Abstract
Mucosa‐associated invariant T (MAIT) cells are unconventional T lymphocytes defined by their innate‐like characteristics and broad antimicrobial responsiveness. Whether MAIT cells are part of the tissue‐resident defense in the oral mucosal barrier is unknown. Here, we found MAIT cells present in the buccal mucosa, with a tendency to cluster near the basement membrane, and located in both epithelium and the underlying connective tissue. Overall MAIT cell levels were similar in the mucosa compared to peripheral blood, in contrast to conventional T cells that showed an altered representation of CD4+ and CD8+ subsets. The major mucosal MAIT cell subset displayed a tissue‐resident and activated profile with high expression of CD69, CD103, HLA‐DR, and PD‐1, as well as a skewed subset distribution with higher representation of CD4–/CD8– double‐negative cells and CD8αα+ cells. Interestingly, tissue‐resident MAIT cells had a specialized polyfunctional response profile with higher IL‐17 levels, as assessed by polyclonal stimulus and compared to tissue nonresident and circulating populations. Furthermore, resident buccal MAIT cells were low in perforin. Together, these data indicate that MAIT cells form a part of the oral mucosal T cell compartment, where they exhibit a tissue‐resident‐activated profile biased toward IL‐17 production.
Keywords: Buccal mucosa, IL‐17, MAIT cells, MR1, Oral mucosa, Tissue residency
Introduction
Mucosa‐associated invariant T (MAIT) cells are nonclassical innate‐like T cells that recognize microbial antigens presented by the MHC‐Ib‐related protein 1 (MR1) 1, 2. MR1 displays an extraordinary level of evolutionary conservation among mammals 2, 3, 4, and was shown to present microbial vitamin B2 (riboflavin) metabolites from a wide range of microbes that carry the riboflavin biosynthesis pathway 5, 6. MAIT cells express a semi‐invariant T cell receptor (TCR), including the Vα7.2 segment coupled with Jα33, Jα12, or Jα20, and limited TCR β‐chain diversity 7, 8. This TCR repertoire endows MAIT cells with the capacity to respond to the MR1‐restricted riboflavin derivatives produced by diverse microbes 9, 10. MAIT cells respond to antigenic stimulus with an innate‐like speed and produce proinflammatory cytokines including TNF, IFN‐γ, and IL‐17 9, 11, 12, 13, 14. They can furthermore kill cells infected by microbes expressing the riboflavin pathway 15, 16 and inhibit intracellular microbial growth 17.
Human MAIT cells are also defined by their high expression of CD161, the IL‐18 receptor α subunit (IL‐18Rα), and the transcription factor ZBTB16 18, also known as promyelocytic leukemia zinc finger protein (PLZF) 9, 19. The majority of MAIT cells are CD8+, with some being CD4–/CD8– double‐negative (DN), and a minor CD4+ population 9, 11, 12, 19. Human MAIT cells acquire innate‐like antimicrobial activity in the fetal intestinal mucosa prenatally, prior to the establishment of the commensal microflora 14. In adults, MAIT cells are highly abundant in mucosal tissues, liver, and peripheral blood 1, 2, 20, 21, 22.
Studies of the murine oral immune system revealed diverse populations of DCs 23, 24, and provided insight into oral T‐cell function 25, but the immunology of the human oral mucosa remains relatively little studied. However, Dutzan et al. recently performed a broad characterization of the immune cell network at the gingival interface and identified T cells as the dominant immune cell population in both the buccal and gingival mucosa 26. CD4+ T cells, mainly of the CD45RO+ "memory" phenotype, were found to be the largest subset at around half of CD3+ cells, whereas smaller subsets of CD8+ and γδ T cells were also present 26. The same study also identified tissue‐resident CD4+ T cells as major producers of IL‐17 in periodontitis. IL‐17 is a cytokine central to the regulation of immune activity at mucosal surfaces 27, 28. Although IL‐17 is implicated in a variety of functions, its role in the regulation of mucosal immunity hinges on three core effects: maintenance of mucosal integrity via regulation of tight junction proteins, induction of antimicrobial molecule production by epithelial cells, and recruitment of neutrophils to sites of infection.
Given the capacity of MAIT cells to recognize and respond to several commensal and disease causing microbes occurring in the oral cavity, and the ability of these cells to locate to mucosal sites, one can hypothesize a role for MAIT cells in the oral mucosal barrier. However, the immunobiology of MAIT cells at this mucosal site has to date not been explored. In the present study, we therefore investigated the presence, location, characteristics, and function of MAIT cells in the human buccal mucosa. The findings demonstrate that MAIT cells are part of the healthy buccal mucosal immune defense with specialized tissue‐resident and nonresident subpopulations exhibiting distinct functional profiles, where in particular, the CD69+CD103+ MAIT cell population show strong IL‐17 production. We discuss these findings in the context of contemporary knowledge of immune homeostasis and defense at this important barrier site.
Results
Identification of MAIT cells in the buccal mucosa
Given the broad antimicrobial reactivity of MAIT cells, we were interested in investigating the potential role of these cells in oral mucosa, a site continuously exposed to a broad array of microbial stimuli. Buccal biopsies and matched peripheral blood samples were collected from healthy donors (Supporting Information Table 1), and cells isolated after tissue processing were analyzed by flow cytometry. All subjects were examined for their oral health and showed no oral mucosal disease, active dental caries, infection, or periodontitis. Using antibody combinations allowing identification of MAIT cells in blood, buccal MAIT cells were defined as CD45+ CD3+ CD161hi Vα7.2+ cells (Fig. 1A). To ensure that cells defined by this gating strategy were bona fide MAIT cells, their MR1 restriction was confirmed by staining with the MR1 5‐(2‐oxopropylideneamino)‐6‐D‐ribitylaminouracil (5‐OP‐RU) tetramer (Fig. 1B). The percentage of MAIT cells in the mucosa ranged between 0.1 and 7% of total T cells (Fig. 1C). Overall, these levels were not significantly different from those in matched blood, although in any given individual, the levels seen at the two sites were often rather divergent. Interestingly, major conventional T‐cell subsets were present in the buccal mucosa at frequencies different from those in the blood. Levels of CD4+ T cells were lower than in blood, whereas CD8+ T cells and CD4–CD8– T cells were enriched in buccal mucosa as compared to blood (Fig. 1C). These data demonstrate that MAIT cells are part of the buccal mucosa T‐cell compartment.
Figure 1.
