Abstract
Background:
In resource-limited settings, HIV-1 drug resistance testing to guide antiretroviral therapy (ART) selection is unavailable. We retrospectively conducted genotypic analysis on archived samples from Nigerian patients who received targeted viral load testing to confirm treatment failure and report their drug resistance mutation patterns.
Methods:
Stored plasma from 349 adult patients on non-nucleoside reverse transcriptase inhibitor (NNRTI) regimens were assayed for HIV-1 RNA viral load and samples with >1,000 copies/ml were sequenced in the pol gene. Analysis for resistance mutations utilized the IAS-US 2011 Drug Resistance Mutation list.
Results:
175 samples were genotyped; majority subtypes were G (42.9%) and CRF02_AG (33.7%). Patients were on ART for a median of 27 months. 90% had the M184V/I mutation, 62% had ≥ 1 thymidine analogue mutations, and 14% had the K65R mutation. 97% had a NNRTI resistance mutation, and 47% had ≥ 2 etravirine associated mutations. In multivariate analysis tenofovir based regimens were less likely to have ≥ 3 nucleoside reverse transcriptase inhibitor (NRTI) mutations after adjusting for subtype, previous ART, CD4 and HIV viral load (p<0.001, OR 0.04). 70% of patients on tenofovir based regimens had at least 2 susceptible NRTIs to include in a second-line regimen compared with 40% on zidovudine based regimens (p=0.04, OR=3.4).
Conclusions:
At recognition of treatment failure, patients on tenofovir based first-line regimens had fewer NRTI drug resistant mutations and more active NRTI drugs available for second-line regimens. These findings can inform strategies for ART regimen sequencing to optimize long-term HIV treatment outcomes in low resource settings.
Keywords: HIV-1, drug resistance, tenofovir disoproxil fumarate, non-B subtype, thymidine analogue mutations, second-line, resource-limited settings
INTRODUCTION
Unprecedented scale up of antiretroviral therapy (ART) for over 2.4 million HIV positive patients in resource limited countries has been achieved through the support of the President’s Emergency Plan For AIDS Relief (PEPFAR) in partnership with the Global Fund to fight AIDS, Tuberculosis and Malaria and national governments [1]. There is mounting evidence that monitoring clinical and immunological parameters, as opposed to HIV-1 RNA viral loads, may increase HIV-1 drug-resistant mutations and therefore limit options for second-line regimens [2,3,4]. New World Health Organization (WHO) guidelines recommend either tenofovir (TDF) or zidovudine (AZT) as the preferred nucleoside reverse transcriptase inhibitor (NRTI) for first-line regimens [5], but there is an increased focus on using TDF because its drug resistance patterns lead to less resistance in alternative thymidine analogue drugs [6]. However, there is limited reported data comparing drug resistance patterns between TDF or AZT containing first-line regimens and their impact on second-line options in low-resource settings.
Nigeria’s HIV-1 prevalence rate of 4.4% combined with a population over 140 million generates the second highest HIV/AIDS burden in the world [7], dominated by subtypes G and CRF02_AG [8,9,10]. In 2002, the government of Nigeria began providing subsidized ART [11] which expanded in 2005 through PEPFAR supported HIV/AIDS treatment services, to treat over 300,000 Nigerians by the end of 2009 [12]. There is evidence that with ART exposure, non-B subtypes may have genetic differences contributing to their pattern of drug resistance [13,14,15,16], and thus, mutations arising in Nigeria’s diverse subtype population could have cross-resistance to second-line options. In this study, we report antiretroviral drug-resistant mutations from Nigerian patients failing different first-line regimens and predict second-line NRTI options for effective long-term HIV treatment.
METHODS
We conducted a retrospective cross-sectional study performing genotypic sequencing analysis on pre-existing blood samples from patients who received targeted viral load testing to confirm virological failure.
Study Population
The Institute for Human Virology-Nigeria (IHVN), a PEPFAR implementing partner, provides ART to over 60,000 public sector patients through the AIDS Care and Treatment in Nigeria (ACTION) program. This study was conducted at IHVN supported sites, University of Abuja Teaching Hospital (UATH) and National Hospital Abuja (NHA). Patients were included in the present study if they received HIV-1 RNA testing between November 2006 and December 2007; were over 18 years; were on NNRTI based first-line regimens; and had not received any protease inhibitor (PI) based therapy.
This study was approved by the National Health Research Ethics Committee of Nigeria, the University of Abuja Teaching Hospital, National Hospital Abuja, and the University of Maryland Baltimore Institutional Review Board.
Clinic Procedures and Data Collection
Clinical care protocols for eligibility, regimen choice and monitoring followed Nigerian national guidelines [17]. During ACTION, six first-line treatment regimens were prescribed: zidovudine, lamivudine and nevirapine or efavirenz (AZT/3TC/NVP or EFV); stavudine, lamivudine, and nevirapine or efavirenz (d4T /3TC/NVP or EFV); and tenofovir, emtricitabine and nevirapine or efavirenz (TDF/FTC/NVP or EFV). First-line regimens were categorized according to type of NRTI prescribed [AZT, d4T, TDF]. Participants who were prescribed multiple first-line regimens (>1NRTI) were further categorized as being given AZT and d4T sequentially and in either order or substituting a first regimen containing AZT or D4T with TDF. Every 6 months, patients provided blood for CD4 counts and toxicity monitoring and received intensified adherence counseling.
Targeted viral load testing was implemented to guide accurate switching decisions in patients at high risk of virological failure defined by: 1) received ART prior to ACTION enrollment; 2) suspected of poor adherence; or 3) had clinical or immunological failure. Information about previous ART was self-reported as 1) yes or no and 2) date started. ART exposure before and during ACTION were summed for total ART. Poor adherence was defined as >7 days late for a scheduled pharmacy refill for over 20% of visits. Data was abstracted by medical folder review.
