Abstract
Bombesin (Bn) receptor subtype 3(BRS-3) is an orphan G-protein–coupled receptor of the Bn family, which does not bind any natural Bn peptide with high affinity. Receptor knockout studies show that the animals develop diabetes, obesity, altered temperature control, and other central nervous system (CNS)/endocrine/gastrointestinal changes. It is present in CNS, peripheral tissues, and tumors; however, its role in normal physiology/pathophysiology, as well as its receptor localization/pharmacology is largely unknown, in part due to the lack of a convenient, specific, direct radiolabeled ligand. This study was designed to address this problem and to develop and characterize a specific radiolabeled ligand for BRS-3. The peptide antagonist Bantag-1 had >10,000-fold selectivity for human BRS-3 (hBRS-3) over other mammalian Bn receptors (BnRs) [i.e., gastrin-releasing peptide receptor (GRPR) and neuromedin B receptor (NMBR)]. Using iodogen and basic conditions, it was radiolabeled to high specific activity (2200 Ci/mmol) and found to bind with high affinity/specificity to hBRS-3. Binding was saturable, rapid, and reversible. The ligand only interacted with known BRS-3 ligands, and not with other specific GRPR/NMBR ligands or ligands for unrelated receptors. The magnitude of 125I-Bantag-1 binding correlated with BRS-3 mRNA expression and the magnitude of activation of phospholipase C in lung cancer cells, as well as readily identifying BRS-3 in lung cancer cells and normal tissues, allowing the direct assessment of BRS-3 receptor pharmacology/numbers on cells containing BRS-3 with other BnRs, which is usually the case. This circumvents the need for subtraction assays, which are now frequently used to assess BRS-3 indirectly using radiolabeled pan-ligands, which interact with all BnRs.
Introduction
Bombesin (Bn) receptor subtype 3 (BRS-3) is an orphan G-protein–coupled receptor (GPCR) included in the Bn family of receptors (BnRs) because of its high homology (47%–51%) to the mammalian members of this receptor family [i.e., gastrin-releasing peptide (GRP) receptor (GRPR) and neuromedin B (NMB) receptor (NMBR)] (Gorbulev et al., 1992; Fathi et al., 1993; Jensen et al., 2008). BRS-3 is receiving increased attention (Majumdar and Weber, 2012b; Gonzalez et al., 2015b; Xiao and Reitman, 2016) because receptor knockout studies in mice show numerous alterations in both energy metabolism/satiety (Ohki-Hamazaki et al., 1997; Feng et al., 2011; Majumdar and Weber, 2012a; Gonzalez et al., 2015b; Xiao and Reitman, 2016) and glucose/insulin control (Ohki-Hamazaki et al., 1997; Gonzalez et al., 2015b; Xiao and Reitman, 2016). These alterations include the development of obesity, diabetes, hypertension, impaired glucose metabolism, reduced metabolic rates, and altered behavior, including changes in feeding with hyperphagia and decreased movement; and wide expression of hyperleptinemia and altered growth hormone secretion in both the central nervous system (CNS), especially in hypothalamic nuclei, amygdala, and the caudate nucleus, and peripheral tissues, such as the gastrointestinal (GI) tract; is frequently overexpressed in human tumors; and its activation has a growth-promoting effect on tumors, similar to GRPR and NMBR (Fathi et al., 1993; Moody et al., 2003; Sancho et al., 2011; Gonzalez et al., 2015b; Moreno et al., 2016; Yamada et al., 2000a,b).
The exploration of the role of BRS-3 in normal physiology as well as in pathologic processes has been hindered by the fact it remains an orphan receptor, by the lack of availability of selective agonists/antagonists until recently, and by confusion in its pharmacology, including the localization of the receptors in both the CNS and peripheral tissue (Jensen et al., 2008; Gonzalez et al., 2015b; Xiao and Reitman, 2016). The latter problem is due to the lack of a convenient specific BRS-3 ligand that can be used for receptor localization/characterization in different tissues as well as the lack of agreement on the reliability of BRS-3 antibodies, with one recent review (Xiao and Reitman, 2016) concluding that existing BRS-3 receptor antibodies are of uncertain specificity. This is a particular issue with the CNS, allowing the correlation of BRS-3 location with the results of recent studies attempting to define the exact CNS areas responsible for many of the changes in energy homeostasis, food intake, metabolic rate, body temperature control, and body weight (Gonzalez et al., 2015b; Xiao and Reitman, 2016; Maruyama et al., 2017; Xiao et al., 2017; Pinol et al., 2018).
The current study was an attempt to address this latter problem by developing and characterizing a radiolabel ligand that could be readily available; had high selectivity for the BRS-3 receptor over the closely related GRPR/NMBR, which are frequently coexpressed on the same cells (Fathi et al., 1993; Jensen et al., 2008; Moreno et al., 2016); and could be used for BRS-3 pharmacological characterization and localization, both in the CNS and peripheral tissues. In this study, evidence is presented that this was accomplished by preparing iodinated Bantag-1 [Boc-Phe-His-4-amino-5-cyclohexyl-2,4,5-trideoxypentonyl-Leu-(3-dimethylamino)benzyl amide-N-methyl ammonium trifluoroacetate)], a BRS-3 antagonist that had a >10,000 selectivity over GRPR/NMBR for the BRS-3, demonstrating that it had a high degree of saturable binding to BRS-3 with rapid kinetics; interacted only with BRS-3 reported ligands, thus demonstrating high specificity for BRS-3; and could be used to directly assess the presence or number of BRS-3 receptors in normal or tumoral tissues. The availability of this ligand should markedly facilitate the ability to study the presence and pharmacology of this increasing important receptor in both normal physiologic and pathophysiological conditions.
Materials and Methods
Materials
BALB 3T3 (mouse fibroblast) cells; NCI-N417 and NCI-H69 (human small cell lung cancer cells); and NCI-H520, NCI-H460, NCI-H720, and NCI-H727 (human non–small cell lung cancer cells) were purchased from the American Type Culture Collection (Gaithersburg, MD); Dulbecco’s minimum essential medium (DMEM), RPMI 1640, PBS, FBS, Dulbecco’s phosphate-buffered saline, trypsin-EDTA 1×, penicillin streptomycin, and GENETICIN-selective antibiotic (G418 sulfate) were from Invitrogen (Carlsbad, CA); GRP and NMB were from Bachem (Torrance, CA); the Bn analogs [D-Tyr6, β-Ala11, Phe13, Nle14]Bn-(6-14) (Peptide-1) (Mantey et al., 1997; Pradhan et al., 1998; Moreno et al., 2013) and other BnR ligands were obtained from Dr. David H. Coy, Tulane University (New Orleans, LA); Bantag-1, a BRS-3 antagonist (Guan et al., 2010; Feng et al., 2011; Nakamura et al., 2016) was from Sigma-Aldrich (Allentown, PA); MK-5046 ([(2S)-1,1,1-trifluoro-2-[4-(1H-pyrazol-1-yl)phenyl]-3-(4-[[1-(trifluoromethyl)cyclopropyl]methyl]-1H-imidazol-2-yl)propan-2-ol]), a BRS-3 agonist (Sebhat et al., 2010; Moreno et al., 2013), was from MedChi Express (Monmouth Junction, NJ); and iodine-125 radionuclide [125I] (10 mCi) (378 MEq) was from Perkin Elmer Life Science (Boston, MA).
Methods
Stable Transfection.
Human BRS-3 (hBRS-3), human GRPR (hGRPR), and human NMBR (hNMBR) stably transfected into BALB 3T3 cells were grown as described previously (Benya et al., 1992, 1993, 1994, 1995; Ryan et al., 1998a). BALB 3T3 cells were grown overnight at 37°C in 10-cm tissue culture plates at a density of 2 × 106 cells per plate. Twenty-four micrograms of hBRS-3, hGRPR, or hNMBR plasmid DNA was transfected into the BALB 3T3 cells by the cationic lipid-mediated method using 60 µl of LipofectAMINE 2000 in OPTI-MEM 1–reduced serum medium for 3 hours at 37°C. At the end of the incubation period, the medium was replaced with regular growth medium. Three days after transfection, cells were split into very dilute concentrations, and 800 mg/l selection antibiotic G418 (Geneticin, Thermo Fisher Scientific) was added to the regular growth medium. In about 2 weeks, single colonies were selected and expanded. The best clones, as determined by binding, were further expanded, frozen, and used for our experiments.
Cell Culture.
BALB 3T3 cells stably expressing hGRPR, hNMBR, and hBRS-3 were grown in DMEM supplemented with supplemented 10% FBS and 300 mg/l G418 sulfate. Lung cancer cells NCI-N417, H69, H520, H460, and H727, which natively express BnRs including BRS-3 (Moreno et al., 2018), were grown in RPMI 1640 supplemented with 10% FBS, and NCI-H720 was grown in DMEM/F12 Medium supplemented with 5% FBS. All the cells were grown in their respective propagation medium supplemented with supplemented 10% FBS and incubated at 37°C in a 5% CO2 atmosphere.
Preparation of 125I-Labeled Peptides.
125I-Bantag-1, with specific activity of 2200 Ci/mmol, was prepared by a modification of methods described previously (Jensen et al., 1983; von Schrenck et al., 1990; Mantey et al., 1993, 1997). Briefly, 0.8 µg of IODO-GEN (in 0.02 µg/ml chloroform) was transferred to a 5-ml glass test tube, dried under a stream of nitrogen, and washed with 100 µl of 0.5 M KH2PO4 (pH 7.4). To the reaction tube, 20 µl of 0.5 M KH2PO4 (pH 8.0), 10 µg of peptide in 5 µl of water, and 2 mCi (20 µl) Na 125I were added, mixed gently, and incubated at room temperature for 6 minutes. The incubation was stopped by the addition of 300 µl of distilled water. The free 125I was separated by applying the reaction mixture onto a Sep-Pak and washing with 5 ml of distilled water and 5 ml of 0.1% trifluoroacetic acid. The radiolabeled peptide was eluted with 60% acetonitrile in 0.1% trifluoroacetic acid (10 × 200 µl sequential elutions). The radioligand was purified by taking the two peaks with the highest radioactivity and applying them to a reverse-phase high-performance liquid chromatography using a μBondapak column (0.46 × 25 cm) from Waters. The column was eluted with a linear gradient of acetonitrile in 0.1% trifluoroacetic acid from 16% to 64% acetonitrile in 60 minutes, with a flow of 1.0 ml/min, and collecting 1-ml fractions. Samples of the fractions were counted, and the highest peaks were tested for radioligand binding to hBRS-3–transfected cells (Ryan et al., 1998a; Moreno et al., 2013). The best binding fractions were stored with 0.5% BSA at −20°C. 125I-Peptide-1 (125I-[D-Tyr6, β-Ala11, Phe13, Nle14] Bn-(6-14)), with a specific activity of 2200 Ci/mmol, was prepared as described previously (Mantey et al., 1997; Moreno et al., 2013; Nakamura et al., 2016).