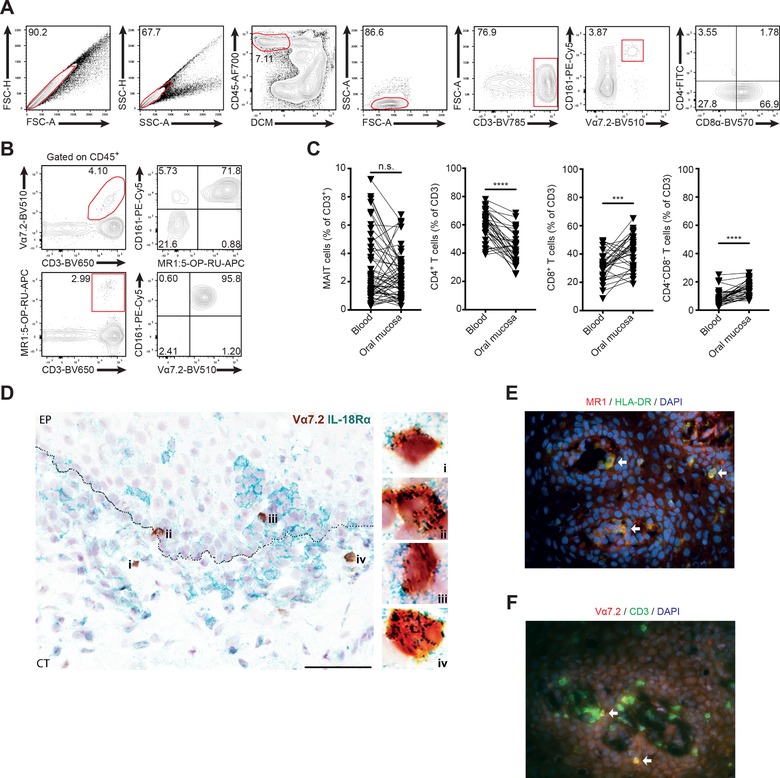
MAIT cells are part of the oral buccal mucosa T cell compartment. (A) Gating strategy to identify T cell populations in buccal mucosa by flow cytometry. (B) Representative staining of identification of MR1:5‐OP‐RU tetramer binding CD161+ MAIT cells within the CD3+Vα7.2+ buccal mucosal cell population (upper panels), and identification of CD161+Vα7.2+ MAIT cells within the MR1:5‐OP‐RU tetramer binding CD3+ buccal mucosal cell population (lower panels). (C) Comparison of T‐cell subset frequency in peripheral blood and buccal mucosa. All numbers given as percentage of CD3+ lymphocytes (n = 55 [MAIT], 32 [others]). (D) Location of MAIT cells within the buccal mucosa. Representative bright field image of buccal mucosal tissue section, from a healthy donor, stained for Vα7.2 in brown (DAB) and IL‐18Rα in green (Vina Green) showing that the MAIT cells are localized close to the basement membrane (n = 6). The images on the right, collected with a 100× objective, are magnified view of the region indicated in the box in the image to the left, collected with a 40× objective. The scale bar represents 60 μm, image including epithelium (EP) and connective tissue (CT). Representative immunofluorescence images of buccal mucosal tissue sections, from a healthy individual, stained for (E) MR1 (red) and HLA‐DR (green) and stained for (F) Vα7.2 (red) and CD3 (green). Double‐positive cells appear as yellow, arrows point to double‐positive cells for each marker combination. DAPI (blue) was used as a counterstain for visualization of cell nuclei. The images were collected with 40× objective. For each panel, "n" indicates the number of individual human donors tested for each dataset. Statistical significance determined using Wilcoxon matched pairs test for paired data. ***p <0.001, ****p <0.0001.
Detection of buccal mucosal MAIT cells in situ
To determine MAIT cell location within the buccal mucosa, we performed in situ staining for Vα7.2 and IL‐18Rα in oral mucosal sections (Fig. 1D). In mucosa, staining for IL‐18Rα can be used in lieu of CD161, as we have previously shown for genital mucosa 22. Using the Vα7.2+ IL‐18Rα+ definition, MAIT cells were readily identified in buccal mucosal sections, and they tended to locate close to the basement membrane (Fig. 1D). MAIT cells were present in the epithelial layer, just above the basal membrane (distance from basal membrane: median = 14.6 μm; range = 1.0–75.9 μm), as well as in the underlying connective tissue, just beneath the basal membrane (median = 20.7 μm; range = 2.8–292.2 μm). Non‐MAIT T cells, as well as MR1‐expressing HLA‐DR+ antigen‐presenting cells, were also found in buccal mucosa as determined by immunofluorescence staining (Fig. 1E and F). MAIT cell location around the basement membrane is consistent with a possible role as innate‐like T cell sentinels to detect microbial epithelial barrier breach.
Altered subset distribution and Jα‐TCR usage in mucosal MAIT cells
In peripheral blood, MAIT cells are predominantly CD8+, with some CD4–CD8– cells and a very small CD4+ subpopulation. In the buccal mucosa, MAIT cells were enriched in CD4–CD8– and CD4+ subsets, and had significantly lower levels of the CD8+ subset (Fig. 2A). Within the CD8+ subset, similar to blood MAIT cells, the mucosal MAIT cells exhibited an increased frequency of CD8αα+ cells compared to non‐MAIT T cells (Fig. 2B and C). Notably, MAIT cells represented almost half of all CD8αα+ T cells in the buccal mucosa, with significant donor variability (Fig. 2D).
Figure 2.
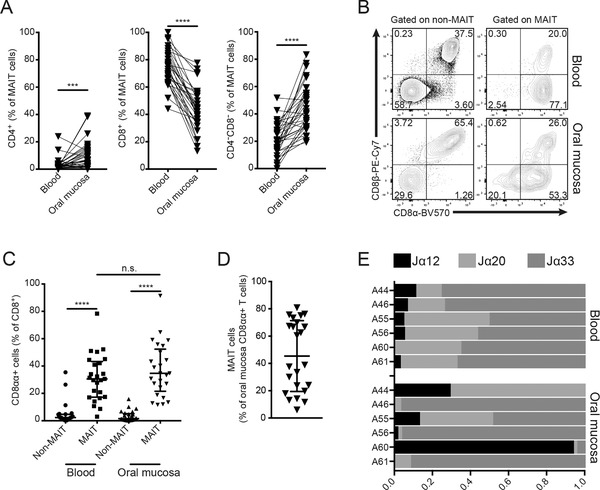
Co‐receptor and TCR‐Jα usage differences between blood and buccal mucosal MAIT cells. (A) Representation of MAIT cell subsets defined by CD4+ and CD8+ expression. All numbers given as percentage of Vα7.2+CD161+ cells (n = 32). (B) Representative flow cytometric plots of CD8α and CD8β expression in T cell subsets in blood and mucosa. (C) Comparison of CD8αα expression in CD8+ T cell subsets in blood and mucosa. All numbers given as percentage of CD8+ cells (n = 25). (D) Percentage MAIT cells among the mucosa CD8αα+ T‐cell population (n = 23). In C and D, medians and interquartile ranges are indicated. (E) MAIT cell TCR Jα segment usage in paired blood and mucosal samples from healthy donors (n = 6). For each panel, "n" indicates the number of individual human donors tested. Statistical significance determined using Wilcoxon matched pairs test for paired data. ***p <0.001, ****p <0.0001.
The distribution of Jα‐TCR usage within the MAIT cell populations in the buccal mucosa and blood was compared using quantitative real‐time PCR (qRT‐PCR; Fig. 2E). We quantified the relative expression of three Jα previously shown to be represented within the MAIT cell TCR repertoire, the Jα12, Jα20, and Jα33 8, 29, 30. As expected, blood MAIT cells displayed a consistent pattern with dominant Jα33 usage, with smaller Jα20 and Jα12 usage. However, in matched mucosa, the pattern was more diverse, with different Jα segments dominating in different donors. Together, these data support a model of compartmentalization where the subset representation and TCR‐Jα usage in buccal MAIT cell population are partly different that of the circulating MAIT cells found in blood.
