Abstract
Advances in genome engineering in the last decade, particularly in the development of programmable nucleases, have made it possible to edit the genomes of most cell types precisely and efficiently. Chief among these advances, the clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9 system is a novel, versatile and easy-to-use tool to edit genomes irrespective of their complexity, with multiple and broad applications in biomedicine. In this review, we focus on the use of CRISPR/Cas9 genome editing in the context of hematologic diseases and appraise the major achievements and challenges in this rapidly moving field to gain a clearer perspective on the potential of this technology to move from the laboratory to the clinic. Accordingly, we discuss data from studies editing hematopoietic cells to understand and model blood diseases, and to develop novel therapies for hematologic malignancies. We provide an overview of the applications of gene editing in experimental, preclinical and clinical hematology including interrogation of gene function, target identification and drug discovery and chimeric antigen receptor T-cell engineering. We also highlight current limitations of CRISPR/Cas9 and the possible strategies to overcome them. Finally, we consider what advances in CRISPR/Cas9 are needed to move the hematology field forward.
Introduction
Genome engineering is defined as the deliberate modification of an organism’s genetic material. It has been used since the early 1980s to study the impact of DNA mutations in human disease precisely and has helped to unravel the genetic basis of many malignancies and to advance their diagnosis, prevention, and treatment. Genome engineering to introduce defined alterations has traditionally employed homologous recombination strategies to modify a gene of interest (gain- or loss-of-function) using segments of exogenous DNA.1 To achieve homologous recombination in the “pre-nuclease” era, large DNA sequences homologous to the target sequence, containing sequence changes designed to produce the desired modification, were introduced into the nucleus of the receiving cell. This technology depends heavily on chance since the DNA construct is expected to interact with the target and induce gene conversion upon recombination of DNA homology arms. The success rate of this technology was historically extremely low, which together with the complexity in designing targeting vectors, and the time and resources required, put it out of reach of some researchers. However, with the advent of highly-specific chimeric nucleases (which are able to recognize 18 or more base pairs) to induce locus-specific DNA double-strand breaks (DSB), the efficiency of homologous recombination rose substantially (e.g., becoming more than 40,000 times more efficient2), depending on the experimental system. The use of such nucleases has increased in recent years with the development of meganucleases,3 zinc-finger nucleases,4 and transcription activator-like effector nucleases (TALEN),5 opening new horizons for genome manipulation (Figure 1A). Nevertheless, designing the aforementioned nucleases to induce DSB in specific loci relies on predicting protein-DNA interactions, which remains technically challenging, and so these nucleases are not practicable in every laboratory. By contrast, the recent breakthrough of clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated (Cas) technology, which is based on nucleic acid interactions, has enabled specific genome editing in a versatile and uncomplicated manner over previous nucleases, and has revolutionized the field of genome engineering (Figure 1B, Table 1).
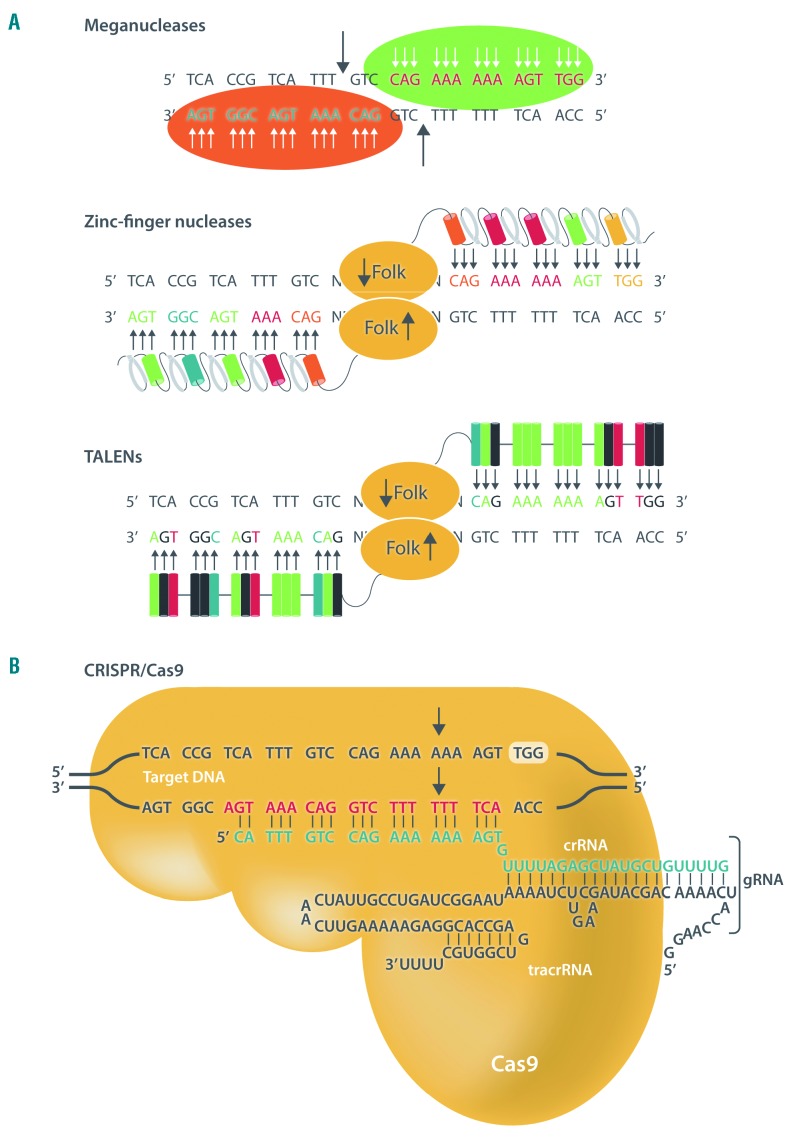
Figure 1.
Nucleases used in genome engineering. (A) Pre-CRISPR nucleases such as meganucleases, zinc-finger nucleases (ZFN) and transcription activator-like effector nucleases (TALEN) are proteins that bind directly to DNA. Meganucleases are naturally occurring restriction enzymes that recognize between 12 to 40 base pair sequences, although they allow for some restricted level of engineering to make them specific to certain loci. Engineered ZFN induce specific double-strand breaks (DSB) acting as dimers. Each monomer is composed of a non-specific cleavage domain from the FokI endonuclease and a zinc-finger protein array where each domain bind three base pairs. ZFN dimers are able to recognize 18–24 base pairs in the target sequence, allowing for highly specific targeting. TALEN are designed combining the same non-specific endonuclease FokI domain and transcription activator-like effector (TALE) proteins. TALE proteins present a central domain responsible for DNA binding, which interacts specifically with just one nucleotide. One of these domains consists of monomers of 34 amino acid residues, two of which are responsible for nucleotide recognition. This makes the design of TALEN very straightforward in principle. (B) In contrast to the nucleases described in (A), the Cas9 endonuclease of the CRISPR/Cas9 system binds to the target DNA thought the guide RNA (gRNA) by Watson-Crick base pairing. The gRNA is composed of two molecules of RNA: (i) the CRISPR RNA (crRNA) (green nucleotides) of which 20 nucleotides [white bold in top panel in (A), black bold in middle and bottom panels in (A)] show strict homology to the target and (ii) the trans-activating crRNA (tracrRNA), which binds to the crRNA and to the Cas9 nuclease (yellow structure). The gRNA brings Cas9 the target sequence, which is always adjacent to a protospacer adjacent motif (PAM) sequence. The PAM sequence for the most used Cas9, isolated from the bacteria Streptococcus pyogenes, is NGG (TGG in the white box). Notes: white arrows in (A) represent hydrogen bonds between amino acids from proteins and DNA base pairs; thick black arrows point to the site of cleavage of the nucleases.
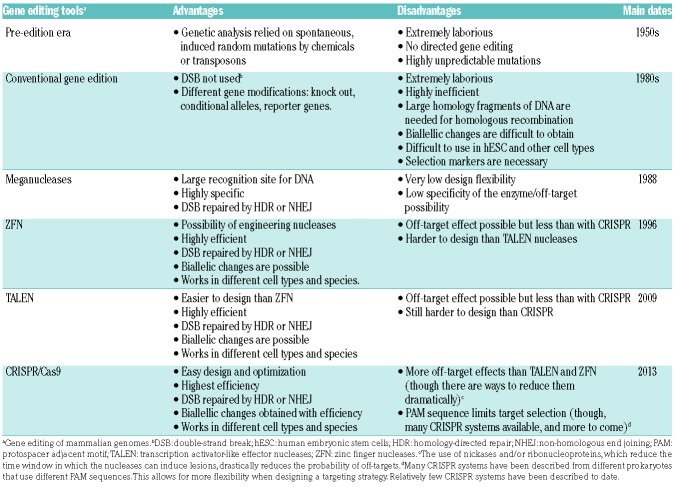
Table 1.