Binding Studies.
hBRS-3/BALB (1 × 106 cells/ml), hGRPR/BALB (0.5 × 106 cells/ml), hNMBR/BALB (0.05 × 106 cells/ml), NCI-N417 (3 × 107 cells/ml), NCI-H69 (6 × 106 cells/ml), NCI-H460 (6 × 106 cells/ml), NCI-H520 (6 × 106 cells/ml), NCI-H720 (3 × 106 cells/ml), NCI-H727 (4 × 106 cells/ml) cells were incubated for 40–60 minutes at 22°C with 125I-labeled ligand and with or without ligand in 300 µl of binding buffer, as described previously (Pradhan et al., 1998; Ryan et al., 1998a,b; Moreno et al., 2013). The standard binding buffer contained 24.5 mM HEPES (pH 7.4), 98 mM NaCl, 6 mM KCl, 2.5 mM KH2PO4, 5 mM sodium pyruvate, 5 mM sodium fumarate, 5 mM sodium glutamate, 2 mM glutamine, 11.5 mM glucose, 0.5 mM CaCl2, 1.0 mM MgCl2, 0.01% (w/v) soybean trypsin inhibitor, 0.2% (v/v) amino acid mixture, 1% (w/v) BSA, and 0.05% (w/v) bacitracin. After the incubation, 100 µl of each sample was removed and added to 300 µl of washing buffer 2% (w/v) BSA in PBS in 400-µl microfuge tubes and centrifuged for 1 minute at 10,000g (Microfuge E; Beckman, Fullerton, CA) to separate the bound radioligand from the unbound radioligand. The supernatant was aspirated, and the pelleted cells were rinsed twice with a washing buffer. The amount of radioactivity bound to the cells was measured in a Wizard 1470 Automatic Gamma Counter (Packard Instruments, Meriden, CT). Total binding was expressed as the percentage of total radioactivity that was associated with the cell pellet. Nonsaturable binding was <15% of the total binding in all experiments. Each point was measured in duplicate, and each experiment was replicated at least four times. The calculation of affinity was performed by determining the IC50 using the curve-fitting program GraphPad Prism 4.0 (GraphPad Software, La Jolla, CA).
Preparation of Membranes from Various Tissues.
Cell membranes were prepared as described previously (Mantey et al., 1993; Tsuda et al., 1997), using a homogenizing buffer containing 50 nM Tris (pH 7.4), 0.2 mg/ml soybean inhibitor, 0.1% bacitracin, and 0.2 mg/ml benzamidine. A concentration of 1 × 107 cell/ml lung cancer cells or rat tissues (cerebellum, cortex, mid-brain, testis, duodenum) were homogenized for 30 seconds with a polytron (Brinkmann Instruments, Inc.) at speed 6 and were centrifuged at 1500 rpm for 10 minutes. The supernatant was removed and recentrifuged at 15,000 rpm to pellet the membranes. The membranes were suspended in binding buffer and stored at −70°C until used.
Binding of 125I-Peptide-1 and 125I-Bantag-1 to Membranes.
The standard membrane-binding buffer contained 10 mM HEPES (pH 7.4, 118 mM NaCl, 4.7 mM KCl, 5 mM MgCl2, 1 mM EGTA, 0.2 mg/ml benzamidine, 0.2 mg/ml soybean trypsin inhibitor, 0.1% bacitracin, and 0.2% (w/v) BSA. Binding was as stated above with cells, except that membranes were diluted with binding buffer to a concentration equivalent to 6 × 106 cells/ml and that after incubation membranes were spun through 4% BSA for 2 minutes to pellet and wash.
Dissociation of Bound Ligand from hBRS-3/BALB Membranes and Cells.
Dissociation studies were performed in both hBRS-3 BALB cells and membranes. Membranes were incubated with 50 pM radioligand (i.e., 125I-Bantag-1 or 125I-Peptide-1) at room temperature for 60 minutes with or without 1 μM peptide-1 in binding buffer as described above. hBRS-3/BALB (1 × 106 cells/ml) cells were incubated for 60 minutes at 37°C with 50 pM 125I-labeled ligand (125I-Bantag-1 or 125I-Peptide-1) and with or without 1 μM peptide-1 in binding buffer, as described above. Then a final concentration of 1 μM peptide-1 was added, and the membranes or cells were incubated at room temperature or 37°C, respectively. At various times, 100-µl aliquots were taken and spun through 4% BSA for 2 minutes to pellet. The pellet was washed, and the amount of radioactivity saturably bound was determined with a gamma counter.
Internalization of 125I-Peptide-1 and 125I-Bantag-1.
Internalization of ligands was determined as described previously (Benya et al., 1993; Mantey et al., 1993). Briefly, after incubation of hBRS-3/BALB cells with 50 pM 125I-Peptide-1 or 125I-Bantag-1 for the indicated times and temperatures, samples (100 µl) were added to acid-stripping buffer [0.2 M acetic acid with 0.5 M NaCl (pH 2.5)] to remove the surface-bound radioligand or were added to standard washing buffer to assess total saturable bounding. After 5 minutes, at 4°C, the cells were centrifuged and washed two times with washing buffer, and the amount of saturable radioactivity left was determined on the gamma counter.
Measurement of [3H]-Inositol Phosphates.
For lung cancer cells not previously reported (Moreno et al., 2018) (H838,A549,H1299), the ability of stimulation of BRS-3 to activate phospholipase C resulting in [3H]-inositol phosphate (IP) production was determined as described previously (Rowley et al., 1990; Moreno et al., 2018). Briefly, all adherent cells were subcultured into 24-well plates (5 × 105 cells/ml) in regular propagation media and then incubated for 24 hours at 37°C in a 5% CO2 atmosphere. Then, the cells were incubated with 3 μCi/ml myo-[2-3H] inositol in growth media supplemented with 2% FBS for an additional 24 hours. After the incubation, the 24-well plates were washed by incubating for 30 minutes at 37°C with 1 ml/well PBS (pH 7.0) containing 20 mM lithium chloride. Cells that grew in suspension were centrifuged to removed RPMI 1640 medium and incubated (2 × 106cell/ml) directly with myo-[2-3H] inositol in RPMI 1640 with 2% FBS for 24 hours (Moreno et al., 2018). The cells were then centrifuged to remove the myo-[2-3H]-inositol, washed with wash buffer containing 20 mM LiCl for 10 minutes at 4°C, recentrifuged to removed wash buffer, and distributed to 5-ml tubes, where they were incubated without (control) or with peptides in 500 μl of IP assay buffer for 60 minutes at 37°C. In both types of cells, the incubation was terminated by the addition of 1 ml of ice-cold 1% (v/v) hydrochloric acid in methanol. Total [3H]IP was isolated by anion exchange chromatography, as described previously (Rowley et al., 1990; Ryan et al., 1998a; Uehara et al., 2011; Moreno et al., 2018).
Cytosolic Ca2+
For lung cancer cells not previously reported (Moreno et al., 2018) (H838, A549, H1299), the ability of stimulation of BRS-3 to stimulate cytosolic Ca2+ was investigated with cells with adherent growth treated with trypsin-EDTA and harvested (2.5 × 106cells/ml). After centrifugation, the cells were resuspended in SIT-medium (RPMI-1640 containing 3 × 10−8 M sodium selenite, 5 μg/ml insulin, and 10 μg/ml transferrin) (Ryan et al., 1998a,b; Moreno et al., 2018) containing 1 μM Fura-2/acetoxymethyl ester (Calbiochem, La Jolla, CA) at 37°C for 30 minutes. Subsequently, the cells were centrifuged at 1000g for 5 minutes, resuspended, and changed in cytosolic Ca2+, assessed as described previously (Moreno et al., 2018).
Statistical Analysis.
The results are the mean and S.E.M. from at least three separate experiments. The Ki (kinetic constant) values were calculated from the IC50 values obtained from competitive inhibition binding curves using different Bn analogs and antagonists (50 pM 125I-Peptide-1 or 50 pM 125I-Bantag-1) using the equation of Cheng and Prusoff (1973). IC50 values from the binding data were obtained by curve fitting using GraphPad Prism 7.0 (nonlinear curve fitting). An analysis of variance was used to determine the statistical significance of differences in the affinity of each Bn agonist/antagonist with Bantag-1 with changes showing a ≥2-fold difference. In all experiments, cell concentrations were set such that <15% of total radioactivity was bound and the amount of saturable ligand bound was similar for all compounds, and, therefore, in the statistical analysis only two variables (i.e., each Bn agonist/antagonist and its own control, binding a similar amount of ligand) were analyzed. Correlation coefficients were calculated using GraphPad Prism 7.0.
Results
Time Course of Association and Dissociation of Binding of the Radiolabeled Nonselective Bn Analog Agonist (125I-Peptide-1) and the Radiolabeled BRS-3–Selective Peptide Antagonist 125I-Bantag-1
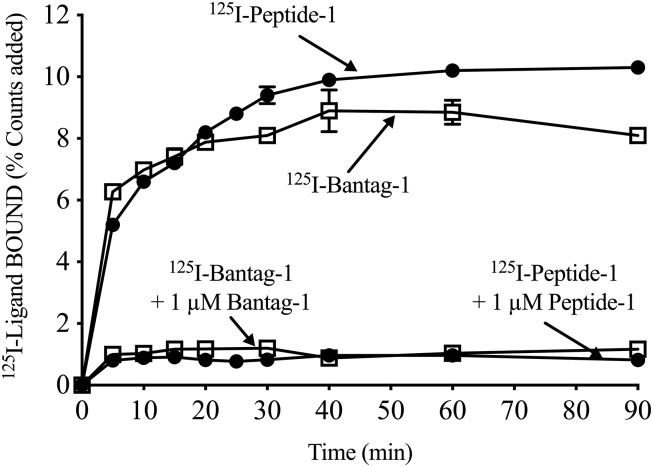
The binding association of both radioligands 125I-Peptide-1 and 125I-Bantag-1 to hBRS-3 BALB 3T3 cells was time dependent, with a rapid increase, reaching a maximal increase after 30 minutes (125I-Peptide-1: 9.24% ± 0.27% of control; 125I-Bantag-1: 8.10% ± 0.23% of control) (Fig. 1) and was maintained for up to 90 minutes (125I-Peptide-1: 10.30% ± 0.05% of control; 125I-Bantag-1: 8.10% ± 0.10% of control) (Fig. 1). Although both radioligands bound well to hBRS-3/BALB cells, the universal ligand 125I-Peptide-1 demonstrated 12% greater steady-state binding than was seen with the peptide antagonist 125I-Bantag-1 (Fig. 1). When each radioligand was incubated with a saturating concentration of Peptide-1 or Bantag-1, the total amount of binding was reduced by 91% and 85%, respectively (At 30 minutes: 125I-Peptide-1 + 1 µM Peptide-1: 0.83% ± 0.05% of control; 125I-Bantag-1 + 1 µM Bantag-1: 1.20% ± 0.11% of control (Fig. 1).
Fig. 1.