Laboratory Evaluation
Plasma HIV-1 RNA viral load (VL) testing was conducted using Roche Amplicor MONITOR 1.5 (Roche, Nutley, New Jersey, USA) assay (limit of detection: 400 copies/ml) at IHVN Asokoro Laboratory Training Center in Abuja. Laboratory quality assurance programs included verification of selected samples with FASCalibur (Beckton-Dickinson, Franklin Lakes, New Jersey, USA) and blinded sample panels testing. Samples eligible for genotyping (VL >1,000 copies/ml) were transported at −70°C to the Institute of Human Virology, University of Maryland School of Medicine Baltimore, USA.
Genotypic analysis of the HIV pol region was performed through nested PCR of protease (codons 1–99) and the amino terminus (codons 1–242) using methods described previously [18]. Amplification products were sequenced with Applied Biosystems 3130 automated sequencer (Applied Biosystems, Foster City, CA), assembled using Sequencher 4.2.2 (Gene Codes Corporation, Ann Arbor, MI), and aligned with standard subtype references (MacGDE). Phylogenetic analysis was conducted using Neighbor-joining and maximum parsimony bootstrap computation [19,20]. Viral sequences outside subtype clusters were analyzed using Simplot, v3.4 for inter–subtype recombination [21].
Drug Resistance Mutation Analysis
Frequencies of drug-resistant mutations were estimated using ViroScore v8.1 [22] and categorized as NRTI and NNRTI associated mutations according to the International AIDS Society-USA 2011 list [23]. Thymidine analog mutations (TAMs) included M41L, D67N, K70R, L210W, T215F/Y, K219E/Q) that were further designated as TAM 1 (M41-L210-T215Y) or TAM 2 (D67-K70-T215F-K219) [24]. Etravirine resistance was calculated using weighted scores [23], where L100I, K101P, Y181C/I/V result in the greatest impaired clinical response. The number of patients with zero, single, or multiple mutations was categorized within each first-line regimen. Phenotypic resistance patterns, susceptible (S), intermediate (I) or resistant (R), were predicted from a sum of scored mutations according to the Stanford Database Algorithm v1.2 [25]. Protease sequences were analyzed for amino acid substitutions at positions previously reported to be associated with protease inhibitor (PI) resistance in subtype B virus [26].
Statistical Analysis
Descriptive analyses were conducted using χ2 and Fisher’s exact test and Student’s t-test and Mann-Whitney test where appropriate. Univariate and multivariate logistic regressions were performed to identify factors associated with mutation patterns. Factors associated at the p<0.2 level with the outcome and potential confounders were included in multivariate models. Akaike’s Information Criterion (AIC) was used for model selection and the Hosmer-Lemeshow test for goodness of fit. Statistical analyses were performed using SAS (9.1.3).
RESULTS
The study population consisted of a cross-sectional sample of 349 patients who received targeted viral load testing. Of the 205 samples designated for genotyping, 30 failed to amplify (n=175). At viral load testing, 52% of patients were female, mean age was 38 years, median time on ART was 27 months and median CD4 count was 128 cells/mm3 (IQR:60–229).
Of those patients genotyped, 14% were on AZT, 21% on d4T, 13% on TDF, and 52% on more than one NRTI. For those with multiple NRTIs, 26% were prescribed AZT and d4T sequentially and in either order. The remaining 25% were prescribed AZT or d4T initially, before substituting either of them with TDF. There was variation in HIV-1 subtypes with most patients harboring subtype G virus and CRF02_AG. Fewer than 6% had subtype A, B and C virus. Those who were genotyped were significantly more likely to have prior ART, more than 24 months of ART exposure, and lower CD4 counts at viral load testing [Table 1]. There were no significant differences between patients whose samples did and did not amplify (data not shown).
Table 1.
Characteristics of NNRTI-based first-line regimen users with or without viral suppression
| No. (%) of NNRTI |
||||
|---|---|---|---|---|
| Characteristic | Overall (N=349) | Genotyped ≥1000 copies/ml (N=175) | Not Genotyped <1000 copies/ml (N=144) | p-valuea |
| NNRTI Regimen | 0.1 | |||
| AZT-3TC-NVP/EFV | 59 (17) | 25 (14) | 32 (22) | |
| d4T-3TC-NVP/EFV | 85 (25) | 36 (21) | 40 (28) | |
| TDF-FTC-NVP/EFV | 46 (13) | 23 (13) | 20 (14) | |
| >1 first-line regimen | 157 (45) | 90 (51) | 51 (36) | |
| Gender | 0.3 | |||
| Male | 166 (48) | 89 (51) | 64 (44) | |
| Female | 183 (52) | 86 (49) | 80 (56) | |
| Age in years (Mean, SD) | 37.9 (8.2) | 38.1 (8.5) | 37.6 (8.3) | 0.6 |
| Site | 0.7 | |||
| UATH | 153 (44) | 75 (43) | 66 (46) | 0.7 |
| NHA | 196 (56) | 100 (57) | 78 (54) | |
| Previous ART | <0.001 | |||
| Yes | 208 (60) | 126 (72) | 61 (42) | |
| No | 141 (40) | 49 (28) | 83 (58) | |
| Length of Total ART | ||||
| Median, IQR | 27 (19–43) | 32 (20–44) | 22 (16–33) | <0.001 |
| ≤24 months | 155 (44) | 62 (35) | 86 (60) | |
| >24 months | 182 (52) | 109 (62) | 53 (37) | |
| Adherent to ART | 0.4 | |||
| Yes | 227 (65) | 109 (62) | 96 (67) | |
| No | 122 (35) | 66 (38) | 48 (33) | |
| Baseline CD4 Count (cells/ml3) | 0.5 | |||
| <100 | 118 (39) | 63 (41) | 47 (37) | |
| 100–199 | 100 (33) | 53 (35) | 40 (32) | |
| ≥200 | 87 (29) | 6 (24) | 40 (32) | |
| CD4 Count at VL (± 3 months) | <0.001 | |||
| <100 | 104 (38) | 71 (50) | 26 (25) | |
| 100–199 | 83 (30) | 43 (30) | 32 (30) | |
| ≥200 | 87 (32) | 29 (20) | 48 (45) | |
| Viral load (copies/ml) | ||||
| Median (log10), IQR | 3.8 (2.6–4.9) | 4.7 (4.1–5.4) | ||
| <1000 | 144 (41) | |||
| 1,000–4,999 | 24 (7) | 19 (11) | ||
| 5,000–99,999 | 103 (30) | 85 (49) | ||
| ≥100,000 | 78 (22) | 71 (41) | ||
| Subtype | ||||
| G | 75 (42.9) | |||
| CRF02_AG | 59 (33.7) | |||
| CRF06_cpx | 9 (5.1) | |||
| A | 6 (3.4) | |||
| C | 2 (1.1) | |||
| B | 1 (0.6) | |||
| URFs | 23 (13.1) | |||
Note. Certain data were missing for selected patients. AZT, zidovudine; d4T, stavudine; TDF, tenofovir; 3TC, lamivudine; FTC, emtricitabine; NVP,nevirapine; EFV, efavirenz; UATH, University of Abuja Teaching Hospital; NHA, National Hospital Abuja; URFs, unique recombinant forms; VL, viral load; IQR, interquartile range.
