Abstract
Self‐organization is a process by which interacting cells organize and arrange themselves in higher order structures and patterns. To achieve this, cells must have molecular mechanisms to sense their complex local environment and interpret it to respond accordingly. A combination of cell‐intrinsic and cell‐extrinsic cues are decoded by the single cells dictating their behaviour, their differentiation and symmetry‐breaking potential driving development, tissue remodeling and regenerative processes. A unifying property of these self‐organized pattern‐forming systems is the importance of fluctuations, cell‐to‐cell variability, or noise. Cell‐to‐cell variability is an inherent and emergent property of populations of cells that maximize the population performance instead of the individual cell, providing tissues the flexibility to develop and maintain homeostasis in diverse environments. In this review, we will explore the role of self‐organization and cell‐to‐cell variability as fundamental properties of multicellularity—and the requisite of single‐cell resolution for its understanding. Moreover, we will analyze how single cells generate emergent multicellular dynamics observed at the tissue level ‘travelling’ across different scales: spatial, temporal and functional.
Keywords: cell‐to‐cell variability, crossing‐scales technologies, development, emergent properties, multicellularity, organoids, pattern formation, regeneration, self‐organization, symmetry‐breaking
Abbreviations
- ESCs
embryonic stem cells
- PI
phosphatidylinositol
- PSM
presomatic mesoderm
Self‐organization during tissue formation, homeostasis and regeneration
Multicellular organisms are composed of cells and tissues with identical genomes but different properties and functions. They all develop from one cell toward multicellular structures of great complexity. On a series of carefully organized steps in space and time, different cell types, architectures, and functions are formed during embryogenesis and development. In adult life, maintaining tissue homeostasis, via periodical tissue renewal and regenerative processes, also requires spatio‐temporal coordination of cells to ensure tissue function and integrity. Moreover, the malfunction of these coordinated behaviours during embryogenesis is the cause of many congenital disorders and their deregulation during adult life in actively proliferating and regenerating tissues, such as the intestine, is the basis of many cancers 1, 2.
These spatio‐temporally organized processes in multicellular organisms are known as collective behaviours. In the early steps of embryogenesis, cells in a seemingly symmetric embryo reorganize themselves collectively into a patterned arrangement giving rise to primitive tissue specification 3, 4. Later into organogenesis, the majority of multipotent dividing cells commits to differentiation and acquires specific functionalities, while only a fraction retains stemness 5. Once development is complete, cells in a tissue must be able to sense organ size and functionality to stop proliferating, and yet, in some cases, retain a minimal level of stemness for homeostasis and a potential to regenerate upon injury 4, 6.
Although the molecular machineries governing these processes are determined genetically, thus making them precise and reproducible, the genome alone does not encode for the complex cellular interactions required to keep them robust and contextual in dynamic environments. To understand biological processes such as development and regeneration, we must understand how a group of individual cells organize themselves into patterns and tissues 4, 7, 8. In many developing organisms, the patterns are driven and maintained by concentration gradients of signals, named morphogens, and each individual cell in the tissue senses its position along the morphogen gradient and responds accordingly 8. Generally, morphogens are released from a local, but dynamic source, and the gradient shape is determined by the flux from the source, the spreading of the morphogen (e.g., composition of the extracellular matrix and transcytosis) 9 and its degradation in the target tissue. Interestingly, morphogen gradients, downstream signaling, and the activity of cell‐intrinsic gene networks respond dynamically to the local environment by sensing complex extracellular cues 8. This means that the precision and robustness of pattern‐forming systems requires not only pre‐existing morphogens but also spatio‐temporally coordinated self‐organized processes (Fig. 1A).
Figure 1.
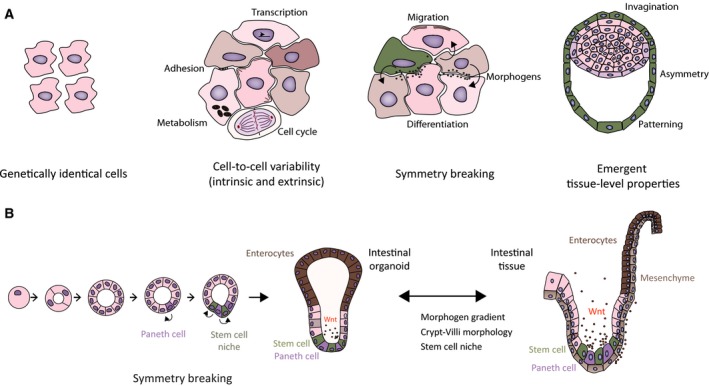
From a population of single cells to tissues. (A) Heterogeneous population of cells where each single cell senses a combination of cell‐intrinsic and cell‐extrinsic cues, ultimately driving tissue patterning. (B) Spatio‐temporal variability and symmetry breaking in intestinal organoids. An intestinal stem cell develops into a symmetrical cyst and undergoes symmetry breaking with the appearance of Paneth cell. This Paneth cell defines the position of the nascent crypt where the stem cell niche will reside. The intestinal organoid develops into a self‐organized structure containing different cell types distributed in a zonated manner recapitulating part of intestinal patterning.
Self‐organization is a process in which interacting entities organize and order themselves in global and larger scale patterns 10, 11. Order appears not because it has been planned by a central controller but because local interactions between individual cells generate complex functional patterns such as tissues and organisms. At the individual level, no cell knows the complexity of the overall structure. Self‐organization is not restricted to developmental processes. In adult organisms, the regeneration of tissues is also an emergent self‐organized property of cells. After an injury, local interactions between different cells drive the healing and repair of the tissue without any single cell knowing how the final tissue should look like at the global scale. To achieve these coordinated and self‐organized process, each individual cell has molecular mechanisms to sense its local environment and respond correctly to injuries, recreating a healthy tissue 10.
In this review, we explore the role of self‐organization and cell‐to‐cell variability as fundamental properties of multicellularity. We discuss cellular mechanisms by which single cells sense their local environment in a multicellular system driving collective behaviours during developmental and regenerative processes. Finally, we provide an overview on current technologies that cross‐scales at the spatial, temporal, and functional level to bridge the gap between single cells and organized tissues.
Sensing mechanisms in a population of interacting single cells
An important question during development is how does a single cell in a tissue sense its complex environmental cues and take individual cellular decisions generating robust and reproducible emergent properties at the population and tissue level?
Number of cells and cell‐packing effects
Eukaryotes evolved many different ways to sense the number of individual cells in a population including global and local mechanisms. One mechanism to generate a defined number of cells in a population is to count the number of cell divisions, as described in Mid Blastula transition in Xenopus laevis 12 and in mammalian hematopoietic stem cells 13. Another way is to rely on chemical information, such as a signal that is secreted locally and sensed globally by other individuals in the population. Morphogen gradient is a conserved strategy in different animals: from the simple counting peptide in Dictyostelium discoideum (secreting a ‘cell‐counting’ factor) 14, toward Dpp gradient in wing tissue development in flies 15, 16 and Wnt3a gradient along the mouse intestinal stem cell niche 17. In addition, a classical environment sensing mechanism that operates at local scale is contact inhibition. MDCK cells, in vitro, have been shown to compute local information on cell density, motility, and cell division rates to trigger contact inhibition 18. Many other mechanisms have been identified in regulating contact inhibition such as increased Clusterin secretion 19 and E‐cadherin‐mediated control of cell proliferation via cell‐cell contact 20.
Cells can also sense and transduce extrinsic physical cues from the microenvironment such as cell‐packing effects. This mechanosensing capability relies on membrane tension sensing pathways such as Yap1 21, 22, Piezo 23, 24 and Misshapen‐Yorkie pathway 25. Understanding how single cells perceive tissue size and function is also essential for growth termination and for successful completion of developmental and regenerative processes. Tissue damage triggers activation of stem cell division and differentiation to replenish lost cells, but this activation must be timely repressed once tissue integrity is restored to limit tissue hyperplasia 26, 27.
Localized signals
Lumen formation is also an important mechanism that cells use to measure their environment by locally restricting and, thereby, coordinating communication between selected groups of cells. In zebrafish lateral line development, the formation of microlumens in a population of migrating cells restricts and enhances FGF signaling only in cells limiting the lumen 28. This increased signaling halts migration and leads to the formation of stable organ precursors 28.
The environmental sensing machineries exemplified above are subjected to cell‐intrinsic cellular states. For example, some cells may present maximal responsiveness to extracellular signals depending on their cell cycle position, rather than to an increased exposure to the signals 29. This regulates how the cell responds to extrinsic cues determining individual cellular behaviours such as secretion of molecules 30, apical constriction 5, counting proliferation rounds 13, symmetric and asymmetric cell divisions 31, adhesion 32, migration 33, and differentiation34. The combination of intrinsic and extrinsic cues establishes positive and negative feedback loops that move the entire population to a new state generating complex architectures. For example, neuronal development requires a period of extensive proliferation of progenitor cells followed by a switch to asymmetric division and differentiation when the population of progenitors has reached the correct size 31, 35, 36. The balance between these two processes in different regions of the nervous system and in different organisms gives rise to differential growth, cellular diversification, and diverse brain structures during evolution 37. Interestingly, in mammals, cell cycle length 38 (particularly the length of G1 phase 39, 40, 41), influences the decision to terminally differentiate. However, it is still unclear if this could be a mechanism of sensing the population size and how progenitor cells would regulate their cell cycle length. Also, during regeneration, especially in Hydra and Planarians, a minimum tissue size and a certain minimum cell number is shown to be required for regeneration and patterning 42, 43.
The mechanisms by which single cells sense their local environments and implement it at the population level are the driving forces of self‐organization and collective behaviours during development, tissue remodeling and regenerative processes.
Symmetry breaking
A defining step during self‐organization and pattern formation is the first moment when initially identical cells in a developing organism or tissue differentiate, and lineage segregation is established. For the mechanistic understanding of this step, symmetry breaking is a key concept. Precisely, the symmetry‐breaking event occurs when, despite all cells being exposed to a uniform growth‐promoting environment, only a fraction of cells becomes activated, differentiates and acquires new functions. This process is called symmetry ‘breaking’ because the transitions usually bring the system from a symmetric, but disordered and variable state, into one or more defined, less variable and asymmetric states (e.g. differentiated states) 44. Symmetry breaking correlates with functional specialization 45 and diversification across different scales: from molecular assemblies, to cell type specification, tissue organization and whole body axis formation.
At the single‐cell level in a tissue, a multitude of reactions and signaling pathways takes place continuously. Each step (e.g., cell cycle progression, metabolism, adhesion and migration), is decided based on an integrated response of computed signals (Fig. 1A). Once a given combinatorial threshold of cell‐intrinsic and cell‐extrinsic signals is reached, a substantial change in behaviour is observed. This change in behaviour can be defined as a newly acquired functionality: a cell which differentiates, starts secreting a molecule or changes its shape, triggers a cascade of effects which moves the entire population to a new state.
Often, cell polarity is the initial building block determining asymmetries at the tissue and body levels. At early stages of mouse embryogenesis, for example, it was demonstrated that the trophectoderm fate is based on differential inheritance of a cell's apical domain 46, 47. Also, the left‐right axis in vertebrates is determined by the polarization and orientation of nodal cilia and molecularly dictated by the chiral nature of molecular motor 48. The directionality of the cilia rotation induces a specific flow of extracellular fluid that, in turn, determines the left‐right body axis properties such as the positioning of the heart 49, a conserved feature of fish 50, frog 51, mouse 52 and humans 53. Similarly, an epithelial cell undergoing apical constriction acts as a nucleator for a pattern of negative straining at the tissue surface driving invagination in embryogenesis or development 5, 54, 55. An essential step during Drosophila development is the ventral furrow formation. It results from apical constriction of a few cells along the ventral side of the embryo leading to invagination and movement of mesoderm into the embryo during gastrulation 3, 56. The initial polarity events occur at the molecular and cellular level and trigger the upcoming patterns at the whole tissue level.
