Myeloproliferative disorders (MPN) are a heterogeneous group of diseases characterized by aberrant proliferation of one or more of the myeloid lineages.1 BCR-ABL1 negative MPN include polycythemia vera (PV), essential thrombocythemia (ET), and primary myelofibrosis (PMF), which have overlapping clinical features and a common molecular basis.1 Molecular mutations frequently involve genes encoding for specific kinases, leading to constitutively activated signal transduction pathways that cause the abnormal proliferation. The most common acquired somatic mutation in BCR-ABL1 negative MPN affects the kinase JAK2 (V617F), and is present in approximately 95% of cases with PV and in approximately 60% of patients with ET or PMF.2 Further frequent mutations in MPN involve MPL (leading to activation of the JAK-STAT pathway similar to JAK2 mutations) and CALR (calreticulin), which is mutated in approximately 20-30% of patients with ET and in 25-35% of PMF cases.3,4 Thus, the vast majority of MPN patients (97%) carries mutations (mut) in one of these three genes in a nearly mutually exclusive manner.4,5 The CALR gene codes for a developmentally highly conserved and multifunctional protein playing a role as a Ca2+ binding chaperone in the endoplasmic reticulum lumen ensuring proper (glyco-) protein folding, thus preventing protein aggregation. One of its targets is represented by major histocompatibility complex (MHC) class I molecules that mediates its assembly and cell surface expression.6–8 For JAK2, copy neutral loss of heterozygosity (CN-LOH) of 9p24 leading to homozygosity of JAK2V617F has been described in a number of studies and was associated with a distinct phenotype and with disease progression.2,9 By contrast, high mutational loads (≥60%) are seen less frequently in CALR mutated cases, and mutated CALR homozygosity by CN-LOH has so far only rarely been described in MPN,3,10 although it was found that homozygous mutations are associated with acquired myeloperoxidase deficiency.11 Thus, the aim of the present study was to identify cases with CALR CN-LOH in a cohort of MPN cases with the help of the parameters high CALRmut loads and/or progressive disease, as well as cytogenetic and molecular genetic characterization of these cases in comparison to CALRmut patients without CN-LOH.
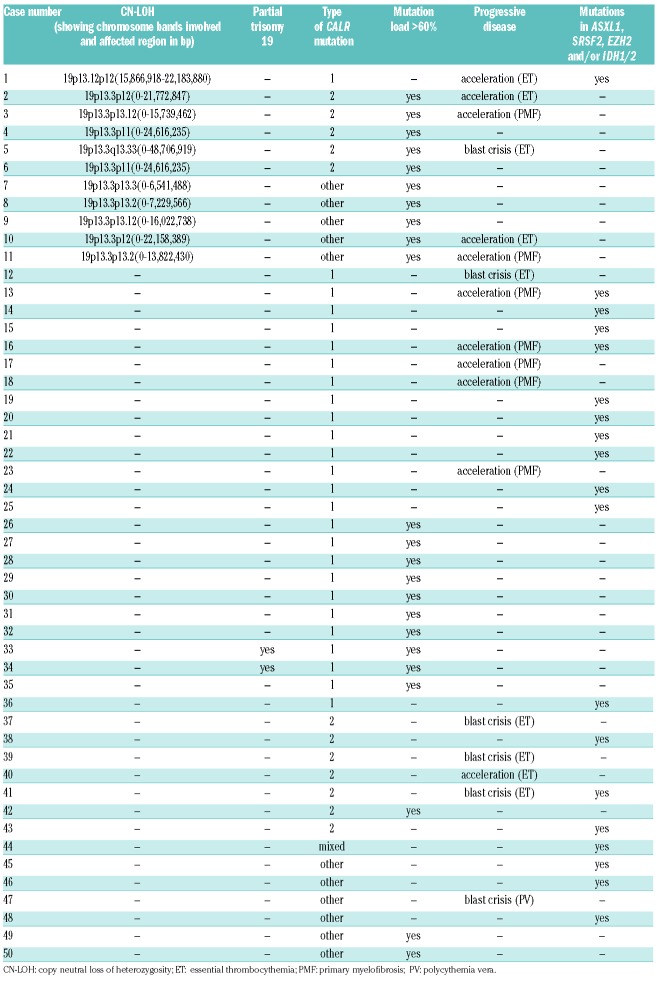
Overall, 50 cases with a CALRmut were included in this study. Bone marrow and/or peripheral blood samples had been sent for diagnosis to the MLL Munich Leukemia Laboratory in the period January 2007 and February 2016. Patients agreed to the use of laboratory data for research studies. The study was carried out in accordance with the Declaration of Helsinki. To detect cases with a CALR CN-LOH, cases were selected according to two different criteria. First, cases with progressive disease, postulating a similar correlation as for JAK2, as homozygosity of JAK2V617F is known to be associated with disease progression.2,9 Cases with progressive disease were defined by: i) accelerated phase; ii) blast crisis; or iii) the presence of at least one mutation in ASXL1, SRSF2, EZH2 and/or IDH1/2 as markers for progression. Second, cases with a high CALRmut load, which could potentially be caused by: i) a CALR CN-LOH; ii) a deletion of the CALR wild-type (wt) allele with a concomitant CALRmut in the other allele; or iii) at