Abstract
Objectives
Early melanoma detection decreases morbidity and mortality. Early detection classically involves dermoscopy to identify suspicious lesions for which biopsy is indicated. Biopsy and histological examination then diagnose benign nevi, atypical nevi, or cancerous growths. With current methods, a considerable number of unnecessary biopsies are performed as only 11% of all biopsied, suspicious lesions are actually melanomas. Thus, there is a need for more advanced noninvasive diagnostics to guide the decision of whether or not to biopsy. Artificial intelligence can generate screening algorithms that transform a set of imaging biomarkers into a risk score that can be used to classify a lesion as a melanoma or a nevus by comparing the score to a classification threshold. Melanoma imaging biomarkers have been shown to be spectrally dependent in Red, Green, Blue (RGB) color channels, and hyperspectral imaging may further enhance diagnostic power. The purpose of this study was to use the same melanoma imaging biomarkers previously described, but over a wider range of wavelengths to determine if, in combination with machine learning algorithms, this could result in enhanced melanoma detection.
Methods
We used the melanoma advanced imaging dermatoscope (mAID) to image pigmented lesions assessed by dermatologists as requiring a biopsy. The mAID is a 21‐wavelength imaging device in the 350–950 nm range. We then generated imaging biomarkers from these hyperspectral dermoscopy images, and, with the help of artificial intelligence algorithms, generated a melanoma Q‐score for each lesion (0 = nevus, 1 = melanoma). The Q‐score was then compared to the histopathologic diagnosis.
Results
The overall sensitivity and specificity of hyperspectral dermoscopy in detecting melanoma when evaluated in a set of lesions selected by dermatologists as requiring biopsy was 100% and 36%, respectively.
Conclusion
With widespread application, and if validated in larger clinical trials, this non‐invasive methodology could decrease unnecessary biopsies and potentially increase life‐saving early detection events. Lasers Surg. Med. 51:214–222, 2019. © 2019 The Authors. Lasers in Surgery and Medicine Published by Wiley Periodicals, Inc.
Keywords: melanoma, dermoscopy, artificial intelligence, machine learning, hyperspectral imaging
INTRODUCTION
The detection of melanoma clinically can be visually challenging and often relies on the identification of hallmark features including asymmetry, irregular borders, and color variegation to identify potentially cancerous lesions. In clinical practice, a substantial number of unnecessary biopsies are performed, as only 11% of all biopsied, suspicious lesions are actually melanomas 1. The dermatoscope aids in detection of melanoma by providing magnified and illuminated images. However, even among expert dermoscopists, the sensitivity of detecting small melanomas (<6 mm) is as low as 39% 2.
Despite evidence that early detection decreases mortality, considerable uncertainty surrounds the effectiveness of state‐of‐the‐art technology in routine melanoma screening 3. Clinical melanoma screening is a signal‐detection problem, which guides the binary decision for or against biopsy. Physicians screening for melanoma prior to the (gold standard) biopsy may be aided or, in some cases, outperformed by artificial‐intelligence analysis 2, 4, 5. However, deep‐learning dermatology algorithms cannot show a physician how a decision was arrived at, diminishing enthusiasm in the medical community 6. In melanoma detection, there is an unmet need for clinically interpretable machine vision and machine learning to provide transparent assistance in medical diagnostics. Improved clinical screening may prevent some of the roughly 10,000 annual deaths from melanoma in the United States 3.
The first computer‐aided diagnosis system for the detection of melanoma was described in 1987 7. Since then, a variety of non‐invasive in vivo imaging methodologies have been developed, including digital dermoscopy image analysis (DDA), total body photography, laser‐based devices, smart phone‐based applications, ultrasound, and magnetic resonance imaging 8. The primary challenge with clinical application of these technologies is obtaining a near perfect sensitivity, as a false negative, or Type II melanoma screening error, can have a potentially fatal outcome.
The most widely employed technology is dermoscopy, in which a liquid interface or cross‐polarizing light filters allow visualization of subsurface features, including deeper pigment and vascular structures. Dermoscopy has been shown to be superior to examination with the naked eye; however, it remains limited by significant inter‐physician variability and diagnostic accuracy is highly dependent on user experience 9, 10, 11. Studies using test photographs and retrospective analyses report increased diagnostic accuracy with the addition of dermoscopy criteria (Menzies, CASH, etc.) 12, 13. In one study, dermatologists with at least 5 years of experience using dermoscopy showed a 92% sensitivity and 99% specificity in detecting melanoma, but this dropped to 69% and 94% with inexperienced dermatologists (less than 5 years of experience), respectively 14. Even more concerning, the use of dermoscopy by inexperienced dermatologists may result in poorer performance compared to examination with the naked eye 9, 10, 15.