Pros and cons of genome engineering tools in mammalian systems.
CRISPR sequence repeats were first reported in Escherichia coli6 and were later characterized in Haloferax mediterranei, an archaeon isolated from a hypersaline environment in Alicante (Spain).7 Soon after, these sequence repeats were identified as a part of a primitive adaptive immune system in prokaryotes.8,9 In 2012, Doudna and Charpentier demonstrated the first use of CRISPR/Cas9 to introduce site-specific DSB in target DNA based on the ability of a single guide RNA (gRNA) to direct sequence-specific Cas9 double-stranded DNA cleavage,10 illustrating the wide-ranging application of CRISPR as a genome-editing technology.11 Indeed, the CRISPR/Cas9 system was first successfully used in human cells in 2013.12–14 The essential components of this technology include a gRNA that binds specifically to a 20-base pair sequence of interest, and the Cas9 enzyme – an endonuclease that introduces a DSB. Additionally, a conserved dinucleotide-containing protospacer adjacent motif sequence upstream of the gRNA-binding site is required by the endonuclease to recognize and cleave the sequence. In the case of Cas9 isolated from Streptococcus pyogenes, the most widely used nuclease, the protospacer adjacent motif sequence is NGG (Figure 2A). If these conditions are fulfilled, CRISPR/Cas9 can be directed to cleave any genomic sequence. Subsequently, the DSB (in eukaryotic cells) triggers endogenous cellular DNA-repair pathways that can be exploited either to generate gene knock-outs based on the introduction of insertions or deletions (indels) at the DSB by non-homologous end joining (NHEJ)15, or for genome editing, by introducing an engineered template DNA via homology-directed repair (HDR)16 (Figure 2A). In contrast to the protein-DNA interactions of other nuclease editing systems, CRISPR relies on Watson-Crick pairing between RNA and DNA. Thus, researchers keen to perform gene editing require only a basic knowledge of molecular biology to design a targeting system against a locus of choice. Here, we will focus mainly on work done with the Cas9 nuclease, although it is worth mentioning that the CRISPR/Cas system can include many other enzyme variants with numerous functions that are suitable for applications beyond gene editing17 (Table 2).
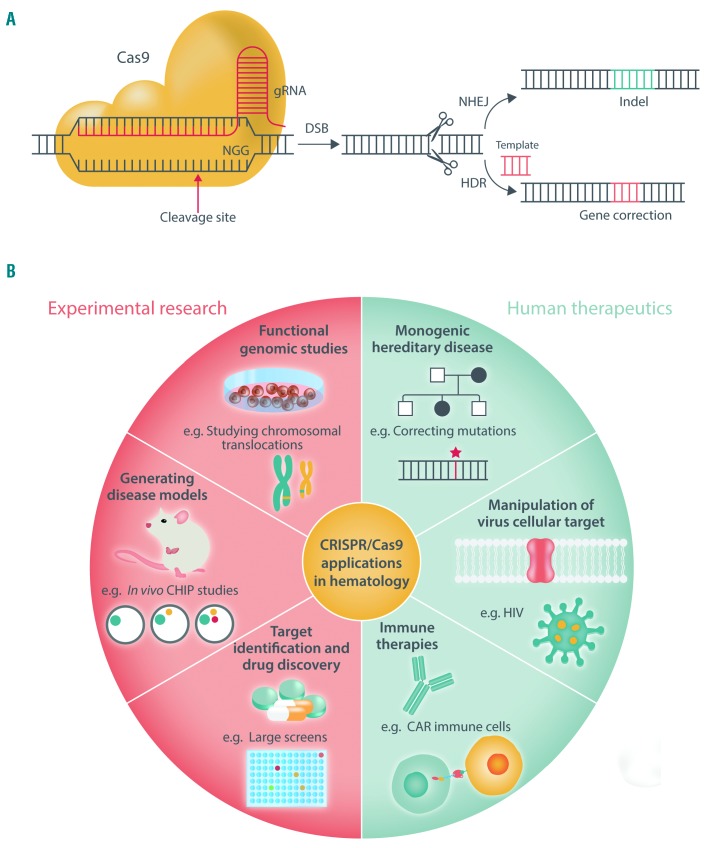
Figure 2.
CRISPR/Cas9-mediated genome editing in hematology. (A) Illustration of the CRISPR/Cas9 system. Site-specific DSB are produced by CRISPR/Cas9 and are either repaired by NHEJ, introducing indels that provoke gene disruption, or by HDR that, in the presence of a DNA template, creates insertions, translocations, or point mutations. gRNA: guide RNA; DSB: double-strand break; NHEJ: non-homologous end joining; HDR: homology-directed repair; indel: insertions and deletions. (B) Applications of CRISPR/Cas9 technology in hematology research and human therapy. HIV: human immunodeficiency virus; CAR: chimeric antigen receptor; CHIP: clonal hematopoiesis of indeterminate potential.
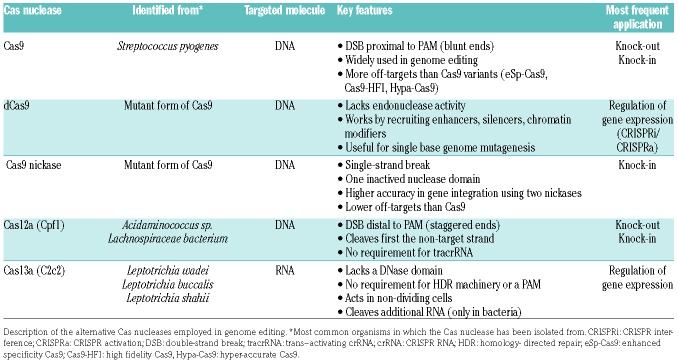
Table 2.
Comparison of the most widely used Cas nucleases.
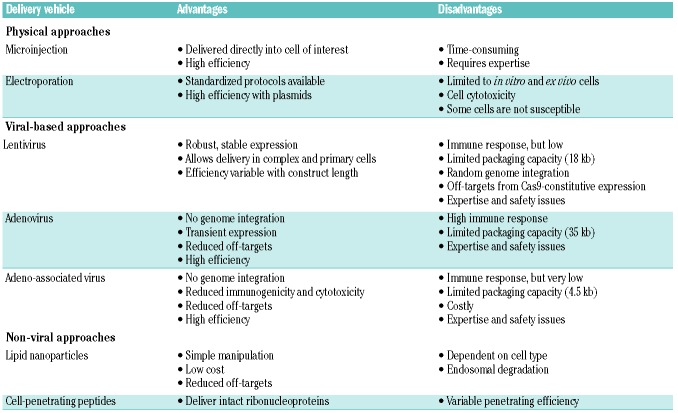
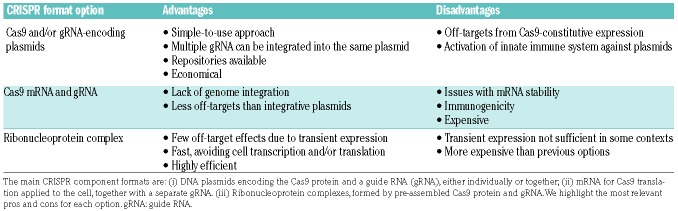
In comparison with engineered nucleases, CRISPR/Cas9 is an easy-to-use genome-editing tool, and several different CRISPR/Cas9-component delivery methods are available for in vitro, ex vivo and in vivo applications18 (Table 3). Generally, Cas9 and gRNA can be introduced into cells in several formats, such as plasmid DNA, lentiviral vectors, mRNA, or more recently by using pre-assembled ribonucleoprotein complexes (Table 4). Indeed, ribonucleoprotein complexes are perhaps the best choice for clinical applications given their high efficiency and short window of action, which reduces the duration of nuclease exposure and, consequently, the possibility of undesired off-target effects. In the hematopoietic setting, CRISPR/Cas9 gene editing has been applied both in research and in clinical translation studies (Figure 2B). In disease modeling, CRISPR/Cas9 technology coupled to next-generation genomics allows researchers to faithfully recapitulate the genetic mutations seen in patients with clonal hematopoiesis or leukemia.19 In the clinical setting, the main goal is to employ CRISPR/Cas9 to treat diseases of the blood and immune system. With this view, several biotechnology companies have pipelines to develop and evaluate therapeutic CRISPR/Cas9 gene editing to correct mutations in pathologies such as autoimmune diseases and multiple myeloma.