Another example of symmetry breaking happens during intestinal organoid formation, where a single intestinal LGR5+ stem cell cultured in Matrigel in the presence of Wnt3a, EGF, Noggin and R‐spondin 57 is able to generate a fully grown organoid (Fig. 1B). First, a symmetrical cyst‐like structure is formed. Then, the first Paneth cell emerges showing hallmarks of active Wnt signaling and determining the future crypt budding sites 58. The emergence of a Paneth cell is the first symmetry‐breaking event. After that, Wnt3a removal from the medium generates local gradients of Wnt3a around the activated Paneth cells within the cyst, inducing the formation of the stem cell niche and, later on, of the intestinal crypt that maintains itself due to positive feedback mechanisms. The paradoxical initial stage, where all cells in the growing cyst are exposed to a uniform growth‐promoting environment but only a fraction becomes activated and differentiates into Paneth cells is still poorly explored 59.
Defining which combination of signals determines the behaviour and interactions of individual cells is important to understand how self‐organized patterns emerge. To model and built theoretical frameworks of such complex mechanisms we need to have access to multivariate information of single cells, including cell cycle phase, signaling pathways, metabolic status, and mechanical properties. The complexity of such multidimensional molecular and cellular interactions has made it difficult to explain, which signals underlie symmetry breaking in a given cell and how complex behaviours emerge.
Cell‐to‐cell variability
Given the fundamental importance of self‐organization, symmetry breaking, and pattern formation in multicellular systems 11, 60, several experimental and theoretical frameworks have been used such as Turing's reaction‐diffusion systems 61, 62, Notch/Delta lateral inhibition and agent‐based models 63, 64. A unifying property of these pattern‐forming theories is the importance of fluctuations, cell‐to‐cell variability and feedback loops 65, 66, 67, 68, 69. In all these systems, an initial cell‐to‐cell variability in a population of cells is then amplified and stabilized by positive and negative feedback loops. The extent of the initial variability and the boundary conditions can confer different steady‐state patterns and robustness to the system. In this review, we will not discuss in length the different pattern‐forming theories, but we will explore the role of the initial heterogeneity in the system.
Cell‐to‐cell variability is an inherent and emergent property of populations of cells. It refers to the phenomenon that no two genetically identical cells behave and look identical 70, 71, 72. This difference may arise from the inherently stochastic and discrete nature of intracellular biochemical reactions, especially when these reactions involve low numbers of molecules. Generally though, robustness in molecular mechanisms 73, 74, 75, 76 can buffer the intrinsic stochasticity of molecular processes 77 while other factors, such as the cell cycle and the microenvironment, can explain the cellular heterogeneity, especially in eukaryotes 78, 79. One major extrinsic factor determining cell‐to‐cell variability is the microenvironment of individual cells. Even in environmentally controlled cell culture conditions, a growing population of adherent cells will continuously experience changing microenvironments as a consequence of an increase in cell number combined with cell adhesion and migration 79. Another significant source of cell‐to‐cell variability in an unsynchronized population of cells is cell cycle phase 29. In fact, when considering cell cycle and microenvironment, much of the unexplained variability in different molecular readouts can be deconvoluted and correlated with local population contexts such as cells being in same cell cycle phases or whether a cell is in a more or less crowded environment 79, 80, 81, 82.
Extensive cell‐to‐cell variability has been shown for key molecular components involved in embryogenesis in the early mouse embryo 83 and in embryonic stem cells 84. For example, Nanog, a key transcription factor for the maintenance of pluripotency, exhibits large variability between cells in the early mouse embryo 85 and populations of undifferentiated embryonic stem cells 86. This variability in Nanog expression has been linked to cell cycle phase, reaching highest expression during G1/S transition 87. Another example is the extensive heterogeneity in expression of Oct4 target genes at the 4‐cell embryonic stage 88, 89. This variability might confer an initial metastable state to a subpopulation of cells with a fluctuating transcriptome that drives the reversible priming of pluripotent cells toward different cell fate decisions. In fact, if populations of pluripotent cells would be uniform in cellular activities, we would expect an ‘all‐or‐none’ response in a homogenous environment, with a single critical threshold below which all cells remain undifferentiated and above which all cells differentiate. A graded response is conceivable only in the presence of initial cell‐to‐cell variability making it possible to control the rate of differentiation at low homogenous stimuli concentrations 67, 90 (Fig. 2). Cell‐to‐cell variability in key molecular components confers to a small fraction of cells an increased probability to break the symmetry and transition to an activated or differentiated state, making stemness and pluripotency not a property of a single cell but a global and statistical property of a population of cells that are able to self‐renew and differentiate 91, 92, 93. This is a particularly important concept to understand the dynamics of stem cell populations 94. A heterogeneous population of cells with different potencies to perform as stem cell is clearly advantageous, as it provides flexibility and easier adaptability to changing environmental conditions. Variability helps to maximize the population performance instead of that of the individual cell. And finally, it is the control of the stemness potential of a given population which provides tissues the flexibility to maintain homeostasis 92.
Figure 2.
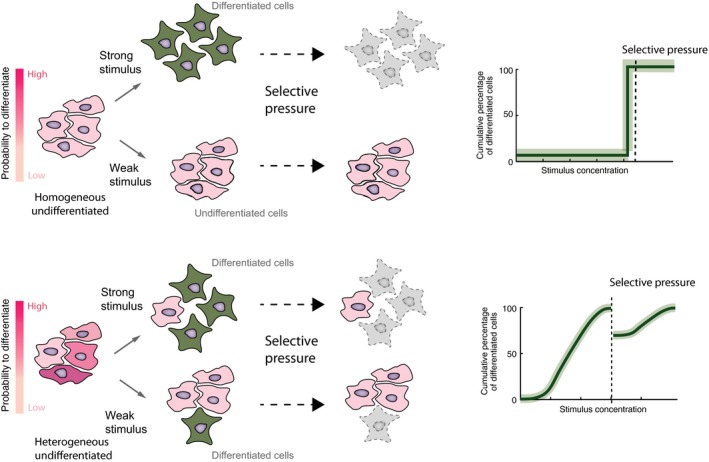
Cell‐to‐cell variability is an advantageous property of a population of stem cells. A population of stem cells which are uniform in their cellular activities, respond to a stimuli in an ‘all‐or‐none’ manner, with a critical threshold below which all cells remain undifferentiated and above which all cells differentiate (upper panel). A graded response is observed in the presence of cell‐to‐cell variability making it possible to control the rate of differentiation according to stimuli concentrations (lower panel). Variability provides adaptability to selective pressure (right side). In a homogeneous scenario, an environmental challenge results in poor population performance, while a heterogeneous population is more robust to the selective pressure, allowing the survival of some individual cells.
Measuring single‐cell behaviour beyond high abundant genes in transcriptomics for whole cell populations in vivo is still challenging. Hence, comprehensive understanding of the extent and sources of cell‐to‐cell variability for different cellular processes and how variability affects in vivo self‐organization, patterning and multicellular programming of cells is sparse 39, 95, 96. One important question is: what is the minimal amount of information required at the single‐cell level to understand molecularly an emergent pattern at the tissue level? It is probably not necessary to follow every single molecular player of every cell over the course of hours or days to describe emergent properties at a higher scale such as development or regeneration processes. With sufficient single cell data of key signaling pathways, gene regulatory networks and positional information, we might be able to predict interactions and infer causal relations between fluctuating cellular activities and the emergence of a pattern over time 44, 79, 80, 81, 89, 97, 98, 99. Ultimately, understanding self‐organization and symmetry breaking in multicellular systems is a problem across scales. To explain with sufficient detail the multicellular dynamic interactions that govern a self‐organized process, the field is moving into developing technologies across scales which combine three essential elements: single cell resolution, temporal resolution, and tissue functionality.
Scale‐crossing technologies
To quantitate and model the population‐level properties of a large group of interacting cells, such as in organogenesis and tissue regeneration, and understand how such properties arise from single cells, we need an experimental framework combining multivariate single‐cell techniques and traceability of spatio‐temporally dynamical problems. Therefore, to explain with sufficient details the multicellular dynamic interactions that govern a self‐organized process, we need scale‐crossing technologies linking three essential elements: multiple simultaneous measurements at single‐cell resolution, temporal resolution accommodating short and long responses, and distinctive quantifiable emergent tissue functionalities (Fig. 3).
Figure 3.
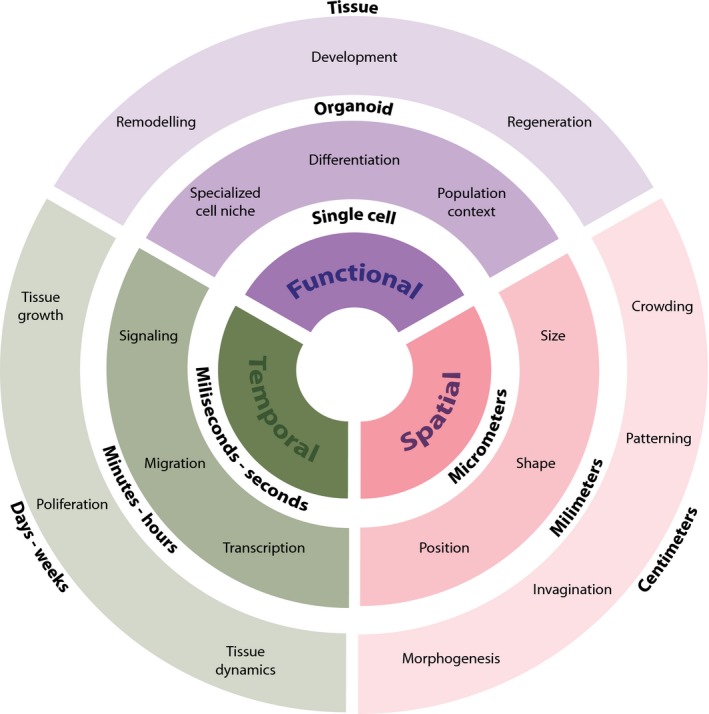
Scale‐crossing technologies required for understanding self‐organization. Different experimental frameworks are required to quantitate and model the population‐level properties of a large group of interacting cells during self‐organized processes. Scale‐crossing technologies described in the text are able to link functional, spatial and temporal scales. Detailed information at each level of these scales, from single cells to tissues, will help to explain the multicellular dynamic interactions that govern a self‐organized process.
An all‐inclusive tool capable of multiplexing single‐cell measurements on a spatio‐temporally resolved scale is still unavailable. We must rely on combinations of advanced imaging, single‐cell ‘omics’ and functional assays as complementary approaches for describing population dynamics at the cellular level. In this final section, we present the available technologies to gain quantitative understanding on the pursuit of self‐organization and emergent properties in multicellular arrangements.