A digital melanoma imaging biomarker is a quantitative metric extracted from a dermoscopy image by simple computer algorithms that is high for melanoma (1) and low for a nevus (0). These imaging biomarkers measure features that are associated with pathological and normal features. They can then be used by more complex machine learning algorithms to create classifiers that are diagnostic. Examples of melanoma imaging biomarkers include symmetry, border, brightness, number of colors, organization of pigmented network pattern, etc. In our previous report, we described two types of imaging biomarkers: single color channel imaging biomarkers derived from gray scale images extracted from individual color channels, that is, Red, Green, Blue (RGB), and multi‐color imaging biomarkers that were derived from all color channels simultaneously 4. An example of a multi‐color imaging biomarker would be the number of dermoscopic colors contained in the lesion, since the definition of a color includes relative levels of intensity for the red, green, and blue channels. These melanoma imaging biomarkers are spectrally dependent in RGB color channels, with the majority of imaging biomarkers showing statistical significance for melanoma detection in the red or blue color channels 4.
Figure 1 shows two imaging biomarkers on one sample lesion. We provide these two imaging biomarkers as a function of wavelength as evidence that a machine learning algorithm utilizing a range of wavelengths has the potential to achieve higher sensitivity and specificity compared to RGB equivalent values. Imaging biomarkers are quantitative features extracted from images that are higher for melanoma than for a nevus. So, for example, if the image that the imaging biomarkers in Figure 2 were derived from was a nevus, the optimum imaging biomarker value for imaging biomarker A (cyan) would be the lowest value (global minimum), which would be in the ultraviolet. Meanwhile, the global minimum of imaging biomarker B (magenta) would be in the infrared. In the case of a melanoma, the optimum imaging biomarker value for imaging biomarker A (cyan) would be the highest value (global maximum), which would be in the red color channel. Meanwhile, the global maximum of imaging biomarker B (magenta) would be in the ultraviolet. Thus, the optimum imaging biomarker values in these examples would not be captured with RGB imaging alone. Further, diagnostic utility may be derived from image heterogeneity measures in the ultraviolet range since ultraviolet light interacts with superficial cytological and morphological atypia, targeting superficial spreading melanoma.
Figure 1.
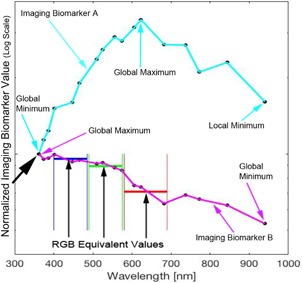
This figure highlights two imaging biomarkers on one sample lesion. The two most diagnostic RGB melanoma imaging biomarkers (from our previous study 4) were evaluated on each hyperspectral gray scale image of a pigmented lesion. They illustrate two classes of hyperspectral imaging biomarkers where the trend is either (A) the maximum is within the spectral range or (B) a constant decrease with a maximum in the ultraviolet (UV) and minimum in the infrared (IR). The data show that hyperspectral melanoma imaging biomarkers can be evaluated at a wavelength where they have values outside the visible (RGB) values (illustrated for imaging biomarker B in magenta).
Figure 2.
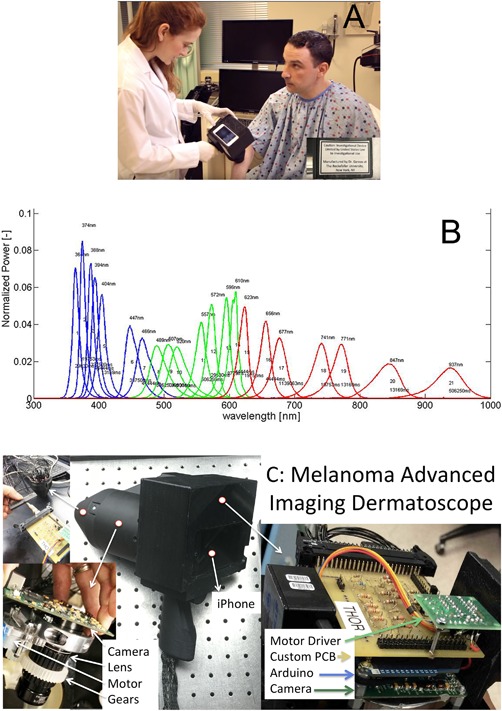
Hyperspectral imaging camera (A) and spectra of the individual imaging wavelengths normalized so the area under the curve equals unity (B). (C) Shows the inner components of the melanoma advanced imaging dermatoscope.
The purpose of this study was to use the same melanoma imaging biomarkers previously described 4, but over a wider range of wavelengths (350–950 nm) to determine if, in combination with machine learning algorithms, this could result in enhanced melanoma detection.
MATERIALS AND METHODS
The Study Device
The Melanoma Advanced Imaging Dermatoscope (mAID) is a non‐polarized light‐emitting diode (LED)‐driven hyperspectral camera (Fig. 2A) that illuminates the skin with 21 different wavelengths of light (Fig. 2B), ranging from the ultraviolet (UV)A (350 nm) to the near infrared (IR) (950 nm) and collects images using a high sensitivity gray scale charge‐coupled device (CCD) array (Mightex Inc., Toronto, Ontario, CA). A detailed schematic of the device is shown in Figure 2C. There is glass overlaid on the device, similar to a dermatoscope. In comparison to a standard digital camera, which captures light at three relatively broad wavelength bands of light (RGB), the mAID device achieves about five times better spectral resolution as well as widened spectral range.
