Abstract
Mitochondria house the metabolic machinery for cellular ATP production. The mitochondrial network is sensitive to perturbations (e.g., oxidative stress and pathogen invasion) that can alter membrane potential, thereby compromising function. Healthy mitochondria maintain high membrane potential due to oxidative phosphorylation (Ly et al., 2003). Changes in mitochondrial function or calcium levels can cause depolarization, or a sharp decrease in mitochondrial membrane potential (Bernardi, 2013). Mitochondrial depolarization induces opening of the mitochondrial permeability transition pore (MPTP), which allows release of mitochondrial components like reactive oxygen species (mtROS), mitochondrial DNA (mtDNA) or intermembrane space proteins into the cytosol (Martinou and Green, 2001; Tait and Green, 2010; Bronner and O'Riordan, 2014). These contents trigger inflammation, and can lead to cell death (West et al., 2011). Both mtROS and cytosolic mtDNA contribute to the activation of inflammasomes, multiprotein complexes that process the proinflammatory cytokines, IL-18 and IL-1β. Studies indicate that cytosolic mtDNA in particular can bind two different inflammasome sensors, AIM2 and NLRP3, leading to inflammasome activation (Burckstummer et al., 2009; Hornung and Latz, 2010). In this protocol, you will be able to specifically extract cytosolic mtDNA and quantify the amount using a qPCR assay.
Part I. Extraction and purification of cytosolic mtDNA
Materials and Reagents
-
For extraction (see Figure 1)
1.5 ml microcentrifuge tubes (Denville Scientific Inc., catalog number: C2170)
Gloves
6 well-plates, tissue culture-treated (Corning, catalog number: 3506)
Cell lifter (Biologix Group Limited, catalog number: 70-2180)
Murine immortalized bone marrow derived macrophages (Bernardi, 2013)
Thapsigargin, 97%, ACROS Organics™ (Thermo Fisher Scientific, Fisher Scientific™, catalog number: AC328570010) (Bronner et al., 2015)
