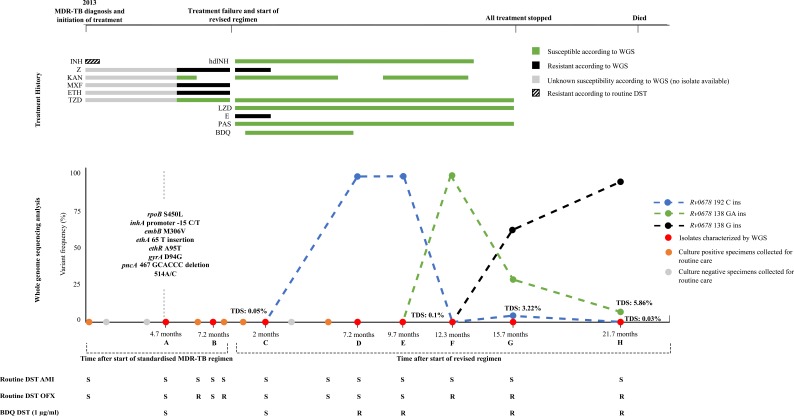
Bedaquiline improves survival among individuals with multidrug-resistant tuberculosis (MDR-TB).1 We report a 65-year old HIV-negative South African male diagnosed in 2013 with MDR-TB (resistant to rifampicin and isoniazid; phenotypically susceptible to a fluoroquinolone and amikacin). Baseline X-ray showed bilateral TB disease with left apex cavitation. He initiated standardised treatment including moxifloxacin, pyrazinamide, kanamycin, ethionamide, isoniazid and terizidone. After initial sputum culture conversion (month 3) and clinical improvement, the patient reconverted to culture positive and developed bilateral cavitation. Following detection of phenotypic ofloxacin resistance (month 6), treatment was revised (month 8) to include high-dose isoniazid, ethambutol, pyrazinamide, terizidone, linezolid, para-aminosalicylic acid and kanamycin (Figure 1). Bedaquiline was added 22 days later and administered for 6 months.2 The patient remained culture positive (treatment failure) and treatment was stopped 15 months after revision of the regimen. The patient died 7 months later.
Figure 1.
Chronology of the diagnosis and treatment of the case study
Summary of treatment provision, genotypic drug resistance (based on whole genome sequencing, WGS), phenotypic bedaquiline drug susceptibility testing (DST, MGIT), targeted deep sequencing and treatment monitoring during standardised treatment and a subsequent individualised bedaquilinecontaining regimen. Overall, eight isolates (A-H) collected 4.7 months after initiation of standard treatment regimen until 6 months after all TB treatment was stopped underwent WGS, targeted deep sequencing of Rv0678 and phenotypic bedaquiline DST. The patient was initially diagnosed with MDRTB with low-level isoniazid resistance using Genotype MTBDRplus, and treated with a standardised MDR-TB treatment regimen but remained culture positive. As per guidelines, subsequent isolates were phenotypically characterized for ofloxacin and amikacin susceptibility. Ofloxacin resistance was first noted 6 months after treatment initiation. All isolates remained susceptible to second-line injectables. At 8.1 months a revised regimen was initiated with the subsequent addition of bedaquiline (22 days after initiation of revised regimen) and withdrawal of pyrazinamide and ethambutol (2 months after initiation of revised regimen). Bedaquiline was administered for 6 months. The patient refused kanamycin at month 6 of the revised regimen for a duration of 2.4 months. The individualized regimen was continued until the outcome of treatment failure at 15 months. Phenotypic DST showed that all isolates with a variant frequency of >1% in Rv0678 were resistant to bedaquiline at 1µg/ml in MGIT.