The LEDs were chosen such that each LED is separated from its’ spectral neighbor by a spectral distance that is approximately the full‐width at half‐maximum of the LED spectrum. This scenario leads to LED spectra that, when normalized to have an area of unity, overlap at the half maximum point. Therefore, we achieve approximately Nyquist sampling and have an appropriate number of LEDS as to not over sample spectrally. There are between one and eight LEDS per wavelength: four for UV wavelength, eight for IR wavelength, and one for most of the visible wavelengths. The number of LEDs per wavelength was empirically determined by evaluating image brightness. Of note, there is no fluorescence as there is no filter to block the reflected UVA light, which is stronger than the fluorescent emission. It is possible that there is unwanted fluorescence, but it is small compared to the reflectance signal, and therefore negligible. There is no photobleaching as the irradiance incident on the skin is several fold less than sunlight and one second of sunlight does not cause photobleaching. Other notable features of the device include a 28 mm imaging window and a mobile phone embedded in its back surface to display a live, “in‐line” view of the target skin lesion. The mobile phone is not used for processing, but is connected to the device via the TwoMon App (DEVGURU Co. Ltd, Seoul, South Korea) to create a secondary display to help align the device properly with the target lesion. In terms of safety, the total light dose is less than one second of direct sunlight exposure and the mAID holds an abbreviated investigational device exemption from the FDA.
The protocol for imaging with the mAID device includes placing the imaging head directly onto the skin after applying a drop of immersion media such as hand sanitizer. After automated focusing, the device sequentially illuminates the skin with 21 different wavelengths of light. Video S1 (https://onlinelibrary.wiley.com/page/journal/10969101/homepage/lsm-23055video001.htm?pbEditor=true) shows four examples of hyperspectral images. The entire process takes less than four minutes including set up and positioning, with the collection of images requiring 20 seconds. In addition, there is no discomfort for the patient. The mAID device automatically encrypts and transfers hyperspectral images from the clinical site of imaging to the site of analysis over a secure internet connection.
Clinical Study
This study was approved by the University of California, Irvine Institutional Review Board. After obtaining informed consent, 100 pigmented lesions from 91 adults 18 years and over who presented to the Department of Dermatology at the University of California, Irvine from December 2015 to July 2018 underwent imaging with the mAID hyperspectral dermatoscope prior to removal and histopathological analysis. All imaged lesions were assessed by dermatologists as suspicious pigmented lesions requiring a biopsy. After obtaining the final histopathologic diagnoses, 30 lesions were excluded from analysis due to their non‐binary classification (i.e., not a melanoma or nevus). These categories included atypical squamous proliferation (1), basal cell carcinoma (9), granulomatous reaction to tattoo pigment (1), lentigo (4), lichenoid keratosis (1), melanotic macule (1), seborrheic keratosis (9), splinter (1), squamous cell carcinoma (2), and thrombosed hemangioma (1). Seventy mAID hyperspectral images then underwent automated computer analysis, adapted as described below following previously published methods, to create a set of melanoma imaging biomarkers. These melanoma imaging biomarkers were derived using hand‐coded feature extraction in the matlab programming environment 4. Images from 52 of the total 70 pigmented lesions were successfully processed. The remaining 18 images were excluded due to one or more of the following errors in processing: bubbles in the imaging medium, image not in focus, camera slipped during imaging, or excessive hair was present in the image obscuring the lesion. In our machine learning, ground truth was the histopathological diagnosis of melanoma or nevus that was accessed automatically during learning. The machine learning, with the melanoma imaging biomarkers as inputs, was trained to output a risk score which was the likelihood of a melanoma diagnosis. In this way, the machine learning created the best transformation algorithm to arrive at the result of the invasive test but using only the noninvasive images acquired prior to the biopsy. A summary of the melanoma classification algorithms used is listed in Table 1. The derivation of melanoma imaging biomarkers and corresponding methods of image analysis have been previously described 4. The extension of single color channel imaging biomarkers to hyperspectral imaging entailed calculating 21 values for each imaging biomarker per hyperspectral image—one for each of the 21 color channels in the hyperspectral image. Using these quantitative metrics, the algorithm generated an overall Q‐score for each image—a value between zero and one in which a higher number indicates a higher probability of a lesion being cancerous. Images were also processed by spectral fitting to produce blood volume fraction (BVF) and oxygen saturation (O2sat), which are candidate components in identifying metabolic and immune irregularity in melanomas (Figure 3) 16.
Table 1.
Melanoma Classification Algorithms
| Method | Description |
|---|---|
| LoR | Logistic regression within the framework of Generalized Linear 18, 19, 20 Models (3) (4) (5) |
| NN | Feed‐forward neural networks with a single hidden layer 21 |
| SVM (linear and radial) | Support vector machines 22, 23 |
| DT | C5.0 decision tree algorithm for classification problems 24 |
| RF | Random Forests 25 |
| LDA | Linear discriminant analysis 26 |
| KNN | K‐nearest neighbors algorithm developed for classification 27 |
| NB | Naive Bayes algorithm 40 |
DT, decision tree; KNN, K‐nearest neighbors; LDA, linear discriminant analysis; LoR, logistic regression; NB, naïve bayes; NN, neural network; RF, random forest; SVM, support vector machine.