Time course of binding of the radiolabeled nonselective Bn analog agonist (Peptide-1) and the BRS-3 peptide antagonist Bantag-1. BALB 3T3 cells transfected with hBRS-3 were incubated with 50 pM 125I-Bantag-1 or 125I-Peptide-1 alone or with 1 µM unlabeled ligand at 22°C for the indicated times. Results are expressed as the percentage of the added counts bound. In each experiment, each value was determined in duplicate, and the results given are from at least four separate experiments.
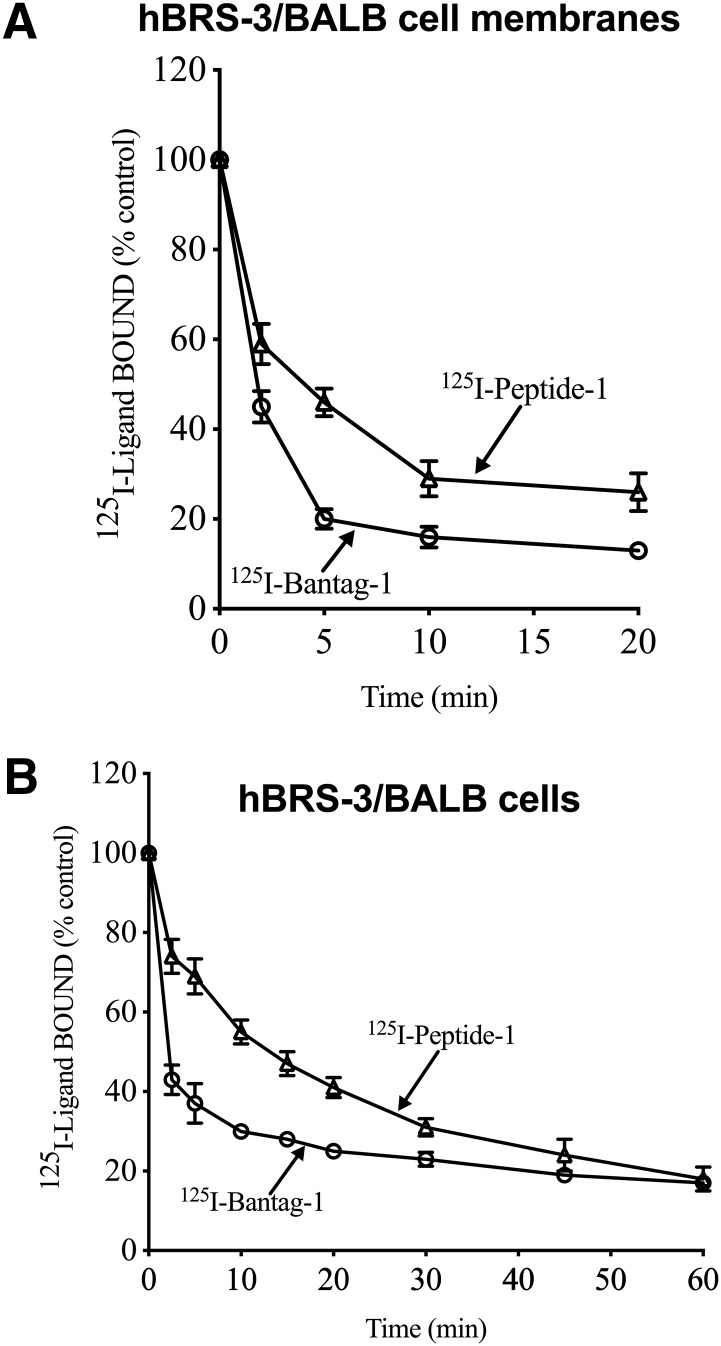
The time course of dissociation for each ligand was similar for membranes and cells. With both membranes and cells, 125I-Bantag-1 binding dissociated more rapidly than 125I-Peptide-1 binding with 70%–80% of 125I-Bantag-1 dissociated by 10 minutes, whereas only 40%–50% of 125I-Peptide-1 was dissociated by this time (Fig. 2).
Fig. 2.
Time course of dissociation of the radiolabeled nonselective Bn analog agonist (Peptide-1) and the BRS-3 peptide antagonist Bantag-1 from hBRS-3/BALB 3T3 cell membranes (top) or intact cells (bottom). hBRS-3/BALB (1 × 106 cells/ml) were incubated for 60 minutes at 37°C with 50 pM 125I-labeled ligand (125I-Bantag-1 or 125I-Peptide-1) and with or without unlabeled 1 μM peptide-1 in binding buffer. Membranes were incubated with 50 pM radioligand (i.e., 125I-Bantag-1 or 125I-Peptide-1) at room temperature for 60 minutes with or without unlabeled 1 μM peptide-1 ligand in binding buffer. Then a final concentration of 1 μM peptide-1 was added, and the membranes or cells were incubated at room temperature or 37°C, respectively. At various times, 100-µl aliquots were taken and saturable binding determined as described in Methods. In each experiment, each value was determined in duplicate, and results are from at least four separate experiments.
Ability of Peptide-1 or Bantag-1 to Interact with BRS-3s, GRPRs, or NMBRs
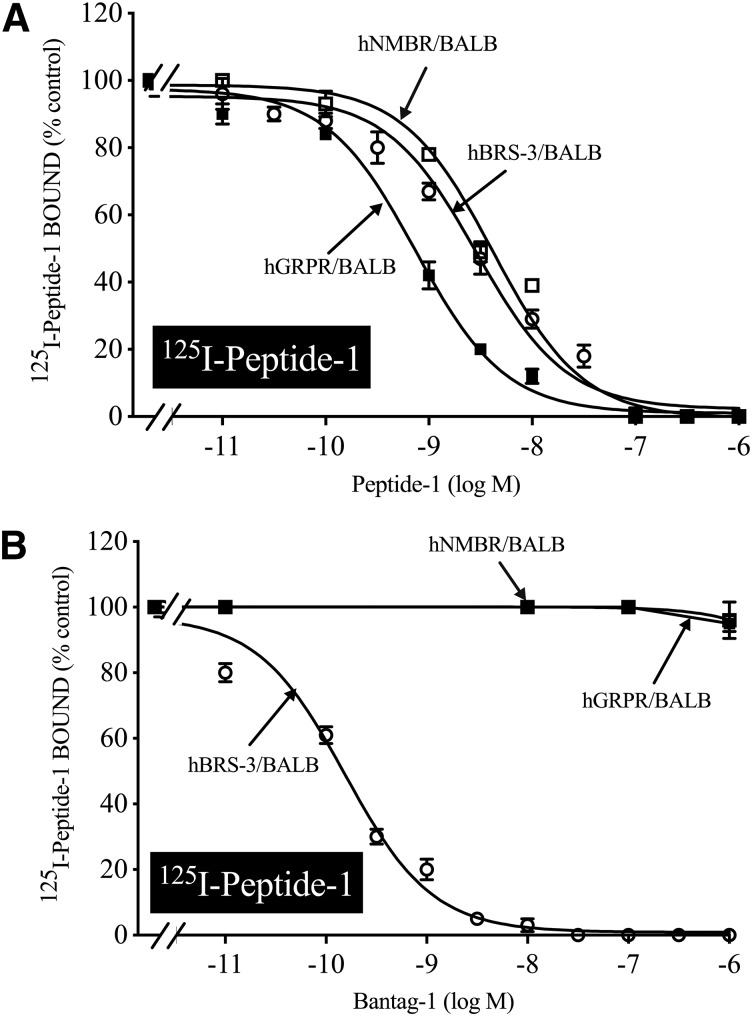
In separate BALB 3T3 cells containing hBRS-3, hGRPR, or hNMBR, 125I-Peptide-1 bound to all three receptors with high affinity and the universal ligand Peptide-1 had a high affinity for the three BnRs (Ki: 1.88, 0.67, and 3.66 nM, respectively) (Fig. 3A; Table 1), similar to reported previously (Mantey et al., 1997; Pradhan et al., 1998; Sancho et al., 2010). However, the peptide antagonist Bantag-1 only had high affinity for hBRS-3/BALB (Ki, 0.72 nM) (Fig. 3B; Table 1) and did not interact with hGRPR/BALB or hNMBR/BALB, even at concentrations of 3000 nM (Ki, >10,000 nM) (Fig. 3B; Table 1). These results demonstrate that Peptide-1 binds with high affinity with all three human BnRs and is therefore nonselective, whereas Bantag-1 has >10,000-fold higher affinity for BRS-3 and, under these experimental conditions, only interacts with hBRS-3.
Fig. 3.
The ability of the nonselective Bn analog agonist (Peptide-1) (A) and the BRS-3–selective peptide antagonist Bantag-1 (B) to inhibit the binding of either ligand to cells containing BRS-3, GRPR, or NMBR. BALB 3T3 cells stably transfected with hBRS-3 (1 × 106 cells/ml), hGRPR (0.5 × 106 cells/ml), or hNMBR (0.05 × 106 cell/ml) were incubated with 50 pM 125I-Peptide-1 for 60 minutes at 22°C, and the saturable binding was determined as described in Materials and Methods. The results are expressed as the percentage of saturable binding without unlabeled peptide added (percentage control). The results are the mean and S.E.M. from at least three separate experiments, and in each experiment the data points were determined in duplicate. BALB, BALB 3T3 cells. For other abbreviations, see the Fig. 1 legend.
TABLE 1.
Comparison of the affinities of the peptide agonist Peptide-1 and the BRS-3 peptide antagonist Bantag-1 for members of the Bn receptor family (hBRS-3, hGRPR, and hNMBR)
Transfected cells hBRS-3 (1 × 106 cells/ml), hNMBR (0.05 × 106 cells/ml), or hGRPR (0.5 × 106 cells/ml) were incubated with 50 pM 125I-Peptide-1 for 60 minutes at 22°C. In each experiment, each value was determined in duplicate, and values given are the mean and S.E.M. from at least three separate experiments. Data are calculated from dose-inhibition curves shown in Fig. 1 for each peptide using a nonlinear regression curve-fitting program (Prism). Each value is the mean ± S.E.M. from at least three experiments.
| Ligand |
Ki
|
||
|---|---|---|---|
| hBRS-3 | hGRPR | hNMBR | |
| nM | |||
| Peptide-1 | 1.58 ± 0.09 | 0.67 ± 0.04 | 3.66 ± 0.02 |
| Bantag-1 | 0.72 ± 0.01 | >10,000 | >10,000 |
Comparison of the Ability of the Radiolabeled BRS-3 Antagonist, 125I-Bantag-1, and the Radiolabeled Nonselective Agonist 125I-Peptide-1 to Bind with Native BALB 3T3 Cells or BALB 3T3 Cells Stably Transfected with hBRS-3, hGRPR, or hNMBR
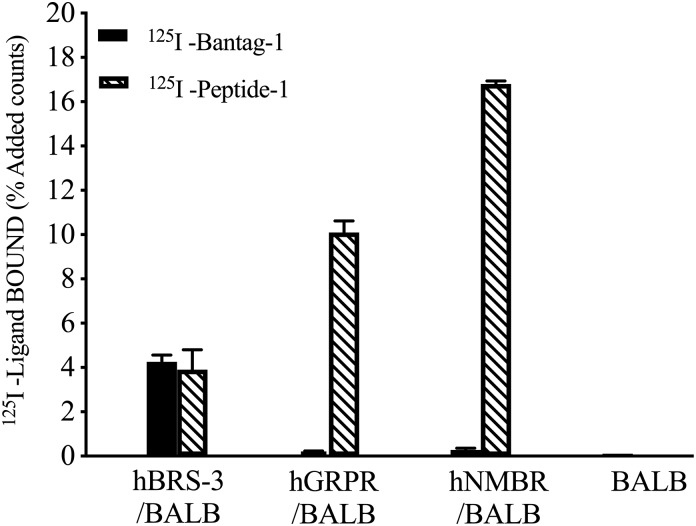
To demonstrate directly by binding studies that, similar to unlabeled Peptide-1 and Bantag-1, the radiolabel agonist 125I-Peptide-1 and the radiolabel antagonist 125I-Bantag-1, also differed in their abilities to interact with cells containing the different BnRs [hBRS-3/BALB, hGRPR/BALB and hNMBR/BALB, and BALB 3T3 cells], we incubated under similar conditions both radiolabels with the different cells. The agonist 125I-Peptide-1 and the antagonist 125I-Bantag-1 bound to similar extent to hBRS-3/BALB cells (3.9% and 4.3% added counts, respectively) (Fig. 4). However, only 125I-Peptide-1 bound to GRPR or NMBR (Fig. 4). In contrast neither ligand bound to native BALB 3T3 cells, which do not contain BnRs (Fig. 4).
