Abstract
PANTHER Classification System (www.pantherdb.org) is a comprehensive system that combines genomes, gene function classifications, pathways and statistical analysis tools to enable biologists to analyze large-scale genome-wide experimental data. The current system (PANTHER v.14.0) covers 131 complete genomes organized into gene families and subfamilies; evolutionary relationships between genes are represented in phylogenetic trees, multiple sequence alignments and statistical models (hidden Markov models, or HMMs). The families and subfamilies are annotated with Gene Ontology terms and sequences are assigned to PANTHER pathways. A suite of tools has been built to allow users to browse and query gene functions, and analyze large-scale experimental data with a number of statistical tests. PANTHER is widely used by bench scientists, bioinformaticians, computer scientists and systems biologists. Since the protocol to use this tool (v8.0) was originally published in 2013, there have been significant improvements and updates in the areas of data quality, data coverage, statistical algorithms and user experience. This Protocol Update will provide a detailed description of how to analyze genome-wide experimental data in the PANTHER Classification System.
Keywords: Gene function, annotation, enrichment analysis, overrepresentation test, Gene Ontology, pathway
INTRODUCTION
The PANTHER Classification System (www.pantherdb.org) is designed to be a comprehensive platform for the analysis of gene function on a genome-wide scale (1). Although its initial development aimed to classify gene and protein functions (2,3), it has evolved through the years to also serve as an online resource for experimental data analysis (4–6). The easy-to-use user interface and timely user support have made PANTHER one of the most widely used online resources for gene function classification and genome-wide data analysis. Nearly 1600 unique IP addresses access the PANTHER website with over 30,000 page views daily (please note that one IP address can have multiple users so the actually number of users is much larger). According to Google Scholar, over 11,000 publications have cited our work (5000 of them since 2016). We believe that there are two main reasons why PANTHER is able to attract more users and may have an advantage compared to other tools in the field: Firstly, as a Gene Ontology (GO) consortium member, PANTHER is integrated into the GO curation process, especially the phylogenetic annotation effort (7), and provides more up-to-date annotation data (which is updated monthly). Secondly, PANTHER provides support to more genomes than other tools (~1000) in collaboration with the Reference Proteome project (8).
PANTHER was initially released publicly in 2003, and quickly became a popular online resource for genome-wide analysis of gene functions on various experimental data, including gene expression, proteomics and genome sequencing data. Our original Nature Protocol was published in 2013 with detailed background (6). Since then, a number of system-wide updates and improvements have been made in order to meet the needs of the user community (Figure 1). The improvements have been made in four main areas, as described below.
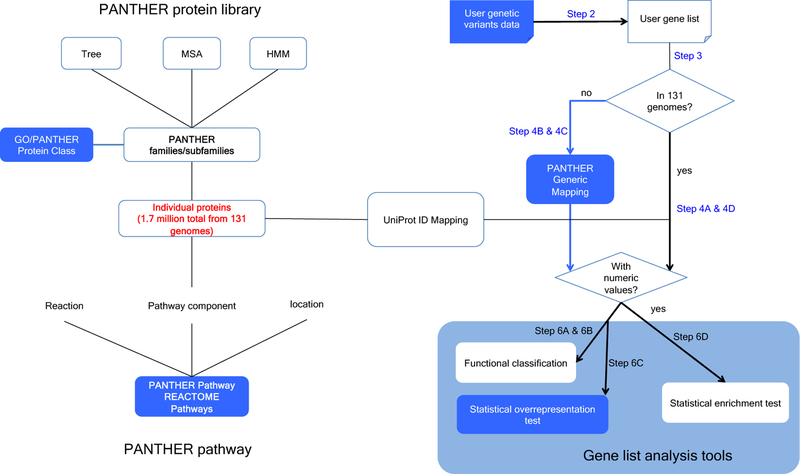
Figure 1.
Overview of PANTHER infrastructure and recent improvements
PANTHER consists of three modules. The core module is the PANTHER protein library (light blue background) that contains a collection of PANTHER families and subfamilies, each of which is represented by a phylogenetic tree, a multiple sequence alignment (MSA) and an HMM. The second module is the pathway module that contains expert-curated pathways from both PANTHER and Reactome (orange background). The pathway components are associated with protein sequences that are also used to build the protein library (dawn pink shade); in this way pathways are also linked to the subfamilies and HMMs. The third module is the tool suite. In this diagram, the gene list analysis tool is used as an example (blue background). Major updates and improvements have been made to the components highlighted in royal blue. Blue arrows surrounding the PANTHER Generic Mapping file indicate a new workflow available for users, that dramatically expands the number of organisms that can be analyzed with PANTHER (see Box 4). There are three types of analysis that can be performed: functional classification, statistical overrepresentation test and statistical enrichment test. Numeric values must be provided in order to use the statistical enrichment test. The corresponding procedure steps are labeled on the arrows.
I. Improved annotation data quality and coverage
All the analyses in PANTHER rely on the accuracy of the annotation data sets. During the past 5 years, improvements have focused on expanding the data coverage and accuracy in the following areas:
The PANTHER GO slim annotation data sets that were reported in the previous protocol now use the data from the Gene Ontology phylogenetic annotation effort (7). These are manually curated annotations to the ancestral nodes on PANTHER family trees based on the experimental annotations on their leaf descendants (extant genes). The annotation can be inferred to other leaf sequences (that have not been tested experimentally) under the annotated ancestral node.
The complete GO annotation data sets (9) have been incorporated into the PANTHER tools for the analysis. They include both experimental and electronic annotations. The data are updated monthly.
In order to expand the coverage of pathway data, Reactome pathways (10) data sets have been added to the system. Our plan is to add more data from the Pathway Commons project (11) in the near future, including pathway databases (e.g., KEGG (12), HumanCyc (13), WikiPathway (14)) and protein-protein interaction databases (e.g., IntAct database (15), BioGRID (16)).
A detailed description of each of the above datasets available in PANTHER analysis tools can be found in Box 1.
Box 1. Annotation data sets.
There are nine annotation data sets, in four general data types, available in PANTHER for users to choose in their analysis. Below is a brief description of each of them.
PANTHER GO-Slim –
GO annotations from the phylogenetic curation effort captured in 3039 GO Slim terms. The annotation data sets use the data from the Gene Ontology phylogenetic annotation effort (7). These are manually curated annotations to the ancestral nodes on PANTHER family trees based on the experimental annotations on their leaf descendants (extant genes). The annotation can be inferred to other leaf sequences (that have not been tested experimentally) under the annotated ancestral node. There are three annotation datasets corresponding to three GO aspects in this data type.
Biological Process (default)
Molecular Function
Cellular Component
Complete GO annotation datasets –
Complete GO annotations including both manually curated and electronic annotations. Electronic annotations are generated by computer algorithm based on sequence similarity, and are usually not reviewed by curators. They are less reliable. There are three datasets corresponding to three GO aspects in this data type..
GO molecular function complete.
GO biological process complete.
GO cellular component complete.
Pathways –
The current PANTHER database includes two pathway datasets from the following two resources.
PANTHER Pathway – PANTHER Pathway consists of over 177, primarily signaling, pathways, each with PANTHER subfamilies and protein sequences mapped to individual pathway components (19).
Reactome Pathways –Reactome is a freely available, open source relational database of signaling and metabolic molecules and their relations organized into biological pathways and processes. The core unit of the Reactome data model is the reaction. Entities (nucleic acids, proteins, complexes, vaccines, anti-cancer therapeutics and small molecules) participating in reactions form a network of biological interactions and are grouped into pathways (10).
PANTHER Protein Class –
The PANTHER Protein Class ontology was adapted from the PANTHER/X molecular function ontology (3), and includes commonly used classes of protein families, many of which are not covered by GO molecular function. There is one corresponding dataset in this data type.