Spatial scale
Spatially, the scales that need to be bridged are from the subcellular resolution (low micrometer range of organelles and cells) to the tissue organization (ranging from millimeters to centimeters) combining multivariate measurements at both scales. Ideally, we would need information on the genome accessibility, mRNA and protein abundance and localization, combined with the phenotypic state of each single cell (such as cell size and shape, cell cycle, signaling, and metabolic state) with spatial localization. At the tissue level, informative measurements of morphological features (size, shape, and curvature), mechanical forces (compactness, pressure, tension, and traction) and functional readouts (morphogen secretion in a niche, organ‐like structures such as hair‐follicle or intestinal crypts) are required as a final outcome of the self‐organized process. Among the different available techniques to obtain spatial information from a tissue at single‐cell resolution, fluorescent light microscopy is the most versatile. With optical sectioning methods such as confocal and light sheet imaging 100 cellular details and general architecture of complex structures can be visualized across the spatial scale: from differential expression of transcripts in neighboring cells 101, toward proteins abundances and specification of different cell types in different organs 102, 103, 104, up to mechanics of tissue folding in development 3, 105, 106. One of the major limitations in tissue and whole animal imaging is sample opacity. Several approaches have been used to overcome it known as tissue clearing methods (for an overview, see 107, 108) and recent developments have enabled whole tissue and animal imaging at the single cell resolution104, 109. Visualizing specific subcellular structures and compartments with fluorescence microscopy has been historically limited by the diffraction limit, the phototoxicity and the number of different fluorophores that can be imaged at once. Now advances on image analysis and antibody multiplexing have recently broadened the spectrum of detection. One of the possibilities is to minimize fluorescence spectral overlap by integrating high‐resolution confocal microscopy with an imaging analysis pipeline which separates up to six fluorophores simultaneously 110. This approach has been applied to mapping dynamic inter‐organelle interactions in live cells with six different organelle‐specific fusion proteins, representing an important tool for investigating organelle spatial organization during different cellular processes. When applied to multicellular 3D structures, it will be necessary to contextualize multiple subcellular readouts with tissue organization. Other alternatives to increase number of readouts in imaging have been developed based on cyclic rounds of antibody staining with chemical inactivation of fluorescence 111, 112 or more recently with sequential antibody elution and stripping, a method called iterative indirect immunofluorescence imaging (4i) 113. The latest allows multiplexing of up to 40 fluorescent molecular readouts inside every single cell in fixed samples with complete sample preservation and unprecedented high spatial resolution. With such an approach, it is possible to monitor different molecular activities on a 3D tissue at single cell level resolution combining, for example, cell cycle reporters, cell type markers, signaling pathways and the cellular microenvironment.
Another method for phenotypic characterization with spatial resolution is imaging mass cytometry 114, 115. It is based on antibodies tagged with metal isotopes allowing quantitation of dozens of proteins simultaneously in individual cells in situ. Because of its high parameterization, it identified heterogeneities at single‐cell level in subpopulations of cells in cancer samples 114, 116. This method, however, relies on tissue sectioning and laser ablation of the sample, and is currently not compatible with a full 3D characterization of the tissue sample.
Spatially resolved data can also be obtained for gene transcript levels by fluorescence in situ hybridization (FISH) and its multiplexed versions such as seqFISH 117, 118 and MERFISH 101. They provide subcellular localization of thousands of RNA species in single cells simultaneously while preserving spatial population context 119. These RNA imaging techniques have identified transcriptionally distinct cells in situ with important applications for characterizing the expression signature of tissues. In the intestine, for example, it has been used to map endogenous markers of intestinal stem cells like Lgr5, Bmi1, and mTert 120 and follow them under different physiological conditions. And by combining spatial information of selected transcripts with whole transcriptome measurements of dissociated cells, it is possible to spatially reconstruct the expression profiles of cells in a tissue coinciding with metabolic cascades and functional zonation as shown in liver 103 and intestine 102.
Besides gene and protein expression, an important aspect to consider during multicellular organization is the metabolic state of cells. Different metabolic identities are adopted during tissue development, homeostasis or disease progression 121, 122, 123, 124. The transition of embryonic stem cells from naïve to primed, for example, is accompanied by a metabolic shift toward a predominantly glycolytic state, and as differentiation progresses, toward a highly respiring mitochondrial state 125. Exploiting a shift in metabolic activities is also observed during organogenesis, where a gradient of glycolytic pattern in the presomitic mesoderm is responsible for coordinating FGF and Wnt signaling during body elongation 126. Later in homeostasis, this has been observed in the intestine, where neighboring cells at the proliferative niche present metabolically distinct identities (yet, complementary functions) with Paneth cells being more glycolytic and providing lactate as a fuel for the oxidative stem cells 127. The relationship between metabolic transitions and morphogenesis seems to hold true also during cancer development, where a zonated glycolytic signature is adopted upon low oxygen input, being a target for therapies 124. High coverage techniques to assess metabolites, like lipidomics and metabolomics, share the same limitations of transcriptomics and proteomics: insufficient spatial resolution and endpoint measurements. An exception being MALDI imaging, where it is possible to identify multiple analytes, including proteins, lipids, and small metabolites, while keeping positional information. However, spatial resolution of MALDI imaging is still limited at the micrometer range 128. Transitions in cellular metabolism are emerging as determinants of cell differentiation and tissue development 122, 129, 130, 131, 132, and learning about the mechanisms driving the onset of these transitions at the single‐cell level certainly will contribute to the understanding of symmetry breaking and self‐organization in multicellularity.
Temporal scale
As discussed before, imaging has the great advantage of combining functional and structural information simultaneously. Self‐organized events leading to multicellular patterns are long‐term dynamic molecular processes occurring over hours, and observing them in living cells in situ requires a temporal layer of information. Again, the different scales are important because we need to record short‐term events such as signaling activation, protein translocations and cell cycle dynamics with long‐term patterning events at tissue scale such as tissue invagination and crypt development (ranging from milliseconds to days). This power to observe dynamical processes over scales enables us to infer causal relationships between molecular mechanisms. Moreover, the ability to use light‐induced manipulation allows to challenge the system and to test experimentally the inferred causal interactions. High‐resolution live cell imaging and optogenetics tools are becoming fundamental in understanding the dynamics of several biological processes and is the current frontier of imaging development 133, 134, 135. Despite number of readouts still being limited by reporters, lasers and filters, the constant advances on temporal and spatial resolution of live fluorescent imaging represents a promising platform for the study of self‐organizing events 136, 137.
Real time
In toto imaging techniques, such as light‐sheet microscopy, enable long‐term live imaging of developing or regenerating tissues and animals with single‐cell resolution 133, 136, 138, 139, 140, 141, opening an extraordinary window to the complexity of living systems. However, observing full development of embryos or having access to complete tissues is not universally applicable to all specimens. Human development, for example, cannot be experimentally studied beyond pre‐implantation stages in vivo 97, 142, 143, 144, an alternative being in vitro culturing in the absence of maternal tissues 97, the study of foetuses 145 or explanted organs 146. Adult model systems, such as mouse, can be fully genetically manipulated but cannot be immobilized for imaging longer than a couple of hours. For the moment, studying self‐organization with in toto imaging during development is performed on embryonic bodies 89, 123, gastruloids 147, organoids 148 or small‐scale animals. Whole animals such as Hydra 42, 149 or C. elegans 150 can be imaged during their entire development, while larger organisms such as D. rerio 151 or D. melanogaster 152 provide fundamental insights into embryonic development. These model systems are highly informative for studying evolutionary conserved mechanisms, despite limitations in their multicellular complexity and tissue functionality. Another aspect to consider is that light sheet microscopy, just as any other fluorescence imaging techniques, suffers from effects of scattering and absorption, which poses technical challenges in deep tissue imaging 133.
Imaging whole animals thorough development is not an absolute requirement for understanding self‐organization in multicellular systems and much can be learned about the molecular mechanisms behind tissue growth and organization, for example, by monitoring the dynamics of live tissue segments. Drosophila wing disc formation and ventral furrow invagination are great models for studying epithelial morphogenesis, where mechanical factors determining cell shape, division rates, and intercellular tension can be assessed with high temporal resolution 3, 16. Similarly, oscillations in the mouse presomatic mesoderm (PSM) can be visualized as wave‐like patterns of signaling and manipulated on the embryos’ tail bud 98, 123, 126, 153 or with PSM‐like tissues 154.
With a high spatio‐temporal resolution and fluorescent reporters, time‐lapse microscopy is a powerful technique to infer causal links between cellular events leading to patterning. In a lineage tracing analysis of stem cell dynamics during epidermal homeostasis 155, a highly coordinated collective behaviour of stem cells during hair‐follicle formation has been shown. Similarly, in intestinal crypts continuous intravital imaging following short‐term dynamics of intestinal stem cells 156confirmed that stemness is a function of a heterogeneous cell population rather than of a single stem cell 157, 158. On the mechanical side, real‐time imaging is an important tool to understand how variability in physical properties of cells in a tissue might drive multicellular patterning 159 and how tissues which are normally robust in their architectures can also be remodelled during regeneration 32, 137.
One important aspect in understanding temporal scales is the ability to manipulate the system in a spatially and temporally controlled manner. Light‐induced manipulations have been largely improved in spatial and temporal control with the advancements of optogenetics 54, 160. This allows, for example, to understand the importance of timing in signaling pathways involved in cell fate specification during tissue formation. Using a light‐induced regulation of phosphatidylinositol(4,5)P2 levels at the plasma membrane of Drosophila embryonic cells, Guglielmi et al. 54 showed that local modulation of cell contractility interferes with tissue contraction and invagination. Also by manipulating the duration of Nodal signaling during zebrafish embryogenesis using a photoactivatable receptor, it has been shown that extended Nodal signaling drives prechordal plate specification at the expense of endoderm differentiation 161 in a process that depends on long‐lasting cell‐to‐cell contacts 162. In tissue morphogenesis, temporal manipulation of signaling pathways which promote cellular contractility, such as RhoA 163, or phosphatidylinositol(4,5)P2 levels at the plasma membrane 54, allows local modulation of mechanical forces at the cellular level.
Pseudotime inference of molecular events
Time‐lapse imaging is currently the main approach by which spatial and temporal scales can be monitored. However, number of samples that can simultaneously be acquired is limited. Temporal information can also be inferred with computational methods using thousands of samples simultaneously. Resolving molecular and cellular processes pseudotemporally can be achieved by reconstructing time‐series of events based on information from numerous fixed time points 80, 164, 165, 166. With single cell transcriptomics, for example, it is possible to resolve differentiation from stem cells to functionally committed progenies 167, 168, 169. The temporal cascade of events can be inferred computationally based on gradual differences within the population capturing a pseudotemporal trajectory. Recently, a whole embryo developmental landscape has been reconstructed based on scRNAseq data 170, 171, describing cells transitioning from pluripotency to different cell types during early zebrafish embryogenesis. This is a useful way to explore high content expression information overcoming the current technical limitations on temporal scale. While differentiation trajectories from transcriptomics can provide a high content molecular picture, imaging and mass cytometry data from fixed samples can also be used to reconstruct trajectories both spatially and temporally 80, 172.
Functional scale
Besides being able to follow single cells in space and time, we require an adequate model system that considers the functional 3D organization of cells in a tissue and has the ability to replicate some of the in vivo environment. Without easy accessibility into live tissues or the possibility of following their long‐term development, scientists over the last decade mostly exploited immortalized cell cultures 18, 173, 174, 175, embryos 4, 5, 97, 98, 144, 176, explanted tissues 16, 98, cocultures 177, as well as whole animal model systems, like Hydra 149, Axolotl 178 and planarians 43. And while much has been learned on fundamental processes, it is clear that each of these systems has a trade‐off between physiological relevance and experimental amenability.