Table 3.
Summary of delivery approaches for CRISPR/Cas9 components.
Table 4.
Comparison of the different formats available for CRISPR/Cas9 components.
Given the exponential output of scientific publications using CRISPR in the last years, there is a clear need to synthesize the latest data on gene editing. With this in mind, here we describe recent advances in the use of CRISPR/Cas9 in hematologic research and clinical translation. We also consider the limitations of CRISPR/Cas9 technology for therapeutic applications, their possible solutions, and how the field of hematology may move forward. While CRISPR/Cas9 gene editing is hoped to be a treatment for many hematologic diseases, large clinical trials are needed to evaluate the efficacy and safety of CRISPR/Cas9 for patients, a promising area that will undoubtedly expand in the near future.
CRISPR/Cas9 in hematology research
In vitro gene editing of blood cells
Hematopoietic cell lines are a robust model for validating gRNA specificity and CRISPR/Cas9 experimental design because of their easy manipulation and expansion, and enrichment of edited cells. In this respect, cell lines have been employed: (i) to analyze gene function upon NHEJ-mediated gene disruption; (ii) to insert a point mutation or a DNA fragment; (iii) to correct a point mutation, and (iv) to create chromosomal translocations. Although valuable as a proof-of-concept approach, editing success cannot always be extrapolated to difficult-to-edit primary cells.
Many studies have reported successful CRISPR/Cas9-mediated gene editing in hematopoietic cell lines. As a consequence of indels introduced by NHEJ-mediated DSB repair, the resulting frameshift or nonsense mutations can give rise to truncated proteins that have a gain- or loss-of-function.20 This is a considerably faster approach than conventional homologous recombination-based gene targeting to create gene knock-outs and, given its simplicity, CRISPR/Cas9 is poised to become the method of choice for knock-out studies in most cases. Moreover, CRISPR/Cas9 is superior to RNA interference approaches for deciphering gene function, since the latter produce hypomorphic phenotypes that do not always mirror the complete loss-of-function that often occurs with genetic mutations.21
CRISPR/Cas9-mediated cleavage followed by HDR has been employed to introduce point mutations or gene fragments into specific loci using donor template DNA flanked by 3′ and 5′ sequences homologous to the target region. However, creating a knock-in allele by homologous recombination of a targeting construct using embryonic stem cells (which could be used to produce a mouse model) or by CRISPR/Cas9 is not so different in terms of cost and time.22 CRISPR/Cas9 gene editing can help to elucidate the role of patients’ mutations by generating cellular models carrying these lesions. Along this line, patients with myelodysplastic syndromes frequently have mutations in splicing genes such as the P95H mutation in serine/arginine factor 2 (SRSF2), which regulates pre-mRNA splicing. Zhang et al. developed an SRSF2/P95H cell line using CRISPR/Cas9-mediated HDR, which resulted in a gain-of-function phenotype and changed its RNA-binding preferences, producing splicing misregulation. This illustrates how a mutation associated with myelodysplastic syndromes alters splicing patterns, some of which are relevant for disease and have therapeutic potential.23 In acute myeloid leukemia, driver mutations can also cause and/or maintain leukemia24 and precise AML models are needed to develop novel, targeted therapies. For instance, the R140Q mutation in the Krebs cycle enzyme isocitrate dehydrogenase 2 (IDH2) endows cells with neomorphic enzyme activity, generating an oncometabolite that interferes with epigenetic cell regulation and contributes to malignant transformation. To study the molecular and functional characteristics of this driver mutation, genome editing was used in K562 cells to introduce the IDH2/R140Q mutation.25 Cells carrying this mutation recapitulated the genetic, epigenetic and functional changes seen in IDH2-mutated patients, offering a suitable model for drug testing.
In addition to modeling disease, CRISPR/Cas9 has been employed to correct mutations in disease-associated genes using single-stranded donor oligonucleotides as DNA donor templates for HDR. For example, a loss-of-function mutation in the Additional sex combs like 1 (ASXL1) gene, frequently mutated in myelodysplastic syndromes, chronic myelomonocytic leukemia, and AML was corrected in a chronic myeloid leukemia cell line.26 Similarly, AML blasts (precursor cells) containing the IDH2R140Q mutation were corrected to restore cell function to wild-type status.25 These results constitute a proof-of-concept that CRISPR/Cas9 gene correction of primary hematopoietic cells is feasible. Beyond hematopoietic cells, CRISPR/Cas9 genome editing of human induced pluripotent stem cells (iPSC) has been used to correct disease-relevant mutations. For example, correction of the HCLS1 associated protein X-1 (HAX1) gene by CRISPR/Cas9-mediated HDR reversed the severe congenital neutropenia phenotype in patient-specific iPSC.27 This is important given that iPSC are excellent platforms to model disease and also hold promise for use in patient-specific, cell-based regenerative therapy. Accordingly, hematopoietic cells carrying a mutation could be isolated from the patient, reprogrammed to iPSC, edited, differentiated to hematopoietic stem cells (HSC) and re-introduced by autologous HSC transplantation. However, the capability of iPSC-derived HSC to reconstitute the blood system in the long-term remains a challenge for clinical translation.28
Most preclinical models of CRISPR/Cas9-based gene repair have been based on precise but relatively poorly efficient HDR. The greater efficiency of NHEJ-based mutation correction in the absence of donor template DNA has been used successfully to repair frame-shift mutations. For example, in a study on X-linked chronic granulomatous disease, which is caused by mutations in the cytochrome b-245 heavy chain (CYBB) gene,29 patient-specific CYBB point mutations were successfully repaired by NHEJ – the dominant DSB-repair pathway in hematopoietic stem and progenitor cells (HSPC) – with gene repair efficiency being between 18-25%. The authors of this study assumed that approximately one-third of NHEJ-mediated indels should re-establish the open reading frame disrupted by the disease mutation, leading to a complete or partial recovery of protein function. Importantly, this high-efficiency approach minimizes the number of reagents required to be introduced into patients’ cells and also circumvents homologous donor template delivery, which might be beneficial for translation of HSPC gene editing to the clinic.
In the context of disease modeling, a more complex scenario would be to recreate the fusion proteins resulting from chromosomal rearrangements, a typical hallmark of some leukemias. CRISPR/Cas9-based editing has been successfully used in human cell lines and human HSC to generate chromosomal translocations resembling those described in acute leukemia, such as t(8;21)/RUNX1-ETO, t(4,11)/KMT2A-AFF1/AFF1-KMT2A and t(11;19)/MLL-ENL.30–32 This achievement is relevant because the modeling of fusion proteins in hematopoietic cells was previously accomplished by viral expression of fusion protein cDNA cloned from patients or by genomic engineering of mouse DNA to create chromosomal rearrangements using recombination systems (e.g., Cre-loxP), which is complicated.33 These aforementioned studies illustrate that CRISPR/Cas9 technology is a reliable and accurate approach to recreate chromosomal translocations, albeit at low efficiencies, providing a powerful tool for cancer studies.
Another application of CRISPR/Cas9 technology that holds great promise is in the arena of functional genomics, in which it has been employed in genome-wide, loss-of-function screens in mammalian cells. Typically, lentiviral gRNA libraries are used in genetic screens for positive and negative selection,34,35 which have advantages over RNA interference-based screening with inherent incomplete gene knockdown. Another advantage of CRISPR/Cas9 is that it can target non-coding genomic regions, including promoters, enhancer elements, and intergenic regions. Positive selection studies screen for perturbations conferring enhanced self-renewal/proliferation/survival potential to the interrogated cells, resulting in cell enrichment over time. By contrast, negative selection studies aim to identify genes essential for survival/proliferation that, when targeted, will cause cell depletion over time. In the context of myeloid malignancies, several high-throughput screens have been performed in drug target discovery applications. Using CRISPR/Cas9 to edit protein domains, Shi et al. identified cancer drug targets by screening 192 chromatin regulatory domains in murine AML cells, validating six known drug targets and also revealing additional dependencies.36 In a study aiming to examine mechanisms of cytarabine drug resistance in AML cell lines, CRISPR/Cas9-based screening identified the deoxycytidine kinase gene as the primary contributor to cytarabine resistance.37 In addition to genome-wide CRISPR screens, targeted panel-based screens of previously selected genes would also allow the interrogation of biological processes, for example, cytokine signal transduction, cancer progression or cell migration, which are suspected to be linked to a disease.