II. Analysis of genotype data
To respond to the increasing requests from our users who use PANTHER to analyze genetic variation data, including single nucleotide polymorphism (SNP) data, from experiments such as genome wide association studies (GWAS) and genome sequencing data, a new feature has been added to support genetic variant data in Variant Call Format (VCF), which is a text file format for storing sequence variation data. Its specification can be found on GitHub at https://samtools.github.io/hts-specs/VCFv4.3.pdf. A sample VCF file can be found in the Supplementary Data 1 for test purposes. Currently, only the overrepresentation test supports the analysis using VCF file format and the human reference genome release GRCh38/hg38 is supported. Each variant is mapped to a gene if it is within the gene region or the flanking region specified by the user. To avoid artifacts, multiple variants in the same gene are only counted once, and a given variant can only be associated with a single gene. The statistical analysis is preformed on the converted gene list the same way as described in Box 2. Users are able to upload both test list(s) and a reference list.
Box 2. Statistical Tests.
Statistical Overrepresentation Test
The input (or test) list is usually a list of genes or variants of your interest. It can be a list of genes that are up-regulated in the gene expression experiment, or a list of significant SNPs from your GWAS experiment, for example. The list is divided into groups based on annotation classification (molecular function, biological process, cellular component, PANTHER Protein Class or pathways). As many as four test lists can be uploaded for each analysis. A reference list, which usually contains all the genes/proteins from which the list was drawn, is divided into groups in the same way. PANTHER provides reference proteome datasets as default reference lists for all 131 genomes, so uploading a reference list is optional. If you work with genomes other than those 131, then you need to prepare and upload a reference list. For each functional category, e.g., protein kinase for GO Molecular Function, cell proliferation for GO Biological Process, or apoptosis signaling pathway for PANTHER Pathway, the statistical test is applied to determine whether there is a statistical over- or under-representation of genes/proteins in the test list relative to the reference list.
P-value calculation in the overrepresentation test
The expected value is the number of genes you would expect in the test list for a particular PANTHER category, based on the reference list. For example, there are 20,000 genes in the reference list (for example, the entire human genome). 440 of these genes map to the GO term induction of apoptosis. Based on this, 2.2% (440 divided by 20,000) of the genes in the reference list are involved in induction of apoptosis. Now a test list that contains 500 genes is uploaded. Based on the reference list, it is expected that 11 genes (500 multiplied by 2.2%) in the test list would be involved in induction of apoptosis.
If for this biological process more genes are observed in the test list than expected, you have an over-representation (+) of genes involved in induction of apoptosis. If fewer genes are observed than expected, you have an under-representation (–). A p-value is calculated then to determine whether the over- or under- representation is significant or not. For example, let’s assume that 21 genes are observed in the test list are involved in induction of apoptosis. Although this is almost twice as the expected value, the p-value is large and not significant (the p-value would be 0.722). Alternatively, if 35 genes are observed, this is very different than the expected value, so you would expect a small, significant p-value (the p-value would be 6.21e-7). This small p-value indicates that the result is non-random and potentially interesting, and worth looking at in closer detail. A p-value cutoff of 0.05 is recommended as a start point.
There two statistical methods used in this test: Fisher’s Exact test and binomial test. They are both standard statistical methods commonly used in the field. The detailed description of the methods can be found at https://en.wikipedia.org/wiki/Fisher%27s_exact_test (for Fisher’s Exact Test) and https://en.wikipedia.org/wiki/Binomial_distribution (for Binomial Distribution Test).
Statistical Enrichment test
The algorithm used in this test is Mann-Whitney Rank-Sum Test (U-Test) (20). The detailed description of the method can also be found at https://en.wikipedia.org/wiki/Mann%E2%80%93Whitney_U_test.
The statistical enrichment test is general enough to handle any numerical data, continuous or discontinuous, generated by experiments such as gene expression, proteomics or GWAS. First, a reference distribution is generated using all values from the input data (blue curve in Figure 8). Although the test would work with any number of input genes, it is statistically meaningful to input all genes from the experiment, which could be the entire genome, or all genes on an array chip, for example. Then the entire list is divided into groups based on annotation classification (molecular function, biological process, cellular component, PANTHER Protein Class or pathways), and the distributions for each group are generated (red curve in Figure 8). For each category in the classification, we calculate the probability that the distribution of input numerical values was drawn randomly from the reference distribution, using the Mann-Whitney Rank-Sum Test (U-Test) (20). A p-value cutoff of 0.05 is recommended as a start point. If the test returns a result with p-value less than 0.05, it means that the distribution of numeric values from the functional category was not drawn randomly from the reference distribution. In another word, the distribution is significant.
III. Improved statistical tests
To meet the current standard in the field, the following improvements have been made.
Fisher’s Exact test (https://en.wikipedia.org/wiki/Fisher%27s_exact_test), with the Benjamini-Hochberg false discovery rate (FDR) correction (17) for multiple testing, has been added as the default algorithm for the overrepresentation test.
FDR correction has been added to the binomial distribution test. The option to use the original settings (Binomial distribution test with Bonferroni correction) is still available in the configuration panel (see Step 6Cii – iv).
FDR correction has been added as one of the multiple testing correction methods to the enrichment test.
More information about these tests can be found in Box 2.
IV. An easier protocol to analyze genomes that are not in the PANTHER database
The current PANTHER (v.14.0) contains 131 of the most commonly researched genomes. One of the most frequent requests from PANTHER users is to include additional genomes beyond these 131. We have now dramatically simplified the steps for analyzing additional genomes. Before now, users would have to download a scoring tool to map their genes of interest to the PANTHER HMMs (hidden Markov models). This proved to be a major obstacle for researchers who are not highly trained in bioinformatics applications. To address this problem, we, in collaboration with InterPro (18) and UniProt Reference Proteomes (8), implemented an easier process to support 877 additional Reference Proteome genomes. We have pre-calculated the PANTHER HMM hits for all of the genes in each Reference Proteome (with UniProtKB identifiers), and stored the results in PANTHER Generic Mapping file format. Users just need to convert their gene list to UniProtKB IDs, and upload to the website. The list of these supported genomes can be found in Supplementary Table 1.
In the following Procedure, we provide an updated step-by-step protocol for using the PANTHER tool, as well as a detailed description of the anticipated results in the subsequent sections.
MATERIALS
Equipment
Laptop or desktop computer with high-speed internet connection is highly recommended.
System Requirements
- Operating System
- Windows XP, Windows 7 or Windows 10 recommended for PC users
- MacOS 10.12 or higher recommended for Mac users
- Minimum of 2GB RAM recommended
- Browser
- Firefox version 59 or higher
- Google Chrome version 67 or higher
- Microsoft Internet Explorer 11 or higher
- Safari version 11.1 or higher
- JavaScript and cookies must be enabled in your browser
PROCEDURE
CRITICAL: Note that sample gene list files can be found in Supplementary Data 2-5 for you to try the tools.
-
1
Access the PANTHER website by entering www.pantherdb.org in your web browser.
-
2Prepare input file(s) according to option A if you are working with one of the 131 genomes in the PANTHER database (see list of genomes at http://pantherdb.org/panther/summaryStats.jsp), option B if you are working with one of the Reference Proteome genomes other than the 131 in the database, option C if you are working with a genome that is not one of the Reference Proteome genomes or one of the 131 genomes in the database, or option D if you are working with genetic variant/SNP data. All input files must be in simple text format (such as .txt or .tab format).
- Working with one of the 131 genomes in the PANTHER database. Timing: 15–30 minutes
- Prepare input file(s) in simple text file with gene or protein identifiers as the first column. If you want to use the Statistical enrichment test, you will also need a second column with a numerical value for each gene. Sample files in the Supplementary Data 2 and 3 can be used for test purposes. Detailed instructions for the file format and supported IDs can be found in Box 3.
- Working with one of the Reference Proteome genomes now also supported in addition to the 131 genomes in the database. Timing: < 15 minutes
- Prepare input file(s) with UniProt IDs in the first column. If you want to use the Statistical enrichment test, you will also need a second column with a numerical value for each gene. For more information about mapping other IDs to UniProt ones, please refer to Box 4. Sample files for this type of input list are available in Supplementary Data 4 and 5.
- Working with a genome that is not one of the 131 geonomes in the database, nor one of the additional Reference Proteome genomes. Timing: Variable, it usually takes 15 minutes to download the data, and 10 minutes to run the script
- Prepare input file(s) in the PANTHER Generic Mapping format by mapping your IDs to the PANTHER HMM IDs using the procedure described in Box 4.
- Working with genetic variant/SNP data.