MAIT cells in buccal mucosa express an activated perforinlow phenotype
To assess the phenotypic characteristics of the buccal MAIT cells in more detail, we next stained isolated mucosal cells with markers of activation, CD38 and HLA‐DR (Fig. 3A). The staining pattern revealed the presence of an activated HLA‐DR+CD38+ MAIT cell population in buccal mucosa (Fig. 3B). Compared to their blood counterparts, the mucosal MAIT cells expressed higher levels of HLA‐DR, whereas CD38 levels appear to be slightly lower (Fig. 3C). In addition, expression of PD‐1 was common in mucosal MAIT cells, whereas PD‐1 was less expressed in peripheral blood MAIT cells (Fig. 3C).
Figure 3.
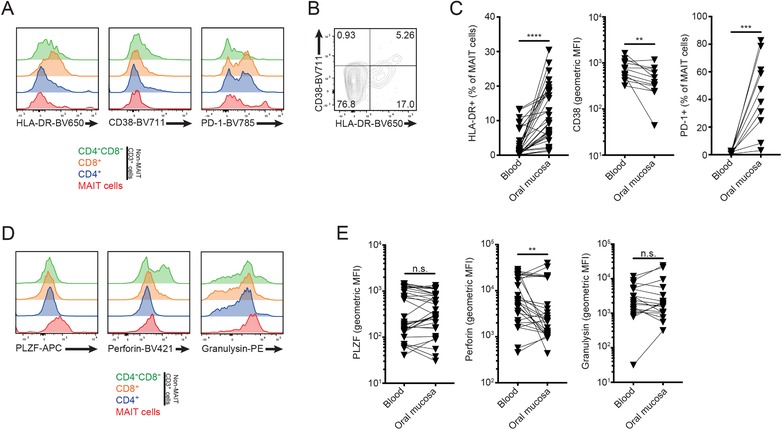
Buccal mucosal MAIT cells express PLZF and exhibit an activated PD‐1+ phenotype. (A) Representative histograms of HLA‐DR and CD38 expression in oral buccal mucosa T cell subsets. (B) Representative FACS plot of CD38 and HLA‐DR coexpression in buccal MAIT cells. (C) Expression of HLA‐DR (n = 34), CD38 (n = 12), and PD‐1 (n = 12) in blood and mucosal MAIT cells, as percentage of or mean fluorescence intensity (MFI) in Vα7.2+CD161+ cells. (D) Intracellular staining histograms of PLZF, perforin and granulysin in buccal T cell subsets. (E) Expression levels of PLZF (n = 32), perforin (n = 29), and granulysin (n = 17) in blood and oral mucosal MAIT cells, as MFI in Vα7.2+CD161+ cells. For each panel, "n" indicates the number of individual human donors tested. Statistical significance determined using Wilcoxon matched pairs test for paired data. **p< 0.01, ***p< 0.001, ****p< 0.0001.
To investigate the phenotype of MAIT cells in more detail, paired blood and mucosal cell samples were stained intracellularly for the expression of the master transcription factor PLZF, as well as the cytolytic effector molecules perforin and granulysin (Fig. 3D). Perforin levels were mostly lower in the buccal MAIT cells than in their blood counterparts (Fig. 3E), whereas expression levels of PLZF and granulysin were similar in both compartments. These findings suggest that MAIT cells in the buccal mucosa are highly activated cells with significant expression of the checkpoint inhibition receptor PD‐1, and display a reduced perforin content as compared to circulating MAIT cells.
Characterization of resident and nonresident MAIT cells in the buccal mucosa
We next investigated the MAIT cell expression of receptors associated with tissue residency in the mucosa. Resident T cells can be distinguished by coexpression of CD69 and CD103 (αE integrin) in a range of tissues 31, 32, 33, 34, 35, including intestinal mucosa 36. Flow cytometric assessment of CD69 and CD103 expression revealed the existence of a major CD69+CD103+ tissue‐resident MAIT cell population in the buccal mucosa (Fig. 4A). However, a minority of MAIT cells were negative for CD69 and CD103, consistent with a tissue nonresident or "passing through" phenotype (Fig. 4A). Compared to peripheral blood MAIT cells, the buccal mucosal MAIT cell population was highly enriched in CD69 and CD103 expression, and CD69 expression was mostly seen in the CD103+ population (Fig. 4B). As the CD69 expression formed a continuum including dim cells of both CD103+ and CD103– character, we compared the four MAIT cell populations defined by CD69 and CD103 with regard to CD4+ and CD8+ expression. CD103+ MAIT cells were mainly CD8+ with a minority CD4–CD8– cells, whereas CD103– MAIT cells had a distinct opposite pattern with a majority CD4–CD8– character (Fig. 4C and D). CD69 expression did not significantly influence this pattern (Fig. 4C). In addition, CD103+ MAIT cells were more activated than their CD103– counterparts, as evidenced by higher expression of HLA‐DR and CD38 (Fig. 4E). In contrast, the CD103+ subpopulation expressed lower levels of the cytolytic effector molecule perforin, whereas PLZF did not vary depending on CD103 (Fig. 4F). These results indicate the existence of two distinct buccal MAIT cell subpopulations where one express the CD103 marker of tissue residency and one lacks CD103 consistent with a nonresident character.
Figure 4.
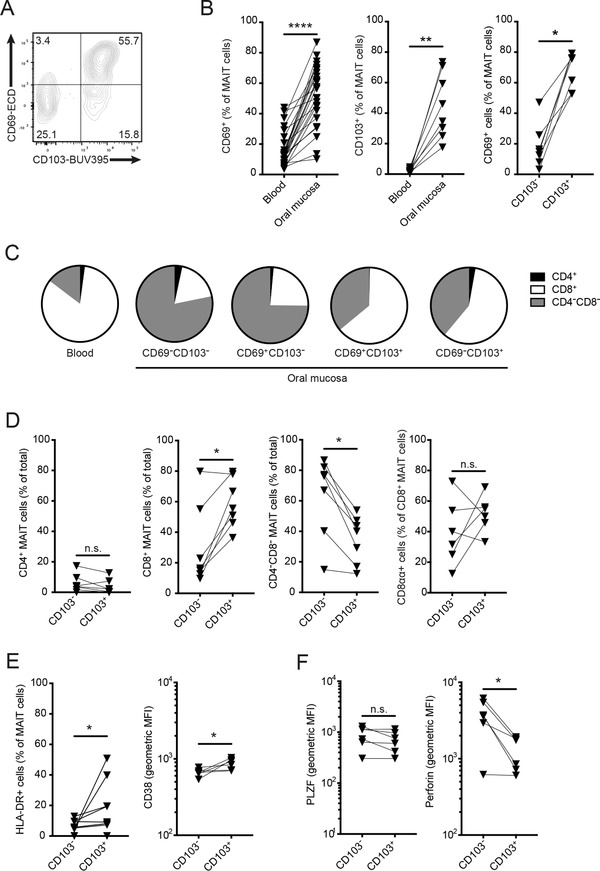
Tissue resident and nonresident MAIT cells in buccal mucosa. (A) Representative plot of CD69 and CD103 expression in buccal MAIT cells. (B) Expression of CD69 (n = 25) and CD103 (n = 8) in blood and mucosal MAIT cells, and expression of CD69 in mucosal CD103+ and CD103– MAIT cells (n = 7). (C) Expression of CD4+ and CD8+ in blood MAIT cells, and subsets of buccal MAIT cells defined by CD69 and CD103. Proportion of each marker given as median value (n = 6). (D) CD4+ and CD8+ expression profile in mucosal CD103+ and CD103– MAIT cells (n = 7). (E) Expression of HLA‐DR (n = 8) and CD38 (n = 6) in mucosal CD103+ and CD103– MAIT cells. (F) Expression of PLZF and perforin in mucosal CD103+ and CD103– MAIT cells (n = 6). For each panel, "n" indicates the number of individual human donors tested. Statistical significance determined using Wilcoxon matched pairs test for paired data. *p <0.05, **p <0.01, ****p< 0.0001.