A powerful model system assessing these limitations are organoids. Organoids are 3D organ‐like structures derived in vitro from primary tissues and adult stem cells, embryonic stem cells (ESCs) or induced pluripotent stem cells (iPSCs) 179, 180, 181, 182, 183, 184, 185, 186, 187. These complex multicellular structures arrange through a self‐organized mechanism requiring no external guidance, only appropriate niche factors and, importantly, a 3D extracellular scaffolding 55, 188, 189. Organoids develop into multicellular structures resembling key aspects of the native organ with differentiated cell types and tissue‐like architecture 190, 191. The cells self‐organize into complex structures from a range of organs such as optic cups 33, neuro‐rosettes 192, 193, cerebral cortex layers 181, intestinal crypts and villi 57, liver 194, lung 195, and kidney 190, 191, 196, 197, 198. Moreover, other powerful systems starting from ES cells have been developed that recapitulate embryo formation 5, 44, 199 and gastrulation 200. Clearly, organoids are simplified models of the complexity of tissue architecture and function. Intestinal organoids, for example, lack important aspects of tissue structure such as stromal cells201, 202, vascularization and enteric nervous system 177. Nonetheless, they deliver powerful means for ex‐vivo modeling of tissue morphogenesis and organogenesis and also represent an opportunity to understand fundamental principles and molecular mechanisms of self‐organized processes.
Organoid cultures combine: (a) advantages of in vitro culture (controlled growth conditions and multi‐parallelized assays possible)203, (b) amenability to chemical and genetic manipulations, (c) single cell accessibility with imaging and genomics techniques in both live and fixed samples 17, 32, 37, 179, 204, (d) temporal resolution, as organoid development can be followed in real time for monitoring short and long‐term events 32, 204, 205, 206, 207, (e) tissue functionality, which ultimately provides cellular information in physiologically unique contexts (such as organ‐specific development, homeostasis, regeneration or disease progression) 208, 209 and finally, (f) organoid cultures are expandable offering the opportunity to reach sample sizes of hundreds or thousands, which is not feasible with explanted tissues.
Organoid cultures allow to question how single cells exposed to a uniform growth‐promoting environment can generate asymmetric structures and how local interactions between single cells give rise to self‐organized patterns visible at the organoid level. Using embryonic kidney cells, for example, it has been shown that after enzymatic dissociation and re‐aggregation in vitro, cells spontaneously recreate the morphological arrangement between epithelial and mesenchymal cells, without prior spatial information 179. This re‐aggregation relies solely on movement of cells and differential cadherin‐based cell‐cell adhesion providing molecular evidence and evolutionary conservation of classical dissociation/re‐aggregation experiments in sponges and differential adhesion hypothesis of Steinberg 210. In an adult intestinal epithelia, the patterning of intermingled progenitors and differentiated cells in the stem cell niche is driven by the higher propensity of elongated cells to intersperse during interkinetic nuclear migration and cell division 207, showing that mechanical properties of cell division are driving forces in tissue patterning 105.
Finally, a recent paper exalted the power of understanding tissue self‐organization for clinical applications. Using hair‐bearing skin organoids from new‐born mice, the authors not only showed successful transplantation and further hair growth in adult nude mice 32, but also dissected the molecular mechanisms and morphological transitions during organoid formation. By combining time‐course transcriptome analysis and immunostaining, they described spatio‐temporally zonated patterns of expression of adhesion molecules and signaling pathways, which allowed experimental manipulation and hair growth restoration. It is therefore clear that understanding self‐organized processes that initiate and propagate regenerative and pathological conditions has also a therapeutic potential in medicine 196. Sensing the tissue environment is essential for a healthy regenerating tissue. In the recent years, the mechanosensors YAP and TAZ have been described as primary sensors of a tissue's physical context 211, 212 and master regulator of tissue regeneration. Moreover, it has been shown previously that engraftment of organoids into a damaged epithelium has a potential regenerative application, such as in ulcerative colitis 213. Because organoids can be derived from human samples, either healthy or diseased, it is now possible to envision personalized strategies 214, 215.
Outcome
Multicellular tissue self‐organization events can now be studied in a quantitative way at single cell resolution. The mechanisms by which single cells sense their local environments and implement it at the population level are the driving forces of self‐organization and collective behaviours during development, tissue remodeling, and regenerative processes. The rapid development of imaging and genomics techniques combined with powerful modeling tools, now, enables us to bridge scales of complexity: spatially, temporally and functionally. With subcellular resolution, we can better understand fundamental concepts of how symmetry‐breaking events occur, which roles cell‐to‐cell variability plays in a biological process and how cellular patterns emerge. From that, we can then move forward to describe the mechanisms behind complex collective behaviours in development and tissue homeostasis, with immediate application in development and regenerative medicine 20, 173.
Acknowledgements
We would like to thanks Prof. Charisios Tsiairis and all Liberali lab members for helpful discussions and comments on the manuscript. We are thankful to Denise Serra and Ilya Lukonin for helping with figures. This study was supported by Swiss initiative in Systems Biology, Systemsx.ch (MorphogenetiX to PL), Swiss National Science Foundation (Foerderungsprofessur grant to PL, POOP3_157531 to PL and Marie‐Heim Vögtlin Grant PMPD3_171365 to AXSS). This project/work has received funding from the European Research Council (ERC) under the European Union's Horizon 2020 research and innovation programme (grant agreement No 758617).
References
- 1. Herr P, Hausmann G & Basler K (2012) WNT secretion and signalling in human disease. Trends Mol Med 18, 483–493. [DOI] [PubMed] [Google Scholar]
- 2. Clevers H & Nusse R (2012) Wnt/beta‐catenin signaling and disease. Cell 149, 1192–1205. [DOI] [PubMed] [Google Scholar]
- 3. Rauzi M, Krzic U, Saunders TE, Krajnc M, Ziherl P, Hufnagel L & Leptin M (2015) Embryo‐scale tissue mechanics during Drosophila gastrulation movements. Nat Commun 6, 8677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wennekamp S, Mesecke S, Nédélec F & Hiiragi T (2013) A self‐organization framework for symmetry breaking in the mammalian embryo. Nat Rev Mol Cell Biol 14, 452–459. [DOI] [PubMed] [Google Scholar]
- 5. Bedzhov I & Zernicka‐Goetz M (2014) Self‐organizing properties of mouse pluripotent cells initiate morphogenesis upon implantation. Cell 156, 1032–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chan CJ, Heisenberg CP & Hiiragi T (2017) Coordination of morphogenesis and cell‐fate specification in development. Curr Biol 27, R1024–R1035. [DOI] [PubMed] [Google Scholar]
- 7. Kicheva A, Cohen M & Briscoe J (2012) Developmental pattern formation: insights from physics and biology. Science 338, 210–212. [DOI] [PubMed] [Google Scholar]
- 8. Rogers KW & Schier AF (2011) Morphogen gradients: from generation to interpretation. Annu Rev Cell Dev Biol 27, 377–407. [DOI] [PubMed] [Google Scholar]
- 9. Gonzalez‐Gaitan M & Julicher F (2014) The role of endocytosis during morphogenetic signaling. Cold Spring Harb Perspect Biol 6, a016881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Werner S, Vu HT & Rink JC (2017) Self‐organization in development, regeneration and organoids. Curr Opin Cell Biol 44, 102–109. [DOI] [PubMed] [Google Scholar]
- 11. Karsenti E (2008) Self‐organization in cell biology: a brief history. Nat Rev Mol Cell Biol 9, 255–262. [DOI] [PubMed] [Google Scholar]
- 12. Jevtić P & Levy DL (2017) Both nuclear size and DNA amount contribute to midblastula transition timing in Xenopus laevis . Sci Rep 7, 7908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bernitz JM, Kim HS, MacArthur B, Sieburg H & Moore K (2016) Hematopoietic stem cells count and remember self‐renewal divisions. Cell 167, 1296–1309. e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Brock DA & Gomer RH (1999) A cell‐counting factor regulating structure size in Dictyostelium . Genes Dev 13, 1960–1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Day SJ & Lawrence PA (2000) Measuring dimensions: the regulation of size and shape. Development 127, 2977–2987. [DOI] [PubMed] [Google Scholar]
- 16. Mao Y, Tournier AL, Hoppe A, Kester L, Thompson BJ & Tapon N (2013) Differential proliferation rates generate patterns of mechanical tension that orient tissue growth. EMBO J 32, 2790–2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Farin HF, Jordens I, Mosa MH, Basak O, Korving J, Tauriello DVF, de Punder K, Angers S, Peters PJ, Maurice MM et al (2016) Visualization of a short‐range Wnt gradient in the intestinal stem‐cell niche. Nature 530, 340–343. [DOI] [PubMed] [Google Scholar]
- 18. Puliafito A, Hufnagel L, Neveu P, Streichan S, Sigal A, Fygenson DK & Shraiman BI (2012) Collective and single cell behavior in epithelial contact inhibition. Proc Natl Acad Sci USA 109, 739–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bettuzzi S, Astancolle S, Guidetti G, Moretti M, Tiozzo R & Corti A (1999) Clusterin (SGP‐2) gene expression is cell cycle dependent in normal human dermal fibroblasts. FEBS Lett 448, 297–300. [DOI] [PubMed] [Google Scholar]