- Gather input file(s) in VCF file format. These files are usually generated by other tools used for genome sequencing and GWAS experiments. See Box 3 for more details about this file format. A sample VCF file (sample_vcf.txt) is available for test purposes in Supplementary Data 1.
Box 3: Input file.
File format
The input file is a tab-delimited text file (.txt or .tab). Only the data in the columns specified below will be used in the analyses. Data in additional columns are ignored. Microsoft Excel format is NOT accepted by the tool. Below are four file types that can be used.
ID list – The first column must be the gene or protein identifiers. See below for the supported IDs. A second column of numerical values is required if a user wants to run the Statistical enrichment test.
Previously exported text search results – Any gene list on the PANTHER site (e.g. generated by a text search, or from a set of uploaded identifiers) can be saved as a text file (see Step 6A in Anticipated Results). This file contains the gene or protein identifiers in the first column. This file type is not associated with numeric values, so it cannot be used for Statistical enrichment test.
- PANTHER Generic Mapping File – For IDs from organisms other than the 112 organisms in the PANTHER database, a user-generated data containing mappings between those IDs and their corresponding PANTHER IDs can be used (see Box 2 for details about mapping). The file must be tab-delimited and contain the following columns:
- The first column can contain a list of unique IDs from the user.
- The second column should be corresponding PANTHER family or subfamily ID (ex: PTHR10078 or PTHR10078:SF1), and is used to look up the association to GO and PANTHER terms (molecular function, biological process, and pathway).
- If you are uploading data for the Statistical enrichment test tool, a third column is required that contains the numeric value of the experiment.
Variant Call Format (VCF) – It is a text file format for storing sequence variation data. It was previously maintained by the 1000 Genomes Project. Currently the group leading the management and expansion of the format is the Global Alliance for Genomics and Health Data Working group file format team. The VCF specification can be found on GitHub at https://samtools.github.io/hts-specs/VCFv4.3.pdf.
!!Critical!! If you are using the Statistical enrichment test, numeric values should be provided to a designated column as described above. No blank or alphabet (eg., N/A) is allowed in that column.
Supported IDs
If the “ID list” file type is used, the IDs in the first column of the file must be from one of the following databases that are supported in the PANTHER system.
Ensembl: Ensembl gene identifier. Example: “ENSG00000126243”
Ensembl_PRO: Ensembl protein identifier. Example: “ENSP00000337383”
Ensembl_TRS: Ensembl transcript identifier. Example: “ENST00000391828”
Gene ID: EntrezGene IDs. Examples: “10203” (for Entrez gene GeneID:10203)
Gene symbol: for example, “CALCA”
GI: NCBI GI numbers. Example: “16033597”
HGNC: HUGO Gene Nomenclature ids. Example: “HGNC:16673”
IPI: International Protein Index ids. Example: “IPI00740702”
- Model Organism Database (MOD)
- MGI: example: “MGI:2444594”
- RGD: example: “1583843”
- ZFIN: example: “ZDB-GENE-060519–23”
- FlyBase: example: “FBgn0039374”
- WormBase: example: “WBGene00001400”
- SGD: example: “S000005863”
- PomBase: example: “SPBC947.14c”
- dictyBase: example: “DDB_G0291918”
- TAIR: example: “AT1G58450”
- Ecoli: EcoGene ID is supported, for example: “EG11161”
UniGene: NCBI UniGene ids. Examples: “Hs.654587”, “At.36040”
UniProtKB: UniProt accession. Example: “O80536”
UniProtKB-ID: UniProt ID. Example: “AGAP3_HUMAN”
The primary IDs used for gene annotations in the PANTHER system are Ensembl gene ID or MODs IDs for genes, and UniProtKB IDs for proteins. All other supported IDs are mapped to those primary IDs using the UniProt “ID mapping” mechanism (https://www.uniprot.org/uploadlists/).
If you are not certain about the ID type in your uploaded gene list, or when you find that your IDs are not mapped to any PANTHER IDs in the result page, you can simply search your ID at NCBI (http://www.ncbi.nlm.nih.gov/) or a search engine website such as Google. You can find the ID type based on the database source on the result page.
Box 4. PANTHER Generic Mapping.
If you are working with a genome that is not one of the 131 in the PANTHER database, you can still use the tool. The back-end mechanism for such analysis is to convert your input list into a PANTHER Generic Mapping file, and then analyze. However, depend on the type of genome you are working on, there are two different approaches. We have pre-calculated the PANTHER Generic Mapping for all the Reference Proteome genomes. Therefore, if you working with one of them, you can submit your list with UniProt IDs and the tool will take care of the rest. If you are working with a genome that is not in the Reference Proteome Project either, you will generate the mapping file using the PANTHER HMM scoring tool. The details of both approaches are described below.
A simple web interface
If you are working with one of the Reference Proteome genomes, you can use this interface to analyze your data with our tools. There are 877 genomes supported by this protocol. The list can be found in the Organism for ID list drop-down menu right below the ID’s from Reference Proteome Genome option on the home page (Figure 2). The details are described in the main text (Steps 2B and 4B).
One crucial requirement is that UniProt IDs must be use in the uploaded list. We recommend that you use the UniProt idmapping tool to convert other IDs to UniProt ones. The tool can be found at https://www.uniprot.org/mapping/.
Score sequences using the PANTHER HMM library
If the genome you are working with is not one of the Reference Proteome genomes, you will need to score your proteins against the PANTHER HMM library using the PANTHER Scoring Tool in order to generate the PANTHER Generic Mapping file.
PANTHER HMM library is a library of HMMER3 models (21) from over 1.70 million training sequences in 131 genomes. There are a total of over 120K models, of which 15.5K are family models, and 104.5K are subfamily models. A subfamily model is built with a subset of genes in a family, often orthologues to each other, that carry out more specific biological functions. Each HMM model is annotated with a name, functions (GO terms) and pathways. The PANTHER Scoring Tool allows users to submit a large number of protein sequences in FASTA file format, and score against the PANTHER HMM library so that the sequence identifiers can be mapped to PANTHER HMM IDs and the functional groups annotated to them, and used in the gene list analysis tools.
UNIX and Perl are required on your computer in order to use the tool. The user needs to have the basic knowledge of using UNIX and Perl in order to complete the procedures described in this Box. If you don’t feel you have adequate knowledge in these areas, you may want to get help from a colleague with the technical expertise and knowledge, such as a bioinformatics support person. You can also send an email to feedback@pantherdb.org for help.
The location to Perl must be defined in your $PATH variable or specified by the users in the arguments. If you have any questions on how to set up $PATH, please contact your UNIX system administrator.
Procedures:
- Download the following scripts and data.
-
pantherScore script (ftp://ftp.pantherdb.org//hmm_scoring/current_release/ ).(Timing: 2 minutes)
-
PANTHER HMM library (ftp://ftp.pantherdb.org/panther_library/current_release/)(Timing: 15 minutes)HMMER3 - Download from http://eddylab.org/software/hmmer3/3.1b2/hmmer-3.1b2.tar.gz(Timing: 5 minutes)
-
Decompress all three downloads. (Timing: 30 minutes)
Define the location of the HMMER binaries in the $PATH variables on your computer. This is usually done quite differently depending on the UNIX shell environment on your computer. Basically, you need to update the $PATH on the UNIX shell files, such as .cshrc (for C shell) or .profile (for Bourne shell). If you are not familiar with $PATH, you need to consult someone with IT knowledge to help you. (Timing: 15 minutes for an expert with bioinformatics training, up to 1–2 hours for others).
Use the following commands:
% tcsh
% cd pantherScore2.1
% source panther.cshrc
% ./pantherScore2.1.pl -l <panther_hmm_library> -D B -V -i <fasta file> -o <output file> -n
or
% ./pantherScore2.1.pl -l <panther_hmm_library> -D B -V -i <fasta file> -o <output file> -n -s
where
-l The path to the PANTHER HMM library downloaded above.
-D display type for results.
Options: B (best hit), A (all hits)
-i input fasta file to score. A sample fasta file is included in the downloaded called test.fasta
-o the output file
-n to display family and subfamily names in the output file.
-s specify using hmmsearch program instead of hmmscan program (default) for scoring large number of input sequences.
CAUTION: If you have a lot of sequences, you can split the fasta file and run the script on multiple computers.