Generating mouse models using the CRISPR/Cas9 system
In vivo mouse models, usually generated by homologous recombination strategies, have been instrumental in deciphering the role of point mutations, translocations, and DNA sequence indels in the context of a whole organism. CRISPR/Cas9 technology can be used to build both germline (heritable) and somatic mouse models in a fast and precise manner.38
Germline CRISPR/Cas9 mouse models
CRISPR/Cas9 has been employed to disrupt the splicing factor ZRSR1 in murine zygotes, resulting in altered erythrocyte function in adult mice, suggesting that ZRSR1-associated minor splicing could have an important role in terminal erythropoiesis.39 More recently, the technology was used for the generation of novel hemophilia mouse models on an immunodeficient NSG (NOD/SCID/IL-2γ −/−) background.40 Hemophilia A and B are congenital, X-linked bleeding disorders caused by mutations in the genes encoding for the blood clotting factor VIII (F8) and factor IX (F9), respectively. CRISPR/Cas9 and gRNA were microinjected into NSG mouse zygotes to generate mice with hemophilia A or hemophilia B. These models should allow the evaluation of the efficacy and safety of novel gene therapy vectors in hemophilia.
Given the importance of reporter mouse lines in biomedical research, it is not surprising that CRISPR/Cas9 technology has been applied in the study of early developmental processes. Recently, a knock-in mouse strain was created for dynamic tracking and enrichment of the MEIS1 transcription factor during hematopoiesis.41 This GFP-HA epitope tag reporter strategy and CRISPR/Cas9 gene editing could be employed to develop new reporter mouse lines to study other transcription factors important for hematopoiesis.
Mice carrying mutations in multiple genes have traditionally been generated by sequential recombination in embryonic stem cells and/or intercrossing of mice with single mutations. CRISPR/Cas9 technology allows the generation of mice bearing different gene mutations in a more affordable, less labor-intensive and time-consuming manner than traditional approaches. Similar to the hemophilia models describe above, mice with bi-allelic mutations in TET1 and TET2 were created by co-injection of targeting gRNA into mouse zygotes, which is a much faster approach compared with traditional techniques and allows one-step generation of animals with precise mutations.42 Accordingly, targeting multiple genes using CRISPR/Cas9 should facilitate, for example, the in vivo study of a family of genes with redundant functions. Indeed, Cas9 mRNA and multiple gRNA targeting B2M, IL2RG, PRF1, PRKDC, and RAG1 genes were microinjected together into mouse embryos to produce different immunodeficient mouse strains,43 thus generating new valuable tools to advance research in human HSPC xeno-transplantation.
There is increasing evidence that the acquisition of somatic mutations in HSC, leading to clonal hematopoiesis, is a cardiovascular risk factor. Indeed, DNMT3A and TET2 somatic mutations are drivers of clonal hematopoiesis of indeterminate potential, a state that predisposes to subsequent development of a hematologic malignancy or cardiovascular death.44 This recent study used CRISPR/Cas9 to inactivate DNMT3A and TET2 genes in HSPC and showed that atherosclerotic plaque size was markedly increased in reconstituted mice.45
Somatic CRISPR/Cas9 mouse models
Mouse models with somatic genome editing can be built by CRISPR/Cas9 modification of ex vivo cells followed by transplantation (murine cells) or xenotransplantation (human cells). For instance, the ability to modulate CRISPR/Cas9 activity has been exploited to perform doxycycline CRISPR/Cas9-inducible Trp53-knockout/mutation. When HSPC isolated from a lymphoma transgenic model (Em-Myc) were transplanted, this resulted in accelerated lymphoma development in vivo. Thus, a highly efficient inducible CRISPR/Cas9 vector system can be used to identify novel gene mutations that drive tumorigenesis or to knock-out essential genes that are required for cell survival in vitro.46
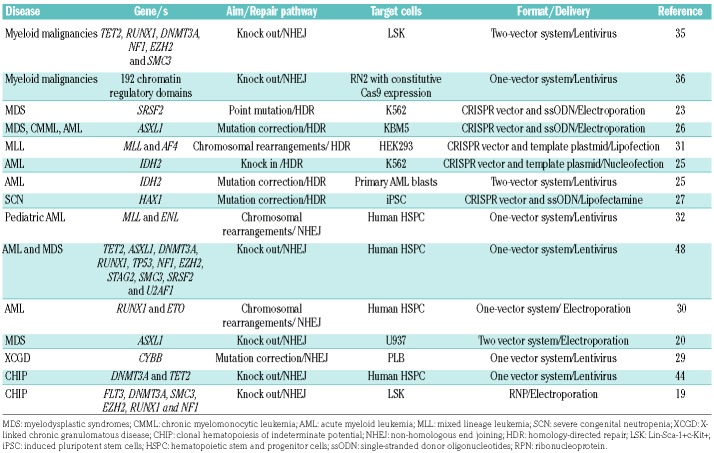
As mentioned in the previous section, one of the unique features of the CRISPR/Cas9 system is its simplicity in enabling simultaneous disruption of several sites in the genome.12 Multiplex CRISPR/Cas9 editing of genes mutated in human leukemias has been demonstrated in mouse and human cells using either lentiviral or ribonucleoprotein approaches. Edited cells were then transplanted into conditioned animals and the identity of the disrupted genes was revealed by next-generation sequencing from clones expanded in sick mice.19,35,47,48 Moreover, ex vivo CRISPR/Cas9 gene editing of HSPC is also useful for studying clonal hematopoiesis of indeterminate potential.19 Multiplex ribonucleoprotein-editing and tracking clonal dynamics by high-throughput sequencing revealed the expansion of mutant clones resembling human clonal hematopoiesis of indeterminate potential, some of which continued to expand and cause death, by hematopoietic failure or AML, in transplanted mice. Accordingly, multiplex CRISPR/Cas9 gene editing is an advantageous tool for functional genomics and for modeling the mutational complexity and co-occurrence patterns observed in hematologic patients at diagnosis, who in the case of AML, carry an average of 2.3 genomic mutations.49 A number of publications on the use of CRISPR/Cas9 gene editing in hematologic research are listed in Table 5.
Table 5.
List of studies on CRISPR/Cas9 gene editing in hematologic diseases.
Gene editing as a therapeutic application in hematologic disorders
Allogeneic HSC transplantation is the frontline treatment for many hematologic disorders; however, this option is only available when a suitable donor exists. Nevertheless, transplanted patients can develop graft-versus-host disease and die of transplant-associated causes. In this scenario, ex vivo gene therapy using viral vectors and ex vivo gene editing by TALEN or zinc-finger nucleases in hematopoietic cells followed by autologous HSC transplantation represent therapeutic alternatives that are currently being investigated in clinical trials.50 However, permanent viral integration into the host genome and/or insertional activation of proto-oncogenes that could lead to secondary leukemia are potential pitfalls related to integrative vector-based gene therapy.51 Site-specific endonucleases, especially CRISPR/Cas9, offer the possibility of delivering non-integrative editing components into target cells, such as mRNA and ribonucleoproteins, constituting a promising approach for HSC gene editing.