Abbreviations: MDR-TB=multi-drug resistant tuberculosis; INH=isoniazid; Z=pyrazinamide; KAN=kanamycin; MXF=moxifloxacin; ETH=ethionamide; TZD=terizidone; hdIND=high dose isoniazid; KAN=kanamycin; LZD=linezolid; E=ethambutol; PAS=para-aminosalicyclic acid; BDQ=bedaquiline; WGS=whole genome sequencing; DST=drug susceptibility testing; ins=insertion; R=resistant; S=susceptible
Overall, eight M. tuberculosis isolates (A-H) underwent whole genome sequencing (WGS), targeted deep sequencing3 of Rv0678 and phenotypic bedaquiline resistance testing. WGS of isolate A collected 4.7 months after standard MDR-TB treatment initiation revealed a Beijing strain with mutations conferring resistance to rifampicin, isoniazid, ethambutol, ethionamide, fluoroquinolones, pyrazinamide and streptomycin (Figure 1). WGS of isolate C, collected 2 months after treatment revision, suggested that bedaquiline (to which the isolate was phenotypically susceptible) was added to a regimen with 5 potentially effective drugs. Targeted deep sequencing of isolate C showed a base pair insertion in Rv06784 at a variant frequency of 0.05% (position 192), which was not present in isolate B taken before bedaquiline treatment. Isolate D, collected after bedaquiline cessation, showed the presence of this insertion in >90% of the bacterial population. The frequency of the Rv0678 192 insertion decreased in subsequent isolates, but two different insertions in Rv0678 emerged (GA and G at position 138, isolates F and G, respectively). The G insertion at position 138 became fixed after all treatment was stopped (isolates G and H). Isolates D, E, F, G, and H were phenotypically resistant to bedaquiline.
This case demonstrates the emergence of bedaquiline resistance despite the presence of five potentially effective drugs and good adherence (based on clinical notes). The emergence of Rv0678 variants, after completion of 6 months of bedaquiline treatment, demonstrates the risk of resistance amplification after cessation of a drug with a long half-life (5.5 months for bedaquiline).5
Supplementary Material
Acknowledgements
This work is supported by the National Research Foundation, the South African Medical Research Council (SA MRC) and the Stellenbosch University Faculty of Medicine Health Sciences. The content is solely the responsibility of the authors and does not necessarily represent the official views of the SA MRC. The work is supported by National Institutes of Health (NIH) grant R01AI131939. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. HC acknowledges support from a Wellcome Trust fellowship (ref 099818/Z/12/Z). MdV, AvR and SL are supported by TORCH funding through the Flemish Fund for Scientific Research (FWO G0F8316N). SL acknowledges support by the Swiss National Science Foundation (P2BSP3_165379). GT acknowledges support from the EDCTP2 programme supported by the European Union (grant number SF1401 – OPTIMAL DIAGNOSIS). The sequencing work was performed at the Translational Genomics Research Institute and supported by the Bill & Melinda Gates Foundation Grant OPP1115887 (M Schito) for the ReSeqTB sequencing platform.
Disclaimer
Use of trade names is for identification only and does not constitute endorsement by the U.S. Department of Health and Human Services, the U.S. Public Health Service, or the Centers for Disease Control and Prevention. The findings and conclusions in this report are those of the authors and do not necessarily represent the views of the Centers for Disease Control and Prevention.
Conflict of interest
The authors declare that there is no conflict of interest regarding the publication of the manuscript.
References
- 1.Schnippel K, Ndjeka N, Maartens G, et al. . Effect of bedaquiline on mortality in South African patients with drug-resistant tuberculosis: a retrospective cohort study. The Lancet Respiratory medicine 2018;6:699-706. [DOI] [PubMed] [Google Scholar]
- 2.Conradie F, Meintjes G, Hughes J, et al. . Clinical access to Bedaquiline Programme for the treatment of drug-resistant tuberculosis. S Afr Med J 2014;104:164-6. [DOI] [PubMed] [Google Scholar]
- 3.Colman RE, Anderson J, Lemmer D, et al. . Rapid Drug Susceptibility Testing of Drug-Resistant Mycobacterium tuberculosis Isolates Directly from Clinical Samples by Use of Amplicon Sequencing: a Proof-of-Concept Study. Journal of clinical microbiology 2016;54:2058-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andries K, Villellas C, Coeck N, et al. . Acquired resistance of Mycobacterium tuberculosis to bedaquiline. PloS one 2014;9:e102135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McLeay SC, Vis P, van Heeswijk RP, Green B. Population pharmacokinetics of bedaquiline (TMC207), a novel antituberculosis drug. Antimicrobial agents and chemotherapy 2014;58:5315-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.