The output file is a tab-delimited file in the following format:
col 1 - sequence ID
col 2 - PANTHER accession (PTHRnnnnn, for family HMMs, PTHRnnnnn:SFnn for subfamilies)
col 3 - PANTHER family or subfamily name
col 4 - HMM E-value score, as reported by HMMER tool.
col 5 - HMM bitscore, as reported by HMMER tool (not used by PANTHER)
col 6 - alignment range of protein for this particular HMM
This file can be used as PANTHER Generic Mapping File for the gene list analysis tool. By default, all results with E-value < 10−3 are included in the file. However, the classification confidence is considered high when E-value < 10−23, and medium when E-value < 10−11. Users should feel free to filter the results depending on the confidence level of their choice.
If the Statistical enrichment test is used, the numeric values need to be inserted in the 3rd column.
!! CRITICAL STEP The input file must be in simple text file format (.txt or .tab). It must also use IDs supported in the PANTHER system and the correct tab delimited format as described in Box 3.
Using online tools TIMING: 10 mins
-
3Upload the gene list to the PANTHER tool system using option A if you have a small list (<200 IDs), option B if you have a large list or VCF file, or option C if you previously saved the list in the Workspace.
- Uploading a small gene list (<200 IDs).
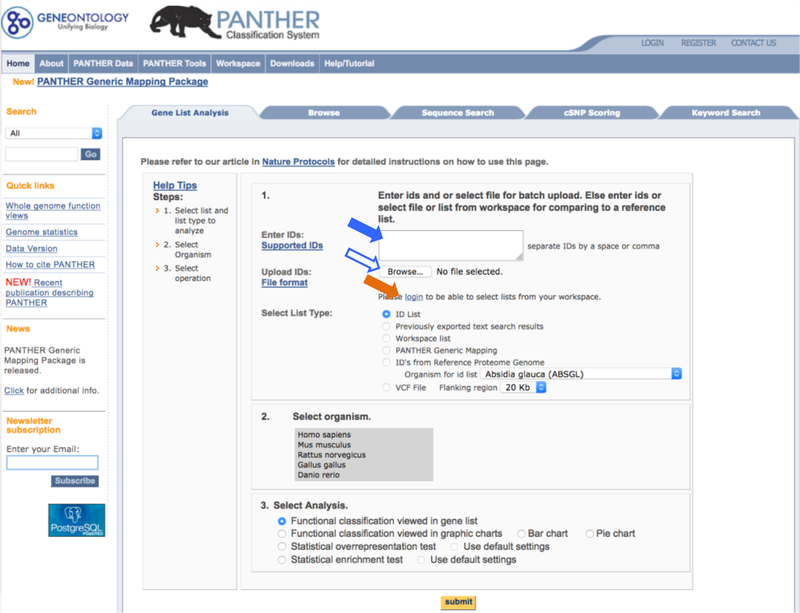
- Paste the ID list prepared in Step 2 into the Enter ID box. Alternatively you can also type IDs, one per line, into the box (solid blue arrow in Figure 2).
- Uploading a large gene list.
- Upload the list file prepared in Step 2 by clicking the Browse button (open blue arrow in Figure 2), and follow the online instructions to locate the file.
- Using a gene list previously saved in the workspace.
- If you have previously saved your list into the Workspace, you can use it by clicking the login link (orange arrow in Figure 2), and follow the online instructions to locate the file in the Workspace. Please note that numeric values cannot be saved in the Workspace, and therefore this type of upload does not support the Statistical enrichment test.
Figure 2.
The PANTHER home page with the Gene List Analysis Tools.
Solid blue arrow points to the Enter IDs box where user can paste the ID list or type IDs, one per line, to upload the gene list. Open blue arrow points to the Browse button where user can upload the list file from the computer. Orange arrow points to the link where user can access saved list in the Workspace.
?Troubleshooting
-
4Select a corresponding list type in order for the tool to work properly. There are four list types that are supported by the tools: ID List, Previously exported text search results, PANTHER Generic Mapping File, and VCF file format. See Box 3 for details of the list types.
- File prepared using Step 2A
- If you prepared your file using Step 2A, select ID List.
- File prepared using Step 2B
- If you prepared your file using Step 2B, select ID’s from Reference Proteome Genomes. You need to also select an organism from the “Organism for ID list” drop-down menu. The genome name used here is a combination of the species name and the organism mnemonic (a 5-letter symbol) to specify the strain. For details of the organism mnemonic, please visit: https://www.uniprot.org/taxonomy/. The list of organisms is also listed in the Supplementary Table 1.
- File prepared using Step 2C
- If you prepared your file using Step 2C, select PANTHER Generic Mapping.
- File prepared using Step 2D
- If you prepared your file using Step 2D, select VCF file format and a flanking region needs be specified. A flanking region is the number of base pairs on either side of the gene on the chromosome, and are usually considered to be associated with the gene function by serving as the regulatory region of the gene. When a SNP is located in the flanking region, it will be mapped to the gene. By default, the tool uses 20Kb on either side of the gene as the flanking region for the analysis.
-
5
Select an organism from the drop-down menu, which lists the 12 “model organisms” first, followed by the remaining organisms ordered alphabetically. Note that Organism selection was done for ID’s from Reference Proteome Genomes option (Step 4B), and is not required for PANTHER Generic Mapping option (Step 4C). Table 1 summarizes the properties of input lists for various types of genomes.
Table 1.
Summary of ID type, List type and organism selection for various types of genomes
| Genome to analyze | One of 131 genomes in PANTHER | One of additional Reference Proteomes | Any other genome |
|---|---|---|---|
| ID type to use in the input file | IDs supported by PANTHER | UniProt | Any |
| Select List Type | ID List | ID’s from Reference Proteome Genome | PANTHER Generic Mapping |
| Select Organism | Required | Required | Not required |
| Reference list for Overrepresentation test | Default or user upload (with IDs supported by PANTHER) | Default or user upload (PANTHER Generic Mapping) | User upload (PANTHER Generic Mapping) |
CAUTION: There are two reasons to select an organism at this point. First, some identifiers, such as gene symbols, are not organism specific. By selecting an organism here, it ensures that the IDs are mapped to those in the organism you are interested in. Second, if the statistical overrepresentation test is selected, the default reference gene list is based on the selected organism.
-
6In the Select Analysis box, select one of the following 4 options (option A for Functional classification viewed in gene list, option B for Functional classification viewed in graphic chart, option C for Statistical overrepresentation test, or option D for Statistical enrichment test) by clicking the radio button, and then click the submit button.
- Functional classification viewed in gene list
-
Select Functional classification viewed in gene list if you want to view the functional classifications of the genes in your list (Figure 3). Once you reach the results page, you can make modifications to the page as described in the steps below to view, customize and export the results?Troubleshooting
- Sort the list – You can always sort the list by clicking on any of the underlined column names. A yellow triangle appears in front of the column name that you choose to sort. The orientation of the triangle indicates the sort is ascending or descending.
- Customize columns – You can click on the “x” button next to the column names to remove the column.
- Customize Gene List – By clicking this link, users can customize what annotation data they want to see on the results page. By default, only PANTHER Protein Class data is displayed.
- Converting a list to another list type – Select the genes you want to convert by clicking the checkboxes. The default is for all genes in the list. Then choose the list type from the drop-down menu after “Convert List to:” at the top of the page.
- Save the list. Select the genes you want to save by clicking the checkboxes. The default is for all genes in the list. You can select one of the following from the pull-down menu as the destination:
- Workspace – You need to register to save data to the workspace. The registration is free. When you make this selection, a pop-up window will ask you to name the list and add any comments. The name and comments can be edited at any time in the future from the Workspace page. Once the gene list is saved in your workspace, it can be returned to at any time. Only the IDs are stored, and they are mapped to the internal PANTHER gene ids, so when you access a list in the future, all information will be updated and current.
- Exporting a list to a file – The list will be exported as a tab-delimited file. You can now import the file into Excel or perform any post-processing you wish.
- View the list as text on the website.
- Use the pie chart view by clicking the colorful pie chart icon (see Glossary for the meaning of the abbreviations). See the next section below for details about how to interpret the pie chart.