Fig. 4.
Comparison of the ability of the radiolabeled BRS-3 antagonist 125I-Bantag and the radiolabeled agonist 125I-Peptide-1 to bind to native BALB 3T3 cells or BALB 3T3 cells stably transfected with hBRS-3, hGRPR, or hNMBR. Cells were incubated with 50 pM 125I-Peptide-1 or 50 pM 125I-Bantag-1 for 60 minutes at 22°C, and the saturable binding was determined as described in Materials and Methods. The results are expressed as the percentage of added saturable counts bound. Cell concentrations were the same as listed in Fig. 3, with 1 × 106 cells/ml BALB cells. The results are the mean and S.E.M. from at least three separate experiments and in each experiment the data points were determined in duplicate. For abbreviations, see the legends of Figs. 1 and 3.
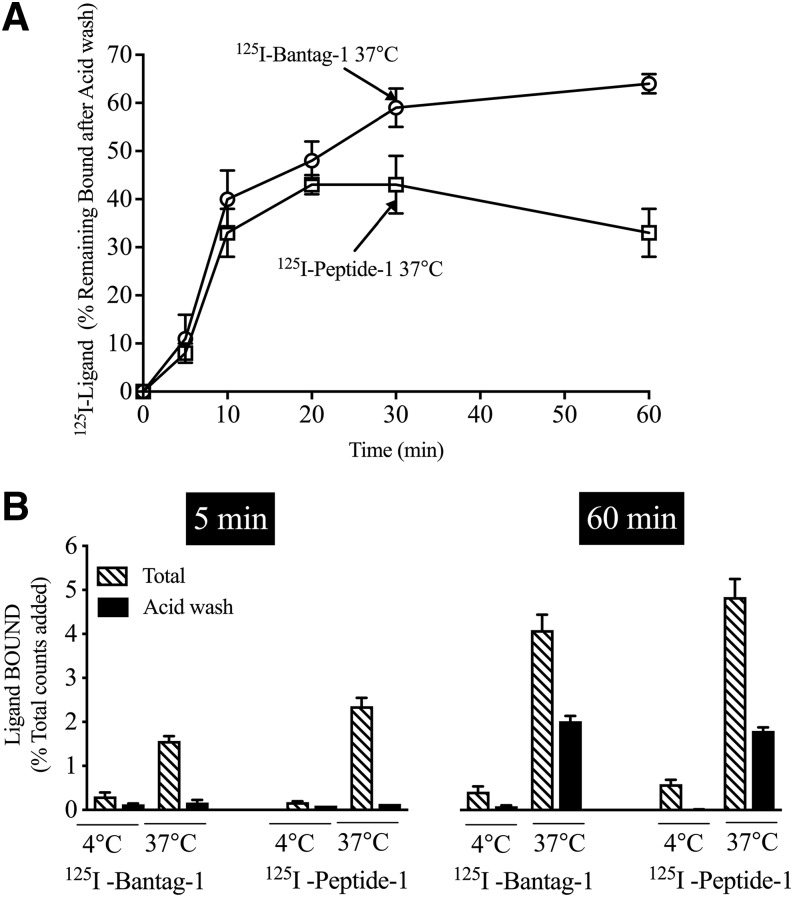
Time Course and Temperature Dependence of Internalization and the Radiolabeled Antagonist 125I-Bantag-1 or Agonist 125I-Peptide 1 by hBRS-3/BALB 3T3 Cells
Previous studies demonstrate that BRS-3, as well as other BnRs, can internalized ligands (Benya et al., 1992, 1993, 1994, 1995; Kumagai et al., 2015). To assess the internalization of each ligand, we performed acid-stripping studies, which have been shown to remove surface-bound ligand (Benya et al., 1992, 1993, 1994, 1995; Kumagai et al., 2015). We found that both 125I-Bantag-1 and 125I-Peptide-1 were internalized in a time-dependent manner, with a significant increase after 10 minutes (40% and 33%, respectively) (Fig. 5A), and maximum internalization occurred after 30 minutes (59% and 43%, respectively) (Fig. 5A). The increase in internalization of 125I-Bantag-1 was maintained in time after 60 minutes (64%) (Fig. 5A). However, the amount of 125I-Peptide-1 internalization decreased slightly over time but was still present after 60 minutes (33%) (Fig. 5A). The internalization of 125I-Bantag-1 and 125I-Peptide-1 was also temperature dependent. At 4°C, no internalization was seen with either of the two radioligands (Fig. 5B). However, both were internalized at 60 minutes at 37°C (Fig. 5B).
Fig. 5.
Comparison of the time course and extent of internalization of the radiolabeled antagonist 125I-Bantag-1 or radiolabeled agonist,125I-Peptide-1 with hBRS-3/BALB 3T3 cells. hBRS-3 stably transfected (top/bottom panels) into BALB 3T3 cells (1 × 106 cells/ml) were incubated with 50 pM 125I-Bantag-1 or 50 pM 125I-Peptide-1 at 37°C or 4°C (bottom panel) for the indicated times. The internalization of bound ligand was determined by assessing the effect of acid-stripping medium (0.2 M acetic acid and 0.5 M NaCl, pH 2.5), which removes surface-bound ligand. “Binding times” refers to the time the cells were incubated with the radioligand before acid stripping. Results are expressed as the percentage of total bound ligand remaining after acid stripping (percentage internalized) and are the mean and S.E.M. from at least three separate experiments, and in each experiment the data points were determined in duplicate. (Top panel) The percentage of total ligand bound that is internalized at the different incubation times is shown. (Bottom panel) Both the total saturable binding and the percentage remaining after acid wash (percentage internalized) are shown after incubation at 4°C or 37°C for the indicated times. For abbreviations, see the Fig. 1 legend.
Specificity of Binding of 125I-Bantag-1 to hBRS-3 Receptor
To assess the specificity of the binding of 125I-Bantag-1 to hBRS-3 receptor, the ability of various ligands that interact preferentially with BRS-3 (Bantag-1, MK-5046) (Sebhat et al., 2010; Moreno et al., 2013), only the other Bn receptors [selective GRPR antagonists [Tyr4,D-Phe12]Bn; [[Leu13, ψ(CH2NH), Leu14]Bn; [D-Phe6, Leu13,Ψ(CH2NH), Cpa14]Bn-(6-14) D-Phe6]Bn-(6-13)methyl ester; D-Phe6]Bn-(6-13)ethyl ester); selective NMBR antagonist PD168368 (PD), selective GRPR (GRP), or NMBR (NMB) agonists] (Heinz-Erian et al., 1987; Coy et al., 1988, 1989; Wang et al., 1990a,b; Ryan et al., 1999; Gonzalez et al., 2009; Uehara et al., 2011; Ramos-Álvarez et al., 2015), ligands of a Drosophila receptor related to mammalian BRS-3 (i.e., CCHamides) (Matsufuji et al., 2014, 2015) or ligands interacting with receptors different from the Bn receptor family, were tested (Tables 2 and 3). At concentrations that cause a maximal effect at their receptors, none of these agents inhibited the binding of 125I-Bantag-1 to hBRS-3–transfected BALB 3T3 cells except for MK-5046, which is a known BRS-3 agonist (Sebhat et al., 2010; Moreno et al., 2013). These results demonstrate the specificity of 125I-Bantag-1 binding.
TABLE 2.
Ability of various agents to alter binding of 125I-Bantag-1 to hBRS-3 receptor
hBRS-3 stably transfected Balb 3T3 cells (1 × 106 cells/ml) were incubated with 50 pM 125I-Bantag-1 either alone or with the indicated concentration of the unlabeled peptide listed. Results are expressed as the percentage of the saturable binding with no unlabeled peptide. Results are the mean ± S.E.M. from at least three experiments, and in each experiment each point was determined in duplicate.
| Agent Added | 125I-Bantag-1 Bound |
|---|---|
| % Control | |
| Peptide-1 (10 µM) | 8.50 ± 1.20 |
| Bantag-1 (0.01 nM) | 80.0 ± 4.1 |
| Bantag-1 (10 µM) | 0 |
| MK-5046 (10 µM) | 62.5 ± 3.4 |
| Bn (10 µM) | 96.0 ± 9.2 |
| CCH Amide I (10 µM) | 87.0 ± 4.3 |
| CCH Amide II (10 µM) | 99.0 ± 5.2 |
| Substance P (10 µM) | 93.0 ± 4.0 |
| CCK-8 (10 µM) | 100.0 ± 3.2 |
| Gastrin-1 (10 µM) | 94.0 ± 2.6 |
| Somatostatin-14 (10 µM) | 100.0 ± 3.3 |
| [Tyr4, D-Phe12]Bn (1 µM) | 98 ± 3.8 |
| [Leu13,Ψ(CH2NH), Leu14]Bn-(6-14) (1 µM) | 100.0 ± 4.1 |
| [D-Phe6, Leu13,Ψ(CH2NH), Cpa14]Bn-(6-14) (10 µM) | 100.0 ± 2.9 |
| [D-Phe6]Bn-(6-13)methyl ester (1 µM) | 96.3 ± 1.2 |
| [D-Phe6]Bn-(6-13)ethyl ester (1 µM) | 100.0 ± 4.2 |
| PD-1676252 (1 µM) | 99.1 ± 3.0 |
CCH, carbamylcholine; CCK, cholecystokin.
TABLE 3.