Distinct polyfunctional responses of mucosal resident, nonresident, and circulating MAIT cells
To investigate the functional profile of MAIT cells in buccal mucosa in comparison with peripheral blood, isolated lymphocytes from the two tissues were activated with PMA/ionomycin and their repertoire of functions evaluated by flow cytometry staining for TNF, IFN‐γ, IL‐2, IL‐17, and granzyme B (Fig. 5A). Overall, both blood and mucosal MAIT cells displayed detectable production of all these effector functions, with some notable tissue‐dependent differences. Peripheral blood MAIT cells produced TNF and IFN‐γ more strongly than their mucosal counterparts, whereas IL‐2 and granzyme B were similar (Fig. 5A and B). In contrast, IL‐17 production was very low in blood MAIT cells, with the exception of one outlier, while this cytokine was clearly more highly expressed in the buccal MAIT cells (Fig. 5A and B). In the mucosal tissue, the resident CD103+ subset and the CD103– nonresident subset were largely similar in their response patterns with two notable exceptions. IL‐17 in particular, and to some extent granzyme B expression, were significantly higher in the CD103+ MAIT cell subset.
Figure 5.
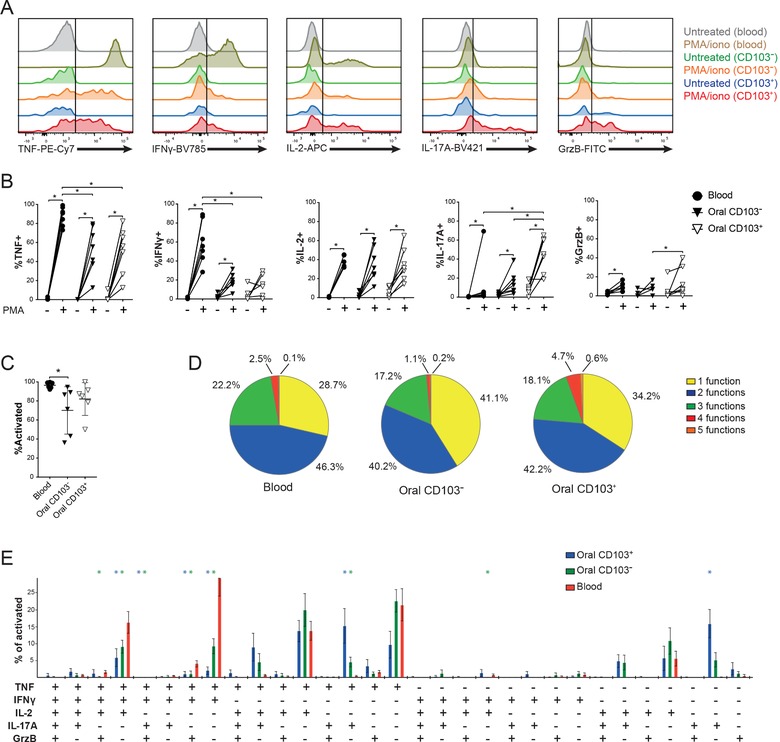
Functional profile of MAIT cells in buccal mucosa and blood. (A) Representative histograms of intracellular staining for TNF, IFN‐γ, IL‐2, IL‐17A, and granzyme B in CD103+ and CD103– MAIT cells from buccal mucosa, in comparison with blood MAIT cells after stimulation with PMA/ionomycin. (B) Expression of cytokines by MAIT cells from blood and buccal mucosa of healthy donors after PMA/ionomycin stimulation (n = 7). (C) Total level of activation of MAIT cells from blood or mucosa after PMA/ionomycin stimulation, defined as percentage of total MAIT cells, which express at least one of the factors TNF, IFN‐γ, IL‐2, IL‐17A, and granzyme B (n = 6). (C) Medians and interquartile ranges are indicated. (D) Overall polyfunctionality of blood, CD103–, and CD103+ mucosal MAIT cells. Percentages represent the mean of values from a set of donors (n = 6). (E) Polyfunctional response profile of activated MAIT cells from blood, as well as CD103– and CD103+ mucosal MAIT cells (n = 6). For each panel, "n" indicates the number of individual human donors tested. Statistical significance determined using Wilcoxon matched pairs test for paired data. *p <0.05, **p <0.01.
To understand the functional specialization of mucosal resident, mucosal nonresident, and circulating MAIT cells in more detail, we analyzed the polyfunctional response profiles of these subsets. First, we determined the percentages of MAIT cells in the three locations that responded with at least one of the five functions measured (Fig. 5C). For all three sites, the vast majority of MAIT cells expressed at least one function. However, the mucosal MAIT cells displayed somewhat lower responsiveness, where in particular, the CD103– MAIT cells were less responsive. When the overall polyfunctionality was analyzed, the ability to express 1, 2, 3, 4, or 5 functions was fairly similar between the different tissue locations, although the CD103+ resident MAIT cells tended to be more 4‐ or 5‐functional (Fig. 5D). Finally, we analyzed the data by Boolean gating to assess which specific polyfunctional profiles were expressed by peripheral blood, CD103+ mucosal resident, and CD103– nonresident MAIT cells (Fig. 5E; Supporting Information Table 2). This analysis brought forward the pattern that peripheral blood MAIT cells were focused primarily on TNF production, either alone or in combinations with IFN‐γ and IL‐2. The mucosal CD103+ resident MAIT cells instead primarily focused on IL‐17 production either alone or in combinations with TNF or IL‐2. The CD103– nonresident MAIT cells had a functional profile intermediate between the circulating and resident populations, and were mostly expressing TNF with or without IL‐2, and less IL‐17. Altogether, these findings indicate that the mucosal resident, the mucosal nonresident, and the circulating MAIT cells deploy different effector cytokine response profiles.