Figure 3.
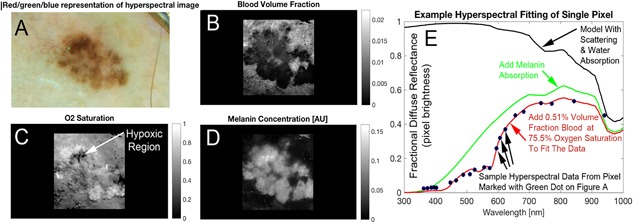
Hyperspectral melanoma imaging biomarkers were derived from spectral analysis (e.g., blood distribution and oxygenation). RGB image (A) shows a visual representative sampling of the hyperspectral image. The correlating blood volume fraction (B), oxygen saturation (C) and melanin factor (D) maps are produced by fitting the spectrum at each pixel. The absorption effect of melanin is added to attenuate the spectrum as shown (green). The final added absorber in our diffuse reflectance spectral simulation is hemoglobin, which we added a reasonable volume fraction and oxygen saturation. The values shown in red text are spectral oximetry biomarkers which are shown as solved for in one pixel of the image but are available in the whole lateral field of view (i.e., at any pixel). These maps, which demonstrate the type of image data hyperspectral melanoma imaging biomarkers are to be derived from, are created by fitting the spectrum (E) produced by the investigational device at each pixel. The spectrum shown (E) is a single pixel in the image (shown in green in A), evaluated across the 21 colors of the LEDs in the hyperspectral camera.
The Theoretical Model
Spectral light transport in turbid biological tissues is a complex phenomenon that gives rise to a wide array of image colors and textures inside and outside the visible spectrum. To help us understand the degree to which different wavelengths interact with tissue at different depths in the skin, we used a Monte Carlo photon transport simulation, adapted from prior work to run at all of the hyperspectral wavelengths 17. Our simulation modeled light transport into and out of pigmented skin lesions. Our modeling involved two steps: (i) 20 histologic sections of pigmented lesions stained with Melan‐A were imaged with a standard light microscope to become the model input; (ii) light transport at 40 wavelengths in the 350–950 nm range was simulated into and out of each input model morphology. First, a digital image of the histology was automatically segmented into epidermal and dermal regions using image processing. Each region was assigned optical properties appropriate for each tissue compartment (i.e., the epidermis had high absorption due to blood and the dermis had an absorption spectrum dominated by hemoglobin but also some melanin). The escaping photons were scored by simply checking, at each propagation step, if they had crossed the boundary of the surface of the skin (all other boundaries were handled with a matched boundary condition). For escaping photons, the numerical aperture of our camera was transformed into a critical angle. If the photons escaped at an angle that was inside the critical angle, their weight at time of escape was added to the simulated pixel brightness at that image point. The positions and directions were scored for each escaping photon as well as the maximum depth of its penetration. Video S2 (https://onlinelibrary.wiley.com/page/journal/10969101/homepage/lsm-23055-video002.htm?pbEditor=true) shows a sample Monte Carlo simulation output.
To generate BVF and O2sat, we started with a tissue phantom composed of scattering collagen, keratin, and melanin. The predicted spectrum is shown in Figure 3 (black) using diffusion theory modified for simulating diffuse reflectance of skin lesions. The spectrum from each pixel was assumed to follow the well‐established diffusion theory of photon transport. However, we know this is not entirely true since, as a consequence of illuminating and detecting from the entire field, illumination occurs both far from detection and on top of the detection points. In the dermis, the absorption coefficient is assumed to be homogenous and contributed to by a fraction of water times the absorption coefficient of water, a fraction of deoxyhemoglobin times the absorption of deoxyhemoglobin, a fraction of oxyhemoglobin times the absorption of oxyhemoglobin. Melanin was modeled in the dermis the same as was the previously mentioned chromophores but with a proportional “extra melanin” factor acting as a transmission filter in the superficial epidermis. This last feature is a departure from simple diffusion theory and it models the dermis as source of diffuse reflectance that transmits through the epidermis, where an extra amount of melanin that is proportional to the dermal melanin (to maintain only one fitting parameter for melanin concentration) attenuates the diffuse reflectance escaping the tissue.
Statistical Analysis
The sensitivity and specificity for detecting melanoma was calculated from the Receiver Operator Characteristic (ROC) curve to assess the overall diagnostic performance as previously described 4. Data analysis was completed using statistical software in the Matlab computing environment 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28.
RESULTS
Our Monte Carlo simulation (Video S2, https://onlinelibrary.wiley.com/page/journal/10969101/homepage/lsm-23055-video002.htm?pbEditor=true) showed that the mean penetration depth of escaping light was a thousand‐fold greater than its wavelength. For example, 350 nm light penetrated 350 μm into the tissue, 950 nm light penetrated 950 μm into the tissue and the relationship was linear at the 40 wavelengths between these two points.