Comparison of affinities of various BnR agonists and antagonists for hBRS-3 containing cells determined from 125I-Peptide-1 or 125I-Bantag-1 binding
hBRS-3 stably transfected into BALB 3T3 cells (1 × 106 cells/ml) (Ryan et al., 1998b; Sancho et al., 2010) were incubated with 50 pM 125I-Peptide-1 or 125I-Bantag-1 for 60 minutes at 22°C. In each experiment, each value was determined in duplicate, and values given are the mean and S.E.M. from at least three separate experiments. Data are calculated from dose-inhibition curves shown in Fig. 5 for each peptide using a nonlinear regression curve-fitting program (Prism). Each value is the mean ± S.E.M. from at least three experiments. Bantag-1 was described in Moreno et al. (2013), Guan et al. (2010), Feng et al. (2011); #1 in Mantey et al. (1997); #2 in Mantey et al. (1997); #6 in Mantey et al. (2004); #7 in Mantey et al. (2004); #15 in Mantey et al. (2001); #34 in Boyle et al. (2005); #54 in Weber et al. (2002, 2003); and #68 in Weber et al. (2002, 2003).
| Peptide No. | Peptide Structure |
Ki
|
|
|---|---|---|---|
| 125I-Peptide-1 | 125I-Bantag-1 | ||
| nM | |||
| Bantag-1 | Boc-Phe-His-4-amino-5-cyclohexyl-2,4,5-trideoxypentonyl-Leu-(3-dimethylamino) benzylamide N-methylammonium trifluoroacetate | 0.40 ± 0.25 | 0.27 ± 0.01 |
| 1 | [D-Tyr6, β-Ala11, Phe13, Nle14]Bn-(6-14) | 1.58 ± 0.09 | 0.68 ± 0.02 |
| 2 | [D-Phe6, β-Ala11, Phe13, Nle14]Bn-(6-14) | 5.50 ± 0.29 | 7.94 ± 0.38 |
| 6 | [D-Tyr6(S), Apa11-4Cl, Phe13, Nle14]Bn-(6-14) | 110 ± 6 | 31.6 ± 0.6 |
| 7 | [D-Tyr6(R), Apa11-4Cl, Phe13, Nle14]Bn-(6-14) | 2.52 ± 0.13 | 2.04 ± 0.06 |
| 15 | [D-Tyr6(S), Apa11, Phe13, Nle14]Bn-(6-14) | 20.0 ± 1.1 | 31.5 ± 1.1 |
| 34 | Ac-Phe, Trp, Ala, His (t-Bzl), Nip, Gly,Arg-NH2 | 162 ± 8 | 74 ± 3 |
| 54 | H-D-Phe, Gln, D-Trp, Phe-NH2 | >1000 | >1000 |
| 68 | 3-phenyl-propionyl-Ala, D-Trp, NH (CH2) C6H5 | >1000 | 467 ± 11 |
| NMB | Neuromedin B | >1000 | >1000 |
| GRP | Gastrin-releasing peptide | >1000 | >1000 |
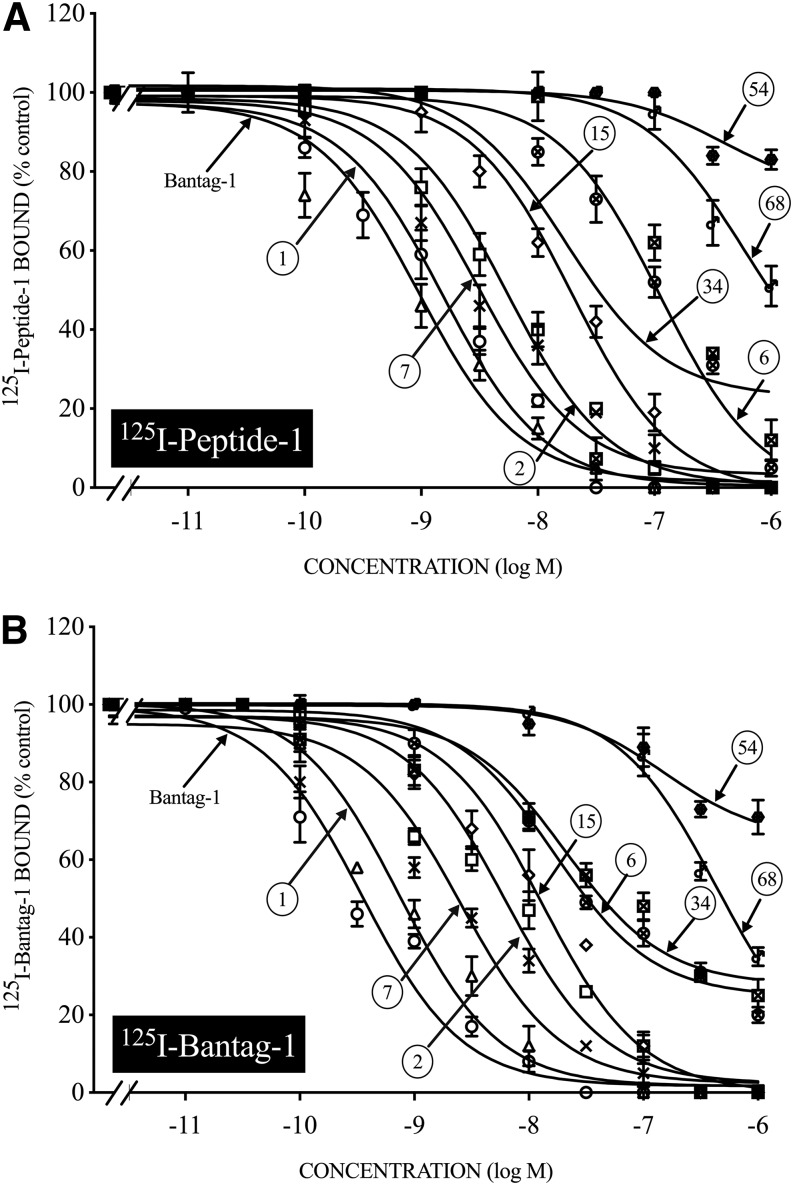
Ability of Various BRS-3 Ligands to Inhibit Binding of 125I-Peptide-1 or 125I-Bantag-1 to hBRS-3/BALB Cells
A number of various selective BRS-3 ligands have been reported recently in various studies using both functional assays and/or various binding studies with 125I-Peptide-1 (Mantey et al., 1997, 2001, 2004; Weber et al., 2002, 2003). To compare the results of the ability of these ligands to interact with BRS-3 receptors directly assessed by binding studies using 125I-Peptide-1 or 125I-Bantag-1 as the radioligand, a number were compared under identical binding conditions (Fig. 6; Table 3). In general, there was a very close comparison between the affinity of each ligand for the hBRS-3 receptor assessed by the two different radioligands (Fig. 5; Table 3). Specifically, Bantag-1 had the highest affinity (0.27–0.40 nM) (Fig. 6; Table 3) for hBRS-3 expressed in BALB 3T3 cells with both radioligands. Peptide-1 had a 2-fold to 4-fold lower affinity than Bantag-1 (Fig. 6A; Table 3), and Peptide-7 had a 7-fold lower affinity than Bantag-1 (Ki, 2.0–2.5 nM) (Fig. 6; Table 3). Peptide-2, which differed from peptide 1 in having a D-Phe6 substitution for D-Tyr6, had a 13-fold to 29-fold decrease in affinity compared with Bantag-1 and Peptide-6, with a similar structure to Peptide-7 but with an (S) configuration, was 28-fold to 117-fold lower than Bantag-1 for both radioligands (Fig. 6A; Table 3). Peptide-15, which differed from Peptide-6 by the removal of a 4-Cl group on Apa11, had a 50-fold to 118-fold lower affinity than Bantag-1 with both radioligands (Ki, 20–31.5 nM) (Fig. 6A; Table 3). The remaining peptides (peptides 34, 54, and 68) had little to no affinity for BRS-3 (>400 nM) assessed by both radiolabeled ligands, as did the selective GRPR agonist GRP or the selective NMBR agonist NMB (Fig. 6; Table 3).
Fig. 6.
The ability of various Bn receptor ligands to inhibit binding of the radiolabeled agonist 125I-Peptide-1 (A) or the radiolabeled antagonist 125I-Bantag-1 (B) to hBRS-3/BALB (1 × 106 cells/ml). Cells were incubated with 50 pM 125I-Peptide-1 or 125I-Bantag-1 with or without increasing the concentrations of various unlabeled Bn receptor ligands for 60 minutes at 22°C. Results are expressed as the percentage of saturable binding with no addition. The results are the mean and S.E.M. from at least three separate experiments, and in each experiment the data points were determined in duplicate. For abbreviations, see the legends of Figs. 1–3 and Table 3.