1% NP-40 (Igepal CA-630) (Sigma-Aldrich, catalog number: I8896) (Bronner and O'Riordan, 2014)
DPBS (Thermo Fisher Scientific, Gibco™, catalog number: 14040-133) (Burckstummer, 2009)
DMEM (Thermo Fisher Scientific, Gibco™, catalog number: 11965-092)
Fetal bovine serum (FBS) (Thermo Fisher Scientific, Gibco™, catalog number: 10437-028)
Medium (see Recipes)
NP-40 (Igepal CA-630) solution (see Recipes)
Thapsigargin stock solution (see Recipes)
-
For purification
Gloves
DNeasy blood & tissue kit, 50 samples (QIAGEN, catalog number: 69504) or 250 samples (QIAGEN, catalog number: 69506)
Ethanol 200 proof (Decon Labs, catalog number: 2716)
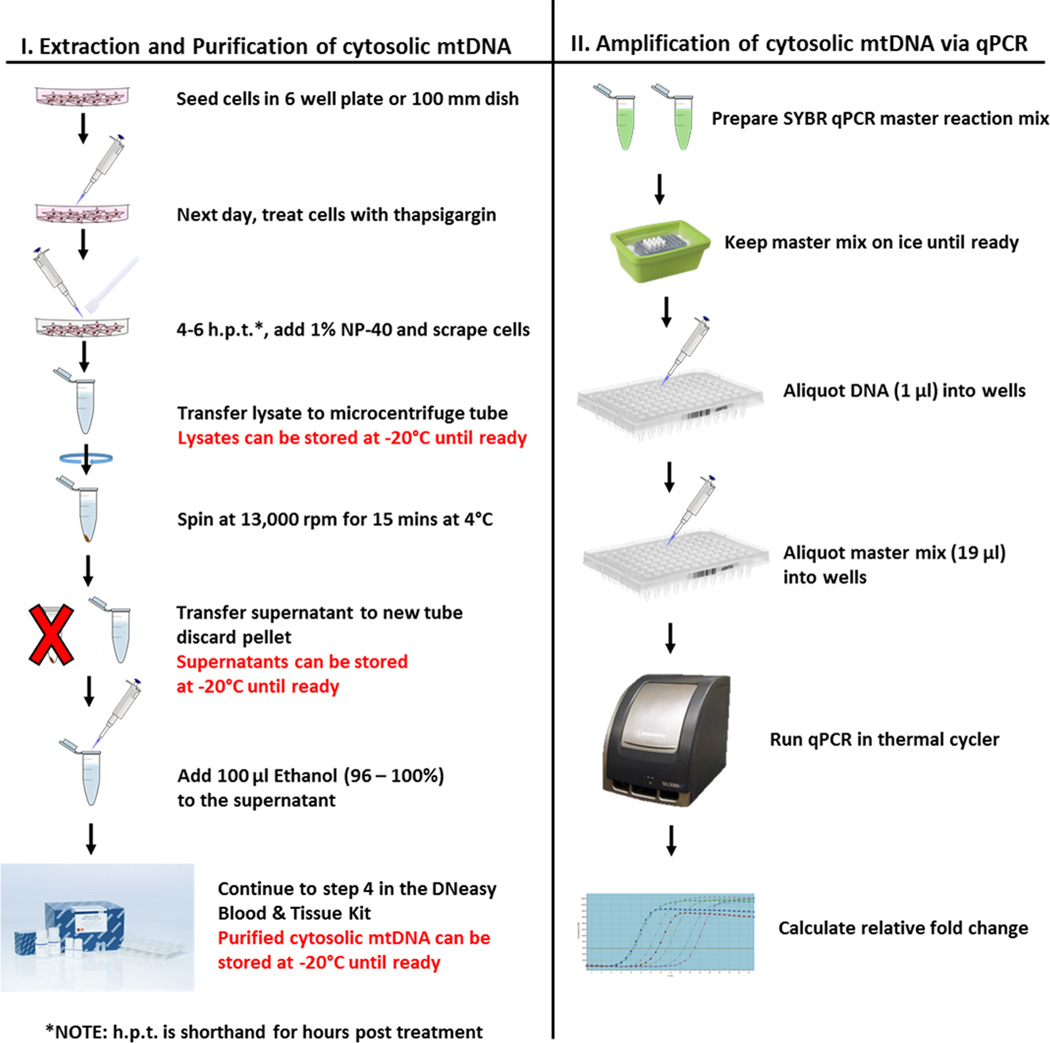
Figure 1. Flowchart for extracting, purifying, and amplifying cytosolic mtDNA.
Equipment
Centrifuge (Thermo Fisher Scientific, Fisher Scientific™, model: accuSpin™ Micro 17)
Procedure
Seed 1 × 106 cells in 2 ml per well into 6-well plate in the appropriate medium.
The following day (~16 h later), aspirate medium and add medium containing thapsigargin (10 µM in 2 ml medium for 4–6 h) (Hornung and Latz, 2010).
At relevant times post treatment (e.g., 2 h, 4 h and 8 h) wash cells with 1× DPBS once then aspirate 1× DPBS.
Add 1% NP-40 (100 µl) to each well and scrape cells.
Place lysates into prelabeled microcentrifuge tubes and incubate on ice for 15 min (Livak and Schmittgen, 2001).
Spin lysates at 13,000 rpm (16,000 × g) for 15 min at 4 °C to pellet the insoluble fraction.
-
Transfer supernatant (the cytosolic fraction) to a new tube and discard the pellet.
Use supernatant in the next step to extract cytosolic mitochondrial DNA.
-
Use DNeasy Blood & Tissue Kit to purify mitochondrial DNA from the cytosolic fraction according to the manufacturer’s instructions (Ly et al., 2003).
Add 100 µl ethanol (96–100%) to the cytosolic fraction and continue to step 4 in the DNeasy Blood & Tissue Kit protocol.
