Abstract
Background: The promising aspect of circulating tumor DNA (ctDNA) is its rapid turnaround and non-invasive nature. According to the American Society of Clinical Oncology (ASCO) and College of American Pathologists joint ctDNA review published in March 2018, there is not sufficient evidence to support the use of ctDNA in practice for GI cancers. However, there were numerous studies presented at ASCO Annual Meeting supporting its value. We aimed to summarize on its role in the management of gastrointestinal cancers based on the studies presented recently, and future directions.
Methods: We limited our search to keywords “ctDNA,” “circulating tumor DNA,” “cell-free DNA (cfDNA)” and/or “liquid biopsy,” at the 2018 ASCO Annual Meeting library abstracts and presentations.
Results: There were 35 studies that revolved around ctDNA as a diagnostic tool, prognostic marker and/or a measure of tumor heterogeneity in gastrointestinal cancers. Depending on the assay used, the results of several studies showed that ctDNA was able to identify relevant mutations or fusions including RAS, HER2/Neu, BRAF, MET, BRCA2, APC, TP53, ALK, ROS1, PTEN, and NF1. The prognosis in terms of tumor mutation burden, objective response rate, metastasis and survival were also estimated by various studies based on ctDNA. The findings showed that higher baseline ctDNA levels and/or increased number of mutations detected in ctDNA were associated with poor survival and multi-site metastasis. Right-sided colon cancer was associated with higher number of mutations in ctDNA than left-sided colon and rectal cancers. Similarly, tubular adenocarcinoma subtype of gastric cancer was more likely to have higher ctDNA levels than signet-ring cell subtype. The feasibility of assessing response to therapy and residual metastatic disease by using ctDNA which was otherwise not detected on imaging was also presented.
Conclusions: The studies presented at ASCO 2018 report on the many ways ctDNA is of value in patients with gastrointestinal malignancies. Experts and discussants at the meeting argued that this may well indeed be ready for prime time for certain GI malignancies including colorectal cancers, especially in the metastatic setting. These findings alongside ongoing studies showing its feasibility into practice would likely lead to revision of the current guidelines for metastatic GI cancers.
Keywords: circulating tumor DNA, ctDNA, liquid biopsy, ASCO, gastrointestinal cancer, colorectal cancer
Introduction
Circulating tumor DNA (ctDNA), considered as cancer biomarker, is the free DNA found predominantly in plasma and derived from tumor cells. Analysis of ctDNA, also referred to as “liquid biopsy,” is a non-invasive and cheaper technique allowing for serial oncologic assessments, though several controversies exist. In March 2018, American Society of Clinical Oncology (ASCO) and the College of American Pathologists published a joint review on ctDNA. They reported on the value in terms of clinical utility and validity for few types of late stage cancers, but not for the majority of late stage cancers and not at all for early stage cancers (1). The consensus statement therefore narrated that routine ctDNA testing is not yet ready to be considered as an integral part of the management of cancers. Since the publication of this review in March 2018, the 54th ASCO Annual Meeting was convened at Chicago, Illinois on June 1–5, 2018. The research abstracts and the discussions, however, at ASCO conference argued otherwise.
In this article we aim to highlight the recent key updates and advances related to ctDNA presented at 2018 ASCO Annual Meeting that was held after the consensus statement. A literature search of 2018 ASCO Annual Meeting library, by using the key words “ctDNA,” “circulating tumor DNA” “cell-free DNA (cfDNA)” or “liquid biopsy” was done; and the most relevant abstracts pertaining to GI malignancies were the ones selected to be summarized/included in this review.
ctDNA as a Diagnostic Tool
Tissue biopsy remains the gold standard test for the diagnosis of cancers. Furthermore, genetic testing done on tissue is what is used to find actionable genes or aberrations. The yield depending on site and procedure often is enough to make the diagnosis but not enough to run additional genetic tests. There were an array of studies reporting on the feasibility and specificity of liquid biopsies aiding and/or corroborating findings noted on tissue biopsies in gastrointestinal cancers including colorectal cancers (CRC) and non-colorectal cancers. Hu et al. investigated the role of ctDNA in CRC patients and reported that the number of DNA mutations detected in tissue biopsy correspond with that found in liquid biopsy (2). Huang et al. did a study on 30 CRC patients who underwent surgery and presented that the majority of patients (83%) had at least one mutation detected in both tissue biopsy as well as liquid biopsy (3). A study conducted on anal cancer patients demonstrated the ability of ctDNA in terms of sensitivity (89%) to help diagnose the cancerous disease (4). An Australian study reported the detection ctDNA in 62.2% pre-operative and 37.1% post-operative plasma samples of patients with early stage pancreatic cancer (5). Mody et al. conducted a study on 104 cholangiocarcinoma patients and reported that at least one gene mutation was identified in ctDNA sample in 77% of patients (median number of mutations per patient = 3; range = 1–15) (6). Another study revealed high mutational concordance between liquid and tissue biopsy for biliary tract cancer (74%) and intrahepatic type (92%) (7). In another study of gastroesophageal cancer patients, researchers were able to detect at least one mutation (median number of mutations per patient = 2; range = 0–15) in ctDNA in 66% of patients (8).
Many clinical trials, mostly ongoing, have demonstrated that the mutations in oncologic genes responsible for carcinogenesis can be identified in ctDNA. These ctDNA mutations correlate well with tissue biopsy results. In the REVERCE phase II trial studying early exposure to regorafenib vs. anti-EGFR in patients with CRC, Tsuji et al. identified mutations in several genes including RAS, BRAF, EGFR, HER2, and MET using ctDNA samples of patients with metastatic CRC (Clinical trial: UMIN000011294) (9). What was interesting was that the arrays of mutations acquired were different depending on the sequence of therapy (Regorafenib anti-EGFR vs. anti-EGFR Regorafenib). Similarly, in the HERACLES study, a phase II trial of trastuzumab and lepatinib in HER-2 positive metastatic CRC, researchers were able to correctly identify the HER-2 amplifications in 96% of samples using ctDNA (10). In another study, researchers reported that HER-2 (ERBB2) amplifications were identified in 61% of gastroesophageal cancer patients using ctDNA samples (11). Iqbal et al. detected a variety of mutations in different genes including HER2, BRCA2, TP53, APC, ROS1, PTEN, KRAS, CCEN1, GNAS, NF1, CTNNB1, PIK3CA, and ARID1A using ctDNA in gastroesophageal adenocarcinoma patients (12). Jia et al. demonstrated the practicality of employing cfDNA for the detection MET amplification in patients with RAS wild-type metastatic CRC (Clinical trial: NCT02008383). They enrolled the patients in two groups i.e., one receiving cabozantinib plus panitumumab and the second receiving cabozantinib alone; and found detectable cfDNA levels and MET amplification in 98 and 18% of patients, respectively (13). In WJOG7112G study, Sukawa et al. studied gastric or gastroesophageal cancer patients who had disease progression despite receiving chemotherapy, and identified HER2 amplifications mutations in ctDNA in 60% of patients (14).
In summary, multiple studies show that GI malignancies in general shed DNA that can be detected and the current technologies available corroborate and correlate well with tissue based genetic testing.
ctDNA as a Prognostic Biomarker
Prediction of Response to Therapy
The behavior of cancer in response to therapy can be predicted by determining the type and number of ctDNA mutations. The research presented at ASCO conference does establish ctDNA as an independent prognostic marker in many cancers. Zhang et al. investigated ctDNA of 43 esophageal squamous cell carcinoma patients, and reported the role of ctDNA in predicting response to therapy. Their results showed that the patients who did not respond to neoadjuvant chemotherapy were associated with higher driver gene molecular mutation burden compared to those who responded well (p < 0.01) (15). Yang et al. studied 88 rectal cancer patients and reported that ctDNA levels became undetectable during neoadjuvant chemoradiotherapy in 65.5% of the patients which were congruous with the imaging and histological changes (16). Another study examined various driver mutational genes detected by ctDNA in CRC patients and reported a noticeable reduction in tumor mutation burden following surgery (3).
There were also studies that reported on ctDNA as a biomarker of the efficacy of specific chemotherapeutic agents in different cancers. Catenacci et al. investigated the response of margetuximab plus pembrolizumab in ERBB2-positive gastroesophageal cancer patients, and demonstrated that the response to therapy was predicted based on ctDNA sample results. They calculated objective response rate and disease control rate using ctDNA which were 57 and 86%, respectively, for their cohort. They also reported that a lot of cancer patients lost their ERBB2 amplifications as detected by ctDNA after receiving trastuzumab (11). Chen et al. conducted a phase II study in China and assessed the clinical rationale of apatinib in chemotherapy-refractory metastatic CRC patients (Clinical trial: NCT03190616). They reported that tumor mutation burden calculated by ctDNA is the main factor determining prognosis (17). In HERACLES study discussed earlier, the researchers reported that ctDNA precisely anticipated the response to HER-2 receptor inhibitor therapy in HER2-positive CRC (10).
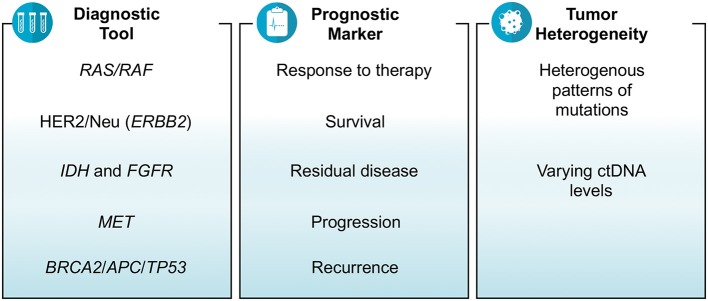
The conclusions one can draw from these studies are that in general if there is a decrease in the variant allele fraction or the number of mutations (or lack of detection of any ctDNA) after receiving therapy, it affirms an improvement in terms of reduction in tumor size. Studies noting a decline in ctDNA as early as 2 weeks could predict response to therapy months later on imaging studies. Unrelated, but utilizing ctDNA testing is also helpful in patients having pseudoprogression in their imaging on immunotherapy. These studies presented at ASCO conference do strongly support ctDNA as a reliable cancer biomarker and favor its use in the management of cancer patients in prognosticating and as a dynamic early biomarker predicting response to therapy (Figure 1).
Figure 1.
Summary of studies reporting on use of ctDNA in GI malignancies at ASCO 2018.
Prediction of Survival
Researchers estimated and reported survival rates of CRC based on the findings of ctDNA. Siveke et al. studied the role of ctDNA in determining the prognosis of metastatic CRC and reported that high baseline ctDNA is associated with multi-site metastasis, increased levels of carcinoembryonic antigen, propensity of having right-sided origin and poor overall performance compared to low baseline ctDNA. They further demonstrated that baseline ctDNA is the predictor of progression free survival (HR = 2.01; 95% CI: 1.25–3.22, p = 0.0033) (18). Poulsen et al. showed the association of ctDNA mutations and overall survival in patients with metastatic CRC. They reported that the patients who received Sym004 (anti-EGFR antibody mixture) and were negative for RAS and BRAF V600E mutation in ctDNA had an overall survival advantage of 3.5 months compared to those who received standard therapy (HR: 0.71; p = 0.134). They further estimated the overall survival of patients who were negative for EGFR mutations in addition to RAS and BRAF V600E mutation in ctDNA (HR: 0.59; p = 0.044) (19). Kehagias et al. investigated 141 patients with advanced CRC in a multicentric trial, and measured cfDNA levels at baseline and at day 14 after starting regorafenib (Clinical trial: NCT01929616). The patients having cfDNA levels ≥1 μg/ml at day 14 were associated with poor progression-free (HR: 2.50, 95% CI: 1.73–3.63) and overall survival (HR: 3.83, 95% CI: 2.52–5.71) compared to those having levels <1 μg/ml (20). A Japanese study showed similar results and reported that the finding of KRAS mutated ctDNA in post-chemotherapy metastatic CRC patients is associated with reduced progression free survival (21). The high levels of ctDNA and high tumor mutation burden correlated with poor survival rates of CRC.
Survival of non-colorectal cancers including esophageal and biliary tract cancers were also estimated based on the numbers of mutations detected in ctDNA. A study of esophageal squamous cell carcinoma patients who underwent radiation therapy reported ctDNA as one of the main factors determining prognosis. Their results revealed that the patients who had mutations in ctDNA after undergoing radiation therapy were associated with poor overall survival (p = 0.005) and a trend of a decrease in disease free survival (p = 0.068) compared to those who did not have any mutations in ctDNA (22). A German study showed the association of variant allele frequency detected by ctDNA and progression-free survival of intrahepatic biliary tract cancer (Spearman, r = −0.5878, p = 0.0288) (7).
Detection of Residual Disease, Progression, and Recurrence
Cancer patients are evaluated after completion of therapy to make sure there is no possible tumor left behind. Imaging and/or tumor markers are usually checked to rule out any residual disease. The feasibility of using ctDNA as a biomarker of residual disease was reported in various studies at ASCO 2018. Tie et al. conducted a study on 95 stage III colon cancer patients and reported that the response to therapy and the residual metastatic disease which is otherwise not identified on imaging can be detected by ctDNA. Their results showed that the finding of positive ctDNA post-surgery (HR: 3.52; p = 0.004) and post-chemotherapy (HR: 7.14; p <0.001) is associated with poor recurrence free survival (23). Murray et al. did a prospective study on 172 post-surgery CRC patients, and demonstrated that ctDNA is a marker of residual disease and recurrence (Clinical trial: 12611000318987). Their results showed that the patients who were positive for ctDNA after surgery were at an increased risk of recurrence (HR: 3.8, 95% CI: 1.5–9.5) (24).
Furthermore, studies have shown the reliability of the use of ctDNA in ascertaining the disease progression in terms of metastasis. A study conducted on CRC patients demonstrated the association of post-surgery detection of ctDNA mutations and increased risk of disease progression. The researchers found that higher proportion of ctDNA mutation positive patients (27.8%) experienced disease progression compared to those who were ctDNA mutation negative (4.4%) after surgery (25). Another study showed the reliability of ctDNA in predicting the response to regorafenib or TAS-102 in patients with metastatic CRC using different PCR based methods (26). Cabel et al. conducted a study on 36 anal cancer patients in France and reported that post-chemoradiotherapy detection of ctDNA is associated with worse outcomes. Their results showed that 17% of the patients had metastatic relapse, and these were the only patients who were positive for ctDNA after receiving chemoradiotherapy (4). Parseghian et al. demonstrated the usefulness of ctDNA in monitoring the decay of clones which were resistant to anti-EGFR agents in metastatic CRC. They calculated the median relative mutant allele fraction (10.5 vs. 10.6%) and the decaying half-life of clones (3.4 vs. 6.9 months) for RAS and EGFR, respectively, while on anti-EGFR therapy (27). Acquisitions of these mutations are known mechanisms of secondary resistance in patients with metastatic CRC. What is indeed intriguing is that these clones can be lost over time allowing for potentially “rechallenging” some of these therapies leading to development of multiple trials employing this strategy.
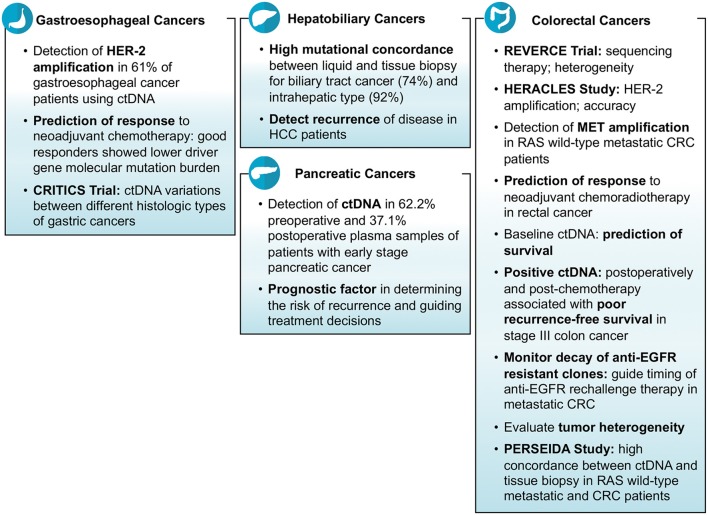
Finally, recurrence of cancers is a common phenomenon and ctDNA was noted in multiple studies to be helpful in predicting the risk of recurrence after being treated for the primary cancer (Figure 2). Murray et al. studied the association of ctDNA and recurrence of CRC, and reported that post-surgery detection of ctDNA is associated with an increased risk of recurrence (HR = 3.8; p = 0.004) (Clinical trial: 12611000318987) (28). A Chinese study, conducted on hepatocellular carcinoma patients who underwent transcatheter arterial chemoembolization, reported that high mutational burden of 10 genes identified in ctDNA might be associated with the recurrence of the disease. The 10 genes detected in their study were NRAS, BRAF, PIK3CA, KRAS, ARID1A, AXIN1, ARID2, TERT, TP53, and CTNNB1 (29). Lee et al. demonstrated that ctDNA is an indicator of prognosis in terms of determining the risk of recurrence and guiding treatment decisions in patients with early stage pancreatic cancer (Clinical trial: ACTRN12612000763842). Their analysis showed that detection of ctDNA both before and after surgery is associated with poor recurrence free and overall survival; and 100% of patients showed recurrence who had measurable ctDNA after surgery despite of being on adjuvant chemotherapy (5).
Figure 2.
Findings of ASCO 2018 studies about the use of liquid biopsy in gastrointestinal cancers.
Tumor Heterogeneity and ctDNA
Gastrointestinal cancers including CRC are heterogeneous in nature in terms of histology, presenting symptoms, progression, response to therapy, and outcomes that can lead to a difference in the management of various types of cancers (30). Tumor heterogeneity can be evaluated by measuring the wide-ranging levels of ctDNA and the identification of diverse patterns of mutations. A study reported that right-sided colon cancer is associated with an increased number of mutations in ctDNA than left-sided colon and rectal cancers (2). Another study demonstrated higher median cfDNA levels (14.2 vs. 8.94 ng/ml) and higher mutation concordance rate identified via liquid and tissue biopsies (94.7 vs. 50%) for colon cancer patients than rectal cancer patients (31). Similarly in another study, researchers identified a heterogeneous pattern of genomic alterations in ctDNA of patients with metastatic CRC including mutations in BRAF, KRAS, NRAS, MAP2K1, PIK3CA, ERBB2, MET, and EGFR genes (19). A Chinese study on patients with rectal cancer indicated that the most common mutations found in ctDNA were of TP53, APC, and KRAS genes (16). A study of cholangiocarcinoma patients reported the detection of mutations in ctDNA in a variety of genes including TP53, KRAS, FGFR2, ARID1A, APC, PIK3CA, BRAF, CCND1, CCND2, CCNE1, CDK4, CDK6, EGFR, ERBB2, FGFR1, MET, MYC, and PDGFRA (6). The same study revealed the finding of FGFR2 fusions and the actionable mutations in 3 and 61% of patients, respectively (6). Clifton et al. did a study for the identification of actionable gene fusions in CRC and reported that the main fusions detected in ctDNA samples were RET, FGFR3, ALK, NTRK1, ROS1, and FGFR2 (32).
Several clinical trials have demonstrated the role of ctDNA in detecting tumor heterogeneity of different gastrointestinal cancers. In CRITICS phase III trial, Leal et al. investigated cfDNA of stage Ib-IVa resectable gastric cancer patients (n = 115) and showed that the level of ctDNA varies between different histologic types of cancer (Clinical trial: NCT00407186). They found higher ctDNA levels for tubular adenocarcinoma (mutant allele fractions: 0.25%) than signet-ring cell subtype (mutant allele fractions: 0.16%) of gastric cancer (33). Yaung et al. demonstrated the ability of ctDNA, by using next-generation sequencing based methods, to detect tumor heterogeneity in a clinical trial (STEAM) evaluating the outcomes of FOLFOXIRI-bevacizumab vs. FOLFOX-bevacizumab as first-line treatment of metastatic CRC (Clinical trial: NCT01765582). Their results showed that high mutant-allele tumor heterogeneity is associated with poor outcomes (34). Okamura et al. examined the ctDNA of gastroesophageal carcinoma patients (n = 55) and found different arrays of genomic alterations (Clinical trial: NCT02478931). The most frequent mutations found in their study were TP53, PIK3CA, ERBB2 and KRAS in 50.9, 16.4, 14.5, and 14.5% of patients, respectively (8). In another clinical trial, the mutated genes on ctDNA in patients with advanced CRC were found to be APC, TP53, KRAS, and PI3KCA in 73, 72, 66, and 23% of ctDNA samples, respectively (20). PERSEIDA study which is an ongoing trial in Spain evaluated the concordance in mutation profile between ctDNA and tissue biopsy. The researchers analyzed the data of tissue biopsy-proven RAS wild type metastatic CRC patients (n = 119) from 20 centers who were later assessed with liquid biopsy, and reported that there was high concordance between the two but new RAS mutations were also detected in ctDNA majority of which were at low mutant allele fraction limit (Clinical trial: NCT02792478) (35).
Future Directions and Conclusion
While the joint statement issued earlier in March 2018 was reasonable, the array of studies presented and discussed at ASCO 2018 argue more in favor of integrating ctDNA in practice for many gastrointestinal malignancies. For some e.g., CRC, gastroesophageal cancers as well as biliary duct cancers, it is helpful in not only identifying relevant actionable mutations but also in helping identifying secondary mechanisms of resistance. Furthermore, it helps in guiding therapy, as the appropriate chemotherapy drugs can be added to the treatment regimen depending on the emerging clones of resistance detected on ctDNA testing. It also aids in monitoring the response to treatment and predicting survival depending on the level of ctDNA detection in serial analyses. While the technology and the methods may not be ready for use in all stages in all cancers, for patients with metastatic GI cancers, particularly CRC, we would argue similar to discussants at colorectal cancer sessions at ASCO that this is indeed ready for primetime because of its feasibility, rapid turn around and non-invasive nature allowing for serial oncologic assessments. Reevaluation of the literature is needed which could result in an update in the current guidelines for use of ctDNA particularly in late stage gastrointestinal cancers.
Author Contributions
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
Conflict of Interest Statement
PK has provided advisory board consultancy to Taiho in January 2017 (to institution) and Ipsen in June 2018 (to institution). The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Special thanks to Kristina K. Greiner, Editor and Designer, at the University of Iowa Design Center for help on the Figures 1, 2.
Footnotes
Funding. This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
References
- 1.Merker JD, Oxnard GR, Compton C, Diehn M, Hurley P, Lazar AJ, et al. Circulating tumor DNA analysis in patients with cancer: American Society of Clinical Oncology and College of American Pathologists Joint Review. J Clin Oncol. (2018) 36:1631–41. 10.1200/JCO.2017.76.8671 [DOI] [PubMed] [Google Scholar]
- 2.Hu Y, Guo H, Chen Y, Chen Y, Lan L, Li J, et al. The mutational landscape of circulating cell free DNA in patients with colorectal adenoma and carcinoma. J Clin Oncol. (2018) 36(15_suppl):e15662 10.1200/JCO.2018.36.15_suppl.e15662 [DOI] [Google Scholar]
- 3.Huang T, Xia L, Xu M, Liu Y, Zhang X, Melançon CE, et al. Dynamic monitoring of driver gene mutation profiles in plasma cfDNA using an 85 gene panel for postoperative prognosis in colorectal cancer. J Clin Oncol. (2018) 36(15_suppl):e15536 10.1200/JCO.2018.36.15_suppl.e15536 [DOI] [Google Scholar]
- 4.Cabel L, Jeannot E, Bieche I, Vacher S, Callens C, Bernard A, et al. Prognostic impact of residual HPV ctDNA detection after chemoradiotherapy for anal canal carcinoma. J Clin Oncol. (2018) 36(15_suppl):3565 10.1200/JCO.2018.36.15_suppl.3565 [DOI] [PubMed] [Google Scholar]
- 5.Lee B, Lipton LR, Cohen J, Tie J, Javed AA, Li L, et al. Circulating tumor DNA as a prognostic biomarker in early stage pancreatic cancer. J Clin Oncol. (2018) 36(15_suppl):e16206 10.1200/JCO.2018.36.15_suppl.e16206 [DOI] [Google Scholar]
- 6.Mody K, Borad MJ, Bekaii-Saab TS, Yang JD, Ramanathan RK, Ahn DH, et al. Real-time circulating tumor DNA profiling of advanced cholangiocarcinoma (CCA). J Clin Oncol. (2018) 36(15_suppl):4089 10.1200/JCO.2018.36.15_suppl.4089 [DOI] [Google Scholar]
- 7.Berger AW, Ettrich TJ, Schwerdel D, Dolnik A, Beuter F, Blätte TJ, et al. Genotyping of circulating tumor DNA in biliary tract cancer reveals diagnostic and prognostic information. J Clin Oncol. (2018) 36(15_suppl):e16147 10.1200/JCO.2018.36.15_suppl.e16147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Okamura R, Kato S, Baumgartner JM, Patel H, Leichman LP, Kelly KJ, et al. Analysis of circulating tumor DNA and clinical correlates in patients with esophageal, gastroesophageal junction and gastric adenocarcinoma. J Clin Oncol. (2018) 36(15_suppl):e16031 10.1200/JCO.2018.36.15_suppl.e16031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tsuji Y, Shitara K, Yamanaka T, Denda T, Shinozaki K, Komatsu Y, et al. REVERCE: Randomized phase II study of regorafenib followed by cetuximab versus the reverse sequence for metastatic colorectal cancer patients previously treated with fluoropyrimidine, oxaliplatin, and irinotecan-Biomarker analysis. J Clin Oncol. (2018) 36(15_suppl):3510. 10.1200/JCO.2018.36.15_suppl.3510 [DOI] [Google Scholar]
- 10.Bardelli A, Siravegna G, Sartore-Bianchi A, Nagy RJ, Mussolin B, Cassingena A, et al. Plasma HER2 (ERBB2) copy number to predict response to HER2-targeted therapy in metastatic colorectal cancer. J Clin Oncol. (2018) 36(15_suppl):3506 10.1200/JCO.2018.36.15_suppl.3506 [DOI] [Google Scholar]
- 11.Catenacci DVT, Park H, Uronis HE, Kang Y-K, Lacy J, Enzinger PC, et al. Margetuximab (M) plus pembrolizumab (P) in ERBB2-amplified PD-L1+ gastroesophageal adenocarcinoma (GEA) post trastuzumab (T). J Clin Oncol. (2018) 36(15_suppl):4030 10.1200/JCO.2018.36.15_suppl.4030 [DOI] [Google Scholar]
- 12.Iqbal M, Kasi PM, Mody K. Feasibility and clinical value of circulating tumor DNA testing in patients with gastroesophageal adenocarcinoma. J Clin Oncol. (2018) 36(15_suppl):e16045 10.1200/JCO.2018.36.15_suppl.e16045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jia J, Niedzwiecki D, Uronis HE, Morse M, Zafar Y, Hsu SD, et al. A phase I/II trial of cabozantinib (C) with or without panitumumab (P) in patients (pts) with RAS wild-type (WT) metastatic colorectal cancer (mCRC): clinical outcomes in pts with MET amplification (amp) detected in blood. J Clin Oncol. (2018) 36(15_suppl):3555 10.1200/JCO.2018.36.15_suppl.3555 [DOI] [Google Scholar]
- 14.Sukawa Y, Makiyama A, Esaki T, Hirano G, Komoda M, Kashiwada T, et al. Biomarker study for trastuzumab continuation beyond progression in a randomized phase II trial of weekly paclitaxel±trastuzumab in patients with HER2-positive advanced gastric or gastro-esophageal junction cancer refractory to trastuzumab combined with fluoropyrimidine and platinum (WJOG7112G). J Clin Oncol. (2018) 36(15_suppl):4029 10.1200/JCO.2018.36.15_suppl.4029 [DOI] [Google Scholar]
- 15.Zhang R, Hu Y, Zhou T, Han W, Chen Y, Liu X, et al. The mutational landscape of circulating cell free DNA in patients with esophageal squamous cell carcinoma response and non-response to neoadjuvant chemotherapy. J Clin Oncol. (2018) 36(15_suppl):e16082 10.1200/JCO.2018.36.15_suppl.e16082 [DOI] [Google Scholar]
- 16.Yang L, Wang Y, Shen L, Wan J, Deng W, Zhu J, et al. Predicting treatment outcome of rectal cancer patients underwent neoadjuvant chemoradiotherapy by ctDNA: the potential use of ctDNA monitoring as organ-sparing approach. J Clin Oncol. (2018) 36(15_suppl):3608 10.1200/JCO.2018.36.15_suppl.3608 [DOI] [Google Scholar]
- 17.Chen X, Li P, Sun J, Wang B, Lin P, Cai X, et al. Apatinib as a salvage treatment for refractory metastatic colorectal cancer. J Clin Oncol. (2018) 36(15_suppl):3556 10.1200/JCO.2018.36.15_suppl.3556 [DOI] [Google Scholar]
- 18.Siveke JT, Lueong S, Hinke A, Herbst A, Tannapfel A, Reinacher-Schick AC, et al. Serial analysis of mutant KRAS in circulation cell-free DNA (cfDNA) of patients with KRAS mutant metastatic colorectal cancer: a translational study of the KRK0207 trial. J Clin Oncol. (2018) 36(15_suppl):e15599 10.1200/JCO.2018.36.15_suppl.e15599 [DOI] [Google Scholar]
- 19.Poulsen TT, Kragh M, Ding C, Pedersen MW, Lindsted T, Blondal T, et al. Circulating tumor DNA profiling to identify patients with metastatic colorectal cancer with improved overall survival following Sym004 treatment. J Clin Oncol. (2018) 36(15_suppl):e15577 10.1200/JCO.2018.36.15_suppl.e15577 [DOI] [Google Scholar]
- 20.Kehagias P, Vandeputte C, Ameye L, Housni HE, Deleporte A, Geboes KP, et al. High levels of cell-free DNA (cfDNA) at baseline (BL) and increase of at least one mutation at day 14 (D14) as independent prognostic biomarkers for patients (pts) with advanced colorectal cancer (aCRC) under regorafenib. J Clin Oncol. (2018) 36(15_suppl):3532 10.1200/JCO.2018.36.15_suppl.3532 [DOI] [Google Scholar]
- 21.Takayama Y, Suzuki K, Fukui T, Ichida K, Watanabe F, Kakizawa N, et al. Monitoring circulating tumor DNA revealed dynamic changes in KRAS status in patients with metastatic colorectal cancer. J Clin Oncol. (2018) 36(15_suppl):e15630 10.1200/JCO.2018.36.15_suppl.e15630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jia R, Li P, Zhao C, Wang Y, Liu R, Chang L, et al. Postradiation ctDNA status as a prognostic factor in locally advanced esophageal squamous cell carcinoma. J Clin Oncol. (2018) 36(15_suppl):e16053 10.1200/JCO.2018.36.15_suppl.e16053 [DOI] [Google Scholar]
- 23.Tie J, Cohen J, Wang Y, Lee M, Wong R, Kosmider S, et al. Serial circulating tumor DNA (ctDNA) analysis as a prognostic marker and a real-time indicator of adjuvant chemotherapy (CT) efficacy in stage III colon cancer (CC). J Clin Oncol. (2018) 36(15_suppl):3516 10.1200/JCO.2018.36.15_suppl.3516 [DOI] [Google Scholar]
- 24.Murray D, Young GP, Pedersen SK, Rabbitt P, Byrne SE, Cornthwaite KJ, et al. A prospective cohort study in colorectal cancer assessing the relationship between post-surgery detection of methylated BCAT1 or IKZF1 ctDNA and risk for residual disease and survival. J Clin Oncol. (2018) 36(15_suppl):3596. 10.1200/JCO.2018.36.15_suppl.359629992492 [DOI] [Google Scholar]
- 25.Li L, Zhou W, Li C, Li P, Gong Y, Guan Y-F, et al. Analysis of circulating tumor DNA to monitor disease status in colorectal cancer after surgery. J Clin Oncol. (2018) 36(15_suppl):e15583 10.1200/JCO.2018.36.15_suppl.e15583 [DOI] [Google Scholar]
- 26.Pereira AAL, Lam M, Marie PK, Raghav KPS, Morris VK, Brown H, et al. Circulating tumor DNA (ctDNA) as an early marker to monitor clinical benefit of regorafenib and TAS-102 in patients with metastatic colorectal cancer (mCRC). J Clin Oncol. (2018) 36(15_suppl):3533 10.1200/JCO.2018.36.15_suppl.3533 [DOI] [Google Scholar]
- 27.Parseghian CM, Loree JM, Morris VK, Pereira AAL, Sanchez EV, Kee BK, et al. Anti-EGFR resistant clones decay exponentially after progression: Implications for anti-EGFR rechallenge. J Clin Oncol. (2018) 36(15_suppl):3511 10.1200/JCO.2018.36.15_suppl.3511 [DOI] [Google Scholar]
- 28.Murray D, Symonds EL, Pedersen SK, Young GP. Detection of ctDNA biomarkers post resection and relationship to recurrent disease in colorectal cancer patients. J Clin Oncol. (2018) 36(15_suppl):e15616 10.1200/JCO.2018.36.15_suppl.e15616 [DOI] [Google Scholar]
- 29.Du X, Li Z, Zhang H. Detection of early recurrence in HCC patients after TACE treatment with plasma cfDNA. J Clin Oncol. (2018) 36(15_suppl):e16146 10.1200/JCO.2018.36.15_suppl.e16146 [DOI] [Google Scholar]
- 30.Wang CB, Merchea A, Shahjehan F, Colibaseanu D, Cochuyt J, Li Z, et al. Impact of tumor location and variables associated with overall survival in patients with colorectal cancer. J Clin Oncol. (2018) 36(15_suppl):e15651 10.1200/JCO.2018.36.15_suppl.e15651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang X, Xia L, Xu M, Qu H, Wen M, Melançon CE, et al. ctDNA sequencing for early detection of colorectal cancers: comparative analysis of colon cancer and rectal cancer data. J Clin Oncol. (2018) 36(15_suppl):e15653 10.1200/JCO.2018.36.15_suppl.e15653 [DOI] [Google Scholar]
- 32.Clifton K, Raymond VM, Dasari A, Raghav KPS, Parseghian CM, Pereira AAL, et al. Actionable fusions in colorectal cancer using a cell-free circulating tumor DNA (ctDNA) assay. J Clin Oncol. (2018) 36(15_suppl):3507 10.1200/JCO.2018.36.15_suppl.3507 [DOI] [Google Scholar]
- 33.Leal A, van Grieken NCT, Phallen J, Palsgrove DN, Adleff V, Warmerdam F, et al. Circulating tumor DNA dynamics in resectable gastric cancer. J Clin Oncol. (2018) 36(15_suppl):4069 10.1200/JCO.2018.36.15_suppl.4069 [DOI] [Google Scholar]
- 34.Yaung S, Tikoo N, Nicholas A, Sommer N, Bendell JC, Hurwitz H, et al. Correlation of measurements of tumor heterogeneity based on next-generation sequencing (NGS) of circulating tumor DNA (ctDNA) with clinical outcomes in STEAM, a prospective, randomized, multicenter study in metastatic colorectal cancer (mCRC). J Clin Oncol. (2018) 36(15_suppl):3545 10.1200/JCO.2018.36.15_suppl.3545 [DOI] [Google Scholar]
- 35.Valladares-Ayerbes M, Alfonso PG, Luengo JM, Cáceres PP, Vieitez JM, Cruz-Hernández JJ, et al. Concordance in RAS mutation status between liquid and solid biopsies in subjects with RAS wild-type (wt) metastatic colorectal cancer (mCRC) in first-line treatment in Spain: PERSEIDA study (NCT02792478). J Clin Oncol. (2018) 36(15_suppl):e15602 10.1200/JCO.2018.36.15_suppl.e15602 [DOI] [Google Scholar]