Determined with 2-sided Pearson’s chi-square test.
NRTI Resistance
Among all patients genotyped, 94% had at least one NRTI mutation and 62% had at least 1 TAM mutation. The average number of NRTI mutations was 3 with a range of 0 to 8. Most (90%) patients harbored the M184V/I mutation and 14% had the K65R mutation. M184V was the only mutation that was associated with a lower average HIV-1 RNA viral load (4.7 vs. 5.1 log10copies/ml, p=0.01).
Patients on TDF based regimens were significantly more likely to have thymidine-sparing mutations (K65R (57%), M184I (30%), Y115F (13%)) as compared to AZT and d4T based regimens (p≤0.02). Conversely, patients on TDF based regimens were less likely to have TAM mutations [Table 2]. The two patients on TDF based regimens with a TAM mutation (K219E) also had the K65R mutation. In multivariate analysis, TDF based regimens were less likely to have three or more NRTI associated mutations after adjusting for subtype, previous ART exposure, CD4 and HIV viral load (OR 0.04, p<0.001). CD4 counts greater than 100 cells/mm3 remained independently protective of multiple NRTI mutations (100–199: OR 0.3, p=0.03; ≥200: OR 0.4, p=0.09). Age, baseline CD4 counts (cells/mm3), adherence, and length of time on ART did not significantly improve the analysis and were not retained in the final model.
Table 2.
Frequency Distribution of Major NRTI Mutations
| No. (%) of NRTI regimen |
|||||||
|---|---|---|---|---|---|---|---|
| All (N=175) | TDF (N=23) | AZT (N=25) | d4T (N=36) | AZT/d4Tb (N=45) | AZT/d4T,TDFb (N=44) | p-valuea | |
|
NRTI mutations | |||||||
| M184V | 144 (83) | 14 (61) | 22 (88) | 29 (81) | 42 (93) | 36 (82) | 0.1 |
| K65R | 25 (14) | 13 (57) | 0 | 2 (6) | 0 | 10 (23) | <0.001 |
| V75I | 17 (10) | 1 (4) | 0 | 7 (19) | 6 (13) | 3 (7) | 0.03 |
| M184I | 12 (7) | 7 (30) | 0 | 1 (3) | 0 | 4 (9) | 0.001 |
| L74V | 5 (3) | 0 | 0 | 2 (6) | 1 (2) | 2 (5) | 0.5 |
| Y115F | 6 (3) | 3 (13) | 0 | 0 | 1 (2) | 2 (5) | 0.02 |
| Q151M | 6 (3) | 0 | 0 | 3 (8) | 1 (2) | 2 (5) | 0.2 |
| A62V | 5 (3) | 0 | 0 | 2 (6) | 2 (4) | 1 (2) | 0.5 |
| F116Y | 4 (2) | 0 | 0 | 2 (6) | 1 (2) | 1 (2) | 0.5 |
| F77L | 4 (2) | 0 | 0 | 1 (3) | 1 (2) | 2 (5) | 1.0 |
| K70E | 3 (2) | 2 (9) | 0 | 0 | 0 | 1 (2) | 0.1 |
| 69i | 1 (1) | 0 | 0 | 1 (3) | 0 | 0 | 1.0 |
|
TAMs | |||||||
| M41L | 53 (30) | 0 | 8 (32) | 11 (31) | 23 (51) | 11 (25) | 0.003 |
| T215Y | 48 (28) | 0 | 8 (32) | 8 (22) | 22 (49) | 10 (23) | 0.01 |
| D67N | 40 (23) | 0 | 9 (36) | 9 (25) | 15 (33) | 7 (16) | 0.002 |
| K70R | 40 (23) | 0 | 10 (40) | 10 (28) | 12 (27) | 8 (18) | 0.001 |
| T215F | 39 (22) | 0 | 7 (28) | 6 (17) | 14 (31) | 12 (27) | 0.01 |
| K219E | 19 (11) | 2 (9) | 3 (12) | 4 (11) | 8 (18) | 2 (5) | 1.0 |
| L210W | 15 (9) | 0 | 1 (4) | 2 (6) | 7 (16) | 5 (11) | 0.8 |
| K219Q | 12 (7) | 0 | 3 (12) | 4 (11) | 2 (4) | 3 (7) | 0.2 |
|
Multiple NRTI mutations | |||||||
| 0 | 10 (6) | 0 | 2 (8) | 4 (11) | 2 (4) | 2 (5) | 0.001 |
| 1 | 29 (17) | 9 (39) | 5 (20) | 5 (14) | 4 (9) | 5 (11) | |
| 2 | 42 (24) | 9 (39) | 3 (12) | 5 (14) | 9 (20) | 15 (34) | |
| 3 | 36 (21) | 5 (22) | 7 (28) | 7 (19) | 7 (16) | 10 (23) | |
| 4+ | 58 (33) | 0 | 8 (32) | 15 (42) | 23 (51) | 12 (27) | |
|
Multiple TAMs | |||||||
| 0 | 66 (38) | 21 (91) | 7 (28) | 13 (36) | 6 (13) | 18 (41) | <0.001 |
| 1 | 26 (15) | 2 (9) | 2 (8) | 3 (8) | 12 (27) | 7 (16) | |
| 2 | 38 (22) | 0 | 8 (32) | 12 (33) | 8 (18) | 10 (23) | |
| 3+ | 44 (25) | 0 | 8 (32) | 8 (22) | 19 (42) | 9 (20) | |
Note. NRTI, nucleoside reverse transcriptase inhibitor; TDF, tenofovir; AZT, zidovudine; d4T, stavudine; TAMs, thymidine analog mutations.