Notes
This protocol is optimized for using immortalized bone marrow derived macrophages (iBMDM) (Bronner et al., 2015). Protocol can be optimized for chosen experimental cell type.
Thapsigargin serves as a positive control for triggering mitochondrial DNA release. Thapsigargin prevents the uptake of calcium into the endoplasmic reticulum (ER) by blocking SERCA channels. Under these conditions, the ER leaks calcium without being able to replenish its calcium stores, and consequently calcium accumulates in the cytosol or mitochondria. Mitochondrial calcium overload triggers depolarization, leading to the release mitochondrial contents into the cytosol.
Make a 10% NP-40 stock solution and dilute to 1% NP-40.
Use DPBS that contains calcium and magnesium to ensure that cells will remain attached during the wash step.
Prelabeled microcentrifuge tubes do not need to be prechilled.
1 × 106 to 1 × 107 (seeded in 100 mm tissue culture dishes) cells will yield 1–20 µg of mtDNA.
Recipes
-
Medium
DMEM
10% Fetal Bovine Serum (FBS)
Add 50 ml heat-inactivated FBS to 450 ml of DMEM.
Notes:
FBS is heat inactivated for 30 min at 55 °C.
This medium has been optimized for iBMDM. Use medium optimized for experimental cell type.
-
NP-40 (Igepal CA-630) solution
For 10% NP-40, add 1 ml of NP-40 to 9 ml of dH2O.
For 1% NP-40, add 1 ml of 10% NP-40 to 9 ml of dH2O.
-
Thapsigargin stock solution
The concentration stated above has been optimized for iBMDM. The concentration used on different cell types must be optimized.
Stock solution of thapsigargin remains usable up to one year after reconstitution and can be aliquoted and stored at −20 °C
Stock solution of thapsigargin is 5 mM (1 mg in 307 µl DMSO). Dilute thapsigargin (4 µl into 2 ml of medium) into medium that will be added to the wells.
Part II. Amplification of mtDNA via qPCR
After extracting DNA from the cytosolic fraction, quantitative PCR is employed to measure cytosolic mitochondrial DNA.
Materials and Reagents
96 well qPCR plate (Denville Scientific Inc., catalog number: C18096) (Bernardi, 2013)
1.5 ml microcentrifuge tubes (Denville Scientific Inc., catalog number: C2170)
Gloves
Brilliant II SYBR® green with low ROX (Agilent Technologies, catalog number: 600830)
Sterilized double distilled water
Primers (see sequences below)
Equipment
qPCR machine (Stratagene MX3000P)
Procedure
-
Prepare the SYBR qPCR master reaction mix as follows in Table 1 for mitochondrial genes of interest and internal control (housekeeping gene for used here is 18S rDNA) in 1.5 ml microcentrifuge tubes:
The following primers were used:
Cytochrome c oxidase I (mt gene)
Forward: 5’-GCCCCAGATATAGCATTCCC-3’
Reverse: 5’-GTTCATCCTGTTCCTGCTCC-3’
18S rDNA (internal control)
Forward: 5’-TAGAGGGACAAGTGGCGTTC-3’
Reverse: 5’-CGCTGAGCCAGTCAGTGT-3’
Mix gently but thoroughly by pipetting.
Incubate reaction mix on ice until ready to use.
Aliquot DNA samples (1 µl) into wells (run in triplicates) (Bronner et al., 2015).
Aliquot corresponding Master Mix (19 µl) into the corresponding wells (Bronner and O'Riordan, 2014).
Use the thermal cycler program as seen below in Table 2:
Once qPCR is complete, calculate relative fold change in cytochrome c oxidase I from the Ct values.
Table 1.
Master Mix recipe for quantifying mtDNA release into cytosol via qPCR.
| Reagents | Amount (µl) |
|---|---|
| SYBR Green with Low Rox Master Mix | 10 |
| Forward primer | 1 |
| Reverse primer | 1 |
| Sterile dH2O | 7 |
| Total | 19 |
Table 2.