-
- Functional classification viewed in graphic chart
- Select Functional classification viewed in graphic chart to get the functional classification of the genes in your list displayed as either a pie chart or a bar chart (Figure 4).
- Statistical overrepresentation test
- Select Statistical overrepresentation test to find functional classes that are statistically over- (or under-) represented in the input list, compared to randomly selected genes.
- The default option uses the PANTHER reference list for the genome as the reference list, and PANTHER GO-Slim Biological Process annotation gene set for the analysis. You can change these (highly recommended) by deselecting the default option and updating the form on the Analysis Summary panel. The tests can be repeated with different annotation data sets and on different user data by updating the parameters on top of the result page.
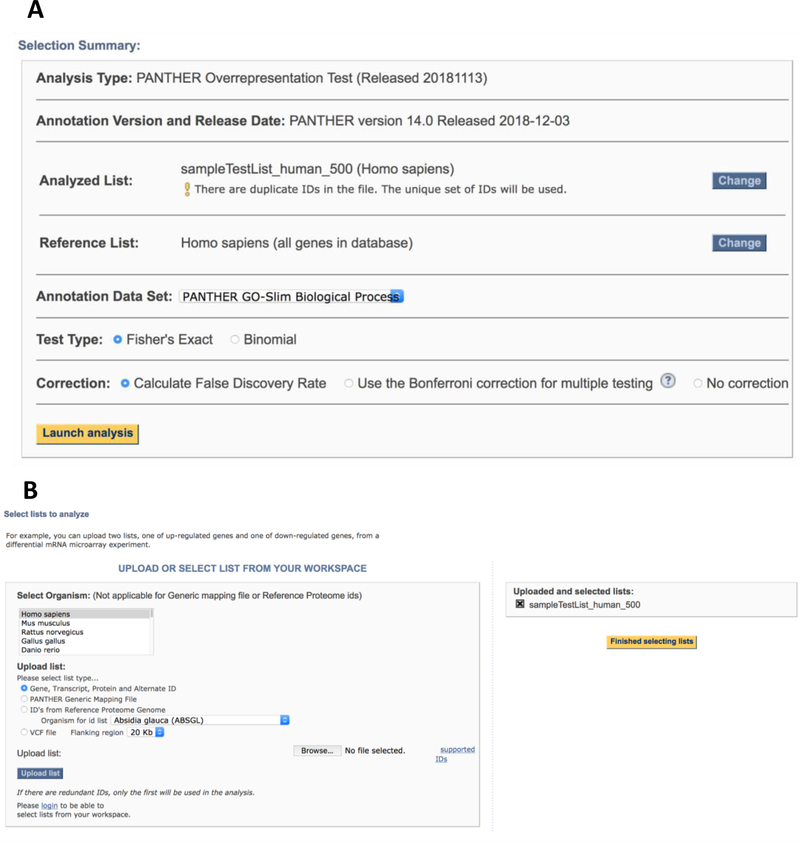
- HIGHLY RECOMMENDED. On the configuration page (Figure 5A), you can change/add the Analyzed List or change the Reference List by clicking the Change button. A new webpage will open and you can upload those lists (Figure 5B). The detailed step-by-step procedure can be found in Box 5. On the configuration page, you can also change the Annotation Data Set from the drop down menu. There are nine data sets to select. See Box 1 for the list and description of the data sets.
- On the configuration page, you can also select the statistical test for the overrepresentation analysis. The default is Fisher’s Exact test with FDR multiple test correction. You can also select the Binomial test with FDR multiple test correction or Bonferroni correction.
- Click the Launch analysis button.
-
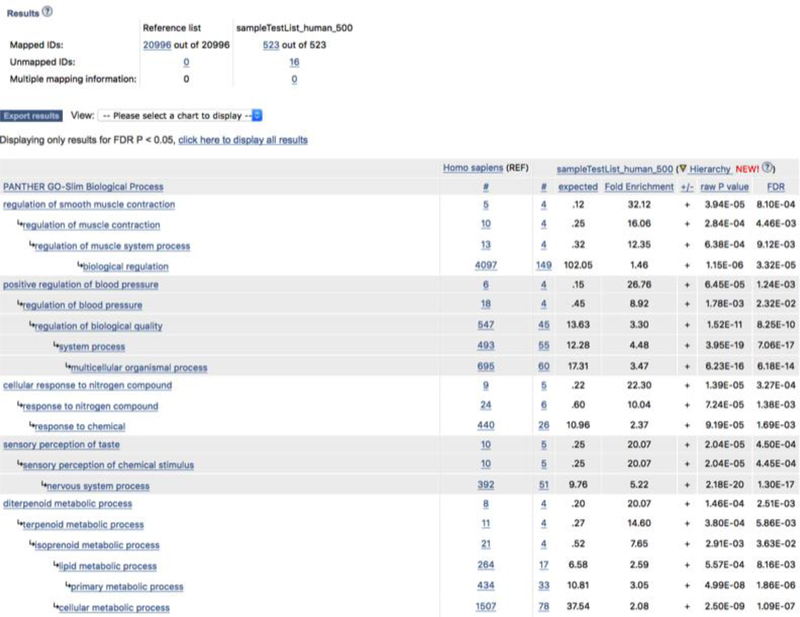
On the results page (Figure 6), you can export the result table in a tab-delimited file by clicking the Export results button, or view the results graphically by using the View drop-down menu and choose one of the annotation class (PANTHER Pathway, PANTHER GO-Slim, or PANTHER Protein Class)?Troubleshooting
- Statistical enrichment test
- Select Statistical enrichment test
- The default setting is to analyze using the PANTHER GO-Slim Biological Process annotation gene set for the analysis. The settings can be changed by deselecting the default option and modifying on the Analysis Summary panel.
- On the configuration page, you can also change the Annotation Data Set from the drop down menu. There are nine data sets available for analysis. See Box 1 for the list and description of the list.
- Select either FDR or Bonferroni multiple test for the analysis.
-
Click the Submit button if you choose to use the default setting from step 6Di, or Launch Analysis button after you modify the settings on the configuration page as in Steps 6Dii-1v.?Troubleshooting
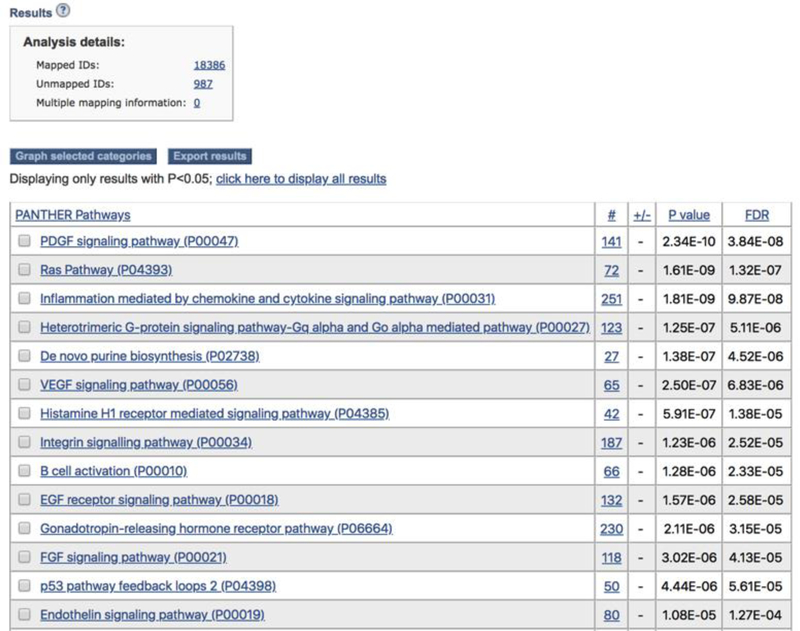
- On the results page (Figure 7), you can export the result table in a tab-delimited file by clicking the Export results button or compare the distribution curve in graph view. To do so, check the box in front of the category or pathway of your interest, and then click the Graph selected categories button (Figure 8).