Inherited diseases
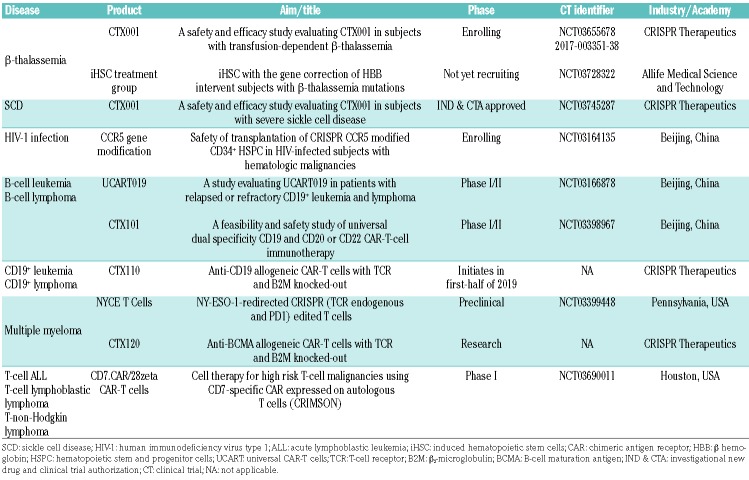
Clinically, CRISPR/Cas9 gene editing holds promise for monogenic hematologic disorders and, thus far, it has been mainly employed in hemoglobinopathies. β-tha-lassemia is caused by mutations in the human hemoglobin beta (HBB) gene and is characterized by reduced β-hemoglobin production, resulting in hemoglobin clumping, hemolytic anemia, and ineffective erythropoiesis. One strategy to remedy this defect using CRISPR/Cas9 is to repair the HBB mutation as has been achieved in iPSC from patients with β-thalassemia.52,53 Another strategy is to reactivate the fetal hemoglobin gene via disruption of the BCL11A gene, an erythroid enhancer regulator of the fetal-to-adult hemoglobin switch and silencer of fetal hemoglobin. BCL11A disruption by CRISPR/Cas9 was shown to facilitate the achievement of threshold levels of functional fetal hemoglobin for treating β-hemoglo-binopathies.54 Likewise, sickle cell disease is caused by a major mutation in the HBB gene, resulting in abnormal hemoglobin and the production of malfunctioning erythrocytes. CRISPR/Cas9-mediated gene editing has been employed to correct one HBB allele in iPSC generated from patients with sickle cell disease,55 and to create the hereditary persistence of a fetal hemoglobin genotype in HSPC, which is suggested as an approach for treating β-thalassemia and sickle cell disease.56,57 In the clinical setting, CTX001, a gene-edited autologous HSC therapy targeting the erythroid-specific enhancer of the BCL11A gene, is entering clinical trials for β-thalassemia (Europe) and sickle cell disease (USA) (Table 6). Specifically, ex vivo edited patients’ cells will be re-infused into patients to produce fetal hemoglobin-containing erythrocytes and overcome the hemoglobin deficiency. These approaches are remarkable because hemoglobinopathies represent a huge cost to healthcare systems as a consequence of frequent transfusions and hospital admissions. Accordingly, novel therapies for these diseases are in high demand.
Table 6.
Current clinical trials in hematology using the CRISPR/Cas9 system.
CRISPR/Cas9 gene editing has recently been employed in Fanconi anemia, a rare genetic disease characterized by progressive bone marrow failure that results in decreased production of all blood cell types. In 80–90% of cases, Fanconi anemia is caused by mutations in FANCA, FANCC or FANCG genes. CRISPR/Cas9-gene editing has successfully corrected a FANCC gene mutation in patient-derived fibroblasts using Cas9 nickase, obtaining a higher correction frequency than Cas9 nuclease.58 As the name might suggest, nickases introduce a single-strand break or “nick” rather than a DSB. Although clinical trials using other nucleases in Fanconi anemia are ongoing,59 to the best of our knowledge CRISPR/Cas9 gene editing to treat this disease has not been employed thus far.
Immunodeficiencies
Primary immunodeficiencies
Primary immunodeficiencies are a heterogeneous group of disorders characterized by variable susceptibility to infections due to hereditary defects in the immune system. One such immunodeficiency, X-linked chronic granulomatous disease, is caused by mutations in the CYBB gene encoding gp91phox, a component of the NADPH oxidase in phagocytes which, when mutated, results in fatal infections. HDR-based therapeutic genome editing (zinc-finger nucleases and CRISPR/Cas9) has been employed to correct a CYBB mutation and restore the functional defect in human HSPC.29 Wiskott-Aldrich syndrome (WAS) is a severe X-linked primary immunodeficiency caused by mutations in the WAS gene and characterized by thrombocytopenia, recurrent infections, tumor development, and autoimmune diseases. Recently, CRISPR/Cas9 gene editing of the WAS locus was reported in a leukemic cell line.60 These preclinical studies hold promise for the clinical translation of CRISPR/Cas9 gene editing therapies for primary immunodeficiencies.
Acquired immunodeficiencies
Human immunodeficiency virus type-1 (HIV-1) infection gives rise to acquired immune deficiency syndrome (AIDS). Several different CRISPR/Cas9 gene-editing strategies have been used to target HIV-1 along its replication cycle.61 For example, disruption of the CCR5 receptor gene, which is present in the host cell and is a co-factor for entry of HIV into the cell, represents a promising strategy to combat the disease, and several studies have reported NHEJ-mediated CRISPR/Cas9 inactivation of CCR5 and other co-factors in lymphocytes. In one study, generation of iPSC homozygous for the naturally occurring CCR5Δ32 mutation through CRISPR/Cas9 genome editing conferred resistance to HIV infection.56 Antiretroviral therapy fails to cure HIV-1 infection because of the persistence of HIV reservoirs harboring integrated HIV DNA. CRISPR/Cas9-mediated deletion or inactivation of proviral DNA could eliminate this source of HIV persistence, thereby being a potentially curative treatment. In preclinical studies, CRISPR/Cas9 efficiently mutated and deactivated HIV proviral DNA in latently infected Jurkat cells.62 However, complete eradication of HIV latent infection is challenging because of the development of mutations resistant to attack by DNA-shearing enzymes.51 Clinically, the safety of transplantation of CRISPR CCR5-modified CD34+ cells in HIV-infected patients with hematologic malignancies is under evaluation in clinical trials (Table 6).
Cancer immunotherapy using chimeric antigen receptor T cells
Cancer immunotherapy can be defined as the induction or enhancement of cancer-specific immune responses against malignant tumors. One approach to this is the ex vivo manipulation of patients’ T cells to express a chimeric antigen receptor (CAR) including an intracellular chimeric signaling domain capable of activating T cells and an extracellular binding domain that recognizes an antigen specific for and strongly expressed on tumor cells. CAR-T cells are re-infused into patients to attack cancer cells in vivo. Currently, CAR-T cells expressing CD19, CD20, CD22, or dual B targeting CAR-T cells are available to treat acute lymphoblastic leukemia, non-Hodgkin lymphoma and chronic lymphocytic leukemia.63 Unfortunately, CAR-T-cell administration can have adverse effects, such as neurotoxicity, cytopenia and cytokine release syndrome, which can be life-threatening. CRISPR/Cas9 can be utilized to complement CAR-T-cell therapy, for example, via disruption of the endogenous T-cell receptor (TCR). Upon its interaction with engineered, transgenic TCR in patients’ cells, endogenous TCR can alter the antigen specificity of CAR-T cells. In a study using CRISPR/Cas9 to knock out endogenous TCR-β, with simultaneous introduction of CAR-T cells, the authors found that this replacement strategy resulted in more robust transgenic anticancer T cells.64 The CRISPR/Cas9 system has also be applied to eliminate other genes that encode inhibitory T-cell surface receptors, such as programmed cell death protein 1 (PD1), to improve the efficiency of T-cell-based immunotherapy.65 To exploit CAR-T-cell therapy beyond the autologous setting, allogeneic universal T cells derived from healthy donors could be engineered by CRISPR/Cas9 upon disruption of TCR to prevent graft-versus-host disease, or beta-2-microglobulin, to eliminate major histocompatibility complex I (MHC I) expression, or by integrating a CAR precisely at the disrupted T-cell receptor α constant (TRAC) locus to improve safety and potency.66–68 Thus, CRISPR/Cas9 technology holds enormous promise to advance the field of cancer immunotherapy and several clinical trials are running to assess the efficacy of CRISPR/Cas9-edited CAR-T cells (Table 6).
Challenges and opportunities for CRISPR/Cas9 therapeutic applications
Delivery of editing tools
Delivery platforms that ensure the access of editing components into a large number of target cells are critical for the clinical development of this technique. Ribonucleoprotein is the cargo format preferred over other transient delivery methods such as mRNA and non-integrating viral vectors because of its hit-and-run mechanism, which reduces the risk of undesired off-target effects, and also because of its ability to efficiently modify cells with low translation rates.47 Nevertheless, the benefit-to-harm ratio of the CRISPR/Cas9 system must be maximized. Possible solutions include the development of novel approaches to integrate ribonucleoprotein and donor template DNA for gene correction in a unique system.