Thermal cycler program for mtDNA release qPCR
| Procedure | # of Cycles | Temperature | Time |
|---|---|---|---|
| Hot start activation | 1 | 95 °C | 10 min |
| Denaturation | 40 | 95 °C | 30 sec |
| Annealing/Extension | 53 °C | 30 sec | |
| Dissociation | 1 | 72 °C | 30 sec |
| 95 °C | 1 min | ||
| 55 °C | 30 sec | ||
| 95 °C | 30 sec |
Representative data
Adapted method for calculating relative fold change (Livak and Schmittgen, 2001) between thapsigargin-treated and untreated Ct values that are represented in Table 3:
Table 3.
Representative values from mtDNA release qPCR
| Untreated | Thapsigargin | |
|---|---|---|
| Cytochrome c oxidase I | 26.16 | 19.15 |
| 18S rDNA | 26.18 | 20.54 |
-
ΔCt Treatment = Target gene - Reference gene
ΔCt Treatment (thapsigargin) = 19.15 – 20.54
ΔCt Control = Target gene - Reference gene
ΔCt Control (untreated) = 26.16 – 26.18
-
ΔΔCt = ΔCt Treatment - ΔCt Control
ΔΔCt = −1.39 – (−0.02)
ΔΔCt = −1.37
-
Relative Fold Change = 2−(ΔΔCt)
Relative Fold Change = 2−(−1.37)
Relative Fold Change = 2.6
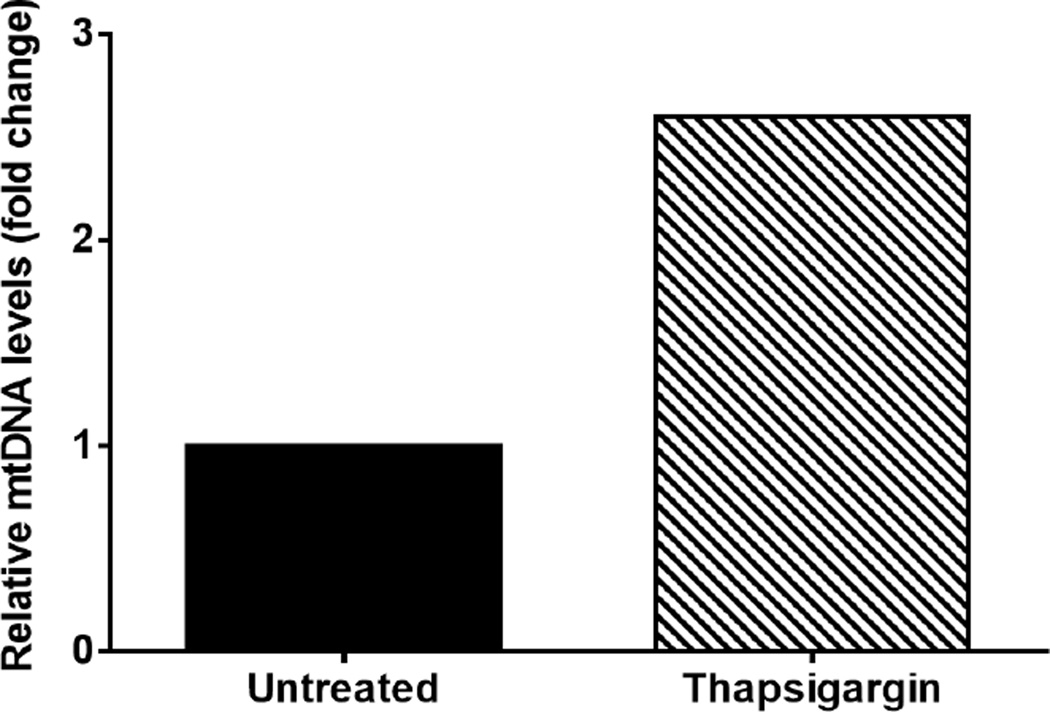
This result indicates that thapsigargin treatment results in a 2.6 fold increase in cytosolic mtDNA compared to untreated (Figure 2).