Of the 52 pigmented lesions that were successfully processed with hyperspectral imaging, 13 (25%) were histologically diagnosed as melanoma and 39 (75%) were diagnosed as nevi. Sensitivity, specificity, and diagnostic accuracy were calculated from the ROC curves (Figure 4). The corresponding confusion matrix is shown in Table 2. These ROC curves demonstrated a sensitivity of 100% and specificity of 36% in detecting melanoma with hyperspectral dermoscopy 4.
Figure 4.
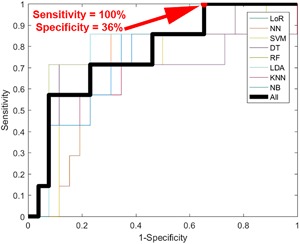
Receiver operator characteristic (ROC) curve for melanoma detection in hyperspectral images. Thin lines represent the individual machine learning approaches used while the thick line represents the “wisdom of the crowds” diagnostic that averaged the risk scores produced by the individual machine learning approaches.
Table 2.
Results Displayed in a Confusion Matrix Table That Correlate to the 100% Sensitivity 36% Specificity Point Indicated in Red on Figure 4
| n = 52 | Negative | Positive | |
|---|---|---|---|
| No disease | TN = 14 | FP = 25 | 39 |
| Disease | FN = 0 | TP = 13 | 13 |
| 14 | 38 |
DISCUSSION
DDA systems have attempted to decrease inter‐physician variability and standardize dermoscopic analysis by incorporating quantitative parameters such as colorimetric and geometric evaluation. There are a variety of proprietary DDA instruments on the market, although none have yet demonstrated a reproducibly high sensitivity and specificity for melanoma detection 29. SolarScan® (Polartechnics Ltd, Sydney, Australia), for example, is an automated digital dermoscopy instrument that extracts lesion characteristics from digital images and then compares them to a database of benign and malignant lesions. In clinical studies, SolarScan® demonstrated a sensitivity of 91% and specificity of 68% for detecting melanoma 14. In the evaluation of another DDA, the FotoFinder Mole‐Analyzer®, a 15‐year retrospective study evaluated diagnostic performance in 1,076 pigmented skin lesions, reporting a low diagnostic accuracy with a sensitivity of 56% and specificity of 74%, which was significantly lower than previous reports with the same system 30, 31. There remains a wide variation in sensitivity and specificity amongst the current DDA systems, ranging from 56% to 100% and 60% to 100%, respectively. Further investigation into the diagnostic accuracy of these DDA systems is needed for more standardized, reproducible results 8.
The benefit of incorporating hyperspectral or multispectral imaging into DDA is that it provides information beyond the visible spectrum regarding variation in tissue oxygenation and melanin distribution, which may help differentiate melanoma from noncancerous lesions. In this work, we continue to explore the biophotonics of pigmented lesion imaging. Figure 1 provides preliminary evidence that melanoma imaging biomarkers have a spectral dependence that goes beyond the visible spectrum and implies that melanoma imaging biomarkers may be tuned to optimize diagnostic significance within a hyperspectral image.
Bringing the basic science of optical biophysics to bear on clinical diagnostic precision is warranted since our preliminary findings indicate that melanoma imaging biomarkers exhibit strong spectral variance. Understanding the biophotonic pathologic contrast mechanisms allows us to target within the spectrum and exercise the rest of the data, which will enable an elegant form of constrained machine learning. In building this approach, we developed a Monte Carlo photon transport simulation that helps us better understand/exploit the optical properties of pigmented lesions for diagnosis. Our result, that the penetration depth is linearly related to the wavelength with a factor of 1,000 relating the two, provides a theoretical basis upon which to understand diagnostic targeting of variously spaced morphologic pathologies. For example, interpreting the images in Video S1 (https://onlinelibrary.wiley.com/page/journal/10969101/homepage/lsm-23055-video001.htm?pbEditor=true) with the relationship that the penetration is roughly 1,000‐fold longer than the wavelength of light is suggestive that the lesion in the top left has a wide area of superficial (<0.5 mm) heterogeneous pigmentation while the upper middle lesion is >1 mm‐deep. The approach of correlating the spectral features with underlying morphology is promising in terms of deepening the scientific understanding and will be a goal of future work.
Hyperspectral imaging has been previously studied for the detection of subclinical lentigo maligna melanoma borders, successfully identifying 18/19 (94.7%) cases in one study 32. Another study evaluating the use of multispectral imaging (450–950 nm) to detect melanoma in 82 pigmented lesions reported a sensitivity and specificity of 94% and 89%, respectively 33. In comparison to previous melanoma spectral imaging systems, which developed direct spectral signatures using select wavelengths, our system images a 21‐wavelength spectral range using relatively inexpensive components.