least a partial trisomy 19 including the CALR gene. For some cases, more than one criterion was present (Table 1). It should be noted that, for large CALR deletions, the mutation load might be over-estimated due to preferential amplification. The selected cases included: 1) n=23 (46% of cases) with a mutation load ≥60% determined by gene scan analysis; 2) n=17 (34%) cases with progressive disease (accelerated phase: n=11, 22%; blast crisis: n=6, 12%) according to cytomorphology; and/or 3) n=19 (38%) cases displaying mutations in ASXL1, SRSF2, EZH2 and/or IDH1/2. All cases were investigated by genomic arrays (SurePrint G3 ISCA CGH+SNP Microarray, Agilent, Waldbronn, Germany). Images were analyzed using the DEVA Software v.1.2.1 (Roche Nimblegen) and Nexus Copy Number 6.1 (Biodiscovery Inc., El Segundo, CA, USA). Aberrations were evaluated in each sample using BioDiscovery’s Fast Adaptive States Segmentation Technique (FASST2) algorithm. In addition, chromosome preparations and banding analysis were performed in all 50 cases according to standard methods, as previously described.12 Amplicon next-generation sequencing was performed to detect mutations in ASXL1, CBL, DNMT3A, EZH2, IDH1/2, KRAS, NRAS, RUNX1, SF3B1, SRSF2, TET2, TP53, and U2AF1 for all cases. The template library was generated with the TruSeq Custom Amplicon Low Input Kit and sequenced with the NextSeq (Illumina, San Diego, CA, USA; sensitivity: 3%). Next-generation sequencing (NGS) data were analyzed using the Sequence Pilot (v.4.1.1 Build 510 for the Illumina platform, JSI Medicalsystems, Kippenheim, Germany). SPSS (v.19.0.0) software (IBM Corporation, Armonk, NY, USA) was used for statistical analysis. All reported P-values are two-sided; P≤0.05 was considered statistically significant. Variants of unknown significance were excluded from statistical analysis.
Table 1.
Description of selected cases with CALR mutations.
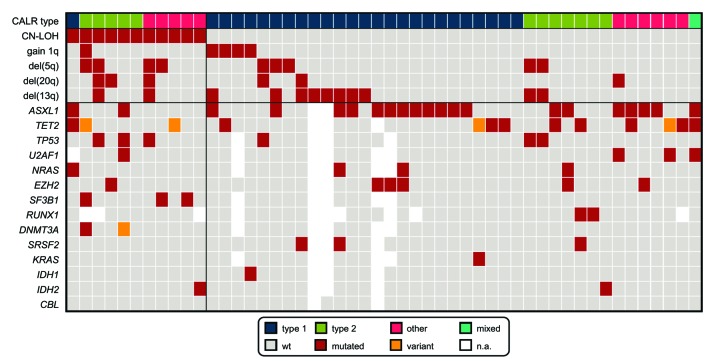
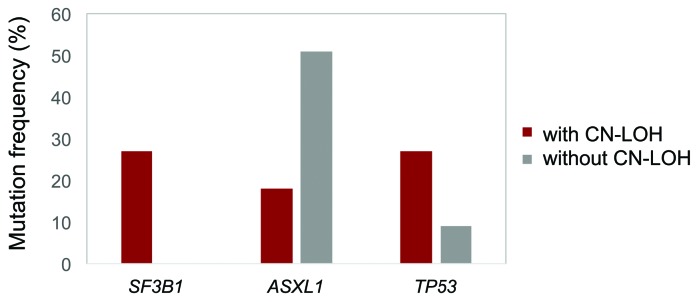
In total, we were able to identify 11 out of 50 cases (22%) with CN-LOH by array CGH, suggesting that CN-LOH of 19p13 is quite a common event in cases with high CALR mutation loads and/or progressive disease. While the type 1 (c.1099_1150del52) CALR mutation was the most frequent mutation type in cases without CN-LOH (25 out of 39, 64%), it was rare in CALR CN-LOH cases (1 out of 11, 9%; P=0.009). Thus, in cases with CN-LOH, it was mainly the type 2 (c.1154_1155insTTGTC) mutation (5 out 11, 45%) and rare mutation types (5 out of 11, 45%) that were detected (Table 1). It can be hypothesized that the observed association of CN-LOH with type 2 CALRmut causes a higher potency of activation of the JAK-STAT signaling pathway. Of note, the only case with CALR CN-LOH showing the type 1 mutation was found to harbor a TET2mut and CN-LOH encompassing 4q23q28 including the TET2 gene. As the CALRmut load was clearly lower compared to the TET2mut load (49% vs. 100%), in this case, the CALRmut might only belong to a subclone. The remaining 10 cases (91%) with CN-LOH showed a CALRmut load of >60% (median: 81%; range 49-100%). In 13 out of 39 cases (33%) without CALR CN-LOH a mutation load >60% was found (median: 48%; range: 3-100%). The reasons for the high mutation load of the majority of these 13 cases without CN-LOH are unclear. A partial trisomy 19 involving the CALR locus was