Safety
Off-target DSB can result from non-specific Cas9 cleavage at unwanted genome sites, which is perhaps the major concern regarding therapeutic CRISPR/Cas9 editing. Accordingly, genome-wide sequencing approaches should be employed to thoroughly examine for modifications at unexpected genome locations, or at anticipated off-target sites indicated by in silico prediction tools. The issue of off-target activity is, nevertheless, controversial since studies have yielded contrasting results.69,70 Along this line, several methods have been developed in the last years to detect CRISPR off-target mutations;71 however, there is a lack of consensus on how to predict which putative off-target sites should be examined via deep targeted sequencing. Additionally, the possibility of Cas9-induced on-target mutagenesis, including large deletions and rearrangements that may have pathogenic consequences, has been highlighted as another safety concern.72 Accordingly, more research is needed for a definitive understanding of the in vivo genomic effects of CRISPR/Cas9. Indeed, the possibility of producing undesired gene modifications raises concerns about the use of the CRISPR/Cas9 system for therapy in humans. For instance, infused gene-edited HSC could have the potential to expand clonally and induce leukemia, and so clinical gene editing might cause panic. Possible solutions include the substitution of Cas9 with a different nuclease, for example, Cas12a (also known as Cpf1), which prohibits mismatches between the 18 nucleotides next to the protospacer adjacent motif.73 Other alternatives include the use of paired nickases, guided by two different gRNA targeting the same locus but on opposite DNA strands, or “base editors” editing nucleotides without inducing a DNA break.17
Efficiency
Suboptimal DNA repair outcomes or insufficient target conversion might prevent an intervention from reaching a critical gene-editing threshold necessary to rescue the genetic defect. Strategies to enhance the frequency of HDR in CRISPR/Cas9-mediated transgenesis have been reported and need to be tested in the clinical context.74 Moreover, it should be considered that efficacy could be reduced if the CRISPR/Cas9-induced mutation is detrimental to the cells, having a negative, non-reversible effect.
Immunogenicity
The immune system reaction to in vivo administration of gene editing reagents or ex-vivo genetically modified cells is also a cause for concern.75 The presence of antibodies against Cas9, mainly isolated from Staphylococcus aureus or Streptococcus pyogenes, is common in neonates and adults. Similarly, T lymphocytes against Staphylococcus aureus Cas9 constitute an obstacle to CRISPR/Cas9 therapeutic gene editing.76 Accordingly, the possible immune response must be examined in depth to ascertain whether it could compromise the efficacy of CRISPR-based treatments. Strategies to minimize/eliminate immunogenicity include the use of nucleases other than Cas9 that have not been exposed to the human immune system, or novel nucleases that do not activate an immune response. Other strategies could be: (i) to design an in silico prediction tool for immunogenic predisposition; (ii) to understand the innate immune mechanism against CRISPR/Cas9 in order to help in vector choice and engineering; (iii) to identify antigenic regions on CRISPR/Cas9 to enable deimmunization and epitope masking; and (iv) to employ immunosuppression by using drugs and/or regulatory T cells to reduce undesired immune reactivity.77
p53-mediated DNA damage response
CRISPR/Cas9 genome editing has recently been shown to induce a p53-mediated DNA damage response in some human cell types,78,79 which is in part responsible for the low targeting efficiencies observed in these cells. Consequently, p53 inhibition may improve the efficiency of genome editing in wild type cells; however, a caveat to this approach would be the increased likelihood of cancerous transformation of cells in which the “guardian” activity of p53 is inhibited. p53 gene sequence and function should, therefore, be monitored closely in cells destined for therapy when developing CRISPR/Cas9 cell-based therapies.
Bioethical regulation
CRISPR/Cas9 gene editing is associated with several ethical issues; for example, its application to humans, embryos or germline cells. While the clinical application in human somatic cells to treat hematologic diseases is generally accepted, there is consensus among geneticists that its application in human embryos and germline cells (except for research purposes), in which genetic changes would be inherited by future generations, should be impermissible. That being said, some alarming news was recently reported about the use of CRISPR/Cas9 in human embryos to inactivate the CCR5 receptor and provoke resistance to HIV infection. The biophysicist He Jiankui presented limited (and non-peer-reviewed) data on the birth of twin girls genetically edited with CRISPR/Cas9. This claim, whether true or not, urgently imposes the establishment of strict regulations on human CRISPR/Cas9 genome editing such that it should only be considered for therapeutic uses, but not for human enhancement or eugenics, although it could be used as a research tool to understand early human development or disease pathogenesis. Thus far, no patients have been clinically treated with in vivo CRISPR-based therapy, whereas patients have been given infusions of ex vivo modified T cells (Table 6).
The ethical and regulatory aspects of therapeutic CRISPR/Cas9 genome editing are very complex.80 Given the proven potential of CRISPR/Cas9 to modify the human genome, there are naturally great expectations for future applications. To discuss all these concerns, a multidisciplinary regulatory committee, composed of geneticists, lawyers, society representatives and clinicians, should be created to define a legislative framework to regulate permission or prohibition of CRISPR applications and any genome engineering technique in the future. Global scientific and biological ethics communities must take the lead and establish standards and procedures that reduce the dangers of these powerful new technologies without forgetting the benefits.
What is necessary to move the hematology field forward
As with any new treatment, safety and efficacy are very important. Efforts should be made to develop novel vector systems to maximize delivery to target cells with minimal side effects. In this sense, platforms for systemic and local administration of CRISPR/Cas9 would be highly desirable, including non-integrative vectors for gene therapy or systems based on nanoparticle technology, ribonucleoproteins or other novel approaches.
In the last decade, next-generation sequencing has led to enormous advances in the molecular diagnostics of hematologic malignancies. Somatic mutations have been revealed in many diseases and some may have important prognostic value. The functional impact of these mutations on tumor initiation and/or maintenance needs to be addressed in the next years, and CRISPR/Cas9-based screens in patient-derived cells will be powerful tools to undertake this endeavor. In addition, some hematologic malignancies are characterized by mutations in epigenetic modifiers, proteins that modify DNA at cytosine residues or cause post-translational histone modifications. Some therapies already exploit epigenetic targets, such as DNA methyltransferase 3A (DNMT3A) or histone deacetylase (HDAC), and hypomethylating agents, including the DNA methyltransferase inhibitors azacytidine and decitabine, are used to treat myelodysplastic syndrome and AML. Accordingly, targeted epigenome editing,81 which is the modification of the epigenome at specific sites as opposed to whole-genome modification, could be an area for research development in hematology – for example, for fine-tuning gene expression by locally modulating DNA methylation or determining the function of specific methylation sites. Because epigenome editing does not involve genetic changes, it may also be less hazardous with respect to off-target effects. The challenges will be how to administer epigenome-editing tools in vivo, to achieve reversible epigenetic modifications at precise sites and to ensure that epigenetic modification is heritable upon cell division.
Last but not least, the contribution of CRISPR/Cas9 to multiplex editing in CAR-T cells to create safer and more effective treatments cannot be underestimated, and it is highly likely that this technique will move forward the field of cancer immunotherapy for personalized T-cell-based therapies. The engineering of novel CAR-T cells by pharmaceutical industries, resulting in costly and unaffordable treatments for the general population, should be accompanied by their production by academia institutions, which could make it easier to tailor CAR-T cells for each patient. Thus, drug regulatory authorities should facilitate their academic production and provide resources for CAR-T- cell manufacturing processes, so that these can be simplified and automated to enable scaling up of these cell products.
Conclusions
CRISPR/Cas9 technology is a revolutionary approach for genome editing with wide applications in molecular biology, genetics, and medicine. It has great potential for dissecting gene function, modeling diseases and editing human genes for curative treatment. The number of publications in this field has doubled every year since its introduction, and the CRISPR/Cas9 system is now more widely used in biotechnology and research laboratories than other, more time-consuming and expensive approaches such as zinc-finger nucleases or TALEN. In hematology, CRISPR/Cas9 can be used to model diseases using cultured cells or model organisms, but perhaps more importantly, it can be a valuable approach to correct ex vivo mutations and chromosomal aberrations in cells from patients with blood disorders for autologous HSC transplantation. However, many pitfalls and challenges need to be overcome for the translation of CRISPR/Cas9 gene editing to the clinic. For example, we do not know the minimum number of edited cells needed to functionally correct a genetic defect or if gene editing can be applied to treat multigenic diseases. Further research is necessary to implement CRISPR/Cas9 in the clinical context, so that genome editing-based treatments are available to patients. In conclusion, the CRISPR/Cas9 revolution brings us a specific, efficient and versatile tool for editing genes. Nowadays, technology is no longer a limitation and scientists can probably do any genetic manipulation they can dream of.