Discussion
Tissue‐resident immune cell populations are increasingly recognized for their importance in defense against invading pathogens. The oral mucosa is continuously exposed to a range of antigens derived from food, microbiota, and pathogens. To maintain barrier immune defenses at this site, while not reacting against nonpathogenic encounters, is a considerable challenge. Recent studies have started to address this research question 37, 38, but no studies to date have addressed the potential role of the MR1‐restricted MAIT cell population in oral mucosa. Here, we observe that MAIT cells are dispersed throughout the buccal mucosa with a preferential location close to the basement membrane. The buccal MAIT cells are biased toward a CD4–/CD8– DN profile, and the CD8+ subset is primarily CD8αα and make up a significant proportion of all CD8αα T cells in the mucosa. The MAIT cell population in this mucosal site is composed of distinct CD69+CD103+ intraepithelial resident and CD69–CD103– nonresident subsets. The CD103+ subset is enriched in CD8+ MAIT cells, is more activated, and has lower cytolytic potential. Finally, the buccal MAIT cells show a distinct functional profile with enhanced IL‐17 production and less TNF and IFN‐γ, as compared to their peripheral blood counterparts, and this pattern was particularly pronounced for the CD103+ resident MAIT cells.
The MR1‐presented riboflavin metabolite antigens recognized by MAIT cells are conserved among many prokaryotic and eukaryotic microbes relevant for mucosal health. Thus, MAIT cells may play a unique role in the oral mucosa with broad innate‐like recognition of such microbes in a manner that does not require the kind of priming which conventional adaptive T‐cell responses do. It is interesting that the MAIT cells to at least some extent locate around the basement membrane, where they may function as innate‐like T‐cell sentinels to detect microbial epithelial barrier breach. Around the similar location, MR1+HLA–DR+ cells are residing and those may be able to present riboflavin metabolite antigens from invading microbes. Together, MAIT cells and MR1+ antigen presenting cells may form a rapid innate response immune unit at the mucosal barrier, capable of responding to pathogens to which the host has no preexisting adaptive immunity.
The CD103+CD69+ mucosal‐resident MAIT cell population is fairly strongly biased toward IL‐17 production, with less TNF and IFN‐γ expression, as compared to the CD103– MAIT cell subpopulation. Resident MAIT cell recognition of an invading microbial pathogen may thus setoff an IL‐17‐mediated inflammatory response with induction of antimicrobial peptides in epithelium and recruitment of neutrophils. Such a pattern would be reminiscent to the situation described for chronic severe periodontitis by Dutzan et al. 26, where IL‐17 produced by presumably adaptive Th17 cells was associated with increased gingival tissue infiltration of neutrophils. Given their innate‐like response characteristics, MAIT cells may be among the first immune cells to detect microbial infiltration and respond with IL‐17 release. Here, it is interesting to note that IL‐17 has emerged as an important mediator and amplifier of immunity against Candida albicans 38. The dimorphic fungus C. albicans expresses the riboflavin biosynthesis pathway and is recognized by MAIT cells in an MR1‐restricted manner 10. Many cells respond to IL‐17 by upregulation of proinflammatory cytokines, such as IL‐6, and chemokines for recruitment of neutrophils including CXCL1, CXCL2, and CXCL5 39. In addition, IL‐17 stimulates production of β‐defensins in epithelial cells 40. MAIT cells are thus well located to initiate mucosal immune responses in oropharyngeal candidiasis.
Peripheral blood MAIT cells are predominantly CD8+ with a minority CD4–CD8– subset. This pattern is reversed in buccal mucosa, such that the CD4–CD8– MAIT cells are more numerous. It is however interesting to note that the CD8+ MAIT subset in buccal mucosa is primarily CD8αα, and that this subset makeup almost half of all CD8αα T cells in the oral mucosa. These CD8αα MAIT cells bear resemblance to the intestinal mucosal CD8αα intraepithelial lymphocytes (IELs) that have been extensively characterized in murine models, but may be less frequent in humans 41, 42. The intestinal IELs of mice are a mix of TCRγδ T cells and TCRαβ T cells with diverse specificities, and the representation of MAIT cells among these IELs in different sites is to our knowledge largely unknown. Our data indicate that the CD103+ MAIT cell population is mostly, but not exclusively, CD8+ and composed of both CD8αα and CD8αβ cells. These findings together suggest that MAIT cells makeup a significant part of the human buccal IEL‐like population.
The human oral mucosal barrier retains a commensal bacterial microbiota that is both varied and unique among other sites 43, 44, dominated by the genera Streptococcus, Haemophilus, Prevotella, and Veillonella. In addition to bacteria, the oral mucosal barrier is home to many species of fungi including C. albicans 45. The oral immune system thus has to manage and tolerate a diverse commensal microbiome, and at the same time guard against conditions arising either from dysfunction of normal oral homeostasis or caused by pathogens normally not present in the oral cavity. T cells are believed to play a role in multiple oral mucosa pathologies including aphthous stomatitis, oral leukoplakia, oral reticular or ulcerative lichen planus, celiac disease, and oral psoriasis 46, 47. Whether MAIT cells have a role in any such conditions in humans remains to be explored.
In summary, we have shown that MAIT cell populations with resident and nonresident characteristics are part of the buccal mucosal immune system in healthy donors and that they have unique functional profiles. Future studies should aim to investigate how these populations respond to commensal and pathogenic microbes, and how they are affected in different disease conditions.
Materials and methods
Tissue donor recruitment and sample collection
A total of 94 volunteers were recruited in two healthy donor groups A and B (Supporting Information Table 1). Inclusion criteria for both groups were: 20–50 years of age, HIV‐negative, non‐smoking, no antibiotics in the last 3 months, and not pregnant. Ethical permission was obtained from the Regional Ethical Review Board in Stockholm in accordance with the Declaration of Helsinki. All participants gave written informed consent. The oral health of all subjects was evaluated using standard dental examination procedures, including inspection of oral mucosa, teeth, and surrounding soft tissues, to ensure the donors had no visible mucosal lesions, no signs of gingivitis, active dental caries, or periodontitis. All donors were instructed to abstain from food or drink for 1 h before tissue collection.
Oral mucosal tissue samples and matched peripheral blood samples were collected from two sets of healthy donors. Donor group A was recruited from the general population. A matched blood sample of 20–30 mL venous blood was taken from each participant on the day of the procedure, prior to the oral sample collection. The blood was stored in heparin tubes at room temperature until analysis. Local anesthesia with xylocain/adrenalin was applied to the left bucca and three punch biopsies (5 mm in diameter, 3–4 mm deep) were taken from the site. For donor group B, tissue samples were taken before wound closure on patients who underwent orthognatic surgery of the mandible. The patients underwent these corrective surgeries due to abnormal position of the mandible in relation to the base of skull, but no buccal disease or inflammation is associated with these conditions. The patients were treated under general anesthesia and local anesthesia was additionally applied. Tissue samples 20 × 10 mm in size were taken bilaterally from the mucosa. Matched blood samples from donor group B were collected at the time of the procedure. After collection, the biopsies were stored at 4°C in serum‐free RPMI (ThermoFisher Scientific, Waltham, MA, USA) supplemented with 50 μg/mL gentamicin (ThermoFisher Scientific) and 100 μg/mL normocin (InvivoGen, San Diego, CA, USA). Biopsies intended for qPCR analysis were stored in RNAlater (QIAgen, Venlo, Netherlands) at −20°C. Biopsies intended for in situ microscopic analysis were submerged in OCT Cryomount (HistoLab, Askim, Sweden), snap‐frozen in liquid nitrogen, and then stored at −80°C.
