Abstract
Serum starvation is a widely used condition in molecular biology experiments. Opti‐MEM is a serum‐reduced media used during transfection of genetic molecules into mammalian cells. However, the impact of such media on cell viability and protein synthesis is unknown. A549 human lung epithelial cell viability and morphology were adversely affected by growing in Opti‐MEM. The cellular protein levels of chloride intracellular channel protein 1, proteasome subunit alpha Type 2, and heat shock 70 kDa protein 5 were dysregulated in A549 cells after growing in serum‐reduced media. Small interfering RNA transfection was done in Dulbecco's modified Eagle's medium (DMEM) with 10% fetal bovine serum, and knockdown efficacy was determined compared with Opti‐MEM. Similar amounts of knockdown of the target proteins were achieved in DMEM, and cell viability was higher compared with Opti‐MEM after transfection. Careful consideration of the impact of Opti‐MEM media during the culture or transfection is important for experimental design and results interpretation.
Keywords: cellular protein expression, Dulbecco's modified Eagle's medium (DMEM), opti‐MEM, serum starvation, transfection
1. INTRODUCTION
Fetal bovine serum (FBS) is one of the most commonly used supplements in eukaryotic cell culture media, but as a complex natural product its composition is poorly defined and may vary between lots from the same manufacturer (Zheng et al., 2006). Maintaining consistent cell growth condition is often very difficult in FBS containing media and can lead to inconsistent or opposing results during bioassays (Krämer, Bouzakri, Holmqvist, Al‐Khalili, & Krook, 2005; Mannello & Tonti, 2007). Thus, serum is often eliminated from the media to remove the known factors to reduce analytical interference and provide more reproducible experimental conditions (Colzani et al., 2009; Lambert & Pirt 1979; Mbeunkui, Fodstad, & Pannell, 2006).
Serum starvation is defined as growing cells in either serum‐free, serum‐reduced, or serum protein‐free medium (Pirkmajer & Chibalin, 2011), which has been used as a tool for molecular mechanism studies, such as autophagy, apoptosis (Bhutia et al., 2010; Terra, Garay‐Malpartida, Wailemann, Sogayar, & Labriola, 2011; Zhao et al., 2010), and cellular stress response (Arrington & Schnellmann, 2008; Levin et al., 2010). Although serum starvation has been performed in hundreds of research studies, the impact of the condition is not well understood (Pirkmajer & Chibalin, 2011).
Serum starvation is often referred to as “environmental stress” (Liu et al., 2010), which reduces basal cellular activity (Codeluppi et al., 2011). Intercellular signaling pathways are widely affected by serum starvation, but the impact is unpredictable and depends on cell types (Oya, Zölzer, Werner, & Streffer, 2003; Pirkmajer & Chibalin, 2011). A proteomic study has showed the divergent responses on different signaling pathways by serum starvation in different tumor cell types (Levin et al., 2010). Several studies have also demonstrated the impact on different aspects of signaling pathways in response to serum‐free conditions (Ching, Rajguru, Marupudi, Banerjee, & Fisher, 2010, Sancho & Fabregat, 2010; Tan, You, Wu, Altomare, & Testa, 2010).
Serum‐free or serum‐reduced media is often recommended for transfection (http://dharmacon.gelifesciences.com/uploadedFiles/Resources/basic‐dharmafect‐protocol.pdf). Transfection is a sophisticated but very common method in molecular biology that is used to deliver nucleic acids (DNA or RNA) into cells for a variety of applications. The efficiency of transfection depends on different factors, such as transfection reagent, nucleic acid dose, cell type, cell density, media composition, and incubation time (https://cellculturedish.com/ask‐the‐expert/transfection‐optimization‐improved‐efficiency‐performance/). Serum proteins in the media also compete with transfection vehicles/viral vectors for cell‐surface receptors for entrance into cells. Serum starvation helps cell cycle synchronization by delaying the G0/G1 stage (Khammanit, Chantakru, Kitiyanant, & Saikhun, 2008).
Opti‐MEM is a serum‐reduced media that contains insulin, transferrin, hypoxanthine, thymidine, and trace elements that compensate for the adverse impact of FBS supplementation reduction with no change in the growth rate or morphology (https://www.thermofisher.com/order/catalog/product/31985070). This media have been used for more than a decade in many research studies for diluting the transfection reagents, sometimes as a transfection media (Chen et al., 2017; Hu et al., 2018; Ivanovic et al., 2018; Zou et al., 2004) and is also recommended for mammalian cell culture (http://dharmacon.gelifesciences.com/uploadedFiles/Resources/basic‐dharmafect‐protocol.pdf).
A549 is a tumor cell line from a human lung carcinoma that has been used in numerous research studies for more than 50 years (Gazdar et al., 2010; Lieber et al., 1976). Serum starvation induces higher expression of E‐cadherin in human lung A549 epithelial cells (Dong et al., 2014). Many studies have performed transfection of A549 cells in Opti‐MEM, but the impact of the media on cell morphology, cell viability, and protein expression is not defined by any of the published literature. In this study we have evaluated cell viability, morphology, and the expression profile of three cellular proteins (proteasome subunit alpha Type 2 [PSMA2], chloride intracellular channel protein 1 [CLIC1], and heat shock 70 kDa protein 5 [HSPA5]) in A549 cells after growing the cells in Opti‐MEM media. Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% FBS was used as a control to determine unaffected expression profiles of the proteins. Small interfering RNA (siRNA)‐transfection efficacy was evaluated in complete media with 10% FBS (DMEM) and compared with serum‐reduced media (Opti‐MEM) to determine if transfection could be performed in DMEM.
2. MATERIALS AND METHODS
2.1. Cells
Human A549 lung epithelial cells were grown in DMEM (GIBCO, Grand Island, NY) after supplementing with 10% FBS (Life Technologies, Thermo Fisher Scientific, Burlington, Ontario, Canada), Na‐pyruvate, l‐glutamine, and nonessential amino acids at 37°C in a humidified incubator with 5% CO2 as described (Coombs et al., 2010). Cells were maintained as monolayers and were passaged by trypsinization three times each week.
2.2. Cell viability
Cell viability was determined using cell proliferation reagent WST‐1 (Roche, Penzberg, Upper Bavaria, Germany) according to the manufacturer's protocol. Briefly A549 cells were grown in 96‐well plates and treated as described below. Nine microliter of WST‐1 reagent (Pierce Biotechnology, Rockford, IL) was added to each well and incubated at 37°C. After 2 hr, colorimetric changes were determined by photodensitometer, and cell viability was calculated compared with positive (no treatment as 100%) and negative (killed as 0%) controls. Each experiment was done in two biological replicates and five technical replicates.
2.3. Protein extraction and quantification
To determine the impact of Opti‐MEM on cellular protein expression, A549 cells were grown in either DMEM with 10% FBS or Opti‐MEM. Cells were harvested every day up to 4 days. To compare transfection efficiency, A549 cells were transfected in DMEM with 10% FBS or Opti‐MEM media and harvested every day up to 4 days. Cells were washed three times in phosphate buffered saline (PBS) and lysed with mammalian protein extraction reagent (MPER) supplemented with 1× HALT protease inhibitor. Insoluble cellular components were removed by centrifugation at 14,000g for 15 min at 4°C. The concentration of protein in each cell lysate was determined by the bicinchoninic acid (BCA) protein assay (Pierce, Rockford, IL) and quantified using bovine serum albumin standards.
2.4. Immunoblotting
Western blot was performed as previously described (Ezzati et al., 2015). Thirty micrograms of protein in cell lysate were separated in 10% sodium dodecyl sulfate‐polyacrylamide gel electrophoresis gels and transferred to 0.2 µm nitrocellulose membranes. Anti‐PSMA2 (Cat. 2455; Cell Signaling Technology, Danvers, MA), anti‐CLIC1 (Cat. MABN46; EMD Millipore, Damstadt, Germany), anti‐HSPA5 (Cat. MABC675; Millipore), anti‐GAPDH (Cat.2118L; Cell Signaling), and anti‐beta actin (Cat. 3700S; Cell Signaling) primary antibodies were used to detect specific proteins. Appropriate secondary horseradish peroxidase–conjugated horse antimouse or antirabbit (cat.7076, cat.7074, respectively; Cell Signaling) were used to detect immune complexes. Bands were developed by enhanced chemiluminescence and imaged with an Alpha Innotech FluorChem Q Multi Image III instrument. Band intensities were quantified with Image J 1.50i (NIH, Bethesda, MD) and graphically presented by the Graphpad Prism software (La Jolla, CA).
2.5. siRNA transfection
A549 cells were grown to 30% to 40% confluency in DMEM media with 10% FBS supplement. Before transfection, cells were washed twice with RNase free PBS and DMEM media with 10% FBS or Opti‐MEM was added into the culture dishes. Smart‐pool siRNA for PSMA2 (25 nM), CLIC1 (50 nM), nontargeting siRNA (Dharmacon) as control, and Dharmafect (Cat. #T‐2001; GE Healthcare Dharmacon, Lafayette, CO) were diluted in Opti‐MEM media following the manufacturer's instructions. The diluted siRNA and Dharmafect were combined, incubated at room temperature for 20 min and added into the A549 cells growing in either Opti‐MEM or DMEM media. Culture dishes were incubated at 37°C in 5% CO2 incubator, and cells were harvested everyday up to 4 days from separate culture dishes.
2.6. Photomicrography
The A549 cells growing in Opti‐MEM and DMEM media, with or without siRNA transfection, were examined every day for 4 days to observe the cytopathic effect of Opti‐MEM media and photographed with a Canon‐A700 digital camera at 200× magnification. Images were imported into power point, and slight adjustments were made in brightness and contrast, which did not alter image context with respect to each other.
2.7. Statistics
Cell viability of A549 cells growing in Opti‐MEM or DMEM, before and after transfection, was compared by unpaired t tests. p < 0.05 was considered statistically significant. All statistical analyses were performed using the Graphpad Prism software.
3. RESULTS AND DISCUSSION
Suppression of protein expression by siRNA transfection often takes 24 to 96 hr depending on the cell type and target protein (McNaughton et al., 2009). Serum free or serum‐reduced media is recommended for transfection to synchronize the cell cycle and reduction of serum protein interference during transport of genetic material into the cells (Khammanit et al., 2008). Understanding the cellular responses to serum starvation conditions is critical for data interpretation and reproducibility. Opti‐MEM is a well‐accepted serum‐reduced media used for transfection experiments. To understand the impact of Opti‐MEM on cell viability and morphology, A549 cells were grown in Opti‐MEM for 4 days, and cell morphology was monitored under the microscope. As a control of optimal growth condition, A549 cells were also grown in DMEM media with 10% FBS. There was no visible change observed in the cells growing in two different media after 24 hr. However, after Day 2, higher numbers of rounded and floating cells were visible in the Opti‐MEM culture dishes compared with the DMEM (Figure 1a). A549 are epithelial cells with characteristic rectangular shape; a rounded morphology is an indication that the cells are under stress and might be dying. Therefore, we monitored the cell viability of A549 cells after growing in two different media. Cell viability of the cells growing in the Opti‐MEM was significantly lower throughout the experiment from Day 1 to Day 4 compared with DMEM (Figure 1b). Serum starvation causes apoptosis‐induced cell death in different human cell lines (Braun et al., 2011; Goyeneche et al., 2006; Huang et al., 2018; Kulkarni and McCulloch, 1994). Serum starvation arrests A549 cells in G1 phase, without inducing apoptosis (Nakhjavani et al., 2016). But in this study, we observed that cell death was increased after growing in serum‐reduced media. The specific cause of the cell death was not determined in this study.
Figure 1.
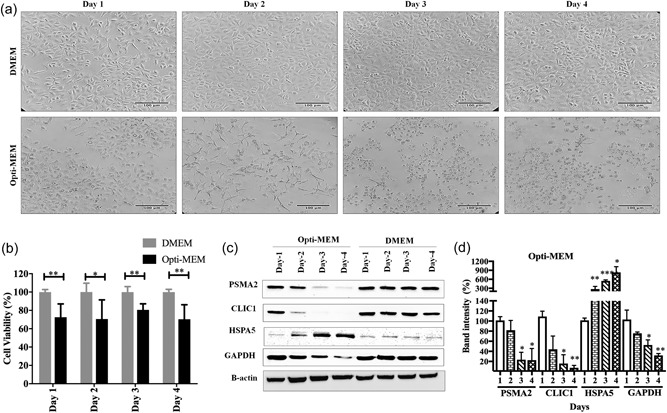
Impact of Opti‐MEM on A549 cells viability, morphology, and protein expression. (a) Observation of cells under microscope in Opti‐MEM and DMEM for 4 days (200×). Scale bar is 100 μm. (b) A549 cell viability in Opti‐MEM and DMEM media, determined by the WST‐1 assay. (c) Expression of cellular proteins in Opti‐MEM and DMEM by western blot. (d) Quantification of PSMA2, CLIC1, HSPA5, and GAPDH expression in Opti‐MEM compared with DMEM from western blot. ***p < 0.001, **p < 0.01, and *p < 0.05. CLIC1: chloride intracellular channel protein 1; DMEM: Dulbecco's modified Eagle's medium; HSPA5: heat shock 70 kDa protein 5; PSMA2: proteasome subunit alpha Type 2
Next, we wanted to know if growing cells in Opti‐MEM could impact cellular protein expression. As part of another ongoing proteomic study, we had noted that several fibronectin‐interacting proteins (CLIC1, PSMA2, and HSPA5) were involved in influenza virus growth. Thus, for the current study, A549 cells were grown in Opti‐MEM for 4 days, and protein expression of CLIC1, PSMA2, and HSPA5 were determined by western blot every day after harvesting cell lysates. Cells were also grown in DMEM media with 10% FBS supplement as a control to determine the usual expression pattern of the proteins in A549 cells. Expression of these three proteins in DMEM was stable throughout the experimental period. The expressions of PSMA2 and CLIC1 were significantly reduced after 3 days in Opti‐MEM, but HSPA5 expression was significantly increased after 2 days of incubation in Opti‐MEM (Figure 1c,d). In addition, a widely‐used loading control, GAPDH, was observed to decrease its expression significantly after Day 3 in Opti‐MEM (Figure 1c,d).
A protein microarray‐based study has shown that many proteins and phosphoproteins involved in EGFR‐MAPK‐Stat, PTEN‐PI‐3 kinase‐AKT, and transcription activator‐polyamine signaling pathways were dysregulated after 24 hr of FBS starvation in glioma and adenocarcinoma cell lines (Levin et al., 2010). Serum starvation can also induce changes in ERK1/2, phospho‐ACC, and phospho‐Akt signaling pathways (Pirkmajer & Chibalin, 2011). Time‐dependent differential expression of GAPDH was also observed after serum starvation in primary human myotubes and HEK293 cells (Pirkmajer and Chibalin, 2011). GAPDH was also found as an unstable reference marker in a colitis mouse model (Eissa et al., 2017). Therefore, a careful consideration of the impact of the media or the experimental condition is very important for choosing the appropriate loading control. Serum starvation dysregulates different cellular signaling pathways, which might affect the expression of different cellular proteins. A proteomic study of serum‐starved cells by mass spectrometry might help us to get a deeper understanding of the impact on cell signaling pathways and cellular functions.
As A549 cell viability and cellular protein expression were affected in Opti‐MEM serum‐reduced media, we wanted to know if similar amounts of knockdown could be achieved by siRNA transfection in complete DMEM media. So, siRNA transfection was performed in A549 cells targeting PSMA2 and CLIC1 genes in complete DMEM media. Knockdown of the target proteins in Opti‐MEM media was used to evaluate the transfection efficacy by side‐by‐side comparison. Nonsilencing siRNA (NSC) was used as a control in both media. Protein expression or knockdown was monitored by western blot everyday up to 4 days after transfection. Compared with NSC, a significant amount of CLIC1 knockdown was achieved by the siRNA in A549 cells 2 days after transfection in both types of media (Figure 2c,d). However, cell viability of the CLIC1 knockdown cells was significantly higher in DMEM compared with Opti‐MEM (Figure 2b). The cell morphology was adversely affected in Opti‐MEM media but was better in DMEM media after CLIC1 KD (Figure 2a). Statistically, there were significant differences in knockdown efficacy on Day 3 and Day 4 between Opti‐MEM and DMEM (Figure 2d), which was because of the reduction of CLIC1 protein expression in control cells. Overall, a stable knockdown of CLIC1 was achieved in DMEM media with less impact on cell viability and morphology compared with Opti‐MEM.
Figure 2.
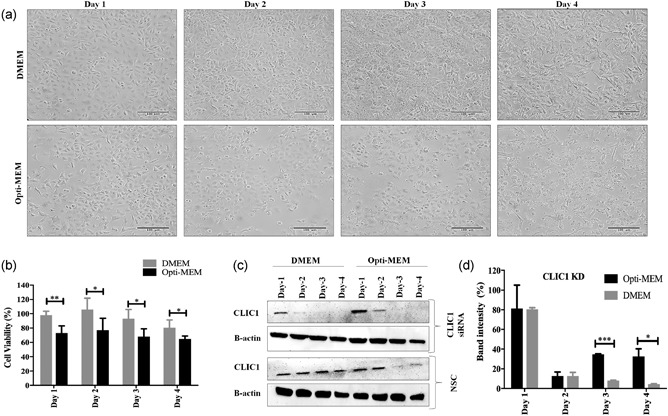
CLIC1 knockdown efficacy in DMEM media compared with Opti‐MEM. (a) A549 cell morphology under microscope (200×) after knockdown of CLIC1 protein. Scale bar is 100 μm. (b) A549 cell viability after CLIC1 knockdown in DMEM and Opti‐MEM media determined by the WST‐1 assay. (c) Knockdown efficacy of CLIC1 siRNA in DMEM and Opti‐MEM media determined by western blot. (d) Quantification of CLIC1 knockdown efficacy siRNA in Opti‐MEM from western blot. ***p < 0.001, **p < 0.01, and *p < 0.05. CLIC1: chloride intracellular channel protein 1; DMEM: Dulbecco's modified Eagle's medium; KD: knockdown; NSC: nonsilencing control (scrambled siRNA); siRNA: small interfering RNA
Transfection of PSMA2 was more efficient in DMEM than in Opti‐MEM media. A significant amount of PSMA2 knockdown was achieved in A549 cells after 2 days of transfection in DMEM. Similar amounts of knockdown were achieved in Opti‐MEM after 4 days of transfection (Figure 3c,d). Cell viability was significantly lower in cells growing in Opti‐MEM than in DMEM after transfection of PSMA2 (Figure 3b). PSMA2 is an important component of the 20S proteasome complex. Knockdown of the protein also affected the cell viability in DMEM after 3 days of transfection but was significantly lower in Opti‐MEM. Cell morphology under the microscope was adversely affected in Opti‐MEM compared with DMEM after PSMA2 transfection (Figure 2a). As with CLIC1 KD, successful knockdown of PSMA2 was achieved in DMEM media with better cell viability and less impact on cell morphology than with Opti‐MEM.
Figure 3.
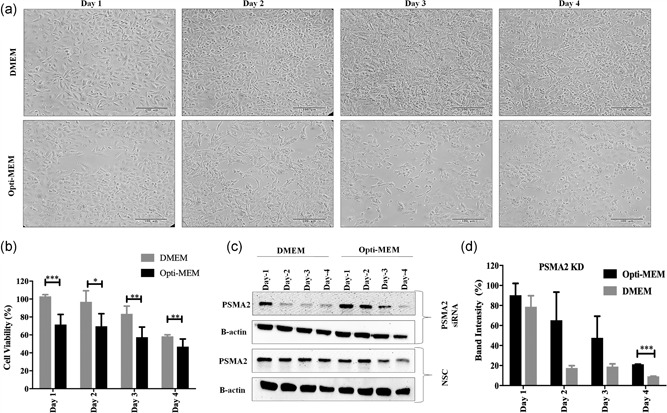
PSMA2 knockdown efficacy in DMEM media compared with Opti‐MEM. (a) A549 cell morphology under microscope (200×) after knockdown of PSMA2 protein. Scale bar is 100 μm. (b) A549 cell viability after PSMA2 knockdown in DMEM and Opti‐MEM media determined by WST‐1 assay. (c) Knockdown efficacy of PSAM2 siRNA in DMEM and Opti‐MEM media determined by western blot. (d) Quantification of PSMA2 knockdown efficacy siRNA in Opti‐MEM from western blot. ***p < 0.001, **p < 0.01, and *p < 0.05. DMEM: Dulbecco's modified Eagle's medium; KD: knockdown; NSC: nonsilencing control (scrambled siRNA); PSMA2: proteasome subunit alpha Type 2; siRNA: small interfering RNA
During any experiment, the adverse impact of media on cellular protein expression and cell viability can lead to misinterpretation of the results. DMEM media supplemented with 10% FBS could be a better alternative for transfection of A549 cells, as cells were growing optimally, expression of proteins was stable as tested with PSMA2, CLIC1, and HSPA5, and transfection efficacy was similar compared with serum‐reduced media.
4. CONCLUSIONS
An appropriate experimental condition is critical for data reproducibility; and an undesirable impact of the media may lead to the misinterpretation of the data. A549 cells were stressed, and cellular protein expression was destabilized after growing in the Opti‐MEM. Consideration of these facts is necessary while using Opti‐MEM as a culture or transfection medium. DMEM media with all nutrients and FBS could be an alternative option as a transfection media for A549 cells. The study was limited by testing only three proteins and only one cell type; thus, we cannot comment about the effects on other cellular proteins or in other cell types.
CONFLICTS OF INTEREST
The authors declare that they have no conflicts of interest.
ACKNOWLEDGMENTS
This study was funded by the Canadian Institutes of Health Research (CIHR) grant no. MOP‐106713. We would also like to thank Ali Zahedi Amiri for his help with data analysis with the Graphpad Prism software.
Rashid M-u, Coombs KM. Serum‐reduced media impacts on cell viability and protein expression in human lung epithelial cells. J Cell Physiol. 2019;234:7718–7724. 10.1002/jcp.27890
References
REFERENCES
- http://dharmacon.gelifesciences.com/uploadedFiles/Resources/basic‐dharmafect‐protocol.pdf. Date 10 August 2018.
- https://cellculturedish.com/ask‐the‐expert/transfection‐optimization‐improved‐efficiency‐performance/. Date 10 August 2018.
- https://www.thermofisher.com/order/catalog/product/31985070. Date 10 August 2018.
- Arrington, D. D. , & Schnellmann, R. G. (2008). Targeting of the molecular chaperone oxygen‐regulated protein 150 (ORP150) to mitochondria and its induction by cellular stress. American Journal of Physiology‐Cell Physiology, 294(2), C641–C650. [DOI] [PubMed] [Google Scholar]
- Bhutia, S. K. , Kegelman, T. P. , Das, S. K. , Azab, B. , Su, Z. , Lee, S. G. , … Fisher, P. B. (2010). Astrocyte elevated gene‐1 induces protective autophagy. Proceedings of the National Academy of Sciences, 107(51), 22243–22248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun, F. , Bertin‐Ciftci, J. , Gallouet, A. S. , Millour, J. , & Juin, P. (2011). Serum‐nutrient starvation induces cell death mediated by Bax and Puma that is counteracted by p21 and unmasked by Bcl‐xL inhibition. PLoS One, 6(8), e23577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, B. , Yoo, K. , Xu, W. , Pan, R. , Han, X. X. , & Chen, P. (2017). Characterization and evaluation of a peptide‐based siRNA delivery system in vitro. Drug Delivery and Translational Research, 7(4), 507–515. [DOI] [PubMed] [Google Scholar]
- Ching, J. K. , Rajguru, P. , Marupudi, N. , Banerjee, S. , & Fisher, J. S. (2010). A role for AMPK in increased insulin action after serum starvation. American Journal of Physiology‐Cell Physiology, 299(5), C1171–C1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Codeluppi, S. , Gregory, E. N. , Kjell, J. , Wigerblad, G. , Olson, L. , & Svensson, C. I. (2011). Influence of rat substrain and growth conditions on the characteristics of primary cultures of adult rat spinal cord astrocytes. Journal of Neuroscience Methods, 197(1), 118–127. [DOI] [PubMed] [Google Scholar]
- Colzani, M. , Waridel, P. , Laurent, J. , Faes, E. , Rüegg, C. , & Quadroni, M. (2009). Metabolic labeling and protein linearization technology allow the study of proteins secreted by cultured cells in serum‐containing media. Journal of Proteome Research, 8(10), 4779–4788. [DOI] [PubMed] [Google Scholar]
- Coombs, K. M. , Berard, A. , Xu, W. , Krokhin, O. , Meng, X. , Cortens, J. P. , … Brown, E. G. (2010). Quantitative proteomic analyses of influenza virus‐infected cultured human lung cells. Journal of Virology, 84(20), 10888–10906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong, S. , Khoo, A. , Wei, J. , Bowser, R. K. , Weathington, N. M. , Xiao, S. , & Zhao, J. (2014). Serum starvation regulates E‐cadherin upregulation via activation of c‐Src in non‐small‐cell lung cancer A549 cells. American Journal of Physiology‐Cell Physiology, 307(9), C893–C899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eissa, N. , Kermarrec, L. , Hussein, H. , Bernstein, C. N. , & Ghia, J. E. (2017). Appropriateness of reference genes for normalizing messenger RNA in mouse 2, 4‐dinitrobenzene sulfonic acid (DNBS)‐induced colitis using quantitative real time PCR. Scientific Reports, 7, 42427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezzati, P. , Komher, K. , Severini, G. , & Coombs, K. M. (2015). Comparative proteomic analyses demonstrate enhanced interferon and STAT‐1 activation in reovirus T3D‐infected HeLa cells. Frontiers in Cellular and Infection Microbiology, 5, 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazdar, A. F. , Girard, L. , Lockwood, W. W. , Lam, W. L. , & Minna, J. D. (2010). Lung cancer cell lines as tools for biomedical discovery and research. Journal of the National Cancer Institute, 102(17), 1310–1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goyeneche, A. A. , Harmon, J. M. , & Telleria, C. M. (2006). Cell death induced by serum deprivation in luteal cells involves the intrinsic pathway of apoptosis. Reproduction, 131(1), 103–111. [DOI] [PubMed] [Google Scholar]
- Hu, R. , Cao, Q. , Sun, Z. , Chen, J. , Zheng, Q. , & Xiao, F. (2018). A novel method of neural differentiation of PC12 cells by using Opti‐MEM as a basic induction medium. International Journal of Molecular Medicine, 41(1), 195–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, Y. , Fu, Z. , Dong, W. , Zhang, Z. , Mu, J. , & Zhang, J. (2018). Serum starvation‐induces down‐regulation of Bcl‐2/Bax confers apoptosis in tongue coating‐related cells in vitro. Molecular Medicine Reports, 17(4), 5057–5064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanovic, R. F. , Viana, N. I. , Morais, D. R. , Silva, I. A. , Leite, K. R. , Pontes‐Junior, J. , … Reis, S. T. (2018). miR‐29b enhances prostate cancer cell invasion independently of MMP‐2 expression. Cancer Cell International, 18(1), 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khammanit, R. , Chantakru, S. , Kitiyanant, Y. , & Saikhun, J. (2008). Effect of serum starvation and chemical inhibitors on cell cycle synchronization of canine dermal fibroblasts. Theriogenology, 70(1), 27–34. [DOI] [PubMed] [Google Scholar]
- Krämer, D. K. , Bouzakri, K. , Holmqvist, O. , Al‐Khalili, L. , & Krook, A. (2005). Effect of serum replacement with plysate on cell growth and metabolismin primary cultures of human skeletal muscle. Cytotechnology, 48(1‐3), 89–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni, G. V. , & McCulloch, C. A. (1994). Serum deprivation induces apoptotic cell death in a subset of Balb/c 3T3 fibroblasts. Journal of Cell Science, 107(5), 1169–1179. [DOI] [PubMed] [Google Scholar]
- Lambert, K. , & Pirt, S. J. (1979). Growth of human diploid cells (strain MRC‐5) in defined medium; replacement of serum by a fraction of serum ultrafiltrate. Journal of Cell Science, 35(1), 381–392. [DOI] [PubMed] [Google Scholar]
- Levin, V. A. , Panchabhai, S. C. , Shen, L. , Kornblau, S. M. , Qiu, Y. , & Baggerly, K. A. (2010). Different changes in protein and phosphoprotein levels result from serum starvation of high‐grade glioma and adenocarcinoma cell lines. Journal of Proteome Research, 9(1), 179–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieber, M. , Todaro, G. , Smith, B. , Szakal, A. , & Nelson‐Rees, W. (1976). A continuous tumor‐cell line from a human lung carcinoma with properties of type II alveolar epithelial cells. International Journal of Cancer, 17(1), 62–70. [DOI] [PubMed] [Google Scholar]
- Liu, H. S. , Hsu, P. Y. , Lai, M. D. , Chang, H. Y. , Ho, C. L. , Cheng, H. L. , … Chow, N. H. (2010). An unusual function of RON receptor tyrosine kinase as a transcriptional regulator in cooperation with EGFR in human cancer cells. Carcinogenesis, 31(8), 1456–1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mannello, F. , & Tonti, G. A. (2007). Concise review: No breakthroughs for human mesenchymal and embryonic stem cell culture: Conditioned medium, feeder layer, or feeder‐free; medium with fetal calf serum, human serum, or enriched plasma; serum‐free, serum replacement nonconditioned medium, or ad hoc formula? All that glitters is not gold!. Stem Cells, 25(7), 1603–1609. [DOI] [PubMed] [Google Scholar]
- Mbeunkui, F. , Fodstad, O. , & Pannell, L. K. (2006). Secretory protein enrichment and analysis: An optimized approach applied on cancer cell lines using 2D LC− MS/MS. Journal of Proteome Research, 5(4), 899–906. [DOI] [PubMed] [Google Scholar]
- McNaughton, B. R. , Cronican, J. J. , Thompson, D. B. , & Liu, D. R. (2009). Mammalian cell penetration, siRNA transfection, and DNA transfection by supercharged proteins. Proceedings of the National Academy of Sciences, 106(15), 6111–6116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakhjavani, M. , Nikounezhad, N. , Ashtarinezhad, A. , & Shirazi, F. H. (2016). Human lung carcinoma reaction against metabolic serum deficiency stress. Iranian Journal of Pharmaceutical Research, 15(4), 817–823. [PMC free article] [PubMed] [Google Scholar]
- Oya, N. , Zölzer, F. , Werner, F. , & Streffer, C. (2003). Effects of serum starvation on radiosensitivity, proliferation and apoptosis in four human tumor cell lines with different p53 status. Strahlentherapie und Onkologie, 179(2), 99–106. [DOI] [PubMed] [Google Scholar]
- Pirkmajer, S. , & Chibalin, A. V. (2011). Serum starvation: Caveat emptor. American Journal of Physiology‐Cell Physiology, 301(2), C272–C279. [DOI] [PubMed] [Google Scholar]
- Sancho, P. , & Fabregat, I. (2010). The NADPH oxidase NOX1 controls autocrine growth of liver tumor cells through up‐regulation of the epidermal growth factor receptor pathway. Journal of Biological Chemistry, 285, 24815–24824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan, Y. , You, H. , Wu, C. , Altomare, D. A. , & Testa, J. R. (2010). Appl1 is dispensable for mouse development, and loss of Appl1 has growth factor‐selective effects on Akt signaling in murine embryonic fibroblasts. Journal of Biological Chemistry, 285(9), 6377–6389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terra, L. F. , Garay‐Malpartida, M. H. , Wailemann, R. A. M. , Sogayar, M. C. , & Labriola, L. (2011). Recombinant human prolactin promotes human beta cell survival via inhibition of extrinsic and intrinsic apoptosis pathways. Diabetologia, 54(6), 1388–1397. [DOI] [PubMed] [Google Scholar]
- Zhao, Y. , Yang, J. , Liao, W. , Liu, X. , Zhang, H. , Wang, S. , … Zhu, W. G. (2010). Cytosolic FoxO1 is essential for the induction of autophagy and tumour suppressor activity. Nature Cell Biology, 12(7), 665–675. [DOI] [PubMed] [Google Scholar]
- Zheng, X. , Baker, H. , Hancock, W. S. , Fawaz, F. , McCaman, M. , & Pungor, E. (2006). Proteomic analysis for the assessment of different lots of fetal bovine serum as a raw material for cell culture. Part IV. Application of proteomics to the manufacture of biological drugs. Biotechnology Progress, 22(5), 1294–1300. [DOI] [PubMed] [Google Scholar]
- Zou, S. Q. , Qu, Z. L. , Li, Z. F. , & Wang, X. (2004). Hepatitis B virus X gene induces human telomerase re verse transcriptase mRNA expression in cultured normal human cholangiocytes. World Journal of Gastroenterology, 10(15), 2259. [DOI] [PMC free article] [PubMed] [Google Scholar]


