The human FAT1 gene was cloned over 20 years ago,1 but there has only been an incremental understanding of its functional role in cancer and developmental disorders.2 FAT1 is highly expressed in a large proportion of cases of T-cell acute lymphoblastic leukemia (T-ALL) and B-cell ALL compared to their normal counterparts suggesting an oncogenic function.3 Conversely, the FAT1 gene is also recurrently mutated in a small subset of TALL cases4,5 and also in chronic lymphocytic leukemia.6 Functionally, the FAT1 cadherin has been implicated in Wnt signaling,7 hippo signaling8 and more recently mitochondrial function9 which together suggests a role outside the classical cadherin function in regulating cell-cell adhesion. Here we show that T-ALL cell lines and clinical samples express a unique N-terminal truncated FAT1 mRNA transcript that generates a protein from a novel transcriptional start site within a retained intronic sequence. This novel transcript is regulated independently of full-length FAT1 and results in the expression of a truncated protein lacking almost the entire extracellular domain of FAT1. Significantly, this truncated protein is a novel biomarker for T-ALL and was found to cooperate with NOTCH in driving T-ALL in vivo, suggesting that in the context of T-ALL, this truncated protein may act as an oncogene.
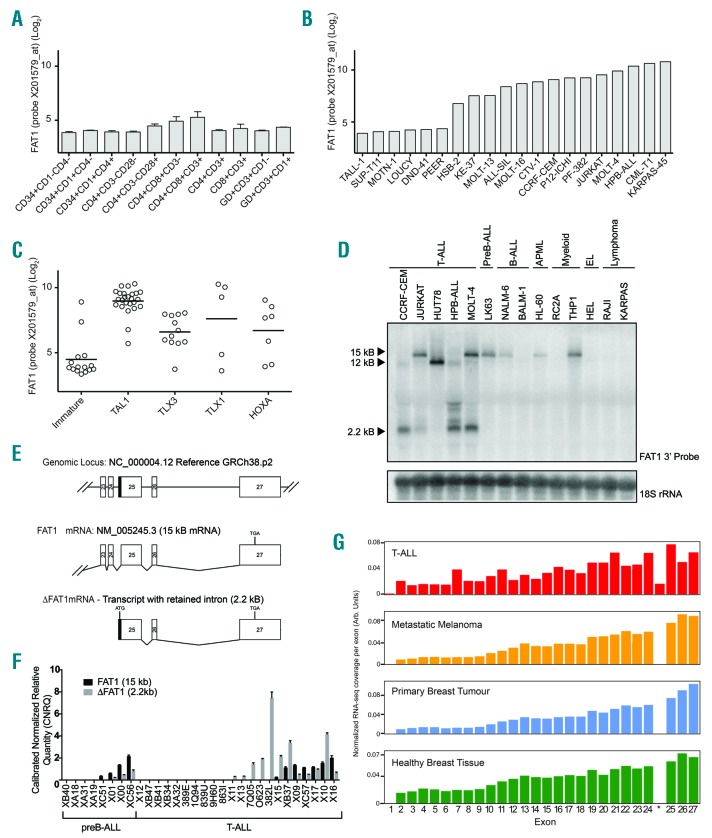
Examining the levels of FAT1 expression in sorted normal healthy hematopoietic and T-cell progenitors revealed that FAT1 is expressed at low levels in all normal T-cell subsets (Figure 1A). In contrast, FAT1 is expressed at significantly higher levels in T-ALL cell lines (Figure 1B). Analysis of FAT1 expression in different molecular-genetic subtypes of human T-ALL10 revealed that FAT1 expression is low or absent in immature T-ALL but expressed at higher levels in TAL1-positive T-ALL cases and variably expressed in TLX1-, TLX3- and HOXA-positive subsets of T-ALL (Figure 1C, Online Supplementary Figure S1).
Figure 1.