Current hyperspectral/multispectral imaging methods on the market include MelaFind and SIAScope. MelaFind® (MELA Sciences, Irvington, New York) is a hand‐held device that images from 430 nm (blue) to 950 nm (near infrared), and was FDA approved in 2011 for melanoma detection. Results from a multi‐center prospective trial in 2011 reported a sensitivity of 98.2% and specificity of 9.5% 34. In a follow‐up study using a test set of 47 lesions to compare MelaFind performance to that of dermatologists in detecting melanoma, the authors report a sensitivity of 96% and specificity of 0.08%. MelaFind recommended biopsy in 44 lesions and no biopsy in 3. In the three lesions that were not biopsied, one was diagnosed as melanoma 31. In a study of 160 board‐certified dermatologists who were asked to evaluate 50 randomly ordered pigmented lesions, the sensitivity and specificity for diagnosing melanoma significantly increased after physicians were provided MelaFind analysis of lesions from 76% to 92% and from 52% to 79%, respectively 35. However, there is still significant debate as to whether MelaFind is a useful tool to guide dermatologists, the concern being that the device almost always recommends biopsy 36.
Spectrophotometric Intracutaneous Analysis (SIAscopy™, Astron Clinica, UK) was first introduced in 2002 as an imaging technology that produces spectrally filtered images in the visible and infrared spectra (400–1000 nm). The first clinical trial with SIAscopy demonstrated a sensitivity of 82.7% and specificity of 80.1% for melanoma in a dataset of 348 pigmented lesions (52 melanomas) 37. However, when implemented in a melanoma screening clinic, the SIAscope did not improve the diagnostic abilities of dermatologists 38. Further studies demonstrated poor correlation between SIAscopy analysis and histopathology in both melanoma and nonmelanoma lesions and worse accuracy than dermoscopy 39, 40. Of note, direct comparison of devices to other systems on the market is limited, as diagnostic performance of a device varies with the difficulty of lesions included in analysis, as well as the proportion of atypical nevi in the benign set 14.
In this study, we demonstrate the improved diagnostic utility of hyperspectral dermoscopy in comparison to standard dermoscopy. By calculating the mean depth of photon penetration at each wavelength, our Monte Carlo simulation (Video S2, https://onlinelibrary.wiley.com/page/journal/10969101/homepage/lsm-23055-video002.htm?pbEditor=true) provided insight regarding which anatomical compartments (e.g., superficial epidermis vs. deeper dermis) the different wavelengths probed. The ability to spectrally target analysis of pagetoid spread and Breslow depth, as examples of diagnostics and prognostics, respectively, shows how our computational theoretical framework can elucidate standard pathological evaluation criteria. In combination with artificial intelligence algorithms, we demonstrated a sensitivity of 100% and specificity of 36% in detecting melanoma. At a high sensitivity, this approach yielded a higher specificity than that estimated in clinical practice 1. Hyperspectral dermoscopy was better able to emphasize sensitivity compared to standard dermoscopy. This was a design feature of our diagnostic algorithm, since false negative, or Type II melanoma screening errors are particularly dangerous. Furthermore, our set of pigmented lesions included suspicious lesions assessed by dermatologists as necessitating a biopsy; thus, our set inherently consisted of more challenging pigmented lesions, as clinically benign lesions were not included.
In addition, it is not entirely clear why melanoma imaging biomarkers are statistically significant in the red and blue color channels and not in the green 4. The blue channel is known to contain information regarding superficial atypia associated with melanoma in situ such as pagetoid spread and junctional atypia at the dermal–epidermal junction. The red channel is thought to contain information about deep pigment so these channels are expected to contain diagnostic value for these reasons. However, we would expect that the green channel would contain atypical features of metabolism since hemoglobin absorbs green light the most. It may be that the green channel in RGB integrates over too much of the spectrum to spectrally resolve the isosbestic points in hemoglobin's absorption spectrum, therefore washing the oximetric information. This washout potentially prevents the differentiation between oxyhemoglobin and deoxyhemoglobin.
One major limitation of the device is the fact that the operator needs to be properly trained. Movement during imaging can lead to a series of laterally sliding positions on the skin, and hence the lesion and its diagnostic morphology will not be spatially coherent. In addition, the presence of hair and bubbles in the imaging medium can interfere with image analysis. This presents a challenge as many of the lesions dermatologists evaluate are in hair bearing regions. Limitations of this study include a small sample size and artificially high melanoma incidence. Further, we analyzed hyperspectral melanoma imaging biomarkers extracted from single gray scale color channels. Evaluating the diagnostic performance of hyperspectral melanoma imaging biomarkers does not yet include imaging biomarkers that used the entire spectrum, such as oxyhemoglobin (HbO2) maps, which are expected to improve the diagnostic results in future work.
CONCLUSION
Our study demonstrated a higher sensitivity in detecting melanoma with hyperspectral dermoscopy compared to standard dermoscopy. Hyperspectral dermoscopy shows promise to noninvasively screen melanoma and guide biopsy. With this novel methodology for evaluating pigmented lesions, dermatologists can harness computational power to aid in standardized evaluation. Over time and with the analysis of more lesions, the computer can gain additional expertise in this area. While our initial results are promising, this technology would need to be validated and results reproduced in larger clinical trials. Continuing work aims to improve the power of the study as well as further investigate the additional diagnostic value of hyperspectral image analysis over RGB image analysis.
Supporting information
Additional supporting information may be found in the online version of this article.
Supporting Video S1.
Supporting Video S2.