Comparison of AZT, d4T and TDF only groups using Pearson’s chi-square and Fisher’s exact test.
Multiple first-line regimens: AZT/d4T refers to switching from AZT to d4T or d4T to AZT; AZT/d4T TDF, refers to switching from AZT or d4T to TDF. One participant switched from TDF to AZT and was not included as a third category for multiple first-line regimens because of the small sample size (n=1).
Forty one percent utilized the TAM 2 pathway and 32% utilized the TAM 1 pathway. Average HIV viral load were lower for TAM 2 as compared to TAM 1 (4.5 vs. 4.9 log10copies/ml, p=0.02). Those who developed TAMs were more likely to be negative for the K65R mutation (70% vs. 12%, p<0.001). D67N and K219E were the TAMs seen with K65R in the three patients who had both. Patients with TAMs had a longer median time on ART (32 months vs. 23 months, p=0.06).
NNRTI and PI Resistance
Among all patients genotyped, 97% patients had at least one NNRTI mutation and 47% had two or more etravirine associated mutations. The most frequent NNRTI mutations were Y181C/V (43%) and K103N (37%). The L100I (36%), K103N (79%), and P225H (21%) mutations were more common in efavirenz based regimens as compared to nevirapine based regimens [Table 3].
Table 3.
Frequency Distribution of Major and Minor NNRTI Mutations
| No. (%) of NNRTI regimen |
|||||
|---|---|---|---|---|---|
| All (N=175) | NVP only (N=131) | EFV only (N=14) | NVP & EFV (N=29) | p-valueb | |
|
NNRTI mutations | |||||
| K103N/S | 67 (38) | 48 (37) | 12 (86) | 6 (21) | 0.001 |
| V108I | 18 (10) | 11 (8) | 2 (14) | 5 (17) | 0.4 |
| Y188L | 6 (3) | 4 (3) | 0 | 2 (7) | 1.00 |
| V106-A/M | 5 (7) | 3 (2) | 1 (7) | 1 (3) | 0.3 |
| P225H | 4 (2) | 0 | 3 (21) | 1 (3) | 0.001 |
|
ETV mutations | |||||
| Y181-C/Vc | 75 (43) | 58 (44) | 4 (29) | 12 (41) | 0.4 |
| G190-A/S | 57 (33) | 39 (30) | 5 (36) | 13 (45) | 0.8 |
| A98G | 40 (23) | 34 (26) | 1 (7) | 5 (17) | 0.2 |
| K101-E/H/P | 36 (21) | 22 (17) | 3 (21) | 11 (38) | 0.7 |
| V90I | 21 (12) | 13 (10) | 2 (14) | 6 (21) | 0.6 |
| E138A/G/K/Q | 16 (9) | 10 (8) | 2 (14) | 4 (14) | 0.3 |
| V106I | 8 (5) | 4 (3) | 0 | 4 (14) | 1.0 |
| L100I | 7 (4) | 0 | 5 (36) | 2 (7) | <0.001 |
| M230L | 4 (2) | 1 (1) | 1 (7) | 2 (7) | 0.2 |
| V179-D/F/T | 0 | 0 | 0 | 0 | |
|
Multiple NNRTI mutations | |||||
| 0 | 6 (3) | 4 (3) | 0 | 2 (7) | 0.1 |
| 1 | 43 (25) | 38 (29) | 1 (7) | 4 (14) | |
| 2 | 79 (45) | 62 (47) | 6 (43) | 10 (34) | |
| 3+ | 47 (27) | 27 (21) | 7 (50) | 13 (45) | |
|
Multiple ETV mutations | |||||
| 0 | 35 (20) | 27 (21) | 2 (14) | 6 (21) | 0.6 |
| 1 | 56 (32) | 46 (35) | 6 (43) | 3 (10) | |
| 2 | 55 (31) | 42 (32) | 3 (21) | 10 (34) | |
| 3+ | 29 (17) | 16 (12) | 3 (21) | 10 (34) | |
|
Phenotypic resistance to second-line NNRTI (ETV)d | |||||
| Susceptible | 27 (15) | 21 (16) | 2 (14) | 4 (14) | 0.03 |
| Intermediate | 131 (75) | 102 (78) | 8 (57) | 20 (69) | |
| Resistant | 17 (10) | 8 (6) | 4 (29) | 5 (17) | |
Note. NNRTI, non-nucleoside reverse transcriptase inhibitor; NVP, nevirapine; EFV, efavirenz; ETV, etravirine.
Bolded mutations signify major mutations for ETV, NVP & EFV.
For comparison between NVP and EFV only groups using Fisher’s exact test.
Y181-I mutation did not occur.
Resistance patterns predicted according to Stanford Database Algorithm v1.2
Three patients had selected IAS PI major mutations (I50V, N83D, I84V and L90M). Secondary mutations related to polymorphisms (I13V (95%), M36I (83%), H69K (82%), V82I (43%) and L63P (28%)) also occurred among those genotyped.