Truncated FAT1 transcript with a retained intron is uniquely expressed in T-ALL. (A) FAT1 is expressed at low levels across a diverse range of normal T-cell subsets. (B) In contrast 14/20 T-ALL cell lines expressed high levels of FAT1 transcript. (C) In clinical T-ALL cases, FAT1 is expressed predominantly within TAL1-positive cases and is negatively correlated with immature cases of T-ALL. (D) Northern blot analysis of T-ALL cell lines identified multiple 5’ truncated FAT1 transcripts present with the majority expressing a band migrating at 2.2 kb. (E) Schematic representation of the 3’ exon structure of the FAT1 gene locus and the 5’ RACE result showing that the 2.2 kb truncated transcript contains a retained intronic sequence directly upstream of exon 25. (F) Real-time polymerase chain reaction analysis of B- and T-ALL patient-derived xenograft samples shows that full-length FAT1 is expressed in both TALL and B-ALL cases but the ΔFAT1 transcript is present at high levels only in T-ALL cases. (G) Analysis of the TCGA and TARGET databases revealed that the retained intronic sequence denoted by * is unique to T-ALL cases when compared to melanoma and breast cancers that also have high levels of FAT1 expression.
In the original paper reporting the molecular cloning of the FAT1 transcript, multiple smaller truncated FAT1 transcripts were described in T-ALL cell lines but were not well characterized.1 To investigate this further we performed Northern blotting with a 3’cDNA probe encoding nucleotides 13091-14751 of FAT1 mRNA. This analysis revealed T-ALL cell lines uniquely express 5’ truncated transcripts of FAT1. These predominantly comprised a high molecular variant migrating at approximately 12 kB in the HuT78 cell line and two smaller variants of approximately 2.2 kB and 2.5 kB in CCRF-CEM, Jurkat, MOLT-4 and HPB-ALL cell lines (Figure 1D). Exon-specific Northern blotting and reverse transcriptase polymerase chain reaction (PCR) showed that the larger 12 kB transcript in HuT78 lacked exons 1 and 2 with no corresponding genomic deletion, while the 2.5 kb transcript lacked exons 1-22 and the 2.2 kb fragment lacked exons 1-24 (Online Supplementary Figure S2A-E).
The smaller 2.2 kb and 2.5 kb transcripts were characterized using 5’ RACE from total RNA isolated from Jurkat cells. Two major transcript clones were sequenced and found to include retained intronic sequences at their 5’ ends (Online Supplementary Table S1). The shorter 2.2 kb transcript had a retained intronic sequence mapping directly upstream of exon 25 (Figure 1E). This transcript is homologous with a previously annotated Ensembl FAT1-004 transcript (OTTHUMT00000360219) and partially aligns with the EST transcript BX362336.2 deposited in Genbank.
The clinical relevance and expression of the FAT1-004 transcript (ΔFAT1 for the remainder of this report) was defined in a panel of primary T-ALL and B-ALL samples (Online Supplementary Table S2) using primer-specific real-time PCR to quantify both full-length FAT1 and ΔFAT1 mRNA levels. This analysis found that 32% (7/22) of the T-ALL samples expressed full-length FAT1 but, significantly, 55% (12/22) expressed the ΔFAT1 mRNA transcript. In five T-ALL cases, only the ΔFAT1 mRNA transcript was present with no full-length FAT1 mRNA. Of the eight preB-ALL cases, four (50%) had full-length FAT1, and three of these cases also had detectable ΔFAT1 transcript albeit always at substantially lower levels compared to those in T-ALL and this was speculated to reflect low levels of pre-mRNA (Figure 1F). Analysis of RNA-sequencing data deposited in the TCGA and TARGET databases found that reads covering the ΔFAT1 retained intronic sequence upstream of exon 25 (indicated by *) were unique to T-ALL and not present in melanoma and breast cancer, previously described as having high levels of FAT1 expression11,12 (Figure 1G).