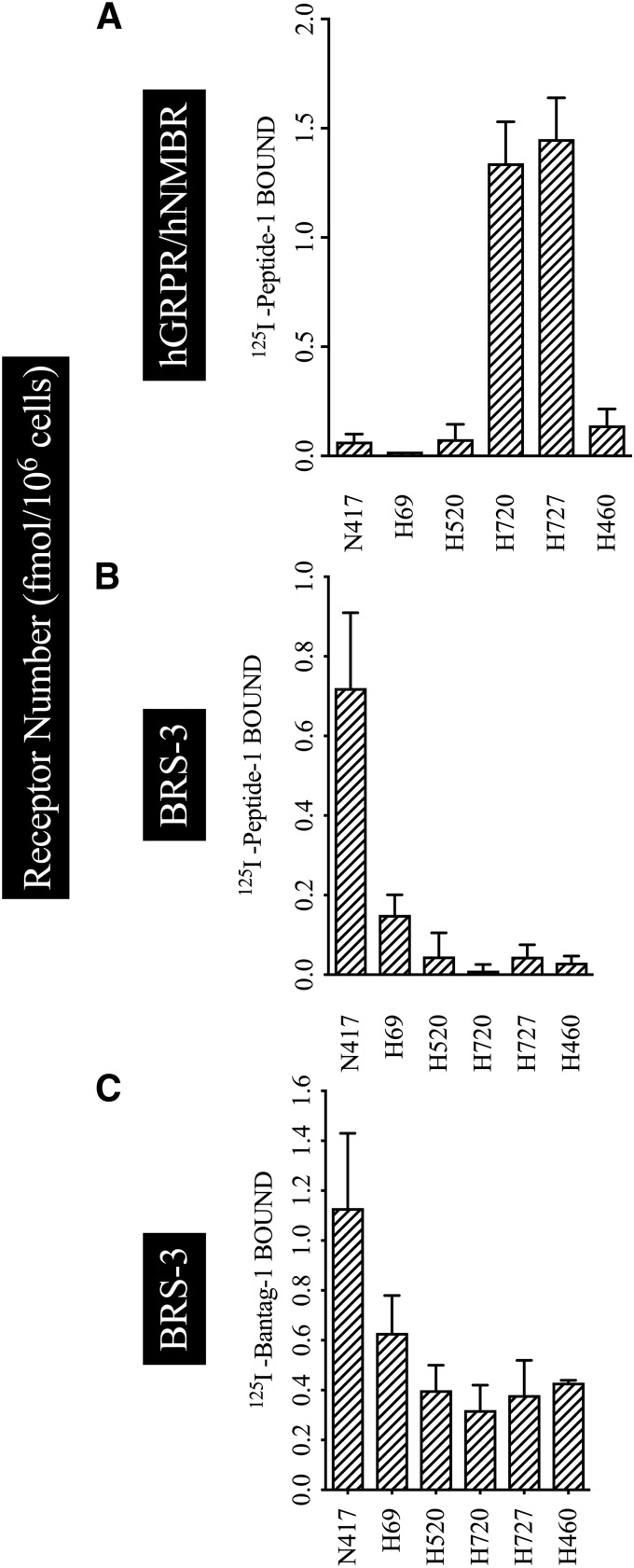
Determination of Relative Receptor Numbers for Various BRS-3 Receptors on Various Cancer Cells Using 125I-Peptide-1 and 125I-Bantag-1
Assessing the presence of BRS-3s is complicating by the occurrence of multiple BnRs on most tissues and the nonselectivity of 125I-Peptide 1 for hBRS-3 receptors, which has been used in most studies (Pradhan et al., 1998; Reubi et al., 2002; Jensen et al., 2008). Until this present study, the only readily available method to directly determine hBRS-3 presence/number on tissues was to use 125I-Peptide-1 with a GRPR- and NMBR-selective ligand to selectively block binding to the NMBR/GRPR receptor, which might also be present in significant numbers to BRS-3-3 (DeMichele et al., 1994; Reubi et al., 2002; Weber, 2009; Moreno et al., 2018). To assess the possible increased usefulness of 125I-Bantag-1, compared with the presently used method, we compared the results of 125I-Bantag-1 binding with the previously generally used subtraction method with 125I-Peptide-1 to identify and quantitate BRS-3 on a number of lung cancer cells, which can contain multiple BnRs (Moreno et al., 2018). We studied six different lung cancer cell lines (N417, H69 are small cell lung cancer cells; and H520, H720, H727, and H460, which are non–small cell lung cancer) because it has been previously reported they contained different amounts of BnRs including hBRS-3 (Fathi et al., 1993; DeMichele et al., 1994; Siegfried et al., 1999; Moreno et al., 2018). To address this study, we compared the binding of saturating concentrations of radioligand 125I-Peptide-1 (2.5 nM) and 125I-Bantag-1 (2.5 nM) (Fig. 7) alone or in combination with a GRPR-selective ligand, methyl ester Bn-(6-13) (ME) (Wang et al., 1990b), and a NMBR-selective ligand, PD (Ryan et al., 1999; Tokita et al., 2001; Gonzalez et al., 2009), to inhibit binding to hGRPR and hNMBR (Fig. 7B).
Fig. 7.
The determination of relative receptor numbers for various Bn receptors on lung cancer cells using the radiolabel agonist 125I-Peptide-1 or the radiolabeled antagonist 125I-Bantag-1. The lung cancer cells (5 × 106 cells/ml) N417, H69, H520, H720, H727, and H460 were incubated with 2.5 nM radioligand with or without 1 µM Peptide-1 or Bantag-1. In (A), the cells were incubated with 125I-Peptide-1, which binds to all three BnRs, in the presence of 1 µM Bantag-1 to block bind to BRS-3, thus showing only the hNMBR/hGRPR cell receptor number. In (B), 125I-Peptide-1 was incubated with 1 µM methyl ester Bn-(6-13) (a selective GRPR ligand) and 1 µM PD (a selective NMBR ligand) to inhibit binding to hGRPR and hNMBR, and this reflects only binding to the BRS-3 receptor. In (C), only 2.5 nM 125I-Bantag-1 was added, which only binds to hBRS-3 and thus directly assesses hBRS-3 receptor number. The results are the mean and S.E.M. from at least three separate experiments, and in each experiment the data points were determined in duplicate. For abbreviations, see the legends of Figs. 1, 3, and 6.
In binding studies with 125I-Peptide-1 and the peptide antagonist Bantag-1 to block the hBRS-3 binding site, the receptor number of hGRPR/hNMBR in H720 and H727 was 8-fold higher than in N417, H69, H520, or H460 (hGRPR/hNMBR receptor numbers: H720, 1.34; H727, 1.45; N417, 0.06; H69, 0.02; H520, 0.07; and H460, 0.14 fmol/106 cells) (Fig. 7A). When these lung cancer cells were incubated with 125I-Peptide-1, and the hGRPR/hNMBR inhibitors ME and PD, coinhibitors that block binding selective to GRPR/NMBR, allowing assessment only of BRS-3-receptor interaction, the receptor number of BRS-3 was 16-fold and 6-fold higher in N417 and H69 than in H520, H720, H727, or H460 (hBRS-3 receptor numbers: N417, 0.720; H69, 0.15; H520, 0.04; H720, 0.01; H727, 0.04; and H460, 0.03 fmol/106 cells) (Fig. 7B). In contrast, when the lung cancer cells were incubated only with the 125I-Bantag-1, N417, H69, and H520 had higher amounts of BRS-3 receptor number than H720, H727, and H460 (hBRS-3 receptor numbers: N417, 1.13; H69, 0.63; H520, 0.40; H720, 0.32; H727, 0.38; and H460, 0.43 fmol/106 cells) (Fig. 7C). The results demonstrate the advantage of the ease of use of the selective BRS-3 ligand 125I-Bantag-1, which does not require the indirect determination by the subtraction of GRPR/NMBR.
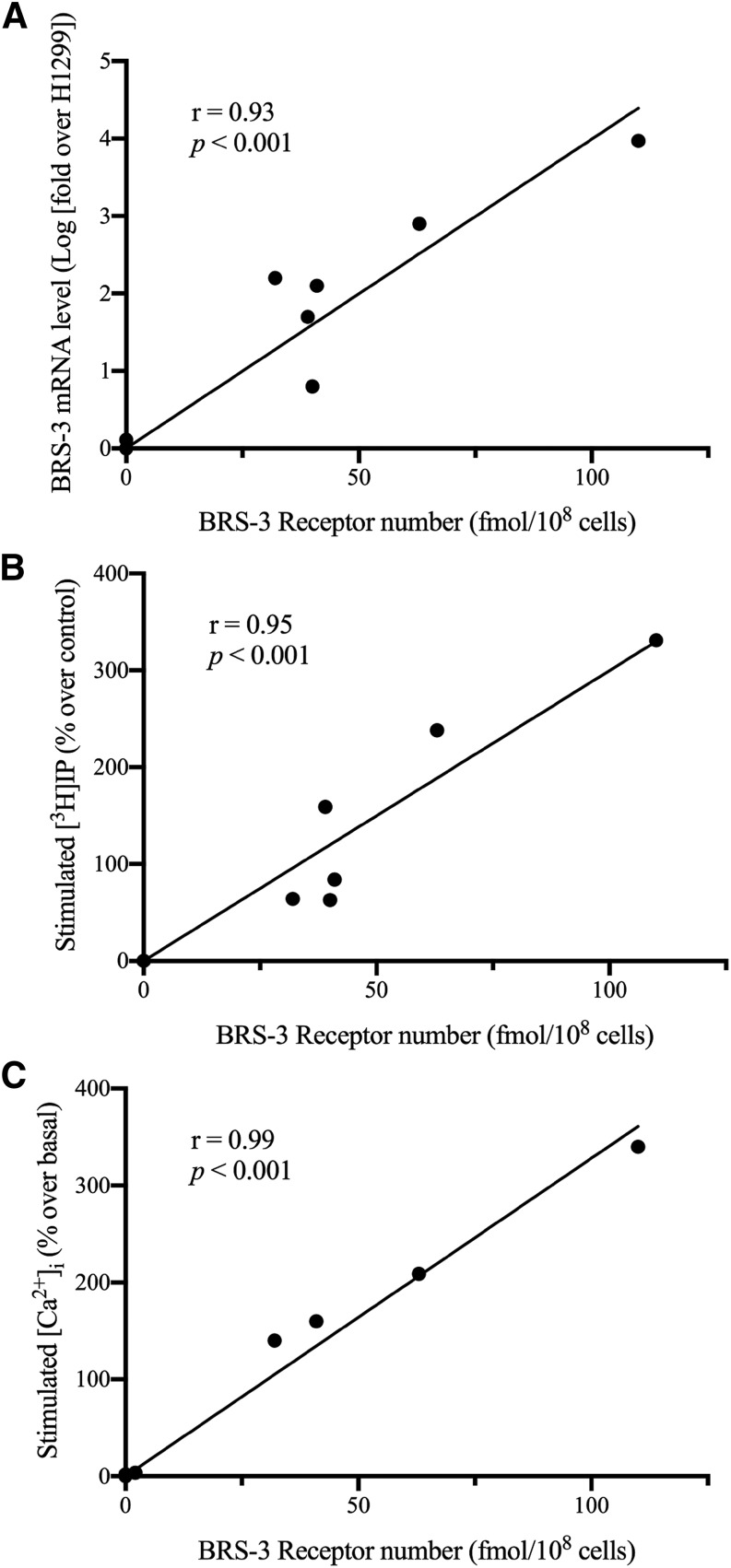
Assess Whether BRS-3 Receptor Number Determined Using 125I-Bantag-1 Correlates with Magnitude of Expression of BRS-3 mRNA, or Maximal Increase in Phospholipase C Activation (in Cytosolic Ca2+ or [3H]IP) in Lung Cancer Cell Lines
In a recent study (Moreno et al., 2018), the BRS-3 agonist MK-5046 was reported to activate phospholipase C, causing increases in cytosolic Ca2+ and IP generation in lung cancer cells possessing BRS-3. To determine whether the BRS-3 receptor number correlated with the magnitude of phospholipase C activation or the mBRS-3 mRNA expression, we correlated BRS-3 receptor number assessed using 125I-Bantag-1 with each of these variables in a number of lung cancer cell lines. As seen in Fig. 8, the BRS-3 receptor number determined in eight lung cancer cell lines correlated closely (r = 93–99, P < 0.001) with the expression level of BRS-3 mRNA determined by quantitative PCR (Moreno et al., 2018) (Fig. 8A), the maximal increase in stimulated [3H]IP (Fig. 8B) and the maximal stimulated increase in cytosolic Ca2+ (Fig. 8C).
Fig. 8.
The correlation of BRS3 receptor number with the ability to activate phospholipase C (Ca2+, [3H]IP) or the magnitude of BRS3 mRNA in various lung cancer cell lines. The magnitude of BRS3 mRNA (A), MK-5046–stimulated increases in [3H]IP (B), or increases in MK-5046–stimulated Ca2+ were correlated with BRS3 receptor number, as determined by saturation binding. The lung cancer cells (5 × 106 cells/ml) N417, H69, H520, H720, H727, A549, H838, and H1299 were incubated with 2.5 nM 125I-Bantag-1 with or without 1 μM Bantag-1, and the total BRS-3 receptor number was calculated using the specific activity. For lung cancer cells not previously reported (Moreno et al., 2018) (H838, A549, H1299), the maximal stimulation of BRS-3 by the BRS-3–specific agonist MK-5046 (10 nM) was determined (Moreno et al., 2018) and correlated with the BRS-3 receptor number, as described in Methods. For the remaining lung cancer cells, the data for change in Ca2+ [3H]IP as well as the BRS-3 mRNA expression determined by quantitative PCR data in all of the cancer cell lines was from Moreno et al. (2018). The results are the mean and S.E.M. from at least three separate experiments, and in each experiment the data points were determined in duplicate.