Effect of Mutation Patterns on Second-line Regimen Options
Overall, 41% of patients genotyped did not have an active NRTI option for a second-line regimen. However, the sensitivity for second-line NRTI options varied depending upon the first-line regimen [Figure 1]. Using the WHO 2010 guidelines, tenofovir was fully active for 52–58% of patients who received AZT or d4T; and 39% who received >1 NRTI. The sensitivity did not differ if the multiple first-line regimens included AZT and d4T (38%) or the regimens included AZT or d4T before TDF (39%). However, patients who had tenofovir as a first-line regimen were 100% sensitive to AZT and 70% sensitive to d4T. Furthermore, 70% of patients on tenofovir based regimens had at least two options of NRTIs to include in a second-line regimen compared with 40% on zidovudine based regimens (OR=3.4, p=0.04).
Figure 1.
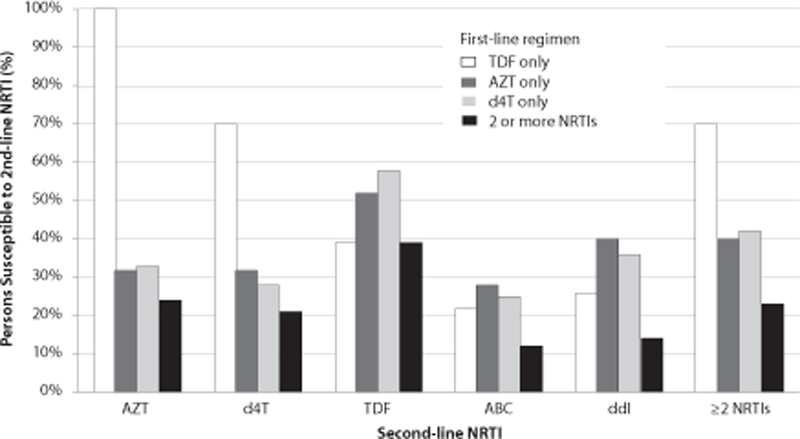
Susceptibility Frequencies of Second-line NRTI Treatment Options.
Overall, 85% of patients genotyped had intermediate to high level resistance to the second-line option, etravirine [Table 3]. For PIs, there was predicted phenotypic resistance to nelfinavir and decreased virological response to tipranavir/ritonavir in 1 patient, as well as predicted intermediate resistance to lopinavir, atazanavir or saquinavir with ritonavir in 2 patients, and to darunavir/ritonavir in 1 patient. There was no documented exposure to PIs in these patients.
DISCUSSION
Tenofovir (TDF) based first-line regimens resulted in significantly fewer NRTI mutations and more fully active NRTI drugs to include in a second-line regimen for this small study of patients at high risk of virologic failure. TDF based first-line regimen retained sensitivity to zidovudine (AZT) and a high proportion retained sensitivity to stavudine (d4T). In comparison, only about half of patients on AZT or d4T retained full sensitivity to their recommended second-line NRTI, TDF because of widespread accumulation of thymidine analogue mutations (TAMs), known to mediate cross resistance to all NRTIs [27].
We report 57% of patients (13/23) on TDF based regimens had K65R, a relatively uncommon mutation (1.7–4%) in viruses of subtype B [28]. K65R is selected by tenofovir, didanosine, stavudine and abacavir [29], but it may be emerging at higher frequencies among non-B subtypes exposed to TDF. Similar prevalences of K65R in TDF based regimens were reported by other studies of non-B subtypes [30,31]. In vitro studies suggest that K65R develops more readily in subtype C because of a site specific pause on the viral template during transcription [32] and this same mechanism may occur for subtypes found in Nigeria. Zidovudine and stavudine remain fully active with K65R, but abacavir and didanosine have reduced activity [33,34]. Tenofovir is partially active and it is enhanced when K65R co-occurs with M184V [35,36]. M184V was the most frequent mutation among TDF based regimens and the only one associated with a lower mean HIV-1 RNA viral load. Viruses with M184V have a decreased replicative capacity and its presence may suggest adherence [37,38]. M184I (7/23) and Y115F (3/23), are relatively uncommon mutations but occurred more often for those on tenofovir. The M184I may eventually switch to M184V and Y115F may be increasing for those on tenofovir, but the small numbers limit our inferences [39,40]. Further studies are needed to evaluate the potential impact of different mutation patterns that are arising in non-B subtypes experiencing selective pressure from tenofovir.
Only 12% of patients with the K65R mutation developed TAMs compared with 70% of those without the K65R mutation. Other studies have shown there is an antagonistic relationship between TAMs and K65R (41, 42). For those with frequent numbers of TAMs, slightly more study patients used the TAM 2 pathway and they had lower average HIV-1 RNA viral loads. Prior studies have shown that the TAM 2 pathway has lower levels of TDF resistance [43], increased sensitivity to AZT with M184V [44], and slower rates of TAM acquisition [45]. Although this augurs well for second-line options, further studies are needed to confirm use of the TAM 2 pathway among non-B subtypes.
High levels of drug resistance in this study mirrors findings in populations who have defined virologic failure with clinical and immunologic parameters rather than viral load testing [45,46,47]. Relying on such measures to define virologic failure increases the number of patients on sub-optimal second-line regimens ultimately leading to poorer treatment outcomes [48,49,50]. It is imperative that viral load testing is conducted in order to maintain effective second-line options in low resource settings.
Limitations of this study include its retrospective nature and its small numbers of patients receiving tenofovir or zidovudine. Larger prospective studies are needed to estimate rates of drug resistance in these first-line regimens. At the time, d4T was a recommended first-line regimen, but PEPFAR and national guidelines promoted substituting tenofovir because of d4T’s associated toxicity issues [17,51,52,53]. This study provided a unique opportunity to evaluate tenofovir as a first-line regimen. Another limitation was relying on predicted resistance patterns from algorithms without confirmation from phenotypic assays [54]. These assays require great resources and may not always be an option in low-resource settings. In addition, there was limited data on baseline mutation patterns, prior ART regimens, adherence, and archived and minority species; all of which could have better described the risk status of this population and whether it biased the mutation patterns observed for tenofovir and the other first-line regimens.