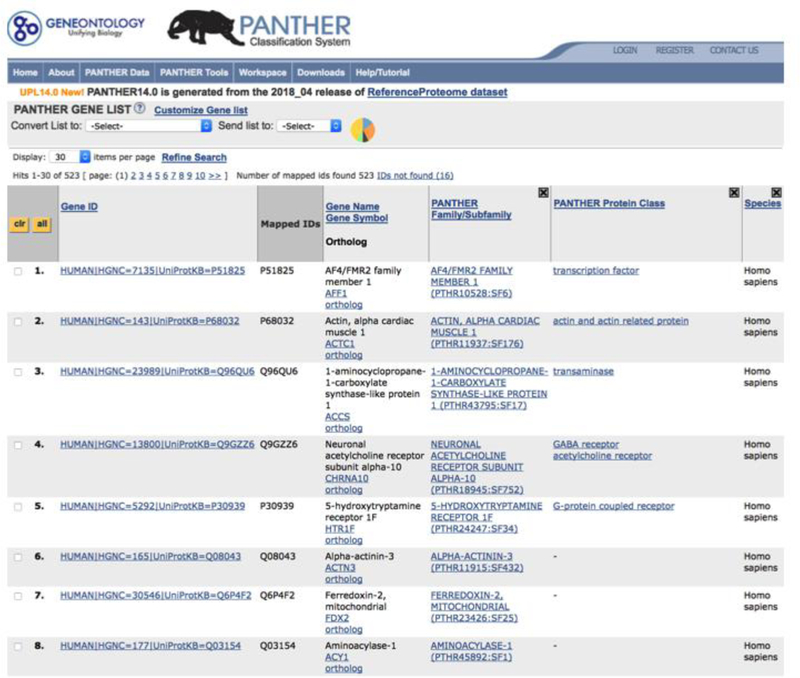
Figure 3.
Results of functional classification displayed as a gene list page. The results are based on the Supplemental Data 3.
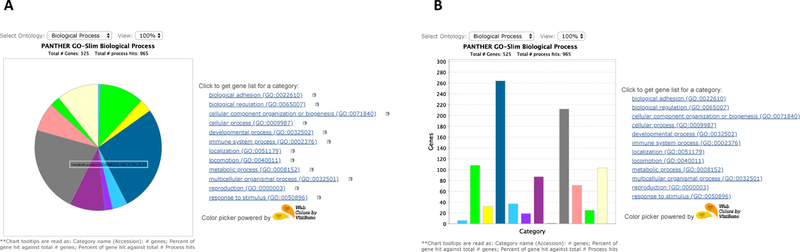
Figure 4.
PANTHER results shown in pie chart (A) or bar chart (B) from the Supplemental Data 3 You can use the Select Ontology drop-down menu to switch to the charts of different ontologies. Click on the chart section to display the child categories. Click on the legends on the right side to retrieve the list of the genes for that category.
Figure 5.
User interface of the statistical overrepresentation test to allow user to configure the analysis criteria. A. The configuration page where user can view the versions of the tool and annotation data set, change/add test lists, change reference list and annotation dataset, and select test type. B. The user interface for users to change/add gene lists.
Box 5. Change/add gene list or reference list in overrepresentation test.
The overrepresentation test allows you to analyze up to four gene lists at a time. You can also upload your own Reference List instead of using the default one. To do so, the default checkbox should be unselected before you click the Submit button.
On the following configuration page (Figure 5A), click the Change button. A new webpage will open (Figure 5B) and you can do the following:
Click Browse button
Select the gene list from your computer
Select the organism. The default is the organism selected when the first gene list is uploaded.
Select the List type. The default is the one selected when the first gene list is uploaded.
Click Upload list.
Steps 1–5 above can be repeated to upload up to 4 analyzed gene lists
It is highly recommended to upload your own reference list file. The reference list should be the list of all the genes from which your smaller analysis list was selected. For example, in a list of differentially expressed genes, the reference list should only contain genes that were detected at all in the experiment, and thus potentially could have been on a list of genes derived from the experiment.
If ID’s from Reference Proteome Genome option is selected, a default reference list is uploaded. If you decide to upload your own reference list, it has to be in the PANTHER Generic Mapping file format.
If PANTHER Generic Mapping file option is selected, a reference list in the same format must be uploaded here.
After all lists are uploaded, click the Finish selecting lists button. The tool will return back to the selection summary panel page displaying the uploaded lists.
Figure 6.
Result from the statistical overrepresentation test. The results are based on the Supplemental Data 3. The summary of the results is displayed in a table. You can export the table in a tab-delimited file by clicking the Export results button. You can also view the results in other views by using the View drop-down menu. If your analysis is done in pathway as shown here, you can click the pathway name and display the pathway diagram. A total of 4 test lists can be analyzed and viewed at the same time.
Figure 7.
The results from the statistical enrichment test. The results are based on the Supplemental Data 2. The output of the tool with a list of P-values for each comparison between a functional category distribution and the reference distribution.
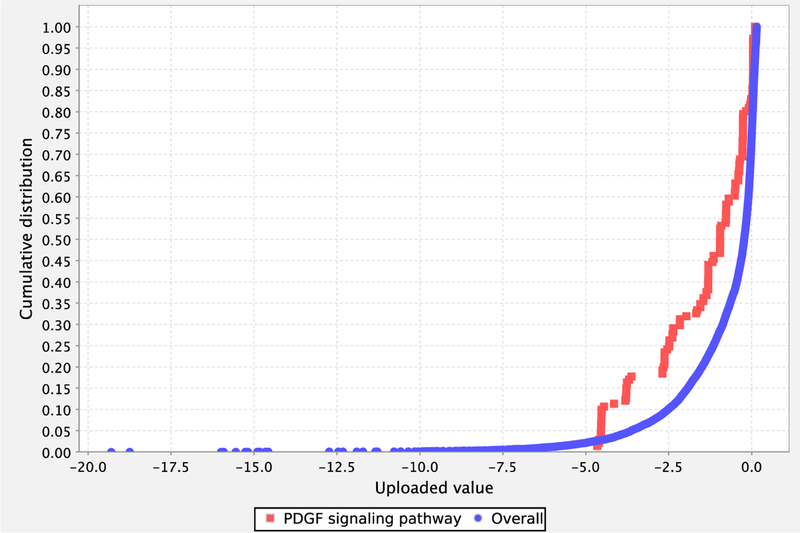
Figure 8.
Graph view of the results from the enrichment test showing a comparison of the distributions from PDGF signaling pathway (red) and reference (blue) in graph view.
CAUTION: In order to use the Statistical enrichment tool, make sure that the uploaded gene list contains a second column with numerical values.
Timing
Step 1, launch website: instant.
Step 2A, prepare input file: 15–30 minutes
Step 2B, prepare UniProt ID input file: < 15 minutes
Step 2C, prepare the PANTHER Generic Mapping file: varies depending on how fast your internet is. It usually takes 15 minutes to download the data, and 10 minutes to run the script.
Step 2D, gather the VCF files: instant.
Steps 3–6, using online tools: 10 minutes.
TROUBLESHOOTING
Table 2 lists some of the common problems and the possible solutions.
Table 2.
Troubleshooting
| Step | Problem | Possible reasons and solutions |
|---|---|---|
| 3 | Fail to upload the file | This is usually because the input file is in the wrong file format. Possible solutions: 1. Make sure that your file is in simple text format (.txt or .tab). 2. If you are uploading a file with numeric values for the enrichment test, make sure that the second column contains only numeric numbers. Any rows with no values should be removed instead of leaving it blank or mark it as “n/a”, etc. 3. Make sure that there are no blank rows in the first column. |
| 6A, 6Cvi, 6Div. | IDs in the uploaded file don’t have a mapped ID in PANTHER | The current PANTHER data is based on the April 2017 release of Reference Proteome Project and its ID mapping. It is possible that a small fraction of the IDs may not map due to the outdated data either in the PANTHER database or your uploaded file. There is no solution to this. If you believe that you are using the current IDs, please do the following: 1. Make sure that the IDs in the uploaded file are supported by PANTHER. Refer to Box 3 for Supported IDs. 2. IDs from certain database may contain a version number at the end of them, e.g., NP_000242.1. Do not include the version number “.1” in the ID, and just use NP_000242. 3. Send feedback to feedback@pantherdb.org. |
| 6Cvi, 6Div | No results returned | By default, only the rows with significant P-value or Q values are returned. Click the Click to display all results link to see all the results. |
ANTICIPATED RESULTS
As illustrated in Figure 1, the tools use the UniProt ID Mapping to map the uploaded IDs to the IDs in the PANTHER annotation data set. Not all IDs can be mapped. This is mainly caused by the outdated IDs used in the uploaded list. The result of the mapping is summarized at the top of the results page (Figures 6 & 7). The list of the genes can be accessed by clicking the counts.