- 20. Kim NG, Koh E, Chen X & Gumbiner BM (2011) E‐cadherin mediates contact inhibition of proliferation through Hippo signaling‐pathway components. Proc Natl Acad Sci USA 108, 11930–11935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yui S, Azzolin L, Maimets M, Pedersen MT, Fordham RP, Hansen SL, Larsen HL, Guiu J, Alves MRP, Rundsten CF et al (2018) YAP/TAZ‐dependent reprogramming of colonic epithelium links ECM remodeling to tissue regeneration. Cell Stem Cell 22, 35–49 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Elbediwy A & Thompson BJ (2018) Evolution of mechanotransduction via YAP/TAZ in animal epithelia. Curr Opin Cell Biol 51, 117–123. [DOI] [PubMed] [Google Scholar]
- 23. Coste B, Xiao B, Santos JS, Syeda R, Grandl J, Spencer KS, Kim SE, Schmidt M, Mathur J, Dubin AE et al (2012) Piezo proteins are pore‐forming subunits of mechanically activated channels. Nature 483, 176–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Piddini E (2017) Epithelial homeostasis: a piezo of the puzzle. Curr Biol 27, R232–R234. [DOI] [PubMed] [Google Scholar]
- 25. Li Q, Li S, Mana‐Capelli S, Roth Flach RJ, Danai LV, Amcheslavsky A, Nie Y, Kaneko S, Yao X, Chen X et al (2014) The conserved misshapen‐warts‐Yorkie pathway acts in enteroblasts to regulate intestinal stem cells in Drosophila . Dev Cell 31, 291–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Marinari E, Mehonic A, Curran S, Gale J, Duke T & Baum B (2012) Live‐cell delamination counterbalances epithelial growth to limit tissue overcrowding. Nature 484, 542–545. [DOI] [PubMed] [Google Scholar]
- 27. Takemura M & Nakato H (2017) Drosophila Sulf1 is required for the termination of intestinal stem cell division during regeneration. J Cell Sci 130, 332–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Durdu S, Iskar M, Revenu C, Schieber N, Kunze A, Bork P, Schwab Y & Gilmour D (2014) Luminal signalling links cell communication to tissue architecture during organogenesis. Nature 515, 120–124. [DOI] [PubMed] [Google Scholar]
- 29. Liu P, Begley M, Michowski W, Inuzuka H, Ginzberg M, Gao D, Tsou P, Gan W, Papa A, Kim BM et al (2014) Cell‐cycle‐regulated activation of Akt kinase by phosphorylation at its carboxyl terminus. Nature 508, 541–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Guisoni N, Martinez‐Corral R, Garcia‐Ojalvo J & de Navascués J (2017) Diversity of fate outcomes in cell pairs under lateral inhibition. Development 144, 1177–1186. [DOI] [PubMed] [Google Scholar]
- 31. Egger B, Gold KS & Brand AH (2010) Notch regulates the switch from symmetric to asymmetric neural stem cell division in the Drosophila optic lobe. Development 137, 2981–2987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lei M, Schumacher LJ, Lai YC, Juan WT, Yeh CY, Wu P, Jiang TX, Baker RE, Widelitz RB, Yang L et al (2017) Self‐organization process in newborn skin organoid formation inspires strategy to restore hair regeneration of adult cells. Proc Natl Acad Sci USA 114, E7101–E7110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Eiraku M, Takata N, Ishibashi H, Kawada M, Sakakura E, Okuda S, Sekiguchi K, Adachi T & Sasai Y (2011) Self‐organizing optic‐cup morphogenesis in three‐dimensional culture. Nature 472, 51–56. [DOI] [PubMed] [Google Scholar]
- 34. Watt FM (2016) Engineered microenvironments to direct epidermal stem cell behavior at single‐cell resolution. Dev Cell 38, 601–609. [DOI] [PubMed] [Google Scholar]
- 35. Borrell V & Calegari F (2014) Mechanisms of brain evolution: regulation of neural progenitor cell diversity and cell cycle length. Neurosci Res 86, 14–24. [DOI] [PubMed] [Google Scholar]
- 36. Noctor SC, Martínez‐Cerdeño V, Ivic L & Kriegstein AR (2004) Cortical neurons arise in symmetric and asymmetric division zones and migrate through specific phases. Nat Neurosci 7, 136–144. [DOI] [PubMed] [Google Scholar]
- 37. Mora‐Bermúdez F, Badsha F, Kanton S, Camp JG, Vernot B, Köhler K, Voigt B, Okita K, Maricic T, He Z et al (2016) Differences and similarities between human and chimpanzee neural progenitors during cerebral cortex development. Elife 5, e18683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hardwick LJ, Ali FR, Azzarelli R & Philpott A (2015) Cell cycle regulation of proliferation versus differentiation in the central nervous system. Cell Tissue Res 359, 187–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Pauklin S & Vallier L (2013) The cell‐cycle state of stem cells determines cell fate propensity. Cell 155, 135–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Coronado D, Godet M, Bourillot PY, Tapponnier Y, Bernat A, Petit M, Afanassieff M, Markossian S, Malashicheva A, Iacone R et al (2013) A short G1 phase is an intrinsic determinant of naïve embryonic stem cell pluripotency. Stem Cell Res 10, 118–131. [DOI] [PubMed] [Google Scholar]
- 41. Carroll TD, Newton IP, Chen Y, Blow JJ & Näthke I (2018) Lgr5(+) intestinal stem cells reside in an unlicensed G1 phase. J Cell Biol 217, 1667–1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Technau U, Cramer von Laue C, Rentzsch F, Luft S, Hobmayer B, Bode HR & Holstein TW (2000) Parameters of self‐organization in hydra aggregates. Proc Natl Acad Sci USA 97, 12127–12131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Reddien PW & Sanchez Alvarado A (2004) Fundamentals of planarian regeneration. Annu Rev Cell Dev Biol 20, 725–757. [DOI] [PubMed] [Google Scholar]
- 44. Chen Q, Shi J, Tao Y & Zernicka‐Goetz M (2018) Tracing the origin of heterogeneity and symmetry breaking in the early mammalian embryo. Nat Commun 9, 1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Anderson PW (1972) More is different. Science 177, 393–396. [DOI] [PubMed] [Google Scholar]
- 46. Korotkevich E, Niwayama R, Courtois A, Friese S, Berger N, Buchholz F & Hiiragi T (2017) The apical domain is required and sufficient for the first lineage segregation in the mouse embryo. Dev Cell 40, 235–247.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Maitre JL, Turlier H, Illukkumbura R, Eismann B, Niwayama R, Nédélec F & Hiiragi T (2016) Asymmetric division of contractile domains couples cell positioning and fate specification. Nature 536, 344–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hirokawa N, Tanaka Y, Okada Y & Takeda S (2006) Nodal flow and the generation of left‐right asymmetry. Cell 125, 33–45. [DOI] [PubMed] [Google Scholar]
- 49. Hirokawa N, Tanaka Y & Okada Y (2009) Left‐right determination: involvement of molecular motor KIF3, cilia, and nodal flow. Cold Spring Harb Perspect Biol 1, a000802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Essner JJ, Amack JD, Nyholm MK, Harris EB & Yost HJ (2005) Kupffer's vesicle is a ciliated organ of asymmetry in the zebrafish embryo that initiates left‐right development of the brain, heart and gut. Development 132, 1247–1260. [DOI] [PubMed] [Google Scholar]
- 51. Schweickert A, Weber T, Beyer T, Vick P, Bogusch S, Feistel K & Blum M (2007) Cilia‐driven leftward flow determines laterality in Xenopus . Curr Biol 17, 60–66. [DOI] [PubMed] [Google Scholar]
- 52. Shiratori H & Hamada H (2006) The left‐right axis in the mouse: from origin to morphology. Development 133, 2095–2104. [DOI] [PubMed] [Google Scholar]
- 53. Okada Y, Takeda S, Tanaka Y, Belmonte J‐C & Hirokawa N (2005) Mechanism of nodal flow: a conserved symmetry breaking event in left‐right axis determination. Cell 121, 633–644. [DOI] [PubMed] [Google Scholar]
- 54. Guglielmi G, Barry JD, Huber W & De Renzis S (2015) An optogenetic method to modulate cell contractility during tissue morphogenesis. Dev Cell 35, 646–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Hughes AJ, Miyazaki H, Coyle MC, Zhang J, Laurie MT, Chu D, Vavrušová Z, Schneider RA, Klein OD & Gartner ZJ (2018) Engineered tissue folding by mechanical compaction of the mesenchyme. Dev Cell 44, 165–178 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Sweeton D, Parks S, Costa M & Wieschaus E (1991) Gastrulation in Drosophila: the formation of the ventral furrow and posterior midgut invaginations. Development 112, 775–789. [DOI] [PubMed] [Google Scholar]
- 57. Sato T, Vries RG, Snippert HJ, van de Wetering M, Barker N, Stange DE, van Es JH, Abo A, Kujala P, Peters PJ et al (2009) Single Lgr5 stem cells build cryptvillus structures in vitro without a mesenchymal niche. Nature 459, 262–265. [DOI] [PubMed] [Google Scholar]
- 58. Sato T, van Es JH, Snippert HJ, Stange DE, Vries RG, van den Born M, Barker N, Shroyer NF, van de Wetering M & Clevers H (2011) Paneth cells constitute the niche for Lgr5 stem cells in intestinal crypts. Nature 469, 415–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Sato T & Clevers H (2013) Growing self‐organizing mini‐guts from a single intestinal stem cell: mechanism and applications. Science 340, 1190–1194. [DOI] [PubMed] [Google Scholar]
- 60. Kondo S & Asai R (1995) A reaction–diffusion wave on the skin of the marine angelfish Pomacanthus . Nature 376, 765. [DOI] [PubMed] [Google Scholar]
- 61. Turing AM (1953) The chemical basis of morphogenesis. Bull Math Biol 52, 153–197; discussion 119‐52. [DOI] [PubMed] [Google Scholar]
- 62. Sugai SS, Ode KL & Ueda HR (2017) A design principle for an autonomous post‐translational pattern formation. Cell Rep 19, 863–874. [DOI] [PubMed] [Google Scholar]
- 63. Thorne BC, Bailey AM & Peirce SM (2007) Combining experiments with multi‐cell agent‐based modeling to study biological tissue patterning. Brief Bioinform 8, 245–257. [DOI] [PubMed] [Google Scholar]
- 64. Van Liedekerke P, Palm MM, Jagiella N & Drasdo D (2015) Simulating tissue mechanics with agent‐based models: concepts, perspectives and some novel results. Computational Particle Mechanics 2, 401–444. [Google Scholar]
- 65. Colman‐Lerner A, Gordon A, Serra E, Chin T, Resnekov O, Endy D, Gustavo Pesce C & Brent R (2005) Regulated cell‐to‐cell variation in a cell‐fate decision system. Nature 437, 699–706. [DOI] [PubMed] [Google Scholar]
- 66. Blum W, Henzi T, Schwaller B & Pecze L (2018) Biological noise and positional effects influence cell stemness. J Biol Chem 293, 5247–5258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Ahrends R, Ota A, Kovary KM, Kudo T, Park BO & Teruel MN (2014) Controlling low rates of cell differentiation through noise and ultrahigh feedback. Science 344, 1384–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Morelli LG, Uriu K, Ares S & Oates AC (2012) Computational approaches to developmental patterning. Science 336, 187–191. [DOI] [PubMed] [Google Scholar]
- 69. Scholes NS, Schnoerr D, Isalan M & Stumpf MPH (2018) Turing patterns are common but not robust. bioRxiv 352302.