Our data suggest that tenofovir may be an optimal first-line regimen because it maintains susceptibility to thymidine analogue drugs in second-line regimens. Tenofovir has an increased tolerability which may foster adherence, limit multi-drug resistance patterns, and promote long term success for HIV/AIDS treatment programs. However, tenofovir is also associated with an increased frequency of the mutation K65R. The long term effects of this mutation are unclear and ongoing monitoring of virologic failure is critical in guiding selection of NRTIs in resource-limited settings.
Acknowledgements
We would like to acknowledge all patients and staff in the ACTION PEPFAR program at University of Abuja Teaching Hospital and National Hospital Abuja which is funded by Centers of Disease Control under the President’s Emergency Plan for AIDS Relief (PEPFAR). We also acknowledge support from the Nigerian National AIDS/STDs Control Program and National Agency for the Control of AIDS. In particular S. Kennedy, C. Ojinna, I. Mamadu and C. Ezeaku were instrumental for data collection. Financial support specifically for this research study was provided by Pfizer Inc. IIR GA001JZ, and NIH Fogarty 2 D43 TW001041–11. The contents are solely the responsibility of the authors and do not necessarily reflect the views of funding institutions or the United States government.
Funding
Funds for performing drug mutation analysis through genotypic sequencing of archived samples were provided by a Pfizer Investigator Initiated Research [IIR GA001JZ] to Dr. Jean Carr for direct total direct costs of $40,000.
Funding Support
Funds for performing drug mutation analysis was supported by a Pfizer Investigator Initiated Research [IIR GA001JZ] to Dr. Jean Carr for direct total direct costs of $40,000.
Footnotes
Sequence Data
Sequences have been deposited in the GenBank Sequence Database under the following accession numbers: HQ843507-HQ843681
REFERENCES
- 1.http://www.pepfar.gov/about/138312.htm. Accessed November 23, 2010.
- 2.Hosseinipour MC, van Oosterhout JJ, Weigel R, et al. The public health approach to identify antiretroviral therapy failure: high-level nucleoside reverse transcriptase inhibitor resistance among Malawians failing first line antiretroviral therapy. AIDS 2009; 23:1127–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sungkanuparph S, Manosuthi W, Kiertiburanakul S, Piyavong B, Chumpathat N, Chantratita W. Options for the second antiretroviral therapy regimen for HIV-infected patients whose initial regimen of fixed-dose combination of stavudine, lamivudine and nevirapine fails. Clin Infect Dis 2007; 44:447–452. [DOI] [PubMed] [Google Scholar]
- 4.Kumarasamy N, Madhavan V, Venkatesh KK, et al. High frequency of clinically significant mutations after first-line generic highly active antiretroviral therapy failure: implications for second-line options in resource limited settings. Clin Infect Dis 2009; 49:306–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.WHO. Antiretroviral therapy for HIV infection in adults and adolescents: Recommendations for a public health approach (2010 version) Geneva: WHO; 2010. [PubMed] [Google Scholar]
- 6.Miller MD. K65R, TAMS and tenofovir. AIDS Rev 2004; 6(1):22–33. [PubMed] [Google Scholar]
- 7.Federal Ministry of Health N: HIV/Syphilis Sero-Prevalence and STD Syndromes Sentinel Survey among PTB and STD Patients in Nigeria Federal Ministry of Health, Abuja, 2007. [Google Scholar]
- 8.McCutchan FE, Carr JK, Bajani M, Sanders-Buell E, Harry TO, Stoeckli TC, et al. Subtype G and multiple forms of A/G intersubtype recombinant human immunodeficiency virus type 1 in Nigeria. Virology 1999; 254(2):226–34. [DOI] [PubMed] [Google Scholar]
- 9.Abimiku AG, Stern TL, Zwandor A, Markham PD, Calef C, Kyari S, et al. Subgroup G HIV type 1 isolates from Nigeria. AIDS Res Hum Retroviruses 1994. November; 10(11):1581–3. [DOI] [PubMed] [Google Scholar]
- 10.Peeters M, Esu-Williams E, Vergne L, Montavon C, Mulanga-Kabeya C, Harry T, et al. Predominance of subtype A and G HIV type 1 in Nigeria, with geographical differences in their distribution. AIDS Res Hum Retroviruses 2000; 16(4):315–25. [DOI] [PubMed] [Google Scholar]
- 11.FMOH/WHO. Situation of Antiretroviral Drug use in Nigeria November 2003.
- 12.http://data.unaids.org/pub/Report/2010/nigeria_2010_country_progress_report_en.pdf. Accessed November 19, 2010.