Sample processing for flow cytometry
All collected samples were processed within 3 h of collection. Buccal biopsies were placed into serum‐free RPMI supplemented with 50 μg/mL gentamicin and 100 μg/mL normocin and then incubated with 1 mg/mL DNase I and collagenase A (both from Roche, Penzberg, Germany) at 37°C with shaking at 800 rpm for 1 h. After incubation, the reaction was stopped by addition of fetal calf serum (FCS; Sigma‐Aldrich, St Louis, MO, USA) to a final concentration of 10%. The biopsies were then pressed through a 100 μm cell strainer in order to detach loose cells. The cells were then washed in PBS and incubated in RBC lysis buffer for 10 min at RT to remove any erythrocyte contamination. The cells were washed again in PBS and stained for flow cytometry.
Donor‐matched PBMCs were isolated from blood samples by Ficoll‐Hypaque density gradient centrifugation using Lymphoprep (Axis‐Shield, Dundee, Scotland). The PBMC layer was washed twice with PBS, and incubated in RBC lysis buffer for 10 minutes at room temperature to remove erythrocyte contamination. The cells were washed again in PBS and 106 cells were stained for flow cytometry.
In situ staining
In situ staining was performed on 8‐μm‐thick sections of frozen biopsies, as previously described 22. Sections were fixed in 2% formaldehyde and stained sequentially. Antibody reagents used for tissue staining are listed in Supporting Information Table 3. The tissue sections stained for Vα7.2 in combination with IL‐18Rα were scanned into digital images using a Pannoramic 250 Flash Slide Scanner (3DHistech, Budapest, Hungary). The distance measurement was performed using the digital microscope application CaseViewer (3DHistech), where the distance from the surface of the double‐positive cells to the basal membrane was measured. The tissue sections stained for HLA‐DR in combination with MR1, and Vα7.2 in combination with CD3 were visualized using DMR‐X microscope (Leica, Weitzlar, Germany) and images were acquired with a Retiga 2000 R camera (Qimaging, Surrey, Canada).
Functional assay
One million lymphocytes in suspension were cultured overnight in RPMI +10% FCS, supplemented with 100 μg/mL normocin and 50 μg/mL gentamicin. After overnight incubation, they were activated using Leukocyte Activation Cocktail (PMA/ionomycin) (BD Biosciences, San Jose, CA, USA) in the presence of GolgiPlug (monensin) for 6 h. At the end of the incubation, the cells were stained and analyzed by flow cytometry.
Flow cytometry and antibodies
Cells in suspension were stained for surface antigens in FACS buffer (PBS with 2 mM EDTA and 2% FCS) for 20 min at 4°C. Afterwards, they were washed in FACS buffer and permeabilized using the transcription factor fixation and permeabilization buffer (BD Biosciences) for 30 min at 4°C. Following permeabilization, cells were washed twice in transcription factor wash and permeabilization buffer (BD Biosciences), and then stained for intracellular antigens in the same buffer for 20 min at 4°C. PE‐conjugated human MR1:5‐OP‐RU tetramer was obtained from the NIH Tetramer Core Facility. For tetramer staining, cells in suspension were incubated with MR1:5‐OP‐RU tetramer for 40 min at room temperature and then washed with FACS buffer prior to surface antibody staining. Monoclonal antibody reagents used in the study are listed in Supporting Information Table 4. After staining, cells were washed and analyzed using an LSRFortessa flow cytometer (BD Biosciences) equipped with 355, 405, 488, 561, and 639 nm lasers. In addition to the antibodies, cells were stained with LIVE/DEAD Fixable Dead Cell Stain (Near‐Infrared or Aqua; ThermoFisher Scientific).
Quantitative RT‐PCR
qRT‐PCR was performed to determine the mRNA expression of TCRs Vα7.2‐Jα33, Vα7.2‐Jα20, and Vα7.2‐Jα12 as well as the constant Cα chain, in healthy oral mucosa and blood. Briefly, total RNA was extracted from buccal mucosa and PBMCs using TRIzol reagent (ThermoFisher Scientific). For reverse transcription, 250 ng of RNA was used in a 15 μL total reaction volume using the IScript cDNA Synthesis kit (BioRad, Hercules, CA, USA). For amplification, a 20 μL supermix reaction was prepared accordingly: 1 μL cDNA, 5 μL SsoAdvanced™ Universal® SYBR Green (BioRad), 1 μL of each primer at a final concentration of 500 nM, and nuclease free H2O. Primer sequences as well as annealing temperatures for each of the primer sets are listed in Supporting Information Table 5. qPCR was performed using 7500 Fast real time PCR system (Applied Biosystems, Waltham, CA, USA) with the following cycling conditions: 1 cycle at 95°C for 5 min, then 40 cycles at 94°C for 10 s, 58°C or 60°C depending on primer for 30 s, and 72°C for 27 s. The relative abundance of each Vα7.2‐Jα was determined as relative to Cα by the comparative ΔΔC T method.
Software and statistical analysis
Analysis of flow cytometric data was performed using FlowJo 10 (FlowJo LLC, Ashland, OR, USA), and microscopic images were processed using Image‐Pro Premier 9.3 (Media Cybernetics, Rockville, MD, USA). Statistical tests were performed using GraphPad Prism version 7.0c for Mac OS X (GraphPad Software, La Jolla, CA, USA). Statistical significance was assessed using Wilcoxon matched pairs test for paired data, and Mann–Whitney U‐test for unpaired data. For data presented in Fig. 5, analysis and presentation of distributions were performed using SPICE version 5.1, downloaded from http://exon.niaid.nih.gov 48. Comparison of distributions was performed using a Wilcoxon matched pairs test as described 48. For all statistical analysis, p‐values below 0.05 were regarded as significant.
Author contributions
J.K.S., E.L., and M.S.C. conceived the original research idea and study design.
M.J.S., H.D., M.M., A.T., J.D., E.L., M.S.C., and J.K.S. designed experiments.
M.J.S., H.D, A.G., and J.E. conducted experiments.
M.J.S., H.D., and A.G. analyzed experimental data.
R.H., S.A., and C.K.W. recruited study volunteers and collected essential human samples.
M.J.S. and J.K.S. wrote the manuscript.
All authors reviewed and commented on the manuscript.
Conflict of interest
The authors declare no commercial or financial conflict of interest.
Abbreviations
- IEL
intraepithelial lymphocyte
- MAIT
mucosal‐associated invariant T
- MR1
MHC‐Ib‐related protein 1
- PLZF
promyelocytic leukemia zinc finger protein
Supporting information
Table 1. Healthy donor demographics
Table 2. Statistical significance obtained using Wilcoxon matched‐pairs statistical test for data presented in Figure 5E. p‐values lower than 0.05 are highlighted.