- 70. Altschuler SJ & Wu LF (2010) Cellular heterogeneity: do differences make a difference? Cell 141, 559–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Elowitz MB (2002) Stochastic gene expression in a single cell. Science 297, 1183–1186. [DOI] [PubMed] [Google Scholar]
- 72. Raj A, Rifkin SA, Andersen E & van Oudenaarden A (2010) Variability in gene expression underlies incomplete penetrance . Nature 463, 913–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Stelling J, Sauer U, Szallasi Z, Doyle FJ & Doyle J (2004) Robustness of cellular functions. Cell 118, 675–685. [DOI] [PubMed] [Google Scholar]
- 74. Macarthur BD, Ma'ayan A & Lemischka IR (2009) Systems biology of stem cell fate and cellular reprogramming. Nat Rev Mol Cell Biol 10, 672–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Barad O, Hornstein E & Barkai N (2011) Robust selection of sensory organ precursors by the Notch‐Delta pathway. Curr Opin Cell Biol 23, 663–667. [DOI] [PubMed] [Google Scholar]
- 76. Ribrault C, Sekimoto K & Triller A (2011) From the stochasticity of molecular processes to the variability of synaptic transmission. Nat Rev Neurosci 12, 375–387. [DOI] [PubMed] [Google Scholar]
- 77. Battich N, Stoeger T & Pelkmans L (2015) Control of transcript variability in single mammalian cells. Cell 163, 1596–1610. [DOI] [PubMed] [Google Scholar]
- 78. Spencer SL, Gaudet S, Albeck JG, Burke JM & Sorger PK (2009) Non‐genetic origins of cell‐to‐cell variability in TRAIL‐induced apoptosis. Nature 459, 428–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Snijder B, Sacher R, Rämö P, Damm E‐M, Liberali P & Pelkmans L (2009) Population context determines cell‐to‐cell variability in endocytosis and virus infection. Nature 461, 520–523. [DOI] [PubMed] [Google Scholar]
- 80. Gut G, Tadmor MD, Pe'er D, Pelkmans L & Liberali P (2015) Trajectories of cell‐cycle progression from fixed cell populations. Nat Methods 12, 951–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Snijder B, Sacher R, Rämö P, Liberali P, Mench K, Wolfrum N, Burleigh L, Scott CC, Verheije MH, Mercer J et al (2012) Single‐cell analysis of population context advances RNAi screening at multiple levels. Mol Syst Biol 8, 579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Frechin M, Stoeger T, Daetwyler S, Gehin C, Battich N, Damm E‐M, Stergiou L, Riezman H & Pelkmans L (2015) Cell‐intrinsic adaptation of lipid composition to local crowding drives social behaviour. Nature 523, 88–91. [DOI] [PubMed] [Google Scholar]
- 83. Ohnishi Y, Huber W, Tsumura A, Kang M, Xenopoulos P, Kurimoto K, Oleś AK, Araúzo‐Bravo MJ, Saitou M, Hadjantonakis AK et al (2014) Cell‐to‐cell expression variability followed by signal reinforcement progressively segregates early mouse lineages. Nat Cell Biol 16, 27–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Dietrich JE & Hiiragi T (2008) Stochastic processes during mouse blastocyst patterning. Cells Tissues Organs 188, 46–51. [DOI] [PubMed] [Google Scholar]
- 85. Dietrich JE & Hiiragi T (2007) Stochastic patterning in the mouse pre‐implantation embryo. Development 134, 4219–4231. [DOI] [PubMed] [Google Scholar]
- 86. Chambers I, Silva J, Colby D, Nichols J, Nijmeijer B, Robertson M, Vrana J, Jones K, Grotewold L & Smith A (2007) Nanog safeguards pluripotency and mediates germline development. Nature 450, 1230–1234. [DOI] [PubMed] [Google Scholar]
- 87. van der Laan S, Golfetto E, Vanacker J‐M & Maiorano D (2014) Cell cycle‐dependent expression of Dub3, Nanog and the p160 family of nuclear receptor coactivators (NCoAs) in mouse embryonic stem cells. PLoS ONE 9, e93663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. White MD, Angiolini JF, Alvarez YD, Kaur G, Zhao ZW, Mocskos E, Bruno L, Bissiere S, Levi V & Plachta N (2016) Long‐lived binding of Sox2 to DNA predicts cell fate in the four‐cell mouse embryo. Cell 165, 75–87. [DOI] [PubMed] [Google Scholar]
- 89. Goolam M, Scialdone A, Graham SJL, Macaulay IC, Jedrusik A, Hupalowska A, Voet T, Marioni JC & Zernicka‐Goetz M (2016) Heterogeneity in Oct4 and Sox2 targets biases cell fate in 4‐cell mouse embryos. Cell 165, 61–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Chang HH, Hemberg M, Barahona M, Ingber DE & Huang S (2008) Transcriptome‐wide noise controls lineage choice in mammalian progenitor cells. Nature 453, 544–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. MacArthur BD & Lemischka IR (2013) Statistical mechanics of pluripotency. Cell 154, 484–489. [DOI] [PubMed] [Google Scholar]
- 92. Clevers H & Watt FM (2018) Defining adult stem cells by function, not by phenotype. Annu Rev Biochem 87, 1015–1027. [DOI] [PubMed] [Google Scholar]
- 93. van Es JH, Sato T, van de Wetering M, Lyubimova A, Yee Nee AN, Gregorieff A, Sasaki N, Zeinstra L, van den Born M, Korving J et al (2012) Dll1 + secretory progenitor cells revert to stem cells upon crypt damage. Nat Cell Biol 14, 1099–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Richard A, Boullu L, Herbach U, Bonnafoux A, Morin V, Vallin E, Guillemin A, Papili Gao N, Gunawan R, Cosette J et al (2016) Single‐cell‐based analysis highlights a surge in cell‐to‐cell molecular variability preceding irreversible commitment in a differentiation process. PLoS Biol 14, e1002585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Connelly JT, Gautrot JE, Trappmann B, Tan DW‐M, Donati G, Huck WTS & Watt FM (2010) Actin and serum response factor transduce physical cues from the microenvironment to regulate epidermal stem cell fate decisions. Nat Cell Biol 12, 711–718. [DOI] [PubMed] [Google Scholar]
- 96. Engler AJ, Sen S, Sweeney HL & Discher DE (2006) Matrix elasticity directs stem cell lineage specification. Cell 126, 677–689. [DOI] [PubMed] [Google Scholar]
- 97. Shahbazi MN, Jedrusik A, Vuoristo S, Recher G, Hupalowska A, Bolton V, Fogarty NNM, Campbell A, Devito LG, Ilic D et al (2016) Self‐organization of the human embryo in the absence of maternal tissues. Nat Cell Biol 18, 700–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Tsiairis CD & Aulehla A (2016) Self‐organization of embryonic genetic oscillators into spatiotemporal wave patterns. Cell 164, 656–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Liberali P, Snijder B & Pelkmans L (2015) Single‐cell and multivariate approaches in genetic perturbation screens. Nat Rev Genet 16, 18–32. [DOI] [PubMed] [Google Scholar]
- 100. Reynaud EG, Peychl J, Huisken J & Tomancak P (2015) Guide to light‐sheet microscopy for adventurous biologists. Nat Methods 12, 30–34. [DOI] [PubMed] [Google Scholar]
- 101. Moffitt JR, Hao J, Bambah‐Mukku D, Lu T, Dulac C & Zhuang X (2016) High‐performance multiplexed fluorescence in situ hybridization in culture and tissue with matrix imprinting and clearing. Proc Natl Acad Sci USA 113, 14456–14461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Moor AE, Harnik Y, Ben‐Moshe S, Massasa EE, Rozenberg M, Eilam R, Bahar Halpern K & Itzkovitz Z (2018) Spatial reconstruction of single enterocytes uncovers broad zonation along the intestinal villus axis. Cell 175, 1156.e15–1167.e15. [DOI] [PubMed] [Google Scholar]
- 103. Halpern KB, Shenhav R, Matcovitch‐Natan O, Toth B, Lemze D, Golan M, Massasa EE, Baydatch S, Landen S, Moor AE et al (2017) Single‐cell spatial reconstruction reveals global division of labour in the mammalian liver. Nature 542, 352–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Tainaka K, Kubota SI, Suyama TQ, Susaki EA, Perrin D, Ukai‐Tadenuma M, Ukai H & Ueda HR (2014) Whole‐body imaging with single‐cell resolution by tissue decolorization. Cell 159, 911–924. [DOI] [PubMed] [Google Scholar]
- 105. LeGoff L & Lecuit T (2016) Mechanical forces and growth in animal tissues. Cold Spring Harb Perspect Biol 8, a019232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Gjorevski N & Nelson CM (2010) The mechanics of development: Models and methods for tissue morphogenesis. Birth Defects Res C Embryo Today 90, 193–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Richardson DS & Lichtman JW (2015) Clarifying tissue clearing. Cell 162, 246–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Susaki EA & Ueda HR (2016) Whole‐body and whole‐organ clearing and imaging techniques with single‐cell resolution: toward organism‐level systems biology in mammals. Cell Chem Biol 23, 137–157. [DOI] [PubMed] [Google Scholar]
- 109. Murakami TC, Mano T, Saikawa S, Horiguchi SA, Shigeta D, Baba K, Sekiya H, Shimizu Y, Tanaka KF, Kiyonari H et al (2018) A three‐dimensional single‐cell‐resolution whole‐brain atlas using CUBIC‐X expansion microscopy and tissue clearing. Nat Neurosci 21, 625–637. [DOI] [PubMed] [Google Scholar]
- 110. Valm AM, Cohen S, Legant WR, Melunis J, Hershberg U, Wait E, Cohen AR, Davidson MW, Betzig E & Lippincott‐Schwartz J (2017) Applying systems‐level spectral imaging and analysis to reveal the organelle interactome. Nature 546, 162–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Lin JR, Fallahi‐Sichani M & Sorger PK (2015) Highly multiplexed imaging of single cells using a high‐throughput cyclic immunofluorescence method. Nat Commun 6, 8390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Lin J‐R, Izar B, Wang S, Yapp C, Mei S, Shah PM, Santagata S & Sorger PK (2018) Highly multiplexed immunofluorescence imaging of human tissues and tumors using t‐CyCIF and conventional optical microscopes. ELife 7, e31657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Gut M, Hermann MD & Pelkmans L (2018) Multiplexed protein maps link subcellular organization to cellular states. Science 361, eaar7042. [DOI] [PubMed] [Google Scholar]
- 114. Giesen C, Wang HA, Schapiro D, Zivanovic N, Jacobs A, Hattendorf B, Schüffler PJ, Grolimund D, Buhmann JM, Brandt S et al (2014) Highly multiplexed imaging of tumor tissues with subcellular resolution by mass cytometry. Nat Methods 11, 417–422. [DOI] [PubMed] [Google Scholar]
- 115. Angelo M, Bendall SC, Finck R, Hale MB, Hitzman C, Borowsky AD, Levenson RM, Lowe JB, Liu SD, Zhao S et al (2014) Multiplexed ion beam imaging (MIBI) of human breast tumors. Nat Med 20, 436–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Schulz D, Zanotelli VRT, Fischer JR, Schapiro D, Engler S, Lun XK, Jackson HW & Bodenmiller B (2018) Simultaneous multiplexed imaging of mRNA and proteins with subcellular resolution in breast cancer tissue samples by mass cytometry. Cell Syst 6, 531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Shah S, Lubeck E, Zhou W & Cai L (2017) SeqFISH accurately detects transcripts in single cells and reveals robust spatial organization in the hippocampus. Neuron 94, 752–758.e1. [DOI] [PubMed] [Google Scholar]
- 118. Shah S, Lubeck E, Zhou W & Cai L (2016) In situ transcription profiling of single cells reveals spatial organization of cells in the mouse hippocampus. Neuron 92, 342–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Chen KH, Boettiger AN, Moffitt JR, Wang S & Zhuang X (2015) RNA imaging. Spatially resolved, highly multiplexed RNA profiling in single cells. Science 348, aaa6090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Itzkovitz S, Lyubimova A, Blat IC, Maynard M, van Es J, Lees J, Jacks T, Clevers H & van Oudenaarden A (2011) Single‐molecule transcript counting of stem‐cell markers in the mouse intestine. Nat Cell Biol 14, 106–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Schell JC, Wisidagama DR, Bensard C, Zhao H, Wei P, Tanner J, Flores A, Mohlman J, Sorensen LK, Earl CS et al (2017) Control of intestinal stem cell function and proliferation by mitochondrial pyruvate metabolism. Nat Cell Biol 19, 1027–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Krejci A & Tennessen JM (2017) Metabolism in time and space ‐ exploring the frontier of developmental biology. Development 144, 3193–3198. [DOI] [PubMed] [Google Scholar]