- 13.Martínez-Cajas J, Pant-Pi N, Klein M, Wainberg M. Differences in resistance mutations among HIV-1 non-subtype B infections: a systematic review of evidence (1996–2008). Journal of the International AIDS Society 2009, 12:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Novitsky V, Wester W, Degruttola V, Bussman H, Gaseitsiwe S, Thomas A, et al. The Reverse Transcriptase 67N 70R 215Y Genotype Is the Predominant TAM Pathway Associated with Virologic Failure among HIV Type 1C-Infected Adults Treated with ZDV/ddI-Containing HAART in Southern Africa. AIDS Res Hum Retroviruses 2007; 23(7): 868–878. [DOI] [PubMed] [Google Scholar]
- 15.Chaplin B, Eisen G, Idoko J, Onwujekwe D, Idigbe E, Adewole I, et al. Impact of HIV Type 1 Subtype on Drug Resistance Mutations in Nigerian Patients Failing First-Line Therapy. AIDS Res and Hum Retroviruses 2010; 26(11): 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Munerato P, Sucupira MC, Oliveros M, Janini L, de Souza DF, Pereira A, et al. HIV Type 1 Antiretroviral Resistance Mutations in Subtypes B, C, and F in the City of Sao Paulo, Brazil. AIDS Res and Hum Retroviruses 2010; 26 (3): 265–273. [DOI] [PubMed] [Google Scholar]
- 17.Federal Ministry of Health. National Guideline for HIV and AIDS Care in Adolescents and Adults 2nd edition 2007. FMOH; Abuja, Nigeria, May 2007. [Google Scholar]
- 18.Eyzaguirre LM, Brouwer KC, Nadai Y, Patterson TL, Ramos R, Firestone Cruz M, et al. First Molecular Surveillance Report of HIV Type 1 in Injecting Drug Users and Female Sex Workers along the U.S.-Mexico Border. AIDS Res Hum Retroviruses 2007; 23(2):331–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kimura M A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 1980; 16:111–120. [DOI] [PubMed] [Google Scholar]
- 20.Saitou N, and Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 1987; 4:406–425. [DOI] [PubMed] [Google Scholar]
- 21.Lole K, Bollinger R, Paranjape R, Gadkari D, Kulkarni S, Novak N, et al. Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J Virol 1999, 73:152–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boulme R, Gonzalez D and Schmit JC.. Storing genotypic resistance data and linking to other clinical information. Poster presented at the XV International AIDS Conference, Bangkok, Thailand, July 11–16, 2004. [Google Scholar]
- 23.Johnson VA, Calvez V, Gunthard HF, Paredes R, Pillay D, Shafer R, et al. 2011 update of the drug resistance mutations in HIV-1. Top Antivir Med 2011; 19(4):156–164. [PMC free article] [PubMed] [Google Scholar]
- 24.Marcelin A, Delaugerre C, Wirden M, Viegas P, Simon A, Katlama C, et al. Thymidine Analogue Reverse Transcriptase Inhibitors Resistance Mutations Profiles and Association to Other Nucleoside Reverse Transcriptase Inhibitors Resistance Mutations Observed in the Context of Virological Failure. J Med Virol 2004; 72:162–165. [DOI] [PubMed] [Google Scholar]
- 25.Liu TF and Shafer RW: Web resources for HIV type 1 genotypic- resistance test interpretation. Clin Infect Dis 2006; 42:1608–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kantor R, Katzenstein D, Efron B, Carvalho AP, Wynhoven B, Cane P, et al. Impact of HIV-1 Subtype and Antiretroviral Therapy on Protease and Reverse Transcriptase Genotype: Results of a Global Collaboration. PLoS Medicine 2005; 2(4):e112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Whitcomb JM, Parkin NT, Chappey C, Hellman NS, Petropoulos CJ. Broad nucleoside reverse-transcriptase inhibitor cross-resistance in human immunodeficiency virus type 1 clinical isolates. J Infect Dis 2003; 188:992–1000. [DOI] [PubMed] [Google Scholar]
- 28.Brenner BG, Coutsinos D. The K65R mutation in HIV-1 reverse transcriptase: genetic barriers, resistance profile and clinical implications. HIV Ther 2009; 3(6):583–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Margot NA, Isaacson E, McGowan I, Cheng AK, Schooley RT, Miller MD. Genotypic and phenotypic analyses of HIV-1 in antiretroviral-experienced patients treated with tenofovir DF. AIDS 2002. ; 16 :1227–1235. [DOI] [PubMed] [Google Scholar]
- 30.Sunpath H, Wu B, Gordon M, Hampton J, Johnson B, Moosa M, et al. High rate of K65R for ART naive patiets with subtype C HIV infection failing a TDF-containing first-line regimen in South Africa. AIDS 2012. ;Epub ahead of print. [DOI] [PMC free article] [PubMed]
- 31.Hawkins CA, Chaplin B, Idoko J, Ekong E, Adewole I, Gashau W, et al. Clinical and genotypic findings in HIV-infected patients with the K65R mutation failing first-line antiretroviral therapy in Nigeria. J Acquir Immune Defic Syndr 2009. ; 52(2) :228–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Coutsinos D, Invernizzi CF, Moisi D, Oliveira M, Martinez-Cajas JL, Brenner BG, Wainberg MA. A template-dependent dislocation mechanism potentiates K65R reverse transcriptase mutation development in subtype C variants of HIV-1. PLoS One 2011; 6(5):e20208 Epub 2011 May 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gu Z, Fletcher RS, Arts EJ, Wainberg MA, Parnaik MA. The K65R mutant reverse transcriptase of HIV-1 cross-resistant to 2’,3’-dideoxycytidine, 2’3’-dideoxy-3’-thiacytidine, and 2’,3’-dideoxyinosine shows reduced sensitivity to specific dideoxynucleoside triphosphate inhibitors in vitro. J Biol Chem 1994. ; 269(45) :28118–22. [PubMed] [Google Scholar]
- 34.White KL, Chen JM, Feng JY, Margot NA, Ly JK, Ray AS, et al. The K65R reverse transcriptase mutation in HIV-1 reverses the excision phenotype of zidovudine resistance mutations. Antir Ther 2006; 11(2):155–63. [DOI] [PubMed] [Google Scholar]
- 35.Gallant JE, Staszewski S, Pozniak AL, DeJesus E, Suleiman JM, Miller MD, et al. Efficacy and safety of tenofovir DF vs stavudine in combination therapy in antiretroviral-naive patients :a 3-year randomized trial. JAMA 2004; 292 :191–201. [DOI] [PubMed] [Google Scholar]