Assess Whether Using 125I-Bantag-1, BRS-3 Receptor Number Can Be Determined in Lung Cancer Cell Line Membranes and How These Values Compare to Results in Various Normal Rat Tissues Reported to Have High BRS-3 Expression of BRS-3 Either in Rats or Other Species
Four different human lung cancer cell lines having different levels of expression of BRS-3 were studied (Table 4). In each case using 125I-Bantag-1, BRS-3 receptors could be detected on the cell membranes (Table 4). However, the BRS-3 receptor densities varied markedly from 1.34 ± 0.81 fmol/mg protein for H838 cells to 64.4 ± 14.2 fmol/mg protein in N417 membranes, which is similar to densities seen in the whole-cell studies shown in Fig. 7.
TABLE 4.
BRS-3 receptor density in selected human lung cancer cell lines and normal rat tissues
Membranes were made from the indicated tissues as described in Methods. The density of BRS-3 binding sites was determined as described in the Fig. 7 legend using 2.5 nM 125I-Bantag-1 binding. Each value is the mean ± S.E.M. from at least three experiments.
| Membrane tissue | BRS-3 |
|---|---|
| fmol/mg protein | |
| Lung cancer cell lines | |
| N417 | 64.4 ± 14.2 |
| H69 | 45.0 ± 10.1 |
| H520 | 7.4 ± 0.6 |
| H838 | 1.34 ± 0.81 |
| Normal rat tissues | |
| Cortex (brain) | 12.0 ± 2.9 |
| Mid-brain | 21.0 ± 2.5 |
| Cerebellum | 15.0 ± 1.5 |
| Testis | 18.1 ± 0.6 |
| Duodenum | 39.1 ± 0.7 |
In rats and other species, high levels of BRS-3 expression are reported by various methods (immunohistochemistry, mRNA studies, and one binding study in the CNS using [3H]-Bag-2) (Fathi et al., 1993; Jennings et al., 2003; Sano et al., 2004; Porcher et al., 2005; Guan et al., 2010; Zhang et al., 2013) in various brain nuclei, testis, and small intestine (especially duodenum). We examined five of these (brain cortex, mid-brain, cerebellum, testis, and duodenum) using 125I-Bantag-1 binding and found that they all possessed BRS-3 receptors (Table 4). In these tissues, the BRS-3 densities overlapped with the levels seen in many of the lung cancer cell lines.
Discussion
The purpose of this study was to attempt to develop and characterize a specific, high-affinity radioligand that could be used for BRS-3 localization and pharmacologic studies in both normal and pathophysiological conditions. This is important for a number of reasons. First, the BRS-3 receptor is receiving increased attention (Gonzalez et al., 2015b; Maruyama et al., 2017; Nio et al., 2017; Xiao et al., 2017; Pinol et al., 2018) because of its prominent role in causing diabetes, hypertension, and obesity by regulating body weight and energy maintenance, body temperature control, and control of metabolic homeostasis, particularly insulin/glucose control, as evidenced from studies of BRS-3 knockout mice (Ohki-Hamazaki et al., 1997; Ladenheim et al., 2008; Majumdar and Weber, 2012a; Lateef et al., 2014; Gonzalez et al., 2015b). Studies using primarily non–ligand receptor localization methods (immunohistochemistry, BRS-3 mRNA methods) (Fathi et al., 1993; Jennings et al., 2003; Sano et al., 2004; Porcher et al., 2005; Guan et al., 2010; Zhang et al., 2013; Maruyama et al., 2017; Xiao et al., 2017) provide evidence that BRS-3 is highly expressed in hypothalamic nuclei associated with metabolic maintenance, and that glutamatergic neurons in this area (Xiao et al., 2017) are particularly important. Furthermore, recent studies (Das et al., 2009; Moreno et al., 2013, 2018) demonstrate that BRS-3 is overexpressed in numerous tumors and has prominent tumoral growth effects, similar to those seen with the other mammalian BnRs (i.e., GRPR, NMBR) (Fathi et al., 1993; Reubi et al., 2002; Ramos-Álvarez et al., 2015; Moreno et al., 2016, 2018). In addition, recent studies demonstrate that the BRS-3 has important roles in a number of different tissues, including regulation of behavioral changes in the CNS (Yamada et al., 2000a,b), the regulation of metabolic changes in skeletal muscle (Ramos-Álvarez et al., 2014; Gonzalez et al., 2015a), important metabolic effects in adipocytes (Nakamichi et al., 2004), the maintenance of bronchial epithelial homeostasis (Liu et al., 2018), and motility effects in the GI tract (Porcher et al., 2005). Second, at present no generally available specific, high-affinity ligand is available to study the expression, location, or pharmacology of the BRS-3 receptor. A ligand, 125I-Peptide-1 (in this study) [(125I-D-Tyr6, β-Ala11, Phe13, Nle14]Bn-(6–14)], which interacts with high affinity with hBRS-3 has been described and used in some receptor localization studies (Fleischmann et al., 2000; Reubi et al., 2002; Waser et al., 2007), but it has a number of disadvantages that limit its usefulness. This ligand also interacts with high affinity with GRPR and NMBR (Mantey et al., 1997; Pradhan et al., 1998), which frequently coexist with BRS-3 on the same cells (Fathi et al., 1993; Ryan et al., 1998b; Moreno et al., 2018). In addition, it has low affinity for mouse and rat BRS-3 and thus is not useful in these commonly used experimental animals (Liu et al., 2002; Uehara et al., 2011; Ramos-Álvarez et al., 2016). The high affinity of this ligand for all three BnRs requires that in receptor studies for BRS that a subtraction approach be used by adding specific ligands for the GRPR and NMBR to block the binding to these receptors (Fleischmann et al., 2000; Reubi et al., 2002; Waser et al., 2007), leaving the BRS-3 binding only; however, this is cumbersome and increases the inaccuracy. In one study (Guan et al., 2010), a tritiated BRS-3 agonist, [3H]-Bag-2, was described, but it is not well characterized and is not generally available. Third, studies, especially in the CNS in nonhypothalamic areas, have shown important differences between the results of possible BRS-3 locations assessed by BRS-3 mRNA studies and BRS-3 antibodies, leading authorities in recent reviews to question the specificity of available BRS-3 antibodies (Gonzalez et al., 2015b; Xiao and Reitman, 2016). Last, even though the endogenous ligand remains unknown at present, nevertheless, because of the important roles of BRS-3 outlined above in energy, glucose/insulin homeostasis, and obesity, a number of pharmaceutical companies/investigators have recently developed selective agonists/antagonists (e.g., MK-5046, chiral diazepines, Bag-1/2, Bantag-1) (Guan et al., 2010; Feng et al., 2011; Matsufuji et al., 2014, 2015; Gonzalez et al., 2015b; Moody et al., 2015; Kiyotsuka et al., 2016; Ramos-Álvarez et al., 2016). These new compounds increasingly are being used to investigate the role of BRS-3 in normal physiologic function as well in pathophysiological conditions. This is already starting to increase the need for specific radioligands to study the location/pharmacology of the BRS-3 in these processes.
A number of our results support the conclusion that in this study we have developed a highly selective, specific ligand, 125-Bantag-1, that should be generally useful for the characterization and study of the pharmacology of the BRS-3 receptor. First, we find that Bantag-1 is highly selective (>10,000-fold) for hBRS-3 over the other closely related mammalian BnRs (i.e., GRPR, NMBR), which is consistent with the results of previous studies (Guan et al., 2010; Moreno et al., 2018). Furthermore, this selectivity is preserved for the radiolabeled Bantag-1, because we find that 125I-Bantag-1 does not bind to either GRPR or NMBR. Second, we find high specificity of 125I-Bantag-1 for BRS-3. This result is supported by findings that ligands of other unrelated GI hormones/neurotransmitters, ligands specific for GRPR/NMBR or ligands of a Drosophila receptor related to mammalian BRS-3 (i.e., CCHamides) (Matsufujiet al., 2014, 2015) all do not alter the interaction of 125I-Bantag-1 with the BRS-3 receptor. Third, dose-inhibition curves and BRS-3 receptor affinities for a number of different ligands reported to interact with the BRS-3, computed from 125I-Bantag-1 binding, show very close agreement with those calculated from the well-characterized panBnR peptide agonist ligand 125I-Peptide-1 (Mantey et al., 1997; Ryan et al., 1998b; Gonzalez et al., 2015b). Fourth, previous studies have demonstrated that the peptide antagonist Bantag-1, in contrast to the peptide agonist Peptide-1, has high affinity for mouse and rat BRS-3 receptors (Guan et al., 2010; Feng et al., 2011). These results demonstrate that 125I-Bantag-1 will also be a valuable tool for assessing BRS-3 in the most commonly used laboratory animals, rats/mice, whereas125I-peptide-1 is not. Fifth, that 125I-Bantag-1 binding studies could be pharmacologically useful for characterizing BRS-3 receptors was shown by the ability to allow clear quantification of BRS-3 receptor expression. This allowed us to establish a close correlation of the BRS-3 receptor number with the amount of BRS-3 mRNA, as well as the maximal magnitude of BRS-3 activation of phospholipase C in lung cancer cells. In addition, it allowed BRS-3 receptor expression to be studied in both normal tissue membranes as well as in those from cancer cells.
In general, in this study the interaction of the radiolabeled antagonist 125I-Bantag-1 with BRS-3 receptors was similar to that seen with the radiolabeled agonist 125I-Peptide 1, which has similarities and differences from other studies comparing the binding of these two classes of ligands on other BnRs, as well as to other GPCRs. This conclusion is supported by the similar kinetics of association of both BRS-3 ligands with the BRS-3; their similar maximal binding under identical conditions to these cells; their similar dissociation kinetics; as well as the similar dose-inhibition curves of various putative BRS-3 ligands with both BRS-3 ligands. This result differs from a previous study (Mantey et al., 1997) comparing a radiolabeled GRPR-radiolabeled peptide antagonist and agonist binding to GRPR receptors, where the kinetics differed between the two, with the antagonist showing more rapid association kinetics, and more rapid and greater time-dependent dissociation than the agonist. However, these GRPR study results were similar to those of the present study in showing that both radiolabeled ligands gave similar affinities for the GRPR (Mantey et al., 1997). The results that similar affinities for BRS-3 for each of the eight putative selective BRS-3 agonists examined were found with both ligands in the present study differs from findings of radiolabeled agonist/antagonist binding with cholecystokinin, gastrin, adrenergic, and luteinizing hormone-releasing hormone receptors (Mantey et al., 1997).