Supplementary Material
Acknowledgments
This work was supported by Ayudas FEDER CIBERONC [CB16/12/00284], Instituto de Salud Carlos III [PI16/01113, PI17/00011, CP16/00011], Conselleria de Educación, Investigación, Cultura y Deporte [PROMETEOII/2015/008, ACIF/2018/255], MINECO [RYC-2015-17534, SAF2017-82171-R], and Beca Leonardo a Investigadores y Creadores Culturales de la Fundación BBVA.
Footnotes
Check the online version for the most updated information on this article, online supplements, and information on authorship & disclosures: www.haematologica.org/content/104/5/881
References
- 1.Mortensen R. Overview of gene targeting by homologous recombination. Curr Protoc Neurosci. 2007;Chapter 4:Unit 4.29. [DOI] [PubMed] [Google Scholar]
- 2.Porteus MH, Baltimore D. Chimeric nucleases stimulate gene targeting in human cells. Science. 2003;300(5620):763. [DOI] [PubMed] [Google Scholar]
- 3.Smith J, Grizot S, Arnould S, et al. A combinatorial approach to create artificial homing endonucleases cleaving chosen sequences. Nucleic Acids Res. 2006;34(22):e149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Urnov FD, Miller JC, Lee YL, et al. Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature. 2005;435(7042):646–651. [DOI] [PubMed] [Google Scholar]
- 5.Miller JC, Tan S, Qiao G, et al. A TALE nuclease architecture for efficient genome editing. Nat Biotechnol. 2011;29(2):143–148. [DOI] [PubMed] [Google Scholar]
- 6.Ishino Y, Shinagawa H, Makino K, Amemura M, Nakata A. Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product. J Bacteriol. 1987;169(12):5429–5433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mojica FJ, Ferrer C, Juez G, Rodriguez-Valera F. Long stretches of short tandem repeats are present in the largest replicons of the Archaea Haloferax mediterranei and Haloferax volcanii and could be involved in replicon partitioning. Mol Microbiol. 1995;17(1):85–93. [DOI] [PubMed] [Google Scholar]
- 8.Mojica FJ, Diez-Villasenor C, Garcia-Martinez J, Soria E. Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J Mol Evol. 2005;60(2):174–182. [DOI] [PubMed] [Google Scholar]
- 9.Mojica FJ, Diez-Villasenor C, Garcia-Martinez J, Almendros C. Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology. 2009;155(Pt3):733–740. [DOI] [PubMed] [Google Scholar]
- 10.Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337(6096):816–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Doudna JA, Charpentier E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346(6213):1258096. [DOI] [PubMed] [Google Scholar]
- 12.Cong L, Ran FA, Cox D, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339(6121):819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mali P, Yang L, Esvelt KM, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339(6121):823–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jinek M, East A, Cheng A, Lin S, Ma E, Doudna J. RNA-programmed genome editing in human cells. Elife. 2013;2:e00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bibikova M, Golic M, Golic KG, Carroll D. Targeted chromosomal cleavage and mutagenesis in Drosophila using zinc-finger nucleases. Genetics. 2002;161(3):1169–1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Capecchi MR. Altering the genome by homologous recombination. Science. 1989;244(4910):1288–1292. [DOI] [PubMed] [Google Scholar]
- 17.Wu WY, Lebbink JHG, Kanaar R, Geijsen N, van der Oost J. Genome editing by natural and engineered CRISPR-associated nucleases. Nat Chem Biol. 2018;14(7):642–651. [DOI] [PubMed] [Google Scholar]
- 18.Lino CA, Harper JC, Carney JP, Timlin JA. Delivering CRISPR: a review of the challenges and approaches. Drug Deliv. 2018;25(1):1234–1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shi X, Kitano A, Jiang Y, Luu V, Hoegenauer KA, Nakada D. et al. Clonal expansion and myeloid leukemia progression modeled by multiplex gene editing of murine hematopoietic progenitor cells. Exp Hematol. 2018;64:33–44.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu ZJ, Zhao X, Banaszak LG, et al. CRISPR/Cas9-mediated ASXL1 mutations in U937 cells disrupt myeloid differentiation. Int J Oncol. 2018;52(4):1209–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hannon GJ, Rossi JJ. Unlocking the potential of the human genome with RNA interference. Nature. 2004;431(7006):371–378. [DOI] [PubMed] [Google Scholar]
- 22.Liu ET, Bolcun-Filas E, Grass DS, et al. Of mice and CRISPR: the post-CRISPR future of the mouse as a model system for the human condition. EMBO Rep. 2017;18(2): 187–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang J, Lieu YK, Ali AM, et al. Disease-associated mutation in SRSF2 misregulates splicing by altering RNA-binding affinities. Proc Natl Acad Sci U S A. 2015;112(34): E4726–4734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Papaemmanuil E, Gerstung M, Bullinger L, et al. Genomic classification and prognosis in acute myeloid leukemia. N Engl J Med. 2016;374(23):2209–2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brabetz O, Alla V, Angenendt L, et al. RNA-guided CRISPR-Cas9 system-mediated engineering of acute myeloid leukemia mutations. Mol Ther Nucleic Acids. 2017;6:243–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Valletta S, Dolatshad H, Bartenstein M, et al. ASXL1 mutation correction by CRISPR/Cas9 restores gene function in leukemia cells and increases survival in mouse xenografts. Oncotarget. 2015;6(42): 44061–44071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pittermann E, Lachmann N, MacLean G, et al. Gene correction of HAX1 reversed Kostmann disease phenotype in patient-specific induced pluripotent stem cells. Blood Adv. 2017;1(14):903–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kaufman DS. Tumor(e) blood cells from human pluripotent stem cells. Blood. 2013;12188):1245–1246. [DOI] [PubMed] [Google Scholar]
- 29.Sürün D, Schwäble J, Tomasovic A, et al. High efficiency gene correction in hematopoietic cells by donor-template-free CRISPR/Cas9 genome editing. Mol Ther Nucleic Acids. 2018;10:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Torres R, Martin MC, Garcia A, Cigudosa JC, Ramirez JC, Rodriguez-Perales S. Engineering human tumour-associated chromosomal translocations with the RNA-guided CRISPR-Cas9 system. Nat Commun. 2014;5:3964. [DOI] [PubMed] [Google Scholar]
- 31.Castaño J, Herrero AB, Bursen A, et al. Expression of MLL-AF4 or AF4-MLL fusions does not impact the efficiency of DNA damage repair. Oncotarget. 2016;7(21):30440–30452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reimer J, Knöß S, Labuhn M, et al. CRISPR-Cas9-induced t(11;19)/MLL-ENL translocations initiate leukemia in human hematopoietic progenitor cells in vivo. Haematologica. 2017;102(9):1558–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rabbitts TH, Appert A, Chung G, et al. Mouse models of human chromosomal translocations and approaches to cancer therapy. Blood Cells Mol Dis. 2001;27(1): 249–259. [DOI] [PubMed] [Google Scholar]
- 34.Zhou Y, Zhu S, Cai C, et al. High-throughput screening of a CRISPR/Cas9 library for functional genomics in human cells. Nature. 2014;509(7501):487–491. [DOI] [PubMed] [Google Scholar]
- 35.Heckl D, Kowalczyk MS, Yudovich D, et al. Generation of mouse models of myeloid malignancy with combinatorial genetic lesions using CRISPR-Cas9 genome editing. Nat Biotechnol. 2014;32(9):941–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shi J, Wang E, Milazzo JP, Wang Z, Kinney JB, Vakoc CR. Discovery of cancer drug targets by CRISPR-Cas9 screening of protein domains. Nat Biotechnol. 2015;33(6):661–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rathe SK, Moriarity BS, Stoltenberg CB, et al. Using RNA-seq and targeted nucleases to identify mechanisms of drug resistance in acute myeloid leukemia. Sci Rep. 2014;4:6048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mou H, Kennedy Z, Anderson DG, Yin H, Xue W. Precision cancer mouse models through genome editing with CRISPR-Cas9. Genome Med. 2015;7(1):53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Horiuchi K, Perez-Cerezales S, Papasaikas P, et al. Impaired spermatogenesis, muscle, and erythrocyte function in U12 intron splicing-defective zrsr1 mutant mice. Cell Rep. 2018;23(1):143–155. [DOI] [PubMed] [Google Scholar]