- 123. Bulusu V, Prior N, Snaebjornsson MT, Kuehne A, Sonnen KF, Kress J, Stein F, Schultz C, Sauer U & Aulehla A (2017) Spatiotemporal analysis of a glycolytic activity gradient linked to mouse embryo mesoderm development. Dev Cell 40, 331–341.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Mathupala SP, Colen CB, Parajuli P & Sloan AE (2007) Lactate and malignant tumors: a therapeutic target at the end stage of glycolysis. J Bioenerg Biomembr 39, 73–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Sperber H, Mathieu J, Wang Y, Ferreccio A, Hesson J, Xu Z, Fischer KA, Devi A, Detraux D, Gu H et al (2015) The metabolome regulates the epigenetic landscape during naive‐to‐primed human embryonic stem cell transition. Nat Cell Biol 17, 1523–1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Oginuma M, Moncuquet P, Xiong F, Karoly E, Chal J, Guevorkian K & Pourquié O (2017) A gradient of glycolytic activity coordinates FGF and Wnt signaling during elongation of the body axis in amniote embryos. Dev Cell 40, 342–353.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Rodríguez‐Colman MJ, Schewe M, Meerlo M, Stigter E, Gerrits J, Pras‐Raves M, Sacchetti A, Hornsveld M, Oost KC, Snippert HJ et al (2017) Interplay between metabolic identities in the intestinal crypt supports stem cell function. Nature 543, 424–427. [DOI] [PubMed] [Google Scholar]
- 128. Rompp A & Spengler B (2013) Mass spectrometry imaging with high resolution in mass and space. Histochem Cell Biol 139, 759–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Dahan P, Lu V, Nguyen RMT, Kennedy SAL & Teitell MA (2018) Metabolism in pluripotency: Both driver and passenger? J Biol Chem, https://doi.org/0.1074/jbc.TM117.000832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Goodman RP, Calvo S & Mootha VK (2018) Spatiotemporal compartmentalization of hepatic NADH and NADPH metabolism. J Biol Chem 293, 7508–7516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Zhang H, Menzies KJ & Auwerx J (2018) The role of mitochondria in stem cell fate and aging. Development 145, dev143420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Mathieu J & Ruohola‐Baker H (2017) Metabolic remodeling during the loss and acquisition of pluripotency. Development 144, 541–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Liu TL, Upadhyayula S, Milkie DE, Singh V, Wang K, Swinburne IA, Mosaliganti KR, Collins ZM, Hiscock TW, Shea J et al (2018) Observing the cell in its native state: Imaging subcellular dynamics in multicellular organisms. Science 360, eaaq1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Royer LA, Lemon WC, Chhetri RK, Wan Y, Coleman M, Myers EW & Keller PJ (2016) Adaptive light‐sheet microscopy for long‐term, high‐resolution imaging in living organisms. Nat Biotechnol 34, 1267–1278. [DOI] [PubMed] [Google Scholar]
- 135. Power RM & Huisken J (2018) Adaptable, illumination patterning light sheet microscopy. Sci Rep 8, 9615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Chen BC, Legant WR, Wang K, Shao L, Milkie DE, Davidson MW, Janetopoulos C, Wu XS, Hammer JA 3rd, Liu Z et al (2014) Lattice light‐sheet microscopy: imaging molecules to embryos at high spatiotemporal resolution. Science.346, 1257998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Chen CH, Puliafito A, Cox BD, Primo L, Fang Y, Di Talia S & Poss KD (2016) Multicolor cell barcoding technology for long‐term surveillance of epithelial regeneration in Zebrafish. Dev Cell 36, 668–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Strnad P, Gunther S, Reichmann J, Krzic U, Balazs B, de Medeiros G, Norlin N, Hiiragi T, Hufnagel L & Ellenberg J (2015) Inverted light‐sheet microscope for imaging mouse pre‐implantation development. Nat Methods 13, 139–142. [DOI] [PubMed] [Google Scholar]
- 139. Keller PJ, Schmidt AD, Wittbrodt J & Stelzer EHK (2008) Reconstruction of zebrafish early embryonic development by scanned light sheet microscopy. Science 322, 1065–1069. [DOI] [PubMed] [Google Scholar]
- 140. Huisken J, Swoger J, Del Bene F, Wittbrodt J & Stelzer EH (2004) Optical sectioning deep inside live embryos by selective plane illumination microscopy. Science 305, 1007–1009. [DOI] [PubMed] [Google Scholar]
- 141. Krzic U, Gunther S, Saunders TE, Streichan SJ & Hufnagel L (2012) Multiview light‐sheet microscope for rapid in toto imaging. Nat Methods 9, 730–733. [DOI] [PubMed] [Google Scholar]
- 142. Martyn I, Kanno TY, Ruzo A, Siggia ED & Brivanlou AH (2018) Self‐organization of a human organizer by combined Wnt and Nodal signalling. Nature 558, 132–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Behjati S, Lindsay S, Teichmann SA & Haniffa M (2018) Mapping human development at single‐cell resolution. Development 145, dev152561. [DOI] [PubMed] [Google Scholar]
- 144. Deglincerti A, Croft GF, Pietila LN, Zernicka‐Goetz M, Siggia ED & Brivanlou AH (2016) Self‐organization of the in vitro attached human embryo. Nature 533, 251–254. [DOI] [PubMed] [Google Scholar]
- 145. Belle M, Godefroy D, Couly G, Malone SA, Collier F, Giacobini P & Chédotal A (2017) Tridimensional visualization and analysis of early human development. Cell 169, 161–173. e12. [DOI] [PubMed] [Google Scholar]
- 146. Regev A, Teichmann SA, Lander ES, Amit I, Benoist C, Birney E, Bodenmiller B, Campbell P, Carninci P, Clatworthy M et al (2017) The human cell Atlas. ELife 6, e27041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. van den Brink SC, Baillie‐Johnson P, Balayo T, Hadjantonakis A‐K, Nowotschin S, Turner DA & Martinez Arias A (2014) Symmetry breaking, germ layer specification and axial organisation in aggregates of mouse embryonic stem cells. Development 141, 4231–4242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.(2018) Method of the year 2017: organoids. Nat Methods 15, 1–1. 10.1038/nmeth.4575 [DOI] [Google Scholar]
- 149. Galliot B (2012) Hydra, a fruitful model system for 270 years. Int J Dev Biol 56, 411–423. [DOI] [PubMed] [Google Scholar]
- 150. Keil W, Kutscher LM, Shaham S & Siggia ED (2017) Long‐term high‐resolution imaging of developing C. elegans Larvae with Microfluidics. Dev Cell 40, 202–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Kaufmann A, Mickoleit M, Weber M & Huisken J (2012) Multilayer mounting enables long‐term imaging of zebrafish development in a light sheet microscope. Development 139, 3242–3247. [DOI] [PubMed] [Google Scholar]
- 152. Kaur P, Saunders TE & Tolwinski NS (2017) Coupling optogenetics and light‐sheet microscopy, a method to study Wnt signaling during embryogenesis. Sci Rep 7, 16636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Masamizu Y, Ohtsuka T, Takashima Y, Nagahara H, Takenaka Y, Yoshikawa K, Okamura H & Kageyama R (2006) Real‐time imaging of the somite segmentation clock: revelation of unstable oscillators in the individual presomitic mesoderm cells. Proc Natl Acad Sci USA 103, 1313–1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Matsumiya M, Tomita T, Yoshioka‐Kobayashi K, Isomura A & Kageyama R (2018) ES cell‐derived presomitic mesoderm‐like tissues for analysis of synchronized oscillations in the segmentation clock. Development 145, dev156836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Rompolas P, Mesa KR, Kawaguchi K, Park S, Gonzalez D, Brown S, Boucher J, Klein AM & Greco V (2016) Spatiotemporal coordination of stem cell commitment during epidermal homeostasis. Science 352, 1471–1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Ritsma L, Ellenbroek SIJ., Zomer A, Snippert HJ, de Sauvage FJ, Simons BD, Clevers H & van Rheenen J (2014) Intestinal crypt homeostasis revealed at single‐stem‐cell level by in vivo live imaging. Nature 507, 362–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Snippert HJ, van der Flier LG, Sato T, van Es JH, van den Born M, Kroon‐Veenboer C, Barker N, Klein AM, van Rheenen J, Simons BD et al (2010) Intestinal crypt homeostasis results from neutral competition between symmetrically dividing Lgr5 stem cells. Cell 143, 134–144. [DOI] [PubMed] [Google Scholar]
- 158. Lopez‐Garcia C, Klein AM, Simons BD & Winton DJ (2010) Intestinal stem cell replacement follows a pattern of neutral drift. Science 330, 822–825. [DOI] [PubMed] [Google Scholar]
- 159. Levayer R & Lecuit T (2013) Oscillation and polarity of E‐cadherin asymmetries control actomyosin flow patterns during morphogenesis. Dev Cell 26, 162–175. [DOI] [PubMed] [Google Scholar]
- 160. Guglielmi G, Falk HJ & De Renzis S (2016) Optogenetic control of protein function: from intracellular processes to tissue morphogenesis. Trends Cell Biol 26, 864–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Sako K, Pradhan SJ, Barone V, Inglés‐Prieto Á, Müller P, Ruprecht V, Čapek D, Galande S, Janovjak H & Heisenberg C‐P (2016) Optogenetic control of nodal signaling reveals a temporal pattern of nodal signaling regulating cell fate specification during gastrulation. Cell Rep 16, 866–877. [DOI] [PubMed] [Google Scholar]
- 162. Barone V, Lang M, Krens SFG, Pradhan SJ, Shamipour S, Sako K, Sikora M, Guet CC & Heisenberg CP (2017) An effective feedback loop between cell‐cell contact duration and morphogen signaling determines cell fate. Dev Cell 43, 198–211.e12. [DOI] [PubMed] [Google Scholar]
- 163. Oakes PW, Wagner E, Brand CA, Probst D, Linke M, Schwarz US, Glotzer M & Gardel ML (2017) Optogenetic control of RhoA reveals zyxin‐mediated elasticity of stress fibres. Nat Commun 8, 15817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Bendall SC, Davis KL, Amir ED, Tadmor MD, Simonds EF, Chen TJ, Shenfeld DK, Nolan GP & Pe'er D (2014) Single‐cell trajectory detection uncovers progression and regulatory coordination in human B cell development. Cell 157, 714–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Setty M, Tadmor MD, Reich‐Zeliger S, Angel O, Salame TM, Kathail P, Choi K, Bendall S, Friedman N & Pe'er D (2016) Wishbone identifies bifurcating developmental trajectories from single‐cell data. Nat Biotechnol 34, 637–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Cannoodt R, Saelens W & Saeys Y (2016) Computational methods for trajectory inference from single‐cell transcriptomics. Eur J Immunol 46, 2496–2506. [DOI] [PubMed] [Google Scholar]