Step 6A. Functional classification tool viewed in gene list page
This tool returns the results as a gene list webpage (Figure 3). The page displays all the IDs from the uploaded gene list and their mapped PANTHER sequence IDs as well as annotation data. The page contains the following information.
Gene ID – This is the identifier for genes in the PANTHER library. The format is as follows: organism|gene database source=gene id|protein database source=protein id. The organism is the 5-letter organism mnemonic code as listed at https://www.uniprot.org/taxonomy/). For example, HUMAN|HGNC=6218|UniProtKB=Q09470 is a human sequence, the gene sequence is from HGNC (Human Gene Nomenclature Committee, https://www.genenames.org/) with ID HGNC:6218, and the protein sequence is from UniProt with id Q09470.
Mapped IDs – IDs from the uploaded gene list that are mapped to the gene ids in the first column.
Gene Name/Gene Symbol – The Entrez gene definition and gene symbol.
PANTHER Family/Subfamily – The name and identifier of the PANTHER family or subfamily where the gene in the first column is in.
PANTHER Protein Class – This is a PANTHER Index terms describing protein classes. The default view only shows PANTHER Protein Class. Other annotation data can be viewed by customizing the columns (see below).
Species – The organism of the gene in Column 1.
B. Functional classification tool viewed in graphical chart
There are two types of charts in this option, pie chart (Figure 4A) and bar chart (Figure 4B). In either chart, the default view displays an overview of all ontology terms at the first (or most general) level within the same ontology. When a slice of the pie chart or a bar in the bar chart, which represents an ontology term, is clicked, a new chart will appear that contains its child ontology terms.
Since one gene can be classified to more than one term, the pie chart is calculated based on the number of “hits” to the terms over the total number of class hits. Class hit means independent ontology terms. For example, if a gene is classified to 2 ontology terms that are not parent or child to each other, it counts as 2 class hits.
For the bar chart, the x-axis is the classification terms (GO terms or pathway terms), and the y-axis is the number of genes annotated to the term.
When you place the computer mouse pointer over a slice or the bar, the category name and a series of counts are displayed. In our example in Figure 4A, the name is a GO term metabolic process (GO:0008152) followed by
the number of genes (212) from the uploaded list that are classified to the term metabolic process;
the percent (40.4%) of genes classified to metabolic process (212) over the total number of genes (525);
the percent (22%) of genes classified to this metabolic process (212) over total number of annotations (965).
The results can be exported as a tab delimited file with Export link on the page.
C. Statistical overrepresentation test
The results using Fisher’s Exact test are displayed in a table (Figure 6). If one test gene list is uploaded, the table contains eight columns of data (seven for binomial distribution test):
The first column contains the name of the PANTHER classification category. If you are doing this analysis using the PANTHER Pathway annotation set, you can click on the pathway name to view the corresponding pathway diagram. For other analyses, the link will take you to more information about that category.
The second column contains the number of genes in the reference list that map to this particular PANTHER classification category.
The third column contains the observed number of genes in your uploaded list that map to this PANTHER classification category.
The fourth column contains the expected value (see Box 2), which is the number of genes you would expect in your list for this PANTHER category, based on the reference list.
The fifth column shows the fold enrichment, which is the ratio of value of column 3 (observed number) over that of column 4 (expected number).
The sixth column has either a + or −. A plus sign indicates over-representation of this category in the analyzed list: you observed more genes than expected based on the reference list (for this category, the number of genes in your list is greater than the expected value). Conversely, a negative sign indicates under-representation, i.e. fewer genes than expected.
For the results from Fisher’s Exact test, this column shows the raw P-values. For binomial distribution test, this column is the P-value as determined by the binomial statistic. In either case, this is the probability that the number of genes you observed in this category occurred by chance (randomly), as determined by your reference list.
The eighth column is the Q-value (adjusted P-value, reflecting the False Discovery Rate) as calculated by the Benjamini-Hochberg procedure (17). By default a critical value of 0.05 is used to filter results, so all results shown are valid for an overall FDR<0.05 even if the FDR for an individual comparison is greater than that value. This value is output when the Fisher’s exact test option is selected.
If more than one test list is uploaded, columns 3 to 6 are repeated for each list.
The default filter has already limited your results to those with statistical significance (overall FDR<0.05, meaning that you can expect that <95% of the enriched classes are true associations). You can view all results regardless of FDR by clicking on the “click here to view all results” link above the table. While this can be useful for troubleshooting, we caution that the additional results are not statistically significant.
By default, the results are sorted “Hierarchically” by to help users understand the hierarchical relations between over-represented or enriched functional classes. Sorting is done only by the most specific subclass first, with its parent terms indented directly below it. These are all related classes in an ontology, and are often interpretable as a group rather than individually. If a term is a parent of more than one term in the results table, it is shown only under its first descendant. You can still sort by a single column (e.g. Fold change or P-value) by clicking on that column header.
From this result page, various statistics can be exported by using the drop-down menu next to the “Export results” button. The list of genes/proteins in any functional group can be viewed by clicking on the listed counts. When PANTHER pathways was chosen as the annotation set, clicking on the pathway name brings up pathway diagrams. The resulting pathway diagram can be exported as an image file (.png) by choosing “Export” function on the page.
D. Statistical enrichment test
The returned results are displayed in a table with four essential columns of data (Figure 7):
If you are doing this analysis using the PANTHER Pathway annotation set, you can click on the pathway name to view the corresponding pathway diagram. For other analyses, the link will take you to more information about that category.
The second column contains the number of genes that map to this particular PANTHER classification category.
The third column has either a + or −. A plus sign indicates that for this category, the distribution of values for your uploaded list is shifted towards greater values than the overall distribution of all genes that were uploaded. A negative sign indicates that the uploaded list is shifted towards smaller values than the overall list.
The fourth column contains the P-value as calculated from the Mann-Whitney U Test (Wilcoxon Rank-Sum test) (Box 2). A large P-value indicates that the genes for this category have a distribution that is similar to randomly choosing genes from the overall distribution. In other words, the values of the uploaded genes for this category have a similar distribution to the overall list of values that were input. A small, significant P-value indicates that the distribution for this category is non-random and different than the overall distribution. A cutoff of 0.05 is recommended as a starting point (note that these are already adjusted for multiple testing using the Bonferroni correction).
If FDR is selected as the multiple test, a fifth column is returned to display the Q-value (adjusted P-value, reflecting the False Discovery Rate) as calculated by the Benjamini-Hochberg procedure (17). By default a critical value of 0.05 is used to filter results, so all results shown are valid for an overall FDR<0.05 even if the FDR for an individual comparison is greater than that value.
By default, only the results with statistical significance are displayed. You can view all results regardless of P-value by clicking on the “click here to view all results” link above the table. While this can be useful for troubleshooting, we caution that the additional results are not statistically significant. Again, the results are sorted “Hierarchically” by default (see details in the previous section).
To have a visual representation of these distributions, select the checkboxes of the categories of interest, and click on the ‘Graph selected categories’ button. The graph will be displayed in a new window (Figure 8). The x-axis is your uploaded value. The y-axis is the cumulative fraction. The blue curve is the overall distribution for all genes. The red curve is the selected functional category. In this case, it is PDGF signaling pathway. If you look at the data point x = −2.5, y is 0.3 for the red curve and 0.1 for the blue curve. This means that 30% of your uploaded genes have a value of −0.25 or smaller, but only 10% of the overall genes have a value of −0.25 or smaller. In other words, it shows that the distribution of the category tends to be smaller than the overall distribution. We find that visualization is essential for interpreting any deviation between the functional category distribution and the overall distribution.
The genes/proteins in each category can also be viewed from the output page by clicking on the listed counts. In addition, for pathways, clicking on the pathway name will bring up the pathway diagram (Figure 8), which can be exported as an image file (.png) by choosing “Export” function from the page.