- 40.Yen CT, Fan MN, Yang YL, Chou SC, Yu IS, Lin SW. Current animal models of hemophilia: the state of the art. Thromb J. 2016;14(Suppl 1):22 eCollection 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xiang P, Wei W, Hofs N, et al. A knock-in mouse strain facilitates dynamic tracking and enrichment of MEIS1. Blood Adv. 2017;1(24):2225–2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang H, Yang H, Shivalila CS, et al. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 2013;153(4):910–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou J, Shen B, Zhang W, et al. One-step generation of different immunodeficient mice with multiple gene modifications by CRISPR/Cas9 mediated genome engineering. Int J Biochem Cell Biol. 2014;46:49–55. [DOI] [PubMed] [Google Scholar]
- 44.Sano S, et al. CRISPR-Mediated gene editing to assess the roles of Tet2 and Dnmt3a in clonal hematopoiesis and cardiovascular disease. Circ Res. 2018;123(3):335–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fuster JJ, MacLauchlan S, Zuriaga MA, et al. Clonal hematopoiesis associated with TET2 deficiency accelerates atherosclerosis development in mice. Science. 2017;355(6327): 842–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Aubrey BJ, Kelly GL, Kueh AJ, et al. An inducible lentiviral guide RNA platform enables the identification of tumor-essential genes and tumor-promoting mutations in vivo. Cell Rep. 2015;10(8):1422–1432. [DOI] [PubMed] [Google Scholar]
- 47.Gundry MC, Brunetti L, Lin A, et al. Highly Efficient Genome Editing of Murine and Human Hematopoietic Progenitor Cells by CRISPR/Cas9. Cell Rep. 2016;17(5):1453–1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tothova Z, Krill-Burger JM, Popova KD, et al. Multiplex CRISPR/Cas9-based genome editing in human hematopoietic stem cells models clonal hematopoiesis and myeloid neoplasia. Cell Stem Cell. 2017;21(4):547–555.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hussaini MO, Mirza AS, Komrokji R, Lancet J, Padron E, Song J. Genetic landscape of acute myeloid leukemia interrogated by next-generation sequencing: a large cancer center experience. Cancer Genomics Proteomics. 2018;15(2):121–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dunbar CE, High KA, Joung JK, Kohn DB, Ozawa K, Sadelain M. Gene therapy comes of age. Science. 2018;359(6372). [DOI] [PubMed] [Google Scholar]
- 51.Zhang H, McCarty N. CRISPR editing in biological and biomedical investigation. J Cell Biochem. 2017;118(12):4152–4162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xie F, Ye L, Chang JC, et al. Seamless gene correction of beta-thalassemia mutations in patient-specific iPSCs using CRISPR/Cas9 and piggyBac. Genome Res. 2014;24(9): 1526–1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Song B, Fan Y, He W, et al. Improved hematopoietic differentiation efficiency of gene-corrected beta-thalassemia induced pluripotent stem cells by CRISPR/Cas9 system. Stem Cells Dev. 2015;24(9):1053–1065. [DOI] [PubMed] [Google Scholar]
- 54.Canver MC, Smith EC, Sher F, et al. BCL11A enhancer dissection by Cas9-mediated in situ saturating mutagenesis. Nature. 2015;527(7577):192–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Huang X, Wang Y, Yan W, et al. Production of gene-corrected adult beta globin protein in human erythrocytes differentiated from patient iPSCs after genome editing of the sickle point mutation. Stem Cells. 2015;33(5):1470–1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ye L, Wang J, Tan Y, et al. Genome editing using CRISPR-Cas9 to create the HPFH genotype in HSPCs: an approach for treating sickle cell disease and beta-thalassemia. Proc Natl Acad Sci U S A. 2016;113(38): 10661–10665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Traxler EA, Yao Y, Wang YD, et al. A genome-editing strategy to treat beta-hemoglobinopathies that recapitulates a mutation associated with a benign genetic condition. Nat Med. 2016;22(9):987–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Osborn M, Lonetree CL, Webber BR, et al. CRISPR/Cas9 targeted gene editing and cellular engineering in Fanconi anemia. Stem Cells Dev. 2016;25(20):1591–1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rio P, Navarro S, Bueren JA. Advances in gene therapy for Fanconi anemia. Hum Gene Ther. 2018;29(10):1114–1123. [DOI] [PubMed] [Google Scholar]
- 60.Gutierrez-Guerrero A, Sanchez-Hernandez S, Galvani G, et al. Comparison of zinc finger nucleases versus CRISPR-specific nucleases for genome editing of the Wiskott-Aldrich syndrome locus. Hum Gene Ther. 2018;29(3):366–380. [DOI] [PubMed] [Google Scholar]
- 61.Wang G, Zhao N, Berkhout B, Das AT. CRISPR-Cas based antiviral strategies against HIV-1. Virus Res. 2018;244:321–332. [DOI] [PubMed] [Google Scholar]
- 62.Zhu W, Lei R, Le Duff Y, et al. The CRISPR/Cas9 system inactivates latent HIV-1 proviral DNA. Retrovirology. 2015;12:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Paczesny S, Pavletic SZ, Bollard CM. Introduction to a review series on emerging immunotherapies for hematologic diseases. Blood. 2018;131(24):2617–2620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Legut M, Dolton G, Mian AA, Ottmann OG, Sewell AK. CRISPR-mediated TCR replacement generates superior anticancer transgenic T cells. Blood. 2018;131(3):311–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Xia AL, He QF, Wang JC, et al. Applications and advances of CRISPR-Cas9 in cancer immunotherapy. J Med Genet. 2019;56(1):4–9. [DOI] [PubMed] [Google Scholar]
- 66.Ren J, Liu X, Fang C, Jiang S, June CH, Zhao Y. Multiplex genome editing to generate universal CAR T cells resistant to PD1 inhibition. Clin Cancer Res. 2017;23(9):2255–2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Eyquem J, Mansilla-Soto J, Giavridis T, et al. Targeting a CAR to the TRAC locus with CRISPR/Cas9 enhances tumour rejection. Nature. 2017;543(7643):113–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Liu X, Zhao Y. CRISPR/Cas9 genome editing: Fueling the revolution in cancer immunotherapy. Curr Res Transl Med. 2018;66(2):39–42. [DOI] [PubMed] [Google Scholar]
- 69.Aryal NK, Wasylishen AR, Lozano G. CRISPR/Cas9 can mediate high-efficiency off-target mutations in mice in vivo. Cell Death Dis. 2018;9(11):1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Iyer V, Shen B, Zhang W, et al. Off-target mutations are rare in Cas9-modified mice. Nat Methods. 2015;12(6):479. [DOI] [PubMed] [Google Scholar]
- 71.Tsai SQ, Joung JK. Defining and improving the genome-wide specificities of CRISPR-Cas9 nucleases. Nat Rev Genet. 2016;17(5):300–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kosicki M, Tomberg K, Bradley A. Repair of double-strand breaks induced by CRISPR-Cas9 leads to large deletions and complex rearrangements. Nat Biotechnol. 2018;36(8): 765–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zetsche B, Gootenberg JS, Abudayyeh OO, et al. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell. 2015;163(3):759–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Devkota S. The road less traveled: strategies to enhance the frequency of homology-directed repair (HDR) for increased efficiency of CRISPR/Cas-mediated transgenesis. BMB Rep. 2018;51(9):437–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Maeder ML, Gersbach CA. Genome-editing technologies for gene and cell therapy. Mol Ther. 2016;24(3):430–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Charlesworth CT, Deshpande PS, Dever DP, et al. Identification of pre-existing adaptive immunity to Cas9 proteins in humans. bioRxiv. 2018;243345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chew WL. Immunity to CRISPR Cas9 and Cas12a therapeutics. Wiley Interdiscip Rev Syst Biol Med. 2018;10(1). [DOI] [PubMed] [Google Scholar]
- 78.Haapaniemi E, Botla S, Persson J, Schmierer B, Taipale J. CRISPR-Cas9 genome editing induces a p53-mediated DNA damage response. Nat Med. 2018;24(7):927–930. [DOI] [PubMed] [Google Scholar]
- 79.Ihry RJ, Worringer KA, Salick MR, et al. p53 inhibits CRISPR-Cas9 engineering in human pluripotent stem cells. Nat Med. 2018;24(7):939–946. [DOI] [PubMed] [Google Scholar]
- 80.Kohn DB, Porteus MH, Scharenberg AM. Ethical and regulatory aspects of genome editing. Blood. 2016;127(21):2553–2560. [DOI] [PubMed] [Google Scholar]
- 81.Kungulovski G, Jeltsch A. Epigenome editing: state of the art, concepts, and perspectives. Trends Genet. 2016;32(2):101–113. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.