ACKNOWLEDGMENTS
The melanoma Advanced Imaging Dermatoscope (mAID) was developed by Dr. Daniel Gareau at the Laboratory for Investigative Dermatology at The Rockefeller University and is his intellectual property. The authors wish to acknowledge Hiroshi Mitsui for allowing access to histology specimens to produce the Monte Carlo simulation shown in Video S2 (https://onlinelibrary.wiley.com/page/journal/10969101/homepage/lsm-23055-video002.htm?pbEditor=true) as well as Michael Deitz and Justin Martin for their work on the mechanical design of the mAID. This work was supported by Paul and Irma Milstein Foundation, Howard and Abby Milstein Foundation.
Conflict of Interest Disclosures: All authors have completed and submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest and none were reported.
REFERENCES
- 1. Salerni G, Terán T, Puig S, et al. Meta‐analysis of digital dermoscopy follow‐up of melanocytic skin lesions: A study on behalf of the International Dermoscopy Society. J Eur Acad Dermatol Venereol 2013;27:805–814. [DOI] [PubMed] [Google Scholar]
- 2. Friedman RJ, Gutkowicz‐Krusin D, Farber MJ, et al. The diagnostic performance of expert dermoscopists vs a computer‐vision system on small‐diameter melanomas. Arch Dermatol 2008;144:476–482. [DOI] [PubMed] [Google Scholar]
- 3. Tripp MK, Watson M, Balk SJ, Swetter SM, Gershenwald JE. State of the science on prevention and screening to reduce melanoma incidence and mortality: The time is now. CA Cancer J Clin 2016;66:460–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gareau DS, Correa da Rosa J, Yagerman S, et al. Digital imaging biomarkers feed machine learning for melanoma screening. Exp Dermatol 2017;26:615–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Haenssle HA, Fink C, Schneiderbauer R, et al. Man against machine: Diagnostic performance of a deep learning convolutional neural network for dermoscopic melanoma recognition in comparison to 58 dermatologists. Ann Oncol 2018;29:1836–1842. [DOI] [PubMed] [Google Scholar]
- 6. Esteva A, Kuprel B, Novoa RA, et al. Dermatologist‐level classification of skin cancer with deep neural networks. Nature 2017;542:115–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cascinelli N, Ferrario M, Tonelli T, Leo E. A possible new tool for clinical diagnosis of melanoma: The computer. J Am Acad Dermatol 1987;16:361–367. [DOI] [PubMed] [Google Scholar]
- 8. Masood A, Ali Al‐Jumaily A. Computer aided diagnostic support system for skin cancer: A review of techniques and algorithms. Int J Biomed Imaging 2013;2013:1–22. Article ID 323268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kittler H, Pehamberger H, Wolff K, Binder M. Diagnostic accuracy of dermoscopy. Lancet Oncol 2002;3:159–165. [DOI] [PubMed] [Google Scholar]
- 10. Vestergaard ME, Macaskill P, Holt PE, Menzies SW. Dermoscopy compared with naked eye examination for the diagnosis of primary melanoma: A meta‐analysis of studies performed in a clinical setting. Br J Dermatol 2008;159:669–676. [DOI] [PubMed] [Google Scholar]
- 11. Bafounta ML, Beauchet A, Aegerter P, Saiag P. Is dermoscopy (epiluminescence microscopy) useful for the diagnosis of melanoma?: Results of a meta‐analysis using techniques adapted to the evaluation of diagnostic tests. Arch Dermatol 2001;137:1343–1350. [DOI] [PubMed] [Google Scholar]
- 12. Henning JS, Dusza SW, Wang SQ, et al. The CASH (color, architecture, symmetry, and homogeneity) algorithm for dermoscopy. J Am Acad Dermatol 2007;56:45–52. [DOI] [PubMed] [Google Scholar]
- 13. Carrera C, Marchetti MA, Dusza S, et al. Validity and reliability of dermoscopic criteria used to differentiate nevi from melanoma. JAMA Dermatol 2016;152:798–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Piccolo D, Ferrari A, Peris K, Daidone R, Ruggeri B, Chimenti S. Dermoscopic diagnosis by a trained clinician vs. a clinician with minimal dermoscopy training vs. computer‐aided diagnosis of 341 pigmented skin lesions: A comparative study. Br J Dermatol 2002;147:481–486. [DOI] [PubMed] [Google Scholar]
- 15. Menzies SW, Bischof L, Talbot H, et al. The performance of SolarScan: An automated dermoscopy image analysis instrument for the diagnosis of primary melanoma. Arch Dermatol 2005;141:1388–1396. [DOI] [PubMed] [Google Scholar]
