Abstract
Background
The tripartite motif-containing protein 59 (TRIM59) is an important member of the TRIM family, which regulates biological processes. However, the relationship between TRIM59 and epithelial ovarian cancer (EOC) is not clear.
Material/Methods
The TRIM59 expression level was detected in EOC tissues and cell lines. CCK-8 assay, Transwell assay, and wound healing assay were performed to determine the effects of TRIM59 on EOC cell proliferation, invasion, and migration. Silencing of the expression of TRIM59 in EOC cells and expression of FAK/AKT/MMP pathway-related protein were detected by Western blot analysis.
Results
Through bioinformatics analysis, TRIM59 was found to be highly expressed in EOC and was correlated with prognosis of patients. TRIM59 was upregulated in EOC tissues and cells. Silencing TRIM59 significantly suppressed EOC cell proliferation, migration, and invasion. In terms of molecular mechanism, silencing TRIM59 inhibited the FAK/AKT/MMP pathway.
Conclusions
TRIM59 is a biomarker for the prognosis of EOC. It is also oncogenic and a potential target for EOC therapy.
MeSH Keywords: Carcinoma, Ovarian Neoplasms, Prognosis
Background
Ovarian cancer is one of the most common malignant tumors in females. Morbidity due to ovarian cancer is the fourth highest after breast, lung, and colorectal cancers [1]. Particularly, epithelial ovarian cancer (EOC) is the most common pathological type, accounting for approximately 80% of all ovarian cancers [2]. Over the past few decades, great advances have been made in surgical procedures and drug development. However, the survival rate of EOC patients remains unsatisfactory, and the 5-year survival rate is less than 40% [3]. At the initial stage of EOC diagnosis, most patients also show uterine, appendage, greater omentum, and pelvic metastases, and patient prognosis is quite poor. Additionally, the molecular mechanism of EOC distant metastasis remains unclear.
The genesis and development of human cancer is a multi-step and complicated process involving the activation of multiple oncogenes and inactivation of various tumor suppressor genes. The tripartite motif-containing protein (TRIM) family is involved in important biological processes such as cell cycle, cell apoptosis, and natural immunity to viruses [4,5]. TRIM proteins mainly contain the RING domain, B-box domain, and coiled-coil region. Among these, the RING domain is associated with E3 ubiquitin ligase activity, which can mediate the ubiquitination modification of target proteins [6]. The B-box structure, which is a unique domain of TRIM proteins, contains conserved cysteine and histidine residues and may play a pivotal role in regulating biological behavior [7]. The coiled-coil region is a super-secondary structure formed by multiple α helices, which promotes the formation of macromolecular polymers [8]. TRIM59 is an important member of the TRIM family that regulates biological processes such as cell proliferation, apoptosis, and tumorigenesis. However, the relationship between TRIM59 and EOC remains unclear. In this study, bioinformatic analysis and experimental results were integrated to evaluate the biological role of TRIM59 in EOC.
Material and Methods
Bioinformatic analysis
The matrix data of GSE66957 chip were downloaded from the Gene Expression Omnibus database (GEO, http://www.ncbi.nlm.nih.gov/geoprofiles/). The GPL15048 was also downloaded concurrently, and the probe matrix was transformed into a Gene Symbol matrix. Differentially expressed genes were identified by further bioinformatic analysis. The Level3 RNA-seq data were downloaded from the cancer genome atlas (TCGA, http://cancergenome.nih.gov/), and 1 file was prepared for each sample for the transcriptome data downloaded from TCGA. The file was merged into a matrix file through script; subsequently, the gene name was converted from an Ensembl ID to the Gene Symbol matrix using the Ensembl database. Finally, the relationship between TRIM59 expression and the prognosis of patients with EOC was analyzed based on TCGA data.
Clinical specimens and cell lines
Cancer tissue and matched para-carcinoma tissue specimens were collected from 15 patients with EOC undergoing surgery at the First Affiliated Hospital of Henan University of Science and Technology. All patients were naïve to neoadjuvant chemotherapy and/or radiotherapy before surgery. This study was approved by the Ethics Committee of the First Affiliated Hospital of Henan University of Science and Technology (Luoyang, China). All patients provided written informed consent to participate in this study.
The EOC cell lines (CAOV3, COV-504, SKOV3, and OVCAR3) were obtained from the Chinese Academy of Sciences Shanghai Cell Bank (Shanghai, China). All cell lines were cultured in RPMI 1640 medium (Gibco, Carlsbad, CA, USA) containing 10% fetal bovine serum (FBS) in an incubator at 37°C with 5% CO2. The culture medium was replaced every 3 days.
Immunohistochemistry and assessment
Paraffin-embedded specimens were prepared, which were then sliced into 4-μm-thick sections, followed by antigen retrieval. The sections were sealed at room temperature for 1 h, incubated with anti-TRIM59 antibody (1: 2000; Abcam, Cambridge, UK) at 4°C overnight, and then incubated with goat anti-rabbit antibody at 37°C for 1 h. Afterwards, the sections were stained with 3,3-diaminobenzidine at room temperature (around 20°C) for 10 min. Images were acquired with an Olympus BX43 microscope (Olympus, Tokyo, Japan).
All immunohistochemical sections were evaluated by 2 pathologists independently. The results were divided as follows according to the staining intensity: 0 point- no staining; 1 point- faint yellow staining; 2 points- claybank staining; and 3 points- brown staining. The sections were also rated from 0% to 100% based on the proportion of positive cells. In this study, high TRIM59 expression was defined as the product of cell staining intensity and the proportion of positive cells >40%; otherwise, the expression was considered as low.
TRIM59 silencing
To examine the role of TRIM59 in EOC, TRIM59 expression in EOC cells was silenced using short interfering RNA (siRNA) technology. EOC cells were inoculated into 24-well plates at a density of 1×105/well. The target sequence of siRNA-TRIM59 was CCCTGAACATTACAGGCAA. Lipofectamine TM 2000 reagent (Invitrogen, Carlsbad, CA, USA) was used for cell transfection, with scrambled siRNA used as the control group. Twenty-four hours later, the RPMI-1640 medium was supplemented, and cells transfected for 48 h were used in subsequent experiments.
RNA extraction and reverse transcription-polymerase chain reaction (RT-PCR)
Total RNA was extracted from tissues and cells using TRIzol Reagent (Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer’s instructions. The purity and concentration of the RNA samples were determined by spectroscopy. The PCR conditions were as follows: 1 cycle of 95°C for 5 min; 40 cycles of denaturation at 95°C for 30 s, annealing at 55°C for 30 s, and extension at 72°C for 30 s; and at 4°C for 60 min. The TRIM59 primers used were as follows: forward primer GTATTCAGCCCCCAAACCAC; and reverse primer GAGCATGGCAGTACACGAGG. Finally, the relative expression of TRIM59 mRNA was calculated according to the 2−ΔΔCt method, with GAPDH used as an internal reference.
Western blotting
The RIPA protein lysis buffer (Beyotime, Shanghai, China) was used to split the proteins in tissues or cells, and the protein concentration was measured using a bicinchoninic acid disodium kit (Beyotime). An equivalent amount of cell lysate was separated on a 10% SDS-PAGE gel and transferred onto polyvinylidene difluoride membranes. The membranes were subsequently sealed, and anti-TRIM59 antibody (1: 5000) was added. GAPDH (1: 10 000; Multisciences, Hangzhou, China) as an internal reference. The membranes were incubated with goat anti-rabbit IgG secondary antibody (1: 5000; Bioworld Technology, lnc., St. Louis Park, MN, USA). The intensity of protein bands was analyzed with Image J software (National Institutes of Health, Bethesda, MD, USA). Other primary antibodies used in this study included anti-FAK antibody, anti-phospho-FAK, anti-AKT, anti-phospho-AKT, anti-MMP2 antibody, and anti-MMP9 antibody, which were all purchased from Abcam or Santa Cruz Biotechnology (Dallas TX, USA).
Cell proliferation assay
Cell proliferation was detected using the cell counting kit-8 (CCK-8) (Sigma-Aldrich, St. Louis, MO, USA). Cells were inoculated into a 96-well plate at a density of 1.5×104/well and were allowed to grow to 75% confluence. Following incubation for 24, 48, 72, and 96 h, 10 μL CCK-8 reagent was added to each well, and the cells were washed 3 times with phosphate-buffered saline (pH 7.4). Finally, the optical density at 450 nm was measured with a Bio-Rad microplate reader (Bio-Rad).
Cell invasion assay
Cell invasion ability was detected in a Transwell assay using cells transfected for 48 h. Cells (1×104) cultured in FBS-free medium were inoculated into the upper Transwell chamber (BD Biosciences, San Jose, CA, USA), and then FBS-containing medium was added. Matrigel (BD Biosciences) was inserted into the Transwell chambers for pre-enveloping. Following incubation for 36 h, the cells were counted, and photos were acquired with a light microscope (Olympus).
Wound healing assay
Cell migration ability was evaluated in a wound healing assay. Briefly, the cells were cultured to 100% confluence and then scratched using the tip of a 200-μL pipette. The isolated cells were then washed with PBS, and photos were acquired. Subsequently, the cells were incubated for another 24 h before imaging. The wound region was analyzed using Image J software, and cell migration was expressed as a percentage of the wound healing/initial wound area.
Statistical analysis
For GEO data, the Wilcox test was employed for differential expression analysis with the filter condition of (|logFC| >2 and P<0.01). For TCGA data, the Survival R software package was used for survival analysis, and Kaplan-Meier analysis was performed using the log-rank method, with P<0.01 as the screening condition. The major outcome indices of survival analysis were disease-free survival and overall survival. Intergroup comparisons were analyzed through the unpaired two-tailed t test. SPSS 18.0 statistics software (SPSS, Inc., Chicago, IL, USA) was used for data analysis. All results were expressed as the means ±SEM, and a difference of P<0.05 was considered as statistically significant.
Results
High TRIM59 expression in EOC
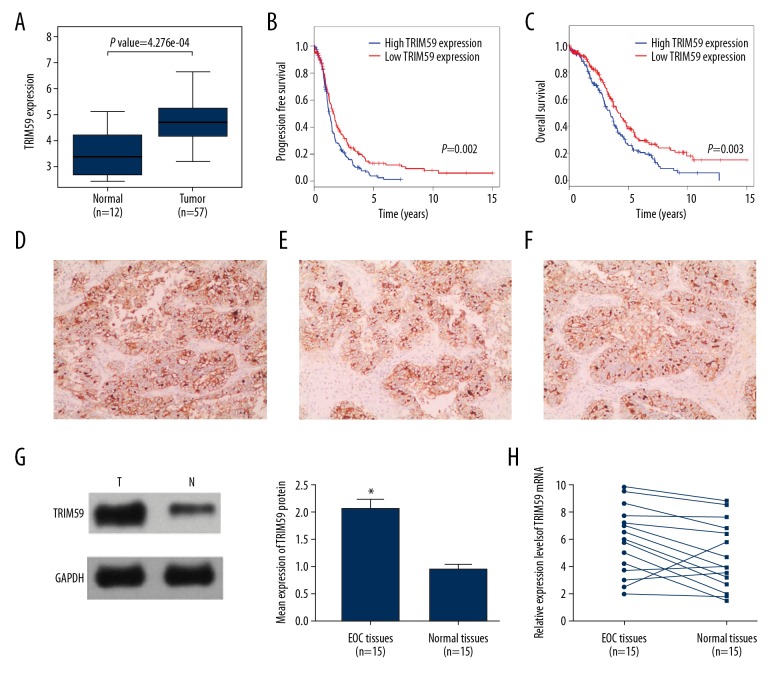
To evaluate TRIM59 expression in EOC, data downloaded from the GEO database were used for differential expression analysis. The results suggested that TRIM59 expression in EOC tissues (n=57) was higher than that in normal tissues (n=12) (Figure 1A). The relationship between patients with EOC and TRIM59 expression was evaluated by Kaplan-Meier analysis based on TCGA data. The results indicated that the prognosis (either disease-free survival or overall survival) of patients with EOC with low TRIM59 expression was better than that of patients with EOC with high TRIM59 expression (Figure 1B, 1C).
Figure 1.
High expression of TRIM59 in EOC and related with prognosis patients. (A) Analysis GEO data (GSE66957) of TRIM59 expression levels in EOC (n=57) and normal tissues (n=12). DFS (B) and OS (C) with different TRIM59 expression status in EOC samples obtained from TCGA were analyzed by Kaplan-Meier survival analysis. (D–F) TRIM59 protein was observed in the cytosol of EOC cells. TRIM59 protein and mRNA was detected in 15 pairs of EOC tissues by Western blotting (G) and RT-PCR (H). * Compared with other groups, P<0.05.
TRIM59 expression in EOC tissues was detected by immunohistochemical staining. Positive TRIM59 expression was mainly observed in the cytoplasm of EOC cells (Figure 1D–1F). Among the 15 EOC tissues, 12 showed high TRIM59 expression. Moreover, TRIM59 protein and mRNA levels in EOC tissues and para-carcinoma tissues were quantitatively analyzed by Western blotting and RT-PCR. The results demonstrated that the TRIM59 protein and mRNA expression levels in EOC tissues were considerably higher than those in para-carcinoma tissues (Figure 1G, 1H).
Silencing TRIM59 expression in EOC cell lines
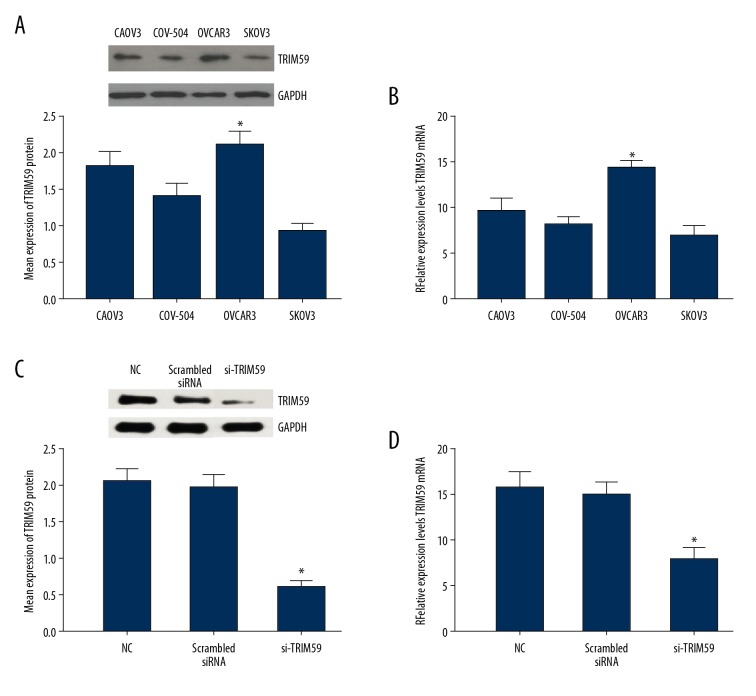
The TRIM59 protein and mRNA expression levels in EOC cell lines CAOV3, COV-504, SKOV3, and OVCAR3 were determined. The Western blotting results suggested that the OVCAR3 cell line had the highest TRIM59 expression levels (Figure 2A). RT-PCR analysis also revealed that the TRIM59 mRNA content was highest in the OVCAR3 cell line (Figure 2B). Therefore, the OVCAR3 cell line was selected for subsequent experiments.
Figure 2.
Expression of TRIM59 in EOC cell lines. TRIM59 protein and mRNA were detected in EOC cell lines CAOV3, COV-504, OVCAR3, and SKOV3 by Western blotting (A) and RT-PCR (B). The results show that the TRIM59 protein and mRNA expression level was highest in EOC cell line OVCAR3. After transfection, the expression levels of TRIM66 protein and mRNA in EOC cells line OVCAR3 was assessed by Western blotting (C) and RT-PCR (D). * Compared with other groups, P<0.05
To further examine the role of TRIM59 in EOC, TRIM59 expression in the OVCAR3 cell line was silenced, which was verified by Western blotting and RT-PCR. Compared to the control group (blank OVCAR3 cells and scrambled siRNA), TRIM59 protein expression in OVCAR3 cells of the siRNA-TRIM59 group was markedly suppressed (Figure 2C); similar results were observed in RT-PCR analysis (Figure 2D). These data indicate that TRIM59 was effectively silenced in the OVCAR3 cell line, which is useful for subsequent studies.
Silencing TRIM59 expression markedly suppresses the proliferation, invasion, and migration of EOC cell lines
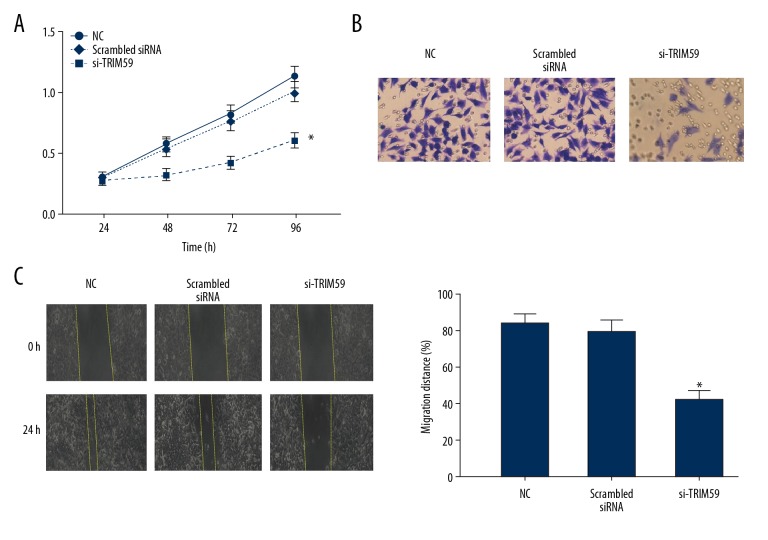
The proliferation ability of EOC cells after TRIM59 silencing was detected by CCK-8 assay. Compared to the scrambled siRNA group, the proliferation ability of OVCAR3 cells in the siRNA-TRIM59 group was significantly inhibited (P<0.05, Figure 3A). Additionally, the results of the Transwell assay suggested that after 36 h of culture, the number of crystal violet-stained cells in the siRNA-TRIM59 group was lower than that in the scrambled siRNA group (P<0.05, Figure 3B), indicating that the invasion ability of EOC cells was dramatically reduced after TRIM59 silencing. The results of the wound healing assay suggested that 24 h after making the scratch, the wound area in the siRNA-TRIM59 group was markedly greater than that in the scrambled siRNA group. These data demonstrate that the migration ability of EOC cells was suppressed after TRIM59 silencing (P<005, Figure 3C).
Figure 3.
The relationship of TRIM59 expression and OVCAR3 cell of biological behavior. (A) CCK-8 assay was used to detect the proliferation of OVCAR3 cell. (B) Transwell assay was used to detect the invasion of OVCAR3 cell. (C) Wound healing assay was used to detect the migration of OVCAR3 cell. * Compared with other groups, P<0.05.
TRIM59 promotes EOC progression through the FAK/AKT/MMP pathway
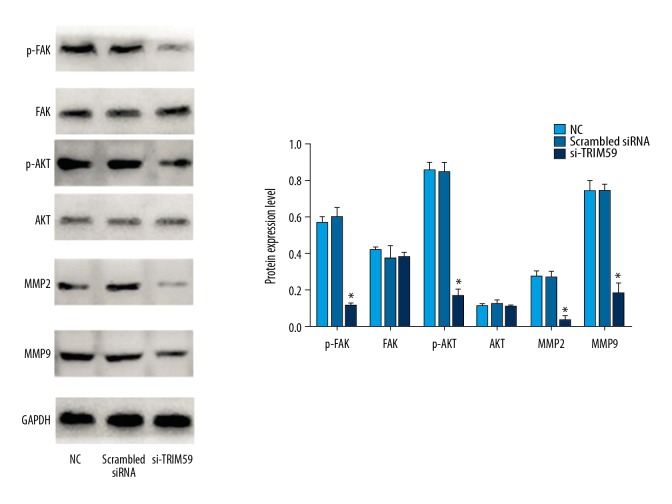
According to previous research and the literature, TRIM59 is closely related to the FAK/AKT/MMP pathway [9–11]. However, the relationship between TRIM59 and the FAK/AKT/MMP pathway in EOC is unclear. To determine the potential mechanism of TRIM59 involved in the proliferation, migration, and invasion of EOC, the expression of FAK/AKT pathway-related proteins (FAK, phospho-FAK, AKT, phospho-AKT, MMP2, and MMP9) was analyzed by Western blotting. The results suggested that silencing of TRIM59 expression in the EOC cell line OVCAR3 did not affect the total FAK and AKT levels. Compared to the scrambled siRNA group, the expression levels of phospho-FAK, AKT, phospho-AKT, MMP2, and MMP9 in the siRNA-TRIM59 groups were reduced (Figure 4).
Figure 4.
After silencing TRIM59 in OVCAR3, the expression levels of phospho-FAK, AKT, phospho-AKT, MMP2, and MMP9 were evidently reduced. * Compared with other groups, P<0.05.
Discussion
TRIM59 is mainly located in the cytoplasm and is composed of 403 amino acids [6]. TRIM59 is involved in numerous biological processes in the body and is a key regulator of processes such as tumorigenesis and development [6,12]. TRIM59 plays an important role in the killing of tumor of cells via bacille Calmette-Guérin (BCG)-activated macrophages [13]. However, most studies have suggested that TRIM59 promotes tumor development. For instance, in gastric cancer cells, TRIM59 negatively regulates p53 to promote tumor cell growth, proliferation, and migration [14]. In a prostate cancer model, TRIM59 was found to affect 2 signaling pathways, RAS and Rb, to accelerate cancer development [15]. Additionally, TRIM59 is highly expressed in lung, breast, and liver cancers [16–18]. However, the relationship between TRIM59 and EOC remains unclear.
In this study, the upregulation of TRIM59 in EOC was predicted using bioinformatic methods based on GEO data. Additionally, TCGA data analysis showed that TRIM59 was the factor responsible for the poor prognosis of patients with EOC. TRIM59 expression in human EOC tissues was determined, and its potential role was examined. Specifically, TRIM59 expression was markedly upregulated in EOC tissues; the positive staining of EOC tissue samples supported this result. To further explore the biological role of TRIM59 in EOC cells, the effects of silencing of TRIM59 on the proliferation, invasion, and migration of EOC cell lines were evaluated. TRIM59 expression levels in different EOC cell lines were detected, with the highest TRIM59 expression observed in OVCAR3 cells; this cell line was further analyzed in vitro. Silencing of TRIM59 was found to suppress the proliferation, invasion, and migration of EOC cells.
The role of TRIM59 in cell migration and invasion has been extensively described, but its underlying mechanism remains unclear. FAK and AKT are important regulators of cell survival, proliferation, migration, and invasion [19–21]. Excessive activation of the FAK/AKT/MMP signaling pathway is related to the malignant biological behaviors of tumors. Additionally, activation of the FAK/AKT/MMP pathway can promote the EOC process. In our study, silencing of TRIM59 suppressed the FAK/AKT/MMP pathway; thus, abnormal activation of the FAK/AKT/MMP signaling pathway plays a key role in TRIM59-related proliferation and invasion. However, the mechanism by which TRIM59 activates the AKT-MMP2/MMP9 pathway and by which molecules mediate the high expression of TIRM59 in EOC warrants further in-depth research and exploration.
Conclusions
We found that TRIM59 expression in human EOC tissues and cell lines was upregulated and that TRIM59 promoted EOC cell proliferation, migration, and invasion through the FAK/AKT/MMP signal transduction pathway. Future studies with larger sample sizes of EOC tissues are needed to analyze the relationships among TRIM59 expression, clinicopathological features, and prognosis of EOC patients. A more comprehensive in vivo investigation is also required.
Footnotes
Source of support: Departmental sources
Conflict of interest
None.
References
- 1.Permuth-Wey J, Sellers TA. Epidemiology of ovarian cancer. Methods Mol Biol. 2009;472:413–37. doi: 10.1007/978-1-60327-492-0_20. [DOI] [PubMed] [Google Scholar]
- 2.Bast RC, Jr, Hennessy B, Mills GB. The biology of ovarian cancer: New opportunities for translation. Nat Rev Cancer. 2009;9(6):415–28. doi: 10.1038/nrc2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mirandola L, Cannon JM, Cobos E, et al. Cancer testis antigens: Novel biomarkers and targetable proteins for ovarian cancer. Int Rev Immunol. 2011;30(2–3):127–37. doi: 10.3109/08830185.2011.572504. [DOI] [PubMed] [Google Scholar]
- 4.Kimsa MW, Strzalka-Mrozik B, Kimsa MC, et al. Differential expression of tripartite motif-containing family in normal human dermal fibroblasts in response to porcine endogenous retrovirus infection. Folia Biol (Praha) 2014;60(3):144–51. doi: 10.14712/fb2014060030144. [DOI] [PubMed] [Google Scholar]
- 5.Hatakeyama S. TRIM family proteins: Roles in autophagy, immunity, and carcinogenesis. Trends Biochem Sci. 2017;21:291–311. doi: 10.1016/j.tibs.2017.01.002. [DOI] [PubMed] [Google Scholar]
- 6.Chu Y, Yang X. SUMO E3 ligase activity of TRIM proteins. Oncogene. 2011;30(9):1108–16. doi: 10.1038/onc.2010.462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Massiah MA, Simmons BN, Short KM, Cox TC. Solution structure of the RBCC/TRIM B-box1 domain of human MID1: B-box with a RING. J Mol Biol. 2006;358(2):532–45. doi: 10.1016/j.jmb.2006.02.009. [DOI] [PubMed] [Google Scholar]
- 8.Peng H, Begg GE, Schultz DC, et al. Reconstitution of the KRAB-KAP-1 repressor complex: A model system for defining the molecular anatomy of RING-B box-coiled-coil domain-mediated protein-protein interactions. J Mol Biol. 2000;295(5):1139–62. doi: 10.1006/jmbi.1999.3402. [DOI] [PubMed] [Google Scholar]
- 9.Gao R, Lv G, Zhang C, et al. TRIM59 induces epithelial-to -mesenchymal transition and promotes migration and invasion by PI3K/AKT signaling pathway in medulloblastoma. Oncol Lett. 2018;15(6):8253–60. doi: 10.3892/ol.2018.8432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shen H, Zhang J, Zhang Y, et al. Knockdown of tripartite motif 59 (TRIM59) inhibits proliferation in cholangiocarcinoma via the PI3K/AKT/mTOR signalling pathway. Gene. 2019;698:50–60. doi: 10.1016/j.gene.2019.02.044. [DOI] [PubMed] [Google Scholar]
- 11.Liu Y, Dong Y, Zhao L, et al. TRIM59 overexpression correlates with poor prognosis and contributes to breast cancer progression through AKT signaling pathway. Mol Carcinog. 2018;57(12):1792–802. doi: 10.1002/mc.22897. [DOI] [PubMed] [Google Scholar]
- 12.Valiyeva F, Jiang F, Elmaadawi A, et al. Characterization of the oncogenic activity of the novel TRIM59 gene in mouse cancer models. Mol Cancer Ther. 2011;10(7):1229–40. doi: 10.1158/1535-7163.MCT-11-0077. [DOI] [PubMed] [Google Scholar]
- 13.Jin Z, Tian Y, Yan D, et al. BCG increased membrane expression of TRIM59 through the TLR2/TLR4/IRF5 pathway in RAW264.7 macrophages. Protein Pept Lett. 2017;24(8):765–70. doi: 10.2174/0929866524666170818155524. [DOI] [PubMed] [Google Scholar]
- 14.Zhou Z, Ji Z, Wang Y, et al. TRIM59 is up-regulated in gastric tumors, promoting ubiquitination and degradation of p53. Gastroenterology. 2014;147(5):1043–54. doi: 10.1053/j.gastro.2014.07.021. [DOI] [PubMed] [Google Scholar]
- 15.Lin WY, Wang H, Song X, et al. Knockdown of tripartite motif 59 (TRIM59) inhibits tumor growth in prostate cancer. Eur Rev Med Pharmacol Sci. 2016;20(23):4864–73. [PubMed] [Google Scholar]
- 16.Zhan W, Han T, Zhang C, et al. TRIM59 promotes the proliferation and migration of non-small cell lung cancer cells by upregulating cell cycle related proteins. PLoS One. 2015;10(11):e0142596. doi: 10.1371/journal.pone.0142596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu Y, Dong Y, Zhao L, et al. TRIM59 overexpression correlates with poor prognosis and contributes to breast cancer progression through AKT signaling pathway. Mol Carcinog. 2018;57(12):1792–802. doi: 10.1002/mc.22897. [DOI] [PubMed] [Google Scholar]
- 18.Sun G, Sui X, Han D, et al. TRIM59 promotes cell proliferation, migration and invasion in human hepatocellular carcinoma cells. Pharmazie. 2017;72(11):674–79. doi: 10.1691/ph.2017.7659. [DOI] [PubMed] [Google Scholar]
- 19.Kim YG, Kim MJ, Lim JS, et al. ICAM-3-induced cancer cell proliferation through the PI3K/Akt pathway. Cancer Lett. 2006;239(1):103–10. doi: 10.1016/j.canlet.2005.07.023. [DOI] [PubMed] [Google Scholar]
- 20.Ren K, Jin H, Bian C, et al. MR-1 modulates proliferation and migration of human hepatoma HepG2 cells through myosin light chains-2 (MLC2)/focal adhesion kinase (FAK)/Akt signaling pathway. J Biol Chem. 2008;283(51):35598–605. doi: 10.1074/jbc.M802253200. [DOI] [PubMed] [Google Scholar]
- 21.Song G, Xu S, Zhang H, et al. TIMP1 is a prognostic marker for the progression and metastasis of colon cancer through FAK-PI3K/AKT and MAPK pathway. J Exp Clin Cancer Res. 2016;35(1):148. doi: 10.1186/s13046-016-0427-7. [DOI] [PMC free article] [PubMed] [Google Scholar]