- 36.Grant PM, Taylor J, Nevins AB, Calvez V, Marcelin AG, Wirden M, Zolopa AR. International cohort analysis of the antiviral activities of zidovudine and tenofovir in the presence of K65R mutation in reverse transcriptase. Antimicrobial Agents and Chemotherapy 2010; 54(4) :1520–1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Castagna A, Danise A, Menoz S, Galli L, Gianotti N, Carini E, et al. Lamivudine monotherapy in HIV-1 infected patients harbouring a lamivudine-resistant virus: A randomized pilot study (E-184V study). AIDS 2006; 20(6):785–803. [DOI] [PubMed] [Google Scholar]
- 38.van Zyl GU, van der Merwe L, Classen M, Zeier M, Preiser W. Antiretroviral resistance patterns and factors associated with resistance in adult patients failing NNRTI-based regimens in the Western Cape, South Africa. J Med Virol 2011; 83:1764–1769. [DOI] [PubMed] [Google Scholar]
- 39.Margot NA, Waters JM, Miller MD. In vitro Human Immunodeficiency Virus Type 1 resistance selections with combinations of tenofovir and emtricitabine or abacavir and lamivudine. Antimicrobial Agents and Chemotherapy 2006; 50(12):4087–4095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wainberg MA, Turner D. Resistance issues with new nucleoside/nucleotide backbone options. J Acquir Immune Defic Syndr 2004; 37:S36–S43. [DOI] [PubMed] [Google Scholar]
- 41.Valer L, Martin-Carbonero L, de Mendoza C, Corral A, Soriano V. Predictors of selection of K65R:tenofovir use and lack of thymidine analogue mutations. AIDS 2004; 18(15):2094–2096. [DOI] [PubMed] [Google Scholar]
- 42.Parikh UM, Barnas DC, Faruki H, Mellors JW. Antagonism between the HIV-1 reverse transcriptase mutation K65R and thymidine analogue mutations at the genomic level. J Infect Dis 2006; 194(5):651–660. [DOI] [PubMed] [Google Scholar]
- 43.Miller MD, Margot N, Lu B, Zhong L, Chen SS, Cheng A, et al. Genotypic and phenotypic predictors of the magnitude of response to tenofovir disproxil fumarate treatment in antiretroviral-experienced patients. J Infect Dis 2004; 189(5):837–46. [DOI] [PubMed] [Google Scholar]
- 44.Luca De, Di Giambenedetto S, Romano L, Gonnelli A, Corsi P, Baldari M, et al. Frequency and treatment related predictors of thymidine analogue mutation patterns in HIV-1 isolates after unsuccessful antiretroviral therapy. J Infect Dis 2006; 193(9):1219–22. [DOI] [PubMed] [Google Scholar]
- 45.Cozzi-Lepri A, Phillips A, Martinez-Picado J, d’Arminio Monforte A, Katlama C, Hansen A, et al. for the EuroSIDA Study Group. Rate of Accumulation of Thymidine Analogue Mutations in Patients Continuing to Receive Virologically Failing Regimens Containing Zidovudine or Stavudine: Implications for Antiretroviral Therapy Programs in Resource-Limited Settings. J Infect Dis 2009; 200: 687–697. [DOI] [PubMed] [Google Scholar]
- 46.Lyagoba F, Dunn DT, Pillay D, Kityo C, Robertson V, Tugume S, et al. ; DART Virology and Trial Team. Evolution of drug resistance during 48 weeks of zidovudine/lamivudine/tenofovir in the absence of real-time viral load monitoring. J Acquir Immune Defic Syndr 2010; 55(2):277–83. [DOI] [PubMed] [Google Scholar]
- 47.Ndembi N, Goodall RL, Dunn DT, McCormick A, Burke A, Lyagoba F, et al. Development of Antiretroviral Treatment in Africa Virology Group and Trial Team. Viral rebound and emergence of drug resistance in the absence of viral load testing: a randomized comparison between zidovudine-lamivudine plus Nevirapine and zidovudine-lamivudine plus Abacavir. J Infect Dis 2010; 201(1):106–13. [DOI] [PubMed] [Google Scholar]
- 48.Doualla-Bell F, Gaolathe T, Avalos A, Cloutier S, Ndwapi N, Holcroft C, et al. Five-year follow up of genotypic resistance patterns in HIV-1 subtype C infected patients in Botswana after failure of thymidine analogue-based regimens. J Int AIDS Soc 2009; 12(1):25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chetchotisakd P, Anunnatsiri S, Mootsikapun P, Kiertiburanakul S, Anekthananon T, Bowonwatanuwong C, et al. ; Study Team. Efficacy and tolerability of a double boosted protease inhibitor (lopinavir + saquinavir/ritonavir) regimen in HIV-infected patients who failed treatment with nonnucleoside reverse transcriptase inhibitors. HIV Medicine 2007; 8(8):529–35. [DOI] [PubMed] [Google Scholar]
- 50.Rodrıguez M, O’Brien D, Humblet P and Calmy A. Second-line antiretroviral therapy in resource-limited settings: the experience of Medecins Sans Frontieres. AIDS 2008, 22:1305–1312. [DOI] [PubMed] [Google Scholar]
- 51.Laurent C, Bourgeois A, Mpoudi-Ngolé E, Ciaffi L, Kouanfack C, Mougnutou R, et al. Tolerability and effectiveness of first-line regimens combining nevirapine and lamivudine plus zidovudine or stavudine in Cameroon. AIDS Res Hum Retroviruses 2008; 24(3):393–9. [DOI] [PubMed] [Google Scholar]
- 52.Hawkins C, Achenbach C, Fryda W, Ngare D, Murphy R. Antiretroviral durability and tolerability in HIV-infected adults living in urban Kenya. J Acquir Immune Defic Syndr 2007; 45(3):304–10. [DOI] [PubMed] [Google Scholar]
- 53.Shepherd J, Habib A, Charurat M, Mondal P, Falajayo K, Abimiku A, Dakum P, et al. ACTION Project Study Group. Efficacy and tolerability of alternative first-line antiretroviral drug regimens in Nigeria. 4th IAS Conference on HIV Pathogenesis, Treatment and Prevention. 22–25 July 2007, Sydney, Australia. [Google Scholar]
- 54.Snoeck J, Kantor R, Shafer RW. Vandamme AM. Discordances between Interpretation Algorithms for Genotypic Resistance to Protease and Reverse Transcriptase Inhibitors of Human immunodeficency Virus are Subtype Dependent. Antimicrobial Agents and Chemotherapy 2006; 50(2): 694–701. [DOI] [PMC free article] [PubMed] [Google Scholar]