- 16. Gareau DS, Truffer F, Perry KA, et al. Optical fiber probe spectroscopy for laparoscopic monitoring of tissue oxygenation during esophagectomies. J Biomed Opt 2010;15:061712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gareau D, Jacques S, Krueger J. In: Choi B, Kollias N, Zeng H, Kang HW, Wong BJF, Ilgner JF, et al., editors. Monte Carlo modeling of pigmented lesions. Proc. SPIE 8926, Photonic Therapeutics and Diagnostics X, 89260V, 2014. 10.1117/12.2040473. [DOI]
- 18. McCullagh P, Nelder JA. Generalized linear models, 2nd edition Boca Raton, Florida: CRC Press; 1989. [Google Scholar]
- 19. Friedman J, Hastie T, Tibshirani R. Regularization paths for generalized linear models via coordinate descent. J Stat Softw 2010;33:1–22. [PMC free article] [PubMed] [Google Scholar]
- 20. Hofner B, Mayr A, Robinzonov N, Schmid M. Model‐based boosting in R: A hands‐on tutorial using the R Package Mboost. Comput Stat 2014;29:3–35. [Google Scholar]
- 21. Haykin SS, Haykin SS. Neural networks and learning machines, 3rd edition. New York: Prentice Hall; 2009. [Google Scholar]
- 22. Cortes C, Vapnik V. Support‐vector networks. Mach Learn 1995;20:273–297. [Google Scholar]
- 23. Scholkopf B, Smola AJ. Learning with kernels: Support vector machines, regularization, optimization, and beyond. Cambridge, MA, USA: MIT Press; 2001. [Google Scholar]
- 24. Quinlan JR. C4.5: Programs for machine learning. San Francisco, CA, USA: Morgan Kaufmann Publishers Inc; 1993. [Google Scholar]
- 25. Breiman L. Random forests. Mach Learn 2001;45:5–32. [Google Scholar]
- 26. Fisher RA. The use of multiple measurements in taxonomic problems. Ann Eugen 1936;7:179–188. [Google Scholar]
- 27. Fix E, Hodges JL. Discriminatory analysis. Nonparametric discrimination: Consistency properties. Int Stat Rev/Revue Internationale de Statistique 1989;57:238–247. [Google Scholar]
- 28. Russell R, Norvig P. Artificial intelligence: A modern approach, 2nd edition Upper Saddle River, New Jersey: Prentice Hall; 1995. [Google Scholar]
- 29. Korotkov K, Garcia R. Computerized analysis of pigmented skin lesions: A review. Artif Intell Med 2012;56:69–90. [DOI] [PubMed] [Google Scholar]
- 30. Rosario del F, Farahi JM, Drendel J, et al. Performance of a computer‐aided digital dermoscopic image analyzer for melanoma detection in 1,076 pigmented skin lesion biopsies. J Am Acad Dermatol 2018;78:927–934.e6. [DOI] [PubMed] [Google Scholar]
- 31. Wells R, Gutkowicz‐Krusin D, Veledar E, Toledano A, Chen SC. Comparison of diagnostic and management sensitivity to melanoma between dermatologists and MelaFind: A pilot study. Arch Dermatol 2012;148:1083–1084. [DOI] [PubMed] [Google Scholar]
- 32. Neittaanmäki‐Perttu N, Grönroos M, Jeskanen L, et al. Delineating margins of lentigo maligna using a hyperspectral imaging system. Acta Derm Venereol 2015;95:549–552. [DOI] [PubMed] [Google Scholar]
- 33. Diebele I, Kuzmina I, Lihachev A, et al. Clinical evaluation of melanomas and common nevi by spectral imaging. Biomed Opt Express 2012;3:467–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Monheit G, Cognetta AB, Ferris L, et al. The performance of MelaFind: A prospective multicenter study. Arch Dermatol 2011;147:188–194. [DOI] [PubMed] [Google Scholar]
- 35. Farberg AS, Winkelmann RR, Tucker N, White R, Rigel DS. The impact of quantitative data provided by a multi‐spectral digital skin lesion analysis device on dermatologists’ decisions to biopsy pigmented lesions. J Clin Aesthet Dermatol 2017;10:24–26. [PMC free article] [PubMed] [Google Scholar]
- 36. Cukras AR. On the comparison of diagnosis and management of melanoma between dermatologists and MelaFind. JAMA Dermatol 2013;149:622–623. [DOI] [PubMed] [Google Scholar]
- 37. Moncrieff M, Cotton S, Claridge E et al. Spectrophotometric intracutaneous analysis: a new technique for imaging pigmented skin lesions. Br J Dermatol 2002;146:448–457. [DOI] [PubMed] [Google Scholar]
- 38. Haniffa MA, Lloyd JJ, Lawrence CM. The use of a spectrophotometric intracutaneous analysis device in the real‐time diagnosis of melanoma in the setting of a melanoma screening clinic. Br J Dermatol 156:1350–1352. [DOI] [PubMed] [Google Scholar]
- 39. Terstappen K, Suurküla M, Hallberg H, Ericson MB, Wennberg A‐M. Poor correlation between spectrophotometric intracutaneous analysis and histopathology in melanoma and nonmelanoma lesions. J Biomed Opt 2013;18:061223. [DOI] [PubMed] [Google Scholar]
- 40. Sgouros D, Lallas A, Julian Y, et al. Assessment of SIAscopy in the triage of suspicious skin tumours. Skin Res Technol 20:440–444. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional supporting information may be found in the online version of this article.
Supporting Video S1.
Supporting Video S2.