only detected by genomic arrays in two of these 13 cases; in the remaining 11 cases no aberration involving chromosome 19 that could, therefore, potentially explain the high mutation loads was found (Table 1). The cases with CALR CN-LOH presented more frequently with MPN in acceleration compared to the cases without CN-LOH (5 out of 11, 45% vs. 6 out of 39, 15%; P=0.048). Almost all of the patients in accelerated phase with CN-LOH (4 out of 5, 80%) showed a mutation load ≥60% (81%, 78%, 94% and 100%, respectively); the remaining case was the patient with concomitant TET2 and CALR CN-LOH. The chromosomal aberrations detected as recurrent abnormalities by array CGH analyses in the total cohort regarded: del(13q) (12 out of 50, 24%), del(5q) (9 out of 50, 18%), del(20q) (6 out of 50, 12%), and gain of 1q (5 out of 50, 10%) (Figure 1). Del(5q) and del(20q) showed a trend for greater frequency in cases with CN-LOH [del(5q): 4 out of 11, 36% vs. 5 out of 39, 13%; P=0.093; del(20q): 3 out of 11, 27% vs. 3 out of 39, 8%; P=0.111]. No differences were observed in the other aberrations between cases with and without CN-LOH: del(13q): 2 out of 11 cases, 18% vs. 10 out of 39 cases, 26%; gain of 1q: 1/11=9% vs. 4 out of 39=10%. With regard to the advanced disease state of the selected cohort, mutation analyses revealed a high frequency of ASXL1mut (44%), followed by mutations in TET2 (19%), EZH2 (13%), TP53 (13%), U2AF1 (9%), NRAS (9%), and SF3B1 (7%) in the total cohort. Interestingly, SF3B1mut were exclusively detected in cases with CALR CN-LOH (3 out of 11, 27% vs. 0 out of 34, 0%; P=0.012), whereas ASXL1mut tended to be more frequent in cases without CN-LOH (19 out of 37, 51% vs. 2 out of 11, 18%; P=0.083); however, this was not statistically significant (Figure 2). TP53mut were detected more often in the subgroup of cases with CALR CN-LOH, although, once again, this was not statistically significant (3 out of 11, 27% vs. 3 out of 35, 9%). In addition, TP53mut showed a significant correlation to del(5q) [TP53mut in cases with del(5q): 5 out of 9, 56% vs. TP53mut in cases without del(5q): 1 out of 37, 3%; P=0.001], an association that is well-known in MDS (myelodysplastic syndrome) cases, where TP53mut are proposed to play a role in disease progression and contribute to transition to acute myeloid leukemia.13,14 However, a correlation of TP53mut with del(5q) has also been described for patients with MPN and MDS/MPN, thus corroborating the present results.15 To analyze if the CALRmut was present in the main clone or in a subclone of the respective patient, its mutation load was compared to the mutation load of accompanying mutations. CALRmut was considered to constitute the main clone if the difference between the loads was >10%: in the vast majority of cases CALRmut is found in the main clone (48 out of 51; 94%). In summary, we can conclude that, in cases with high CALRmut loads and/or progressive disease, CALR CN-LOH is quite common (22%). While cases without CN-LOH frequently show the type 1 CALRmut, while mainly the type 2 mutation and rare mutation types were detected in CALR CN-LOH cases. Moreover, we found that CN-LOH cases show a distinct pattern of chromosomal aberrations and additional molecular mutations, as they are associated with del(5q) and del(20q) and mutations in SF3B1 and TP53, whereas ASXL1mut are less frequent.