Supplementary Material
Supplementary Data 1. sample_vcf.txt
This file contains 206 variants in VCF format. It can be used to test the “VCF File” list type.
Supplementary Data 2. sampleTestList_human
This file contains 19911 human genes, with UniProtKB IDs in the first column and numeric values in the second column. This file can be used as a sample upload file for all tools.
Supplementary Data 3. sampleTestList_human_500
This file contains 555 human genes with UniProtKB IDs, but without numeric values. This file can be used as a sample upload file to test the functional classification tools and the overrepresentation tool.
Supplementary Data 4. sampleTestList_xenopus
This file contains 269 Xenopus laevis genes in UniProtKB IDs. It can be used as a sample upload file to test the “ID’s from Reference Proteome Genomes” list type for function classification tools and the overrepresentation test.
Supplementary Data 5. sampleTestList_xenopus_withValues
This file contains 41561 Xenopus laevis genes in UniProtKB IDs in the first column and numeric values in the second column. It can be used as a sample upload file to test the “ID’s from Reference Proteome Genomes” list type for all tools.
Supplementary Table 1. supported_reference_proteome_genomes.pdf
This file lists 877 Reference Proteome genomes that can be analyzed using the new “ID’s from Reference Proteome Genomes” functionality. The first column of the file is the species name. The second column is the total number of genes. The third column is the number of genes in the PANTHER HMM library.
ACKNOWLDGEMENTS
The authors would like to thank the InterPro team, especially Lorna Richardson and Rob Finn for their support in providing PANTHER matching files that are used to generate PANTHER Generic Mapping files; the Reference Proteome team, especially Maria Martin, for their support in providing up-to-date Reference Proteome datasets; the Gene Ontology Consortium, especially Pascale Gaudet, Marc Feuermann and Suzanna Lewis for their support on providing GO phylogenetic annotation data; and the Reactome team, especially Robin Haw, for their support in providing the Reactome dataset. The authors would also like to thank Arthur Toga and the BDDS (Big Data for Discovery Science) Project for providing the funding to develop the software for supporting genetic variant analysis. This work is supported by NIH/NHGRI U41HG002273 and NSF 1458808 to P.D.T., and NIH U54EB020406 to Arthur Toga. Funding for open access charge: University of Southern California.
Footnotes
COMPETING FINANCIAL INTERESTS
The authors declare that they have no competing financial interests.
DATA AVAILABILITY
All PANTHER data are publically available and can be downloaded at http://www.pantherdb.org/downloads/index.jsp.
CODE AVAILABILITY
Source codes for various PANTHER software, including the PANTHER scoring tool, tree building tool (GIGA), can be downloaded at http://www.pantherdb.org/downloads/index.jsp.
REFERENCES
- 1.Mi H, Muruganujan A, Ebert D, Huang X, Thomas PD. PANTHER version 14: More genomes, a new PANTHER go-slim, and improvements in enrichment analysis tools. Nucleic Acid Research 2019, January 1:In press. [DOI] [PMC free article] [PubMed]
- 2.Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, et al. The sequence of the human genome. Science 2001, February 16;291(5507):1304–51. [DOI] [PubMed] [Google Scholar]
- 3.Thomas PD, Campbell MJ, Kejariwal A, Mi H, Karlak B, Daverman R, et al. PANTHER: A library of protein families and subfamilies indexed by function. Genome Res 2003, September;13(9):2129–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thomas PD, Kejariwal A, Campbell MJ, Mi H, Diemer K, Guo N, et al. PANTHER: A browsable database of gene products organized by biological function, using curated protein family and subfamily classification. Nucleic Acids Res 2003, January 1;31(1):334–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thomas PD, Kejariwal A, Guo N, Mi H, Campbell MJ, Muruganujan A, Lazareva-Ulitsky B. Applications for protein sequence-function evolution data: MRNA/protein expression analysis and coding SNP scoring tools. Nucleic Acids Res 2006, July 1;34(Web Server issue):W645–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mi H, Muruganujan A, Casagrande JT, Thomas PD. Large-scale gene function analysis with the PANTHER classification system. Nat Protoc 2013, August;8(8):1551–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gaudet P, Livstone MS, Lewis SE, Thomas PD. Phylogenetic-based propagation of functional annotations within the gene ontology consortium. Brief Bioinform 2011, September;12(5):449–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.UniProt Consortium T. UniProt: The universal protein knowledgebase. Nucleic Acids Res 2018, March 16;46(5):2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.The Gene Ontology Consortium. Expansion of the gene ontology knowledgebase and resources. Nucleic Acids Res 2017;45(D1):D331–8. doi: 10.1093/nar/gkw1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fabregat A, Jupe S, Matthews L, Sidiropoulos K, Gillespie M, Garapati P, et al. The reactome pathway knowledgebase. Nucleic Acids Res 2018, January 4;46(D1):D649–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cbio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov 2012, May;2(5):401–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kanehisa M, Sato Y, Furumichi M, Morishima K, Tanabe M. New approach for understanding genome variations in KEGG. Nucleic Acids Res 2018, October 13. [DOI] [PMC free article] [PubMed]
- 13.Caspi R, Billington R, Ferrer L, Foerster H, Fulcher CA, Keseler IM, et al. The metacyc database of metabolic pathways and enzymes and the biocyc collection of pathway/genome databases. Nucleic Acids Res 2016, January 4;44(D1):D471–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Slenter DN, Kutmon M, Hanspers K, Riutta A, Windsor J, Nunes N, et al. WikiPathways: A multifaceted pathway database bridging metabolomics to other omics research. Nucleic Acids Res 2018, January 4;46(D1):D661–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kerrien S, Aranda B, Breuza L, Bridge A, Broackes-Carter F, Chen C, et al. The intact molecular interaction database in 2012. Nucleic Acids Res 2012, January;40(Database issue):D841–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chatr-Aryamontri A, Oughtred R, Boucher L, Rust J, Chang C, Kolas NK, et al. The biogrid interaction database: 2017 update. Nucleic Acids Res 2017;45(D1):D369–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. Journal of the Royal Statistical Society. Series B (Methodological) 1995;57(1):289–300. [Google Scholar]
- 18.Finn RD, Attwood TK, Babbitt PC, Bateman A, Bork P, Bridge AJ, et al. InterPro in 2017-beyond protein family and domain annotations. Nucleic Acids Res 2017;45(D1):D190–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mi H, Thomas P. PANTHER pathway: An ontology-based pathway database coupled with data analysis tools. Methods Mol Biol 2009;563:123–40. doi: 10.1007/978-1-60761-175-2_7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Clark AG, Glanowski S, Nielsen R, Thomas PD, Kejariwal A, Todd MA, et al. Inferring nonneutral evolution from human-chimp-mouse orthologous gene trios. Science 2003, December 12;302(5652):1960–3. [DOI] [PubMed] [Google Scholar]
- 21.Eddy SR. A new generation of homology search tools based on probabilistic inference. Genome Inform 2009, October;23(1):205–11. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Data 1. sample_vcf.txt
This file contains 206 variants in VCF format. It can be used to test the “VCF File” list type.
Supplementary Data 2. sampleTestList_human
This file contains 19911 human genes, with UniProtKB IDs in the first column and numeric values in the second column. This file can be used as a sample upload file for all tools.
Supplementary Data 3. sampleTestList_human_500
This file contains 555 human genes with UniProtKB IDs, but without numeric values. This file can be used as a sample upload file to test the functional classification tools and the overrepresentation tool.
Supplementary Data 4. sampleTestList_xenopus
This file contains 269 Xenopus laevis genes in UniProtKB IDs. It can be used as a sample upload file to test the “ID’s from Reference Proteome Genomes” list type for function classification tools and the overrepresentation test.
Supplementary Data 5. sampleTestList_xenopus_withValues
This file contains 41561 Xenopus laevis genes in UniProtKB IDs in the first column and numeric values in the second column. It can be used as a sample upload file to test the “ID’s from Reference Proteome Genomes” list type for all tools.
Supplementary Table 1. supported_reference_proteome_genomes.pdf
This file lists 877 Reference Proteome genomes that can be analyzed using the new “ID’s from Reference Proteome Genomes” functionality. The first column of the file is the species name. The second column is the total number of genes. The third column is the number of genes in the PANTHER HMM library.