Figure 2. Graph of representative data from Table 3.
Graph depicts cytosolic mtDNA relative fold change seen in Thapsigargin treated iBMDM when compared to untreated iBMDM.
Notes
Use plates that are optimized for qPCR machine.
-
The total reaction volume is 20 µl. When adding DNA ensure the amount is consistent between samples.
10 ng and 2 µg represent the minimum and maximum amount of DNA to use for qRT-PCR analysis.
Pipette as accurately as possible during steps 4 and 5, since small variations in volumes can increase variability in the qPCR results.
Acknowledgments
We acknowledge financial support from the University of Michigan Rackham Graduate School (D. N. B.), the UM Genetics Training Program (D. N. B., GM007544). This research was supported by funding from the NIH to M. X. D. O. (AI101777). The funders had no role in study design, data collection, and analysis, decision to publish, or preparation of the manuscript.
We acknowledge that this protocol was adapted (Nakahira et al., 2011) and modified for use with immortalized bone marrow macrophages infected by a bacterial pathogen.
References
- 1.Bernardi P. The mitochondrial permeability transition pore: a mystery solved? Front Physiol. 2013;4:95. doi: 10.3389/fphys.2013.00095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bronner DN, Abuaita BH, Chen X, Fitzgerald KA, Nunez G, He Y, Yin XM, O'Riordan MX. Endoplasmic reticulum stress activates the inflammasome via NLRP3- and Caspase-2-driven mitochondrial damage. Immunity. 2015;43(3):451–462. doi: 10.1016/j.immuni.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bronner DN, O'Riordan MX. A near death experience: Shigella manipulates host death machinery to silence innate immunity. EMBO J. 2014;33(19):2137–2139. doi: 10.15252/embj.201489680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burckstummer T, Baumann C, Bluml S, Dixit E, Durnberger G, Jahn H, Planyavsky M, Bilban M, Colinge J, Bennett KL, Superti-Furga G. An orthogonal proteomic-genomic screen identifies AIM2 as a cytoplasmic DNA sensor for the inflammasome. Nat Immunol. 2009;10(3):266–272. doi: 10.1038/ni.1702. [DOI] [PubMed] [Google Scholar]
- 5.Hornung V, Latz E. Intracellular DNA recognition. Nat Rev Immunol. 2010;10(2):123–130. doi: 10.1038/nri2690. [DOI] [PubMed] [Google Scholar]
- 6.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 7.Ly JD, Grubb DR, Lawen A. The mitochondrial membrane potential (deltapsi(m)) in apoptosis; an update. Apoptosis. 2003;8(2):115–128. doi: 10.1023/a:1022945107762. [DOI] [PubMed] [Google Scholar]
- 8.Martinou JC, Green DR. Breaking the mitochondrial barrier. Nat Rev Mol Cell Biol. 2001;2(1):63–67. doi: 10.1038/35048069. [DOI] [PubMed] [Google Scholar]
- 9.Nakahira K, Haspel JA, Rathinam VA, Lee SJ, Dolinay T, Lam HC, Englert JA, Rabinovitch M, Cernadas M, Kim HP, Fitzgerald KA, Ryter SW, Choi AM. Autophagy proteins regulate innate immune responses by inhibiting the release of mitochondrial DNA mediated by the NALP3 inflammasome. Nat Immunol. 2011;12(3):222–230. doi: 10.1038/ni.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tait SW, Green DR. Mitochondria and cell death: outer membrane permeabilization and beyond. Nat Rev Mol Cell Biol. 2010;11(9):621–632. doi: 10.1038/nrm2952. [DOI] [PubMed] [Google Scholar]
- 11.West AP, Shadel GS, Ghosh S. Mitochondria in innate immune responses. Nat Rev Immunol. 2011;11(6):389–402. doi: 10.1038/nri2975. [DOI] [PMC free article] [PubMed] [Google Scholar]