Figure 1.
Summary of accompanying cytogenetic and molecular genetic aberrations. The result of each analysis is depicted for the total cohort of 50 cases; each column represents one patient. Type 1, type 2, other, and mixed (type 1 and type 2) denote the respective CALR types. wt: wild-type; n.a.: not analyzed.
Figure 2.
Frequency of accompanying molecular mutations in cases with and without CALR copy neutral loss of heterozygosity (CN-LOH). Mutation frequencies of genes that show differences between the two subgroups (with CN-LOH: red; without CN-LOH: gray) are depicted.
Supplementary Material
Acknowledgments
The authors would like to thank all co-workers at the MLL Munich Leukemia Laboratory for approaching together many aspects in the field of leukemia diagnostics and research by their dedicated work. The authors would like to thank all physicians for providing samples and caring for patients as well as collecting data.
Footnotes
Information on authorship, contributions, and financial & other disclosures was provided by the authors and is available with the online version of this article at www.haematologica.org.
References
- 1.Arber DA, Orazi A, Hasserjian R, et al. The 2016 revision to the World Health Organization (WHO) classification of myeloid neoplasms and acute leukemia. Blood. 2016;127(20):2391–2405. [DOI] [PubMed] [Google Scholar]
- 2.Kralovics R, Passamonti F, Buser AS, et al. A gain-of-function mutation of JAK2 inmyeloproliferative disorders. N Engl J Med. 2005; 352(17):1779–1790. [DOI] [PubMed] [Google Scholar]
- 3.Klampfl T, Gisslinger H, Harutyunyan AS, et al. Somatic mutations of calreticulin in myeloproliferative neoplasms. N Engl J Med. 2013; 369(25):2379–2390. [DOI] [PubMed] [Google Scholar]
- 4.Nangalia J, Massie CE, Baxter EJ, et al. Somatic CALR mutations in myeloproliferative neoplasms with nonmutated JAK2. N Engl J Med. 2013;369(25):2391–2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jeromin S, Kohlmann A, Meggendorfer M, et al. Next-generation deep-sequencing detects multiple clones of CALR mutations in patients with BCR-ABL1 negative MPN. Leukemia. 2016;30(4):973–976. [DOI] [PubMed] [Google Scholar]
- 6.Guglielmelli P, Nangalia J, Green AR, Vannucchi AM. CALR mutations in myeloproliferative neoplasms: Hidden behind the reticulum. Am J Hematol. 2014;89(5):453–456. [DOI] [PubMed] [Google Scholar]
- 7.Gold LI, Eggleton P, Sweetwyne MT, et al. Calreticulin: non-endoplasmic reticulum functions in physiology and disease. FASEB J. 2010;24(3):665–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Caramelo JJ, Parodi AJ. Getting in and out from calnexin/calreticulin cycles. J Biol Chem. 2008;283(16):10221–10225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stein BL, Oh ST, Berenzon D, et al. Polycythemia Vera: An Appraisal of the Biology and Management 10 Years After the Discovery of JAK2 V617F. J Clin Oncol. 2015;33(33):3953–3960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tapper W, Jones AV, Kralovics R, et al. Genetic variation at MECOM, TERT, JAK2 and HBS1L-MYB predisposes to myeloproliferative neoplasms. Nat Commun. 2015;6:6691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Theocharides AP, Lundberg P, Lakkaraju AK, et al. Homozygous calreticulin mutations in patients with myelofibrosis lead to acquired myeloperoxidase deficiency. Blood. 2016;127(25):3253–3259. [DOI] [PubMed] [Google Scholar]
- 12.Schoch C, Schnittger S, Bursch S, et al. Comparison of chromosome banding analysis, interphase- and hypermetaphase-FISH, qualitative and quantitative PCR for diagnosis and for follow-up in chronic myeloid leukemia: a study on 350 cases. Leukemia. 2002;16(1):53–59. [DOI] [PubMed] [Google Scholar]
- 13.Fernandez-Mercado M, Burns A, Pellagatti A, et al. Targeted re-sequencing analysis of 25 genes commonly mutated in myeloid disorders in del(5q) myelodysplastic syndromes. Haematologica. 2013;98(12):1856–1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jädersten M, Saft L, Smith A, et al. TP53 mutations in low-risk myelodysplastic syndromes with del(5q) predict disease progression. J Clin Oncol. 2011;29(15):1971–1979. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.