- 167. Grün D, Muraro MJ, Boisset JC, Wiebrands K, Lyubimova A, Dharmadhikari G, van den Born M, van Es J, Jansen E, Clevers H et al (2016) De Novo prediction of stem cell identity using single‐cell transcriptome data. Cell Stem Cell 19, 266–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Lu R, Markowetz F, Unwin RD, Leek JT, Airoldi EM, MacArthur BD, Lachmann A, Rozov R, Ma'ayan A, Boyer LA et al (2009) Systems‐level dynamic analyses of fate change in murine embryonic stem cells. Nature 462, 358–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Klein AM, Mazutis L, Akartuna I, Tallapragada N, Veres A, Li V, Peshkin L, Weitz DA & Kirschner MW (2015) Droplet barcoding for single‐cell transcriptomics applied to embryonic stem cells. Cell 161, 1187–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Briggs JA, Weinreb C, Wagner DE, Megason S, Peshkin L, Kirschner MW & Klein AM (2018) The dynamics of gene expression in vertebrate embryogenesis at single‐cell resolution. Science 360, eaar5780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Farrell JA, Wang Y, Riesenfeld SJ, Shekhar K, Regev A & Schier AF (2018) Single‐cell reconstruction of developmental trajectories during zebrafish embryogenesis. Science 360, eaar3131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. Chevrier S, Levine JH, Zanotelli VRT, Silina K, Schulz D, Bacac M, Ries CH, Ailles L, Jewett MAS, Moch H et al (2017) An immune atlas of clear cell renal cell carcinoma. Cell 169, 736–749.e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. Elia N & Lippincott‐Schwartz J (2009) Culturing MDCK cells in three dimensions for analyzing intracellular dynamics. Curr Protoc Cell Biol 43, 4.22.1–4.22.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174. Montévil M, Speroni L, Sonnenschein C & Soto AM (2016) Modeling mammary organogenesis from biological first principles: cells and their physical constraints. Prog Biophys Mol Biol 122, 58–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175. Liu JS, Farlow JT, Paulson AK, Labarge MA & Gartner ZJ (2012) Programmed cell‐to‐cell variability in ras activity triggers emergent behaviors during mammary epithelial morphogenesis. Cell Rep 2, 1461–1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176. Denker HW (2016) Self‐organization of stem cell colonies and of early mammalian embryos: recent experiments shed new light on the role of autonomy vs external instructions in basic body plan development. Cells 5, 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Puzan M, Hosic S, Ghio C & Koppes A (2018) Enteric nervous system regulation of intestinal stem cell differentiation and epithelial monolayer function. Sci Rep 8, 6313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. Haas BJ & Whited JL (2017) Advances in decoding axolotl limb regeneration. Trends Genet 33, 553–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179. Lefevre JG, Chiu HS, Combes AN, Vanslambrouck JM, Ju A, Hamilton NA & Little MH (2017) Self‐organisation after embryonic kidney dissociation is driven via selective adhesion of ureteric epithelial cells. Development 144, 1087–1096. [DOI] [PubMed] [Google Scholar]
- 180. Lancaster MA & Knoblich JA (2014) Organogenesis in a dish: modeling development and disease using organoid technologies. Science 345, 1247125. [DOI] [PubMed] [Google Scholar]
- 181. Renner M, Lancaster MA, Bian S, Choi H, Ku T, Peer A, Chung K & Knoblich JA (2017) Self‐organized developmental patterning and differentiation in cerebral organoids. EMBO J 36, 1316–1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182. Spence JR, Mayhew CN, Rankin SA, Kuhar MF, Vallance JE, Tolle K, Hoskins EE, Kalinichenko VV, Wells SI, Zorn AM et al (2011) Directed differentiation of human pluripotent stem cells into intestinal tissue in vitro . Nature 470, 105–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183. Workman MJ, Mahe MM, Trisno S, Poling HM, Watson CL, Sundaram N, Chang CF, Schiesser J, Aubert P, Stanley EG et al (2017) Engineered human pluripotent‐stem‐cell‐derived intestinal tissues with a functional enteric nervous system. Nat Med 23, 49–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184. Wells JM & Spence JR (2014) How to make an intestine. Development 141, 752–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185. Tetteh PW, Basak O, Farin HF, Wiebrands K, Kretzschmar K, Begthel H, van den Born M, Korving J, de Sauvage F, van Es JH et al (2016) Replacement of lost Lgr5‐positive stem cells through plasticity of their enterocyte‐lineage daughters. Cell Stem Cell 18, 203–213. [DOI] [PubMed] [Google Scholar]
- 186. Yin X, Farin HF, van Es JH, Clevers H, Langer R & Karp JM (2014) Niche‐independent high‐purity cultures of Lgr5 + intestinal stem cells and their progeny. Nat Methods 11, 106–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187. Forster R, Chiba K, Schaeffer L, Regalado SG, Lai CS, Gao Q, Kiani S, Farin HF, Clevers H, Cost GJ et al (2014) Human intestinal tissue with adult stem cell properties derived from pluripotent stem cells. Stem Cell Reports 2, 838–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188. Gjorevski N, Sachs N, Manfrin A, Giger S, Bragina ME, Ordóñez‐Morán P, Clevers H & Lutolf MP (2016) Designer matrices for intestinal stem cell and organoid culture. Nature 539, 560–564. [DOI] [PubMed] [Google Scholar]
- 189. Meinhardt A, Eberle D, Tazaki A, Ranga A, Niesche M, Wilsch‐Bräuninger M, Stec A, Schackert G, Lutolf M & Tanaka EM (2014) 3D reconstitution of the patterned neural tube from embryonic stem cells. Stem Cell Reports 3, 987–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190. Kretzschmar K & Clevers H (2016) Organoids: modeling development and the stem cell niche in a dish. Dev Cell 38, 590–600. [DOI] [PubMed] [Google Scholar]
- 191. Simian M & Bissell MJ (2017) Organoids: a historical perspective of thinking in three dimensions. J Cell Biol 216, 31–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192. Elkabetz Y, Panagiotakos G, Al Shamy G, Socci ND, Tabar V & Studer L (2008) Human ES cell‐derived neural rosettes reveal a functionally distinct early neural stem cell stage. Genes Dev 22, 152–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193. Ying Q‐L, Stavridis M, Griffiths D, Li M & Smith A (2003) Conversion of embryonic stem cells into neuroectodermal precursors in adherent monoculture. Nat Biotechnol 21, 183. [DOI] [PubMed] [Google Scholar]
- 194. Ramachandran SD, Schirmer K, Münst B, Heinz S, Ghafoory S, Wölfl S, Simon‐Keller K, Marx A, Øie CI, Ebert MP et al (2015) In vitro generation of functional liver organoid‐like structures using adult human cells. PLoS ONE 10, e0139345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195. Dye BR, Hill DR, Ferguson MA, Tsai YH, Nagy MS, Dyal R, Wells JM, Mayhew CN, Nattiv R, Klein OD et al (2015) In vitro generation of human pluripotent stem cell derived lung organoids. ELife 4, e05098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196. Schweiger PJ & Jensen KB (2016) Modeling human disease using organotypic cultures. Curr Opin Cell Biol 43, 22–29. [DOI] [PubMed] [Google Scholar]
- 197. Sachs N & Clevers H (2014) Organoid cultures for the analysis of cancer phenotypes. Curr Opin Genet Dev 24, 68–73. [DOI] [PubMed] [Google Scholar]
- 198. Takasato M, Er PX, Chiu HS, Maier B, Baillie GJ, Ferguson C, Parton RG, Wolvetang EJ, Roost MS, Chuva de Sousa Lopes SM et al (2016) Kidney organoids from human iPS cells contain multiple lineages and model human nephrogenesis. Nature 536, 238. [DOI] [PubMed] [Google Scholar]
- 199. Rivron NC, Frias‐Aldeguer J, Vrij EJ, Boisset JC, Korving J, Vivié J, Truckenmüller RK, van Oudenaarden A, van Blitterswijk CA & Geijsen N (2018) Blastocyst‐like structures generated solely from stem cells. Nature 557, 106–111. [DOI] [PubMed] [Google Scholar]
- 200. Turner DA, Girgin M, Alonso‐Crisostomo L, Trivedi V, Baillie‐Johnson P, Glodowski CR, Hayward PC, Collignon J, Gustavsen C, Serup P et al (2017) Anteroposterior polarity and elongation in the absence of extra‐embryonic tissues and of spatially localised signalling in gastruloids: mammalian embryonic organoids. Development 144, 3894–3906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201. Greicius G, Kabiri Z, Sigmundsson K, Liang C, Bunte R, Singh MK & Virshup DM (2018) PDGFRalpha(+) pericryptal stromal cells are the critical source of Wnts and RSPO3 for murine intestinal stem cells in vivo . Proc Natl Acad Sci USA 115, E3173–e3181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202. Shoshkes‐Carmel M, Wang YJ, Wangensteen KJ, Tóth B, Kondo A, Massasa EE, Itzkovitz S & Kaestner KH (2018) Subepithelial telocytes are an important source of Wnts that supports intestinal crypts. Nature 557, 242–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203. Czerniecki SM, Cruz NM, Harder JL, Menon R, Annis J, Otto EA, Gulieva RE, Islas LV, Kim YK, Tran LM et al (2018) High‐throughput screening enhances kidney organoid differentiation from human pluripotent stem cells and enables automated multidimensional phenotyping. Cell Stem Cell 22, 929–940.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204. Rios AC & Clevers H (2018) Imaging organoids: a bright future ahead. Nat Methods 15, 24–26. [DOI] [PubMed] [Google Scholar]
- 205. Jamieson PR, Dekkers JF, Rios AC, Fu NY, Lindeman GJ & Visvader JE (2017) Derivation of a robust mouse mammary organoid system for studying tissue dynamics. Development 144, 1065–1071. [DOI] [PubMed] [Google Scholar]
- 206. Okkelman IA, Foley T, Papkovsky DB & Dmitriev RI (2017) Live cell imaging of mouse intestinal organoids reveals heterogeneity in their oxygenation. Biomaterials 146, 86–96. [DOI] [PubMed] [Google Scholar]
- 207. McKinley KL, Stuurman N, Royer LA, Schartner C, Castillo‐Azofeifa D, Delling M, Klein OD & Vale RD (2018) Cellular aspect ratio and cell division mechanics underlie the patterning of cell progeny in diverse mammalian epithelia. ELife 7, e36739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208. Tsai Y‐H, Nattiv R, Dedhia PH, Nagy MS, Chin AM, Thomson M, Klein OD & Spence JR (2017) In vitro patterning of pluripotent stem cell‐derived intestine recapitulates in vivo human development. Development 144, 1045–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209. Nusse YM, Savage AK, Marangoni P, Rosendahl‐Huber AKM, Landman TA, de Sauvage FJ, Locksley RM & Klein OD (2018) Parasitic helminths induce fetal‐like reversion in the intestinal stem cell niche. Nature 559, 109–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210. Steinberg MS (1958) On the chemical bonds between animal cells a mechanism for type‐specific association. Am Nat 92, 65–81. [Google Scholar]
- 211. Panciera T, Azzolin L, Cordenonsi M & Piccolo S (2017) Mechanobiology of YAP and TAZ in physiology and disease. Nat Rev Mol Cell Biol 18, 758–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212. Dupont S, Morsut L, Aragona M, Enzo E, Giulitti S, Cordenonsi M, Zanconato F, Le Digabel J, Forcato M, Bicciato S et al (2011) Role of YAP/TAZ in mechanotransduction. Nature 474, 179–183. [DOI] [PubMed] [Google Scholar]
- 213. Yui S, Nakamura T, Sato T, Nemoto Y, Mizutani T, Zheng X, Ichinose S, Nagaishi T, Okamoto R, Tsuchiya K et al (2012) Functional engraftment of colon epithelium expanded in vitro from a single adult Lgr5(+) stem cell. Nat Med 18, 618–623. [DOI] [PubMed] [Google Scholar]
- 214. Dekkers JF, Wiegerinck CL, de Jonge HR, Bronsveld I , Janssens HM, de Winter‐de Groot KM, Brandsma AM, de Jong NW, Bijvelds MJ, Scholte BJ et al (2013) A functional CFTR assay using primary cystic fibrosis intestinal organoids. Nat Med 19, 939. [DOI] [PubMed] [Google Scholar]
- 215. Saini A (2016) Cystic fibrosis patients benefit from mini guts. Cell Stem Cell 19, 425–427. [Google Scholar]