Table 3. Antibodies used for in situ microscopic analysis
Table 4. Monoclonal antibodies used for flow cytometry
Table S5. Primer sequences and annealing temperatures
Peer review correspondence
Acknowledgements
The authors wish to acknowledge the contribution of Sam Chehrehgosha, Angelica Kroonder, Emelie Molin, and Beatrice Wiberg in recruiting study volunteers and gathering oral biopsies. This research was supported by grants to JKS from the Swedish Research Council (2016‐03052), the Swedish Cancer Society (CAN 2017/777), and the US National Institutes of Health (R01DK108350). Grants to M.S.C. were from the Foundation for Odontological Research (OF11211233), and the Swedish Cancer Society (CAN 2016/731). Further support to E.L. was from the Swedish Research Council (2015‐00174) and Marie Skłodowska Curie Actions, Cofund, Project INCA 600398. J.D. was supported by Fundação para a Ciência e a Tecnologia (SFRH/BD/85290/2012, doctoral fellowship), through program QREN‐POPH‐typology 4.1. The MR1 tetramer technology was developed jointly by Dr. James McCluskey, Dr. Jamie Rossjohn, and Dr. David Fairlie, and the material was produced by the NIH Tetramer Core Facility as permitted to be distributed by the University of Melbourne. The authors wish to thank the healthy volunteers that contributed time and effort to sample donation for this study.
References
- 1. Treiner, E. , Duban, L. , Bahram, S. , Radosavljevic, M. , Wanner, V. , Tilloy, F. , Affaticati, P. et al., Selection of evolutionarily conserved mucosal‐associated invariant T cells by MR1. Nature 2003. 423: 1018–1018. [DOI] [PubMed] [Google Scholar]
- 2. Huang, S. , Martin, E. , Kim, S. , Yu, L. , Soudais, C. , Fremont, D. H. , Lantz, O. et al., MR1 antigen presentation to mucosal‐associated invariant T cells was highly conserved in evolution. Proc. Natl. Acad. Sci. USA 2009. 106: 8290–8295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Riegert, P. , Wanner, V. and Bahram, S. , Genomics, isoforms, expression, and phylogeny of the MHC class I‐related MR1 gene. J. Immunol. 1998. 161: 4066–4077. [PubMed] [Google Scholar]
- 4. Tsukamoto, K. , Deakin, J. E. , Graves, J. A. and Hashimoto, K. , Exceptionally high conservation of the MHC class I‐related gene, MR1, among mammals. Immunogenetics 2013. 65: 115–124. [DOI] [PubMed] [Google Scholar]
- 5. Kjer‐Nielsen, L. , Patel, O. , Corbett, A. J. , Le Nours, J. , Meehan, B. , Liu, L. , Bhati, M. et al., MR1 presents microbial vitamin B metabolites to MAIT cells. Nature 2012. 491: 717–723. [DOI] [PubMed] [Google Scholar]
- 6. Corbett, A. J. , Eckle, S. B. , Birkinshaw, R. W. , Liu, L. , Patel, O. , Mahony, J. , Chen, Z. et al., T‐cell activation by transitory neo‐antigens derived from distinct microbial pathways. Nature 2014. 509: 361–365. [DOI] [PubMed] [Google Scholar]
- 7. Reantragoon, R. , Kjer‐Nielsen, L. , Patel, O. , Chen, Z. , Illing, P. T. , Bhati, M. , Kostenko, L. et al., Structural insight into MR1‐mediated recognition of the mucosal associated invariant T cell receptor. J. Exp. Med. 2012. 209: 761–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lepore, M. , Kalinichenko, A. , Colone, A. , Paleja, B. , Singhal, A. , Tschumi, A. , Lee, B. et al., Parallel T‐cell cloning and deep sequencing of human MAIT cells reveal stable oligoclonal TCRbeta repertoire. Nat. Commun. 2014. 5: 3866. [DOI] [PubMed] [Google Scholar]
- 9. Le Bourhis, L. , Martin, E. , Peguillet, I. , Guihot, A. , Froux, N. , Core, M. , Levy, E. et al., Antimicrobial activity of mucosal‐associated invariant T cells. Nat. Immunol. 2010. 11: 701–708. [DOI] [PubMed] [Google Scholar]
- 10. Dias, J. , Leeansyah, E. and Sandberg, J. K. , Multiple layers of heterogeneity and subset diversity in human MAIT cell responses to distinct microorganisms and to innate cytokines. Proc. Natl. Acad. Sci. USA 2017. 114: E5434–E5443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Walker, L. J. , Kang, Y. H. , Smith, M. O. , Tharmalingham, H. , Ramamurthy, N. , Fleming, V. M. , Sahgal, N. et al., Human MAIT and CD8alphaalpha cells develop from a pool of type‐17 precommitted CD8+ T cells. Blood 2012. 119: 422–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Leeansyah, E. , Ganesh, A. , Quigley, M. F. , Sonnerborg, A. , Andersson, J. , Hunt, P. W. , Somsouk, M. et al., Activation, exhaustion, and persistent decline of the antimicrobial MR1‐restricted MAIT‐cell population in chronic HIV‐1 infection. Blood 2013. 121: 1124–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cosgrove, C. , Ussher, J. E. , Rauch, A. , Gartner, K. , Kurioka, A. , Huhn, M. H. , Adelmann, K. et al., Early and nonreversible decrease of CD161++/MAIT cells in HIV infection. Blood 2013. 121: 951–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Leeansyah, E. , Loh, L. , Nixon, D. F. and Sandberg, J. K. , Acquisition of innate‐like microbial reactivity in mucosal tissues during human fetal MAIT‐cell development. Nat. Commun. 2014. 5: 3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Le Bourhis, L. , Dusseaux, M. , Bohineust, A. , Bessoles, S. , Martin, E. , Premel, V. , Core, M. et al., MAIT cells detect and efficiently lyse bacterially‐infected epithelial cells. PLoS Pathog. 2013. 9: e1003681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kurioka, A. , Ussher, J. E. , Cosgrove, C. , Clough, C. , Fergusson, J. R. , Smith, K. , Kang, Y. H. et al., MAIT cells are licensed through granzyme exchange to kill bacterially sensitized targets. Mucosal. Immunol. 2014. 8: 429–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Meierovics, A. , Yankelevich, W. J. and Cowley, S. C. , MAIT cells are critical for optimal mucosal immune responses during in vivo pulmonary bacterial infection. Proc. Natl. Acad. Sci. USA 2013. 110: E3119–E3128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Beaulieu, A. M. and Sant'Angelo, D. B. , The BTB‐ZF family of transcription factors: key regulators of lineage commitment and effector function development in the immune system. J. Immunol. 2011. 187: 2841–2847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Martin, E. , Treiner, E. , Duban, L. , Guerri, L. , Laude, H. , Toly, C. , Premel, V. et al., Stepwise development of MAIT cells in mouse and human. PLoS Biol. 2009. 7: e54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Le Bourhis, L. , Mburu, Y. K. and Lantz, O. , MAIT cells, surveyors of a new class of antigen: development and functions. Curr. Opin. Immunol. 2013. 25: 174–180. [DOI] [PubMed] [Google Scholar]
- 21. Dusseaux, M. , Martin, E. , Serriari, N. , Peguillet, I. , Premel, V. , Louis, D. , Milder, M. et al., Human MAIT cells are xenobiotic‐resistant, tissue‐targeted, CD161hi IL‐17‐secreting T cells. Blood 2011. 117: 1250–1259. [DOI] [PubMed] [Google Scholar]