A somewhat surprising finding in the present study is that the radiolabeled antagonist is internalized to the same degree as the radiolabeled agonist. Generally, GPCR peptide antagonists are poorly and slowly internalized, if at all, whereas agonists are extensively and rapidly internalized (Mantey et al., 1993; Ginj et al., 2006; Moreno et al., 2016; Dalm et al., 2017). The above generalization is also true for BnRs, where numerous studies have reported that most BnR peptide antagonists are poorly internalized, if at all (Mantey et al., 1993; Cescato et al., 2008; Varasteh et al., 2013; Nock et al., 2017; Popp et al., 2017; Cheng et al., 2018). The internalization of radiolabeled Bantag-1 in our studies was not due to the possibility that Bantag-1 was functioning as a partial agonist, because in previous studies it has been reported to have no agonist activity at the BRS-3 receptor for the activation of phospholipase C, as assessed by either stimulation of the generation of phosphoinositides (Moreno et al., 2013) or mobilization of cellular calcium (Guan et al., 2010). Furthermore, Bantag-1 functioned as fully competitive antagonist of the BRS-3 receptor activation for stimulating IP generation (Moreno et al., 2013).
In addition to its use for pharmacological studies of the BRS-3 in normal physiologic and pathologic conditions, radiolabeled Bantag-1 may prove of value in tumor localization and for receptor-directed cytotoxicity for tumors. Recently, numerous studies have demonstrated that a number of common neoplasms overexpress BnRs (i.e., prostate, breast, lung, pancreas, and CNS tumors) (Reubi et al., 2002; Reubi, 2013; Moreno et al., 2016), and in some tumors such as lung cancers, BRS-3 is the most frequently overexpressed and is expressed in the highest numbers (Moreno et al., 2018). A number of recent studies report the ability of BnR radioligands to image various human tumors in humans, particularly breast and prostate, using high-affinity GRPR ligands (Sancho et al., 2011; Moreno et al., 2016; Ferreira et al., 2017; Cheng et al., 2018; Zang et al., 2018), and with the high expression of BRS-3 in lung tumors (Moreno et al., 2018) a similar approach might be used with radiolabeled Bantag-1. Furthermore, using 177Lu- or 90Y-labeled peptides, peptide-radioreceptor therapy has been shown to be effective and has been recently approved for the treatment of neuroendocrine tumors with radiolabeled somatostatin receptor ligands (Strosberg et al., 2016; Ito and Jensen, 2017). Because of this, there is now considerable interest in using a similar approach with radiolabeled GRP agonists/antagonists for a number of cancers overexpressing these receptors, particularly, prostate and breast cancer (Moreno et al., 2016; Reynolds et al., 2016; Maina et al., 2017). This raises the possibility that a similar approach be used for radiolabeled Bantag-1 for lung cancer, because of its frequent high expression in these tumors (Moreno et al., 2018).
In conclusion, we describe and characterize a novel, high-affinity, selective, and specific radiolabeled ligand, 125I-Bantag-1, that can be easily prepared and thus should be generally useful for pharmacological characterization of BRS-3 receptors, both in peripheral tissues and the CNS. The availability of this ligand should be useful in exploring the role of this orphan receptor in energy/insulin-glucose homeostasis, body temperature control, GI motility, and allergic lung diseases, and their possible theranostic use to selectively image and deliver cytotoxic agents to neoplasms overexpressing this receptor.
Abbreviations
- BALB 3T3 or BALB
mouse MIC embryonic fibroblast cells
- Bantag-1
Boc-Phe-His-4-amino-5-cyclohexyl-2,4,5-trideoxypentonyl-Leu-(3-dimethylamino) benzylamide N-methylammonium trifluoroacetate
- Bn
bombesin
- BnR
bombesin receptors
- BRS-3
bombesin receptor subtype 3
- BSA
bovine serum albumin
- CNS
central nervous system
- DMEM
Dulbecco’s minimum essential medium
- GI
gastrointestinal
- GPCR
G-protein–coupled receptor
- GRP
gastrin-releasing peptide
- GRPR
gastrin-releasing peptide receptor
- hBRS-3
human bombesin receptor subtype 3
- hGRPR
human gastrin-releasing peptide receptor
- hNMBR
neuromedin B receptor
- IP
inositol phosphate
- 125I-Peptide-1
125I-[D-Tyr6, β-Ala11, Phe13, Nle14]Bn-(6-14)
- ME
(Bn-(6-13) methyl ester
- MK-5046 [(2S)-1
1,1-trifluoro-2-[4-(1H-pyrazol-1-yl)phenyl]-3-(4-[[1-(trifluoromethyl)cyclopropyl]methyl]-1H-imidazol-2-yl)propan-2-ol]
- NMB
neuromedin B
- NMBR
neuromedin B receptor
- PD
PD168368
Authorship Contributions
Participated in research design: Ramos-Alvarez, Lee, Mantey, and Jensen.
Conducted experiments: Ramos-Alvarez, Lee, Mantey, and Jensen.
Contributed new reagents or analytic tools: Ramos-Alvarez, Lee, Mantey, and Jensen.
Performed data analysis: Ramos-Alvarez, Lee, Mantey, and Jensen.
Wrote or contributed to the writing of the manuscript: Ramos-Alvarez, Lee, Mantey, and Jensen.
Footnotes
This work was partially supported by intramural funds of the National Institutes of Diabetes, Digestive and Kidney Diseases, National Institutes of Health [Grant DK-053200-29].
References
- Benya RV, Fathi Z, Battey JF, Jensen RT. (1993) Serines and threonines in the gastrin-releasing peptide receptor carboxyl terminus mediate internalization. J Biol Chem 268:20285–20290. [PubMed] [Google Scholar]
- Benya RV, Fathi Z, Kusui T, Pradhan T, Battey JF, Jensen RT. (1994) Gastrin-releasing peptide receptor-induced internalization, down-regulation, desensitization, and growth: possible role for cyclic AMP. Mol Pharmacol 46:235–245. [PubMed] [Google Scholar]
- Benya RV, Kusui T, Pradhan TK, Battey JF, Jensen RT. (1995) Expression and characterization of cloned human bombesin receptors. Mol Pharmacol 47:10–20. [PubMed] [Google Scholar]
- Benya RV, Wada E, Battey JF, Fathi Z, Wang LH, Mantey SA, Coy DH, Jensen RT. (1992) Neuromedin B receptors retain functional expression when transfected into BALB 3T3 fibroblasts: analysis of binding, kinetics, stoichiometry, modulation by guanine nucleotide-binding proteins, and signal transduction and comparison with natively expressed receptors. Mol Pharmacol 42:1058–1068. [PubMed] [Google Scholar]
- Boyle RG, Humphries J, Mitchell T, Showell GA, Apaya R, Iijima H, Shimada H, Arai T, Ueno H, Usui Y, et al. (2005) The design of a new potent and selective ligand for the orphan bombesin receptor subtype 3 (BRS3). J Pept Sci 11:136–141. [DOI] [PubMed] [Google Scholar]
- Cescato R, Maina T, Nock B, Nikolopoulou A, Charalambidis D, Piccand V, Reubi JC. (2008) Bombesin receptor antagonists may be preferable to agonists for tumor targeting. J Nucl Med 49:318–326. [DOI] [PubMed] [Google Scholar]
- Cheng S, Lang L, Wang Z, Jacobson O, Yung B, Zhu G, Gu D, Ma Y, Zhu X, Niu G, et al. (2018) Positron emission tomography imaging of prostate cancer with Ga-68-labeled gastrin-releasing peptide receptor agonist BBN7-14 and antagonist RM26. Bioconjug Chem 29:410–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y, Prusoff WH. (1973) Relationship between the inhibition constant (K1) and the concentration of inhibitor which causes 50 per cent inhibition (I50) of an enzymatic reaction. Biochem Pharmacol 22:3099–3108. [DOI] [PubMed] [Google Scholar]
- Coy DH, Heinz-Erian P, Jiang NY, Sasaki Y, Taylor J, Moreau JP, Wolfrey WT, Gardner JD, Jensen RT. (1988) Probing peptide backbone function in bombesin. A reduced peptide bond analogue with potent and specific receptor antagonist activity. J Biol Chem 263:5056–5060. [PubMed] [Google Scholar]
- Coy DH, Taylor JE, Jiang NY, Kim SH, Wang LH, Huang SC, Moreau JP, Gardner JD, Jensen RT. (1989) Short-chain pseudopeptide bombesin receptor antagonists with enhanced binding affinities for pancreatic acinar and Swiss 3T3 cells display strong antimitotic activity. J Biol Chem 264:14691–14697. [PubMed] [Google Scholar]
- Dalm SU, Bakker IL, de Blois E, Doeswijk GN, Konijnenberg MW, Orlandi F, Barbato D, Tedesco M, Maina T, Nock BA, et al. (2017) 68Ga/177Lu-NeoBOMB1, a novel radiolabeled GRPR antagonist for theranostic use in oncology. J Nucl Med 58:293–299. [DOI] [PubMed] [Google Scholar]
- Das M, Mohanty C, Sahoo SK. (2009) Ligand-based targeted therapy for cancer tissue. Expert Opin Drug Deliv 6:285–304. [DOI] [PubMed] [Google Scholar]
- DeMichele MA, Davis AL, Hunt JD, Landreneau RJ, Siegfried JM. (1994) Expression of mRNA for three bombesin receptor subtypes in human bronchial epithelial cells. Am J Respir Cell Mol Biol 11:66–74. [DOI] [PubMed] [Google Scholar]
- Fathi Z, Corjay MH, Shapira H, Wada E, Benya R, Jensen R, Viallet J, Sausville EA, Battey JF. (1993) BRS-3: a novel bombesin receptor subtype selectively expressed in testis and lung carcinoma cells. J Biol Chem 268:5979–5984. [PubMed] [Google Scholar]
- Feng Y, Guan XM, Li J, Metzger JM, Zhu Y, Juhl K, Zhang BB, Thornberry NA, Reitman ML, Zhou YP. (2011) Bombesin receptor subtype-3 (BRS-3) regulates glucose-stimulated insulin secretion in pancreatic islets across multiple species. Endocrinology 152:4106–4115. [DOI] [PubMed] [Google Scholar]
- Ferreira CA, Fuscaldi LL, Townsend DM, Rubello D, Barros ALB. (2017) Radiolabeled bombesin derivatives for preclinical oncological imaging. Biomed Pharmacother 87:58–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleischmann A, Läderach U, Friess H, Buechler MW, Reubi JC. (2000) Bombesin receptors in distinct tissue compartments of human pancreatic diseases. Lab Invest 80:1807–1817. [DOI] [PubMed] [Google Scholar]
- Ginj M, Zhang H, Waser B, Cescato R, Wild D, Wang X, Erchegyi J, Rivier J, Mäcke HR, Reubi JC. (2006) Radiolabeled somatostatin receptor antagonists are preferable to agonists for in vivo peptide receptor targeting of tumors. Proc Natl Acad Sci USA 103:16436–16441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González N, Mantey SA, Pradhan TK, Sancho V, Moody TW, Coy DH, Jensen RