Abstract
The angucyclines form the largest family of polycyclic aromatic polyketides, and have been studied extensively. Herein, we report the discovery of lugdunomycin, an angucycline‐derived polyketide, produced by Streptomyces species QL37. Lugdunomycin has unique structural characteristics, including a heptacyclic ring system, a spiroatom, two all‐carbon stereocenters, and a benzaza‐[4,3,3]propellane motif. Considering the structural novelty, we propose that lugdunomycin represents a novel subclass of aromatic polyketides. Metabolomics, combined with MS‐based molecular networking analysis of Streptomyces sp. QL37, elucidated 24 other rearranged and non‐rearranged angucyclines, 11 of which were previously undescribed. A biosynthetic route for the lugdunomycin and limamycins is also proposed. This work demonstrates that revisiting well‐known compound families and their producer strains still is a promising approach for drug discovery.
Keywords: angucycline, Baeyer–Villiger oxidation, molecular networking, natural product, polyketide
Actinobacteria are Gram‐positive, and often filamentous, bacteria that are a major source of bioactive natural products,1, 2 and most of these are produced by actinomycetes of the genus Streptomyces.2, 3 Despite the increasing difficulty to isolate new molecules, the biosynthetic potential of actinomycetes is far from exhausted.4, 5 Many such molecules are most likely specified by biosynthetic gene clusters (BGCs) that are poorly expressed in the laboratory, generally referred to as cryptic gene clusters.6, 7 However, novel molecules have also been identified via the “one strain—many compounds” (OSMAC) strategy,8 which further supports the notion that actinobacteria harbor significant unexplored chemical diversity. Angucyclines and angucyclinones,9 which bear an unsymmetrically assembled benz[a]anthracene frame, represent the largest family of polycyclic aromatic polyketides from actinomycetes, and exhibit a broad range of biological, predominantly anticancer and antibacterial, activities.9, 10 The minimal polyketide synthase (PKS) forms the initial angucycline or angucyclinone framework that is further modified by a wide array of post‐PKS tailoring enzymes.9
Herein, we report the discovery of a novel angucycline‐derived compound, lugdunomycin (1, Figure 1) with a striking benzaza[4,3,3]propellane‐6‐spiro‐2′‐2H‐naphtho[1,8‐bc]furan backbone, found in Streptomyces sp. QL37. OSMAC, combined with MS/MS‐based molecular networking, characterized 24 other rearranged and non‐rearranged angucyclines 2–25 (Figure 1), 11 of which were new structures featuring unique ring rearrangement, oxidation, reduction, and amidation patterns. The new structural features, and in particular the Baeyer–Villiger oxidative cleavage and expansion of the C‐ring of angucycline, further enrich the existing diversity of angucycline‐ and/or angucyclinone‐type natural products.
Figure 1.
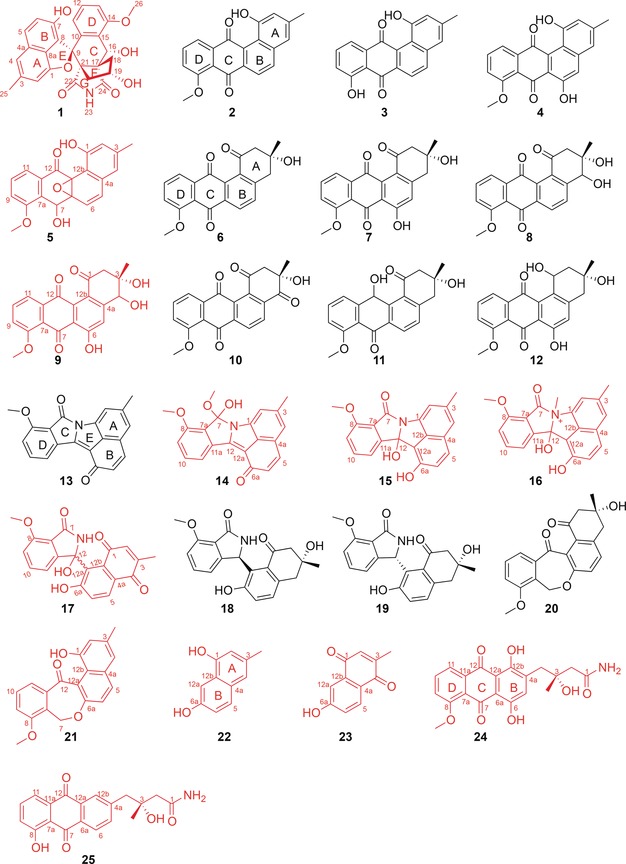
Angucyclines isolated from Streptomyces sp. QL37. Lugdunomycin (1) is a novel angucycline derivative. All the previously undescribed compounds are shown in red.
In our search for novel chemical diversity, seven strains showing distinctive pigmentation were prioritized from our actinomycete strain collection11 because distinctive pigmentation is a beacon for chemical diversity.12 The actinomycetes were grown in six different culture media, and their metabolomes were compared by thin‐layer chromatography (TLC). Of these, Streptomyces sp QL37 yielded a rich metabolic profile, and this strain was therefore subjected to up‐scale (7.5 liter) on MM agar plates. Repeated chromatography of the crude extract (2.3 g) resulted in compounds 1 (0.5 mg), 2 (27 mg), 6/7 (27 mg), 8 (3.4 mg), and 11 (1 mg).
The isolated colorless 1 was called lugdunomycin after Lugdunum batavorum, the Latin name for the city of Leiden. UHPLC‐ToF‐MS analysis of 1 identified an [M+H]+ peak at m/z 474.1553, establishing its molecular composition as C27H23NO7. The deduced chemical formula was corroborated by the attached proton test (APT) that exhibited 27 carbons. The three aromatic rings A, B, and D, were readily assigned based on the proton‐splitting pattern in the 1H NMR spectrum and COSY correlations (Supporting Information, Table S1 and Figure S1). Benzene rings A and B were fused into a naphthalene system by sharing a double bond between C‐4a and C‐8a, based on the HMBC correlations H‐4/C‐8a, H‐5/C‐8a, H‐4/C‐5, H‐5/C‐4, and H‐6/C‐4a. Ring C was identified by HMBC correlations from H‐16 to C‐10, C‐14, and C‐15 of ring D. Key HMBC correlations , such as H‐19/C‐17, H‐19/C‐21, H‐20/C‐17, and H‐20/C‐21, unequivocally showed that the cyclopentanol ring F is joined to ring C by sharing the bond between C‐17 and C‐21. Furthermore, H‐18 and H‐20 showed 3 J CH HMBC correlations with two carbonyl groups at δ C 182.4, and 182.5, respectively, which indicated the presence of another ring fused to two all‐carbon stereocenters,13 C‐17 and C‐21, apart from rings C and F; these two carbonyls are joined by a nitrogen atom in line with both the molecular formula and the chemical shifts, consistent with succinimide ring G. Consequently, rings D/C/F/G constituted a benzaza[4,3,3]propellane motif. Ring systems A/B and D/C/F/G are linked at spiroatom C‐9, as established by key HMBC correlations H‐20/C‐9, H‐11/C‐9, and H‐6/C‐9. An additional five‐membered furan ring (ring E) is formed to make a 2H‐naphtho[1,8‐bc]furan module [7‐methyl‐2H‐naphtho[1,8‐bc]furan‐3‐ol] to join the whole structure and fit in the molecular formula. Taken together, the benzaza[4,3,3]propellane skeleton is adorned with a spirocyclic 2H‐naphtho[1,8‐bc]furan moiety and two all‐carbon quaternary centers embedded within five contiguous stereogenic carbons. The benzaza[4,3,3]propellane‐6‐spiro‐2′‐2H‐naphtho[1,8‐bc]furan architecture is unprecedented.
Single‐crystal X‐ray diffraction confirmed the structure of lugdunomycin (1) (Supporting Information, Table S2). Crystallization of 1 exhibited a centrosymmetric space group P42/n, explicitly supporting a racemic mixture. The configurations of five chiral centers in 1 were determined as 9R*, 16S*, 17R*, 19S*, and 21S* (Figure 2 and Supporting Information, Figure S2). The racemic nature of the obtained lugdunomycin was also suggested by the measured optical rotation of zero.
Figure 2.
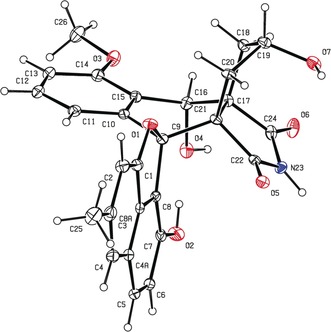
ORTEP drawing of the crystal structure of lugdunomycin. The absolute configurations of the five chiral centers of lugdunomycin are 9R*, 16S*, 17R*, 19S*, and 21S*.
To obtain insight into the diversity of angucycline‐derived molecules produced by Streptomyces sp. QL37, we performed global natural products social (GNPS) molecular networking using MS/MS profiles14, 15 and compared the output to the GNPS database.16 To ensure optimal chemical diversity, Streptomyces sp. QL37 was fermented in 77 different culture media (Supporting Information, Table S3), as described.11 MS/MS‐based molecular networking of QL37 grown on R5 + 0.8 % peptone + 1 % mannitol and MM + 0.5 % mannitol + 1 % glycerol presented the relatedness among metabolites (Supporting Information, Figure S3), including the structurally related angucyclines (Figure 1) and other families of compounds (Supporting Information, Figure S5). This observation not only dereplicated the non‐rearranged angucyclines 2, 6–8, and 11 isolated from MM medium (Supporting Information, Figure S4), but also revealed many nitrogen‐containing angucyclines (Figure 3 B). Especially, the subnetwork for lugdunomycin identified a putative aldehyde analogue of 1 with m/z 502 (Figure 3 A).
Figure 3.
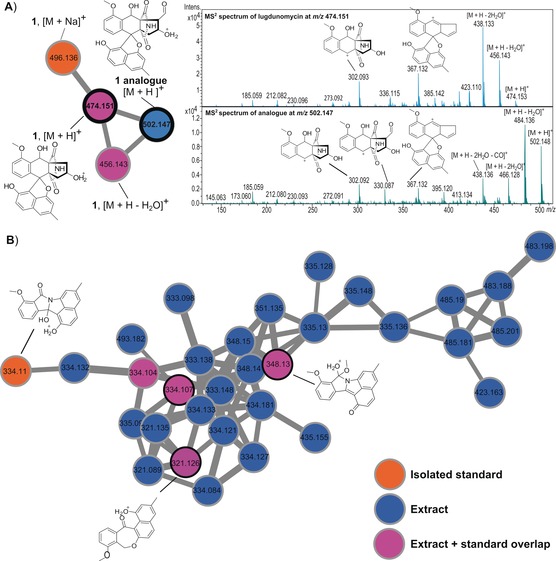
MS2‐based molecular networking of Streptomyces sp. QL37. A) Subnetwork for lugdunomycin reveals an analogue at m/z 502. Comparison of MS2 spectra of 1 at m/z 474.151 and its analogue at m/z 502.147 suggests an additional aldehyde group in the latter; see the fragmentation pattern above the ion peak. B) Rearranged angucyclines. The edge thickness between connecting nodes corresponds to the similarity of the MS/MS spectra. The full GNPS network is presented in Figure S3 in the Supporting Information, and the subnetwork for non‐rearranged angucyclines in Figure S4 in the Supporting Information.
Up‐scale refermentation (20 L) of Streptomyces sp. QL37 in R5 medium followed by extensive systematic isolation, via OSMAC8 identified compounds 2–45, including unrearranged benz[a]anthracene angucyclines (2–12), limamycins17 (13–19), emycins17, 18, 19 (20, 21), naphthalenones (22, 23), anthraquinones (24, 25, Figure 1), nucleosides (26–33), cyclodipeptides (34–41), 4‐quinolones (42, 43), vitamin K (44), and quinazolinone (45, Supporting Information, Figure S5). Of these, compounds 5, 9, 14–17, and 21–25 were new structures. The NMR data assignments of new angucyclines are summarized in Tables S4–S6 in the Supporting Information, and the 2D NMR correlations (HMBC and COSY) are displayed in Figure S1 in the Supporting Information. The 11 new angucyclines featured, among others, unique ring cleavage (ring A or C) and rearrangement (13–25), hydration of double bond Δ12, 12a (15, 16), nucleophilic addition of methanol to the ketone at C‐7 (14), epoxidation of double bond Δ6a, 12b (5), N‐quaternary methylation (16), amidation (13–19, 24, and 25), and reduction of the ketone at C‐7 (20, 21). Though 14 is likely an artifact due to the usage of methanol during chromatographic isolation, all these new structures add further chemical diversity to this important family of polyketides.
In terms of bioactivity, an agar diffusion assay whereby pure compound was spotted on filter on a lawn of the target bacterium, showed that lugdunomycin had antimicrobial activity against the Gram‐positive Bacillus subtilis 168, but not against the Gram‐negative Escherichia coli K12 (not shown). More extensive experiments are needed to assess the bioactivity and mode of action of lugdunomycin.
Based on the structures of the identified molecules (Figure 1), a biosynthetic route of rearranged limamycins and/or non‐rearranged angucyclines is proposed (Figure 4). Versatile post‐PKS oxidations are critical for the rearrangement of the precursors 2 and/or 6. The cleavage at the C‐1/12b bond in the A‐ring of 2 generates the tricyclic anthraquinone scaffold in 24 and 25. Another pivotal Baeyer–Villiger oxidation at the C‐6a/C‐7 bond of the quinone ring C is proposed to initiate the structural rearrangements of 2 and/or 6 to give compounds 13–23. The lactone intermediate 6 a or 2 a is susceptible to hydration resulting in lactone ring opening, followed by the amidation at C‐12 ketone to introduce a nitrogen atom in 6 c or 2 c, which is cyclized to form limamycins17 13–19 (Figure 4 A). Alternatively, 2 a could go through an evolutionary pathway involved in emycin biosynthesis,18 whereby a reduction at the ketone group of C‐7 is essential to generate compounds 20 and 21.
Figure 4.
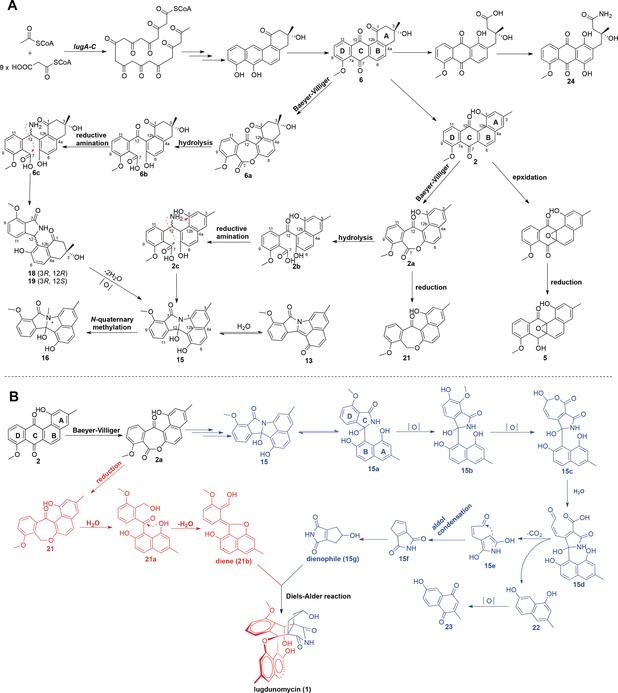
Biosynthetic route to limamycins and proposed pathway towards lugdunomycin. A) Biosynthesis of limamycins. Baeyer–Villiger oxidative cleavage at the quinone ring of compounds 2 and/or 6 is pivotal for the biosynthesis of lugdunomycin and rearranged limamycins 13–19. B) Proposed pathway for lugdunomycin biosynthesis. As possible final step for the assembly of lugdunomycin a Diels–Alder reaction is proposed. As dienophile for the Diels–Alder reaction an isomer of maleimycin (Supporting Information, Figure S6) is a likely candidate. We propose its formation from the oxidative ring contraction in the D‐ring of limamycins. The limamycin and emycin biosynthetic pathways are drawn in blue and red, respectively.
The co‐identification of compounds 2–25 in Streptomyces sp. QL37 and elucidation of the biosynthetic pathway for the nitrogen‐containing limamycins shed light on the biosynthetic logic of lugdunomycin (1, Figure 4 B). We hypothesize that the benzaza[4,3,3]propellane skeleton of lugdunomycin is eventually constructed through a Diels–Alder [4+2] cycloaddition, and the diene and dienophile reagents for the Diels–Alder reaction are provided by the limamycin and emycin biosynthetic pathway, respectively. The putative limamycin 15 a is likely to go through a cascade of oxidative C−C bond cleavage in the D‐ring, followed by decarboxylation and aldol condensation, to give an isomer of maleimycin20 (compound 15 g) that serves as the dienophile for Diels–Alder reaction. In support of this, the identity of compound 22 was spectroscopically confirmed, and MS/MS‐based molecular networking analysis of QL37 confirmed the presence of a mass consistent with that of the maleimycin isomer (15 g) (Supporting Information, Figure S6). The hydroxy‐o‐quinodimethane intermediate 21 b is a candidate diene reagent for the Diels–Alder reaction, which is probably derived from the dehydration of 21 a. The cycloaddition of 21 b and 15 g mimics the reported Diels–Alder trapping of photochemically generated hydroxy‐o‐quinodimethanes by maleimides,21 and this intermolecular Diels–Alder reaction has been explored to construct complex pseudo‐natural polyketides from simple intermediates.22
Genome sequencing of Streptomyces sp. QL37 allowed the identification of a type II PKS gene cluster (lug, Supporting Information, Table S7 and Figure S7). Genetic inactivation of the minimal PKS genes lugA–C completely abolished the production of angucyclines, limamycins, and lugdunomycin. We are investigating the precise function of the various genes of the lug gene cluster.
Future investigation into the genetic and synthetic knowledge underlying lugdunomycin biosynthesis, will not only guide the up‐scale production of lugdunomycin and its potential variants through synthetic biology approaches, but will also offer new opportunities to expand the existing structural diversity of known polyketides. Thus, our work will likely form the basis of new explorations into the exciting chemical space of the angucyclines and other polyketides.
Conflict of interest
The authors declare no conflict of interest.
Supporting information
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Supplementary
Acknowledgements
We are grateful to Hermen Overkleeft for discussions and to Lies Bouwman and Raj Pannu for advice on X‐ray crystallography. C.S.W. was supported by a grant from the China Scholarship Council. A.V.M. and P.C.D. want to acknowledge grants 5P41GM103484, S10RR029121 and GM107550. G.P.vW. acknowledges grant 10467 from the Netherlands Organization for Scientific Research.
C. Wu, H. U. van der Heul, A. V. Melnik, J. Lübben, P. C. Dorrestein, A. J. Minnaard, Y. H. Choi, G. P. van Wezel, Angew. Chem. Int. Ed. 2019, 58, 2809.
References
- 1. Hopwood D. A., Streptomyces in Nature and Medicine: The Antibiotic Makers, Oxford University Press, New York, 2007. [Google Scholar]
- 2. Barka E. A., Vatsa P., Sanchez L., Gaveau-Vaillant N., Jacquard C., Klenk H., Clément C., Ouhdouch Y., van Wezel G. P., Microbiol. Mol. Biol. Rev. 2016, 80, 1–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bérdy J., J. Antibiot. (Tokyo) 2012, 65, 385–395. [DOI] [PubMed] [Google Scholar]
- 4. Kolter R., van Wezel G. P., Nat. Microbiol. 2016, 1, 15020. [DOI] [PubMed] [Google Scholar]
- 5. Cooper M. A., Shlaes D., Nature 2011, 472, 32. [DOI] [PubMed] [Google Scholar]
- 6. Zhu H., Sandiford S. K., van Wezel G. P., J. Ind. Microbiol. Biotechnol. 2014, 41, 371–386. [DOI] [PubMed] [Google Scholar]
- 7. Rutledge P. J., Challis G. L., Nat. Rev. Microbiol. 2015, 13, 509–523. [DOI] [PubMed] [Google Scholar]
- 8. Bode H. B., Bethe B., Höfs R., Zeeck A., ChemBioChem 2002, 3, 619–627. [DOI] [PubMed] [Google Scholar]
- 9. Kharel M. K., Pahari P., Shepherd M. D., Tibrewal N., Nybo S. E., Shaaban K. A., Rohr J., Nat. Prod. Rep. 2012, 29, 264–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Rohr J., Thiericke R., Nat. Prod. Rep. 1992, 9, 103–137. [DOI] [PubMed] [Google Scholar]
- 11. Zhu H., Swierstra J., Wu C., Girard G., Choi Y. H., van Wamel W., Sandiford S. K., van Wezel G. P., Microbiology 2014, 160, 1714–1725. [DOI] [PubMed] [Google Scholar]
- 12. Brady S. F., Chao C. J., Handelsman J., Clardy J., Org. Lett. 2001, 3, 1981–1984. [DOI] [PubMed] [Google Scholar]
- 13. Minko Y., Pasco M., Lercher L., Botoshansky M., Marek I., Nature 2012, 490, 522–526. [DOI] [PubMed] [Google Scholar]
- 14. Watrous J., Roach P., Alexandrov T., Heath B. S., Yang J. Y., Kersten R. D., van der Voort M., Pogliano K., Gross H., Raaijmakers J. M., et al., Proc. Natl. Acad. Sci. USA 2012, 109, 1743–1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Nguyen D. D., Wu C.-H., Moree W. J., Lamsa A., Medema M. H., Zhao X., Gavilan R. G., Aparicio M., Atencio L., Jackson C., et al., Proc. Natl. Acad. Sci. USA 2013, 110, E2611–2620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wang M., Carver J. J., Phelan V. V., Sanchez L. M., Garg N., Peng Y., Nguyen D. D., Watrous J., Kapono C. A., Luzzatto-Knaan T., et al., Nat. Biotechnol. 2016, 34, 828–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fotso S., Mahmud T., Zabriskie T. M., Santosa D. A., Proteau P. J., J. Antibiot. (Tokyo) 2008, 61, 449–456. [DOI] [PubMed] [Google Scholar]
- 18. Gerlitz M., Udvarnoki G., Rohr J., Angew. Chem. Int. Ed. Engl. 1995, 34, 1617–1621; [Google Scholar]; Angew. Chem. 1995, 107, 1757–1761. [Google Scholar]
- 19. Ma M., Rateb M. E., Teng Q., Yang D., Rudolf J. D., Zhu X., Huang Y., Zhao L. X., Jiang Y., Li X., et al., J. Nat. Prod. 2015, 78, 2471–2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Elstner E. F., Carnes D. M., Suhadolnik R. J., Kreishman G. P., Schweizer M. P., Robins R. K., Biochemistry 1973, 12, 4992–4997. [DOI] [PubMed] [Google Scholar]
- 21. Dell'Amico L., Vega-Peñaloza A., Cuadros S., Melchiorre P., Angew. Chem. Int. Ed. 2016, 55, 3313–3317; [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew. Chem. 2016, 128, 3374–3378. [Google Scholar]
- 22. Asai T., Tsukada K., Ise S., Shirata N., Hashimoto M., Fujii I., Gomi K., Nakagawara K., Kodama E. N., Oshima Y., Nat. Chem. 2015, 7, 737–743. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Supplementary