- 22. Gibbs, A. , Leeansyah, E. , Introini, A. , Paquin‐Proulx, D. , Hasselrot, K. , Andersson, E. , Broliden, K. et al., MAIT cells reside in the female genital mucosa and are biased towards IL‐17 and IL‐22 production in response to bacterial stimulation. Mucosal. Immunol. 2017. 10: 35–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Nudel, I. , Elnekave, M. , Furmanov, K. , Arizon, M. , Clausen, B. E. , Wilensky, A. and Hovav, A. H. , Dendritic cells in distinct oral mucosal tissues engage different mechanisms to prime CD8+ T cells. J. Immunol. 2011. 186: 891–900. [DOI] [PubMed] [Google Scholar]
- 24. Hovav, A. H. , Dendritic cells of the oral mucosa. Mucosal. Immunol. 2014. 7: 27–37. [DOI] [PubMed] [Google Scholar]
- 25. Conti, H. R. , Peterson, A. C. , Brane, L. , Huppler, A. R. , Hernandez‐Santos, N. , Whibley, N. , Garg, A. V. , et al., Oral‐resident natural Th17 cells and gammadelta T cells control opportunistic Candida albicans infections. J. Exp. Med. 2014. 211: 2075–2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Dutzan, N. , Konkel, J. E. , Greenwell‐Wild, T. and Moutsopoulos, N. M. , Characterization of the human immune cell network at the gingival barrier. Mucosal. Immunol. 2016. 9: 1163–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cua, D. J. and Tato, C. M. , Innate IL‐17‐producing cells: the sentinels of the immune system. Nat. Rev. Immunol. 2010. 10: 479–489. [DOI] [PubMed] [Google Scholar]
- 28. Abusleme, L. and Moutsopoulos, N. M. , IL‐17: overview and role in oral immunity and microbiome. Oral Dis. 2017. 23: 854–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Tilloy, F. , Treiner, E. , Park, S. H. , Garcia, C. , Lemonnier, F. , de la Salle, H. , Bendelac, A. , et al., An invariant T cell receptor alpha chain defines a novel TAP‐independent major histocompatibility complex class Ib‐restricted alpha/beta T cell subpopulation in mammals. J. Exp. Med. 1999. 189: 1907–1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Reantragoon, R. , Corbett, A. J. , Sakala, I. G. , Gherardin, N. A. , Furness, J. B. , Chen, Z. , Eckle, S. B. , et al., Antigen‐loaded MR1 tetramers define T cell receptor heterogeneity in mucosal‐associated invariant T cells. J. Exp. Med. 2013. 210: 2305–2320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Masopust, D. , Vezys, V. , Wherry, E. J. , Barber, D. L. and Ahmed, R. , Cutting edge: gut microenvironment promotes differentiation of a unique memory CD8 T cell population. J. Immunol. 2006. 176: 2079–2083. [DOI] [PubMed] [Google Scholar]
- 32. Skon, C. N. , Lee, J. Y. , Anderson, K. G. , Masopust, D. , Hogquist, K. A. and Jameson, S. C. , Transcriptional downregulation of S1pr1 is required for the establishment of resident memory CD8+ T cells. Nat. Immunol. 2013. 14: 1285–1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mackay, L. K. , Braun, A. , Macleod, B. L. , Collins, N. , Tebartz, C. , Bedoui, S. , Carbone, F. R. et al., Cutting edge: CD69 interference with sphingosine‐1‐phosphate receptor function regulates peripheral T cell retention. J. Immunol. 2015. 194: 2059–2063. [DOI] [PubMed] [Google Scholar]
- 34. Sathaliyawala, T. , Kubota, M. , Yudanin, N. , Turner, D. , Camp, P. , Thome, J. J. , Bickham, K. L. , et al., Distribution and compartmentalization of human circulating and tissue‐resident memory T cell subsets. Immunity 2013. 38: 187–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Buggert, M. , Nguyen, S. , Salgado‐Montes de Oca, G. , Bengsch, B. , Darko, S. , Ransier, A. , Roberts, E. R. , et al., Identification and characterization of HIV‐specific resident memory CD8(+) T cells in human lymphoid tissue. Sci. Immunol. 2018. 3 10.1126/sciimmunol.aar4526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kiniry, B. E. , Li, S. , Ganesh, A. , Hunt, P. W. , Somsouk, M. , Skinner, P. J. , Deeks, S. G. et al., Detection of HIV‐1‐specific gastrointestinal tissue resident CD8(+) T‐cells in chronic infection. Mucosal. Immunol. 2018. 11: 909–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Moutsopoulos, N. M. and Konkel, J. E. , Tissue‐specific immunity at the oral mucosal barrier. Trends Immunol. 2018. 39: 276–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Conti, H. R. and Gaffen, S. L. , IL‐17‐mediated immunity to the opportunistic fungal pathogen Candida albicans . J. Immunol. 2015. 195: 780–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Shen, F. , Hu, Z. , Goswami, J. and Gaffen, S. L. , Identification of common transcriptional regulatory elements in interleukin‐17 target genes. J. Biol. Chem. 2006. 281: 24138–24148. [DOI] [PubMed] [Google Scholar]
- 40. Conti, H. R. , Shen, F. , Nayyar, N. , Stocum, E. , Sun, J. N. , Lindemann, M. J. , Ho, A. W. , et al., Th17 cells and IL‐17 receptor signaling are essential for mucosal host defense against oral candidiasis. J. Exp. Med. 2009. 206: 299–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Cheroutre, H. , Lambolez, F. and Mucida, D. , The light and dark sides of intestinal intraepithelial lymphocytes. Nat. Rev. Immunol. 2011. 11: 445–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Mayassi, T. and Jabri, B. , Human intraepithelial lymphocytes. Mucosal. Immunol. 2018. 11: 1281–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Human Microbiome Project Consortium , Structure, function and diversity of the healthy human microbiome. Nature 2012. 486: 207–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Aas, J. A. , Paster, B. J. , Stokes, L. N. , Olsen, I. and Dewhirst, F. E. , Defining the normal bacterial flora of the oral cavity. J. Clin. Microbiol. 2005. 43: 5721–5732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ghannoum, M. A. , Jurevic, R. J. , Mukherjee, P. K. , Cui, F. , Sikaroodi, M. , Naqvi, A. and Gillevet, P. M. , Characterization of the oral fungal microbiome (mycobiome) in healthy individuals. PLoS Pathog. 2010. 6: e1000713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wu, R. Q. , Zhang, D. F. , Tu, E. , Chen, Q. M. and Chen, W. , The mucosal immune system in the oral cavity‐an orchestra of T cell diversity. Int. J. Oral Sci. 2014. 6: 125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Fatahzadeh, M. and Schwartz, R. A. , Oral psoriasis: an overlooked enigma. Dermatology 2016. 232: 319–325. [DOI] [PubMed] [Google Scholar]
- 48. Roederer, M. , Nozzi, J. L. and Nason, M. C. , SPICE: exploration and analysis of post‐cytometric complex multivariate datasets. Cytometry A 2011. 79: 167–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table 1. Healthy donor demographics
Table 2. Statistical significance obtained using Wilcoxon matched‐pairs statistical test for data presented in Figure 5E. p‐values lower than 0.05 are highlighted.
Table 3. Antibodies used for in situ microscopic analysis
Table 4. Monoclonal antibodies used for flow cytometry
Table S5. Primer sequences and annealing temperatures
Peer review correspondence