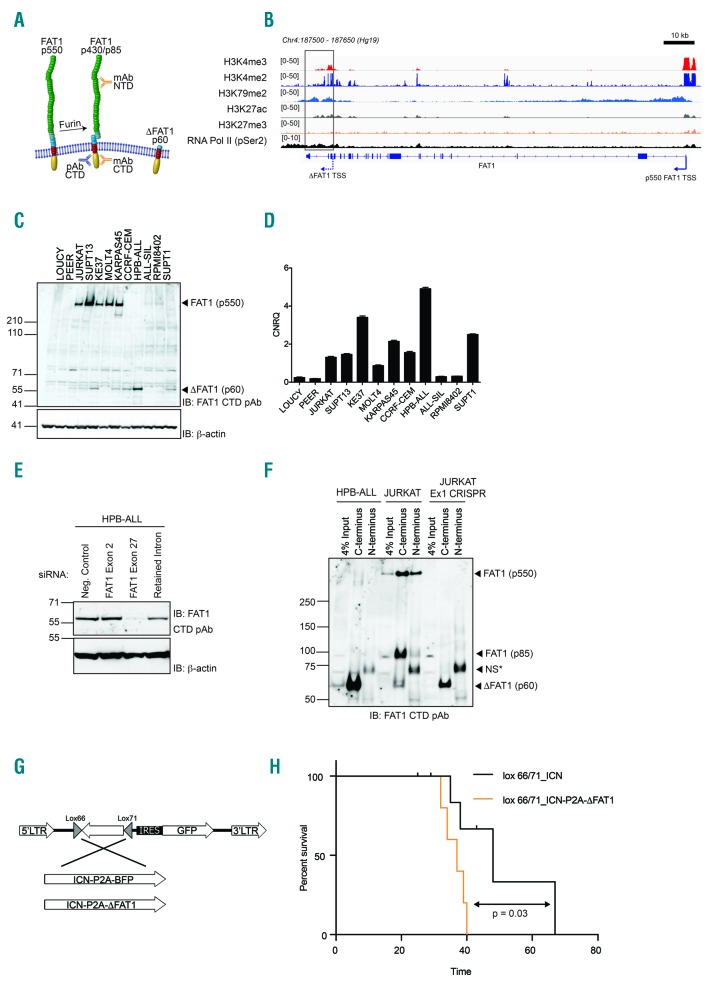
In silico analysis of the retained intronic sequence of the ΔFAT1 transcript revealed the presence of an upstream in-frame ATG (methionine) start codon suggesting that the ΔFAT1 transcript has the potential to generate a protein. This protein would consist of a single extracellular EGF-like domain, a transmembrane region and the entire cytoplasmic tail (Figure 2A), and is predicted to be expressed on the plasma membrane (Online Supplementary Figure S3). The independent regulation of ΔFAT1 is supported by chromatin immunoprecipitation-sequencing data from Jurkat cells that have a strong promotor H3K4me3 signal surrounding the putative start site of the ΔFAT1 transcript together with strong H3K27ac and H3K79me2 marks indicating open chromatin and a transcriptional transition region, respectively, and strong RNA pol II loading (Figure 2B). Moreover, the H3K27Ac mark at the ΔFAT region was consistent among T-ALL cells lines that express the ΔFAT1 transcript and absent from those cell lines that do not express FAT1 (e.g. LOUCY) (Figure 2B) or other cells lines known to express only full-length FAT1 (e.g. MDA-MB-231 and 293T) (Online Supplementary Figure S4).
Figure 2.
The truncated ΔFAT1 transcript produces a protein independent of full-length FAT1. (A) Schematic showing the full-length FAT1 (p550) protein which undergoes furin-mediated cleavage to produce a (p450/p85) heterodimer alongside the predicted truncated ΔFAT protein. (B) Chromatin immunoprecipitation-sequencing data for histone marks and RNA-pol II (pSer2 C-terminal domain) across the FAT1 genetic locus in the Jurkat cell line. The transcriptional start sites (TSS) for FAT1 and ΔFAT1 are highlighted. (C) Western blot analysis of a panel of T-ALL cell lines using an antibody directed against the cytoplasmic tail of FAT1 identified both full-length p550 FAT1 and a specific band migrating at the predicted molecular weight for ΔFAT (p60). (D) Real-time polymerase chain reaction analysis of the levels of ΔFAT1 mRNA transcript reconciles directly with the Western blot analysis. (E) HPB-ALL cells only express the ΔFAT1 protein and siRNA directed toward exon 27 (the cytoplasmic tail) ablated expression while siRNA directed toward exon 2 of FAT1 was equivalent to a negative control. A siRNA directed toward the small intronic sequence also reduced the expression of ΔFAT1. (F) CRISPR/Cas9-mediated knockout of FAT1 using a guide RNA directed toward exon 1 completely removed full-length FAT1 (p550) protein expression but did not reduce ΔFAT1 p60 expression in Jurkat cells. HPB-ALL cells exclusively expressed ΔFAT which was only detected after immunoprecipitation using C-terminal but not N-terminal directed antibodies (*NS = nonspecific band). (G) Schematic representation of the inducible retroviral constructs used to express either ICN or ICN and ΔFAT1 in the presence of Cre-recombinase. (H) Bone marrow transplant survival curve for CD2-Cre-driven expression of either ICN only or ICN and ΔFAT1 in early T-cell progenitors (log-rank, Mantel-Cox test, P=0.03).
Expression of the truncated protein was confirmed by Western blotting of a panel of T-ALL cell lines using a rabbit polyclonal antibody raised against the cytoplasmic tail of FAT1 which revealed a prominent immunoreactive band of the predicted size for ΔFAT1 at ~60 kDa in eight of 12 T-ALL lines (Figure 2C). Protein expression corresponded with levels of ΔFAT1 mRNA (Figure 2C,D). As determined by Northern blotting, HPB-ALL cells exclusively expressed the ΔFAT1 transcript (Figure 1D) and had a strong immunoreactive band at ~60 kDa that was lost through specific short interfering (si)RNA targeting of FAT1 exon 27 but not siRNA targeting of FAT1 exon 2 (Figure 2E). A custom-designed siRNA, albeit with low efficacy parameters due to the short non-optimal intronic sequence, also resulted in reduced expression of the ΔFAT1 protein (Figure 2E).
In solid tumors, FAT1 undergoes furin-mediated post-translational cleavage to generate a p430/p85 heterodimer on the cell surface as well as ADAM10-mediated ectodomain shedding.11 To confirm that the ΔFAT1 protein is neither a splice variant of full-length FAT1 mRNA nor the result of any post-translational cleavage events, immunoprecipitation was carried out using FAT1 antibodies targeting either the extracellular domain or intracellular cytoplasmic domain on HPB-ALL, Jurkat and a CRISPR/Cas9 Jurkat cell line targeting exon 1 of FAT1 to selectively target only the high molecular weight FAT1 (Figure 2F). HPB-ALL cells had a strong enrichment for ΔFAT1 only with the antibody directed toward the cytoplasmic domain and not with the N-terminal targeting anti-FAT1 antibody. In Jurkat cells, there was enrichment for the full length FAT1 with both N-terminal and C-terminal antibodies and ΔFAT1 was only present when immunoprecipitated with the C-terminal domain-directed antibody. When full-length FAT1 was removed by CRISPR/Cas9 guide RNA directed toward FAT1 exon 1 in Jurkat cells, there was no full-length FAT1 present confirming successful loss of FAT1 protein. Significantly, the expression of the novel ΔFAT1 protein remained after immunoprecipitation with the FAT1 C-terminal domain antibody.
The ΔFAT1 truncated protein product was strikingly reminiscent of the Gull mutant allele occurring in Drosophila fat that leads to overgrowth of the wing imaginal disc.13 Similarly, overexpression of ΔFAT1 led to increased cell proliferation and colony formation according to in vitro murine cell-based transformation assays (Online Supplementary Figure S5). However, in an in vivo bone marrow transplant model, ΔFAT1 expression only led to modest B-cell expansion and no overt leukemia within the 200-day observation period (Online Supplementary Figure S6). In the context of T-ALL, chromatin immunoprecipitation-sequencing for activated Notch1 (ICN) shows a specific binding peak upstream of the putative ΔFAT1 transcriptional start site14 suggesting that ICN and ΔFAT1 may cooperate in the development and/or progression of T-ALL. However, in vitro siRNA knockdown of NOTCH1 did not alter levels of ΔFAT1 mRNA after 24 h suggesting that other factors must also regulate ΔFAT1 expression (Online Supplementary Figure S7). However, loss of both NOTCH1 and ΔFAT1 did lead to a decrease in MYC expression in ALL-SIL and CCRF-CEM (Online Supplementary Figure S7). Therefore, to determine the leukemogenic potential of ΔFAT1, a bone marrow transplant model was used to express ΔFAT1 in an inducible manner with ICN using a novel Cre-inducible retroviral construct in conjunction with a CD2-Cre mouse.15 When ΔFAT1 was expressed with ICN using the inducible CD2-Cre model, mice rapidly developed the expected CD4+/CD8+ T-ALL (Online Supplementary Figure S8) with the combination leading to a significant decrease in leukemic latency in vivo revealing that this truncated FAT1 protein acts in an oncogenic manner in the context of NOTCH1-induced T-ALL (Figure 2G,H).
In conclusion, these data show that T-ALL express a unique and novel truncated FAT1 protein that is not only a specific biomarker for T-ALL, but also cooperates with mutant NOTCH1 in driving the disease. This finding urges caution in defining and interpreting the functional role of full-length FAT1 when using recombinant FAT1 constructs that delete the FAT1 extracellular domain7 as these may potentially act as dominant negative for full-length FAT1 signaling, or have de novo protein signaling functions.
Supplementary Material
Footnotes
Information on authorship, contributions, and financial & other disclosures was provided by the authors and is available with the online version of this article at www.haematologica.org.
References
- 1.Dunne J, Hanby AM, Poulsom R, et al. Molecular cloning and tissue expression of FAT, the human homologue of the Drosophila fat gene that is located on chromosome 4q34-q35 and encodes a putative adhesion molecule. Genomics. 1995;30(2):207–223. [DOI] [PubMed] [Google Scholar]
- 2.Sadeqzadeh E, de Bock CE, Thorne RF. Sleeping giants: emerging roles for the fat cadherins in health and disease. Med Res Rev. 2013;34(1):190–221. [DOI] [PubMed] [Google Scholar]
- 3.de Bock CE, Ardjmand A, Molloy TJ, et al. The Fat1 cadherin is over-expressed and an independent prognostic factor for survival in paired diagnosis-relapse samples of precursor B-cell acute lymphoblastic leukemia. Leukemia. 2012;26(5):918–926. [DOI] [PubMed] [Google Scholar]
- 4.Liu Y, Easton J, Shao Y, et al. The genomic landscape of pediatric and young adult T-lineage acute lymphoblastic leukemia. Nat Genet. 2017;49(8):1211–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Neumann M, Heesch S, Schlee C, et al. Whole-exome sequencing in adult ETP-ALL reveals a high rate of DNMT3A mutations. Blood. 2013;121(23):4749–4752. [DOI] [PubMed] [Google Scholar]
- 6.Messina M, Del Giudice I, Khiabanian H, et al. Genetic lesions associated with chronic lymphocytic leukemia chemo-refractoriness. Blood. 2014;123(15):2378–2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Morris LGT, Kaufman AM, Gong Y, et al. Recurrent somatic mutation of FAT1 in multiple human cancers leads to aberrant Wnt activation. Nat Genet. 2013;45(3):253–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ahmed AF, de Bock CE, Lincz LF, et al. FAT1 cadherin acts upstream of Hippo signalling through TAZ to regulate neuronal differentiation. Cell Mol Life Sci. 2015;72(23):4653–4669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cao LL, Riascos-Bernal DF, Chinnasamy P, et al. Control of mitochondrial function and cell growth by the atypical cadherin Fat1. Nature. 2016;539(7630):575–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Soulier J, Clappier E, Cayuela J-M, et al. HOXA genes are included in genetic and biologic networks defining human acute T-cell leukemia (T-ALL). Blood. 2005;106(1):274–286. [DOI] [PubMed] [Google Scholar]
- 11.Sadeqzadeh E, de Bock CE, Zhang XD, et al. Dual processing of FAT1 cadherin protein by human melanoma cells generates distinct protein products. J Biol Chem. 2011;286(32):28181–28191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kwaepila N, Burns G, Leong AS-Y. Immunohistological localisation of human FAT1 (hFAT) protein in 326 breast cancers. Does this adhesion molecule have a role in pathogenesis? Pathology. 2006; 38(2):125–131. [DOI] [PubMed] [Google Scholar]
- 13.Mahoney PA, Weber U, Onofrechuk P, et al. The fat tumor suppressor gene in Drosophila encodes a novel member of the cadherin gene superfamily. Cell. 1991;67(5):853–868. [DOI] [PubMed] [Google Scholar]
- 14.Durinck K, Van Loocke W, Van der Meulen J, et al. Characterization of the genome-wide TLX1 binding profile in T-cell acute lymphoblastic leukemia. Leukemia. 2015;29(12):2317–2327. [DOI] [PubMed] [Google Scholar]
- 15.de Bock CE, Demeyer S, Degryse S, et al. HOXA9 cooperates with activated JAK/STAT signaling to drive leukemia development. Cancer Discov. 2018;8(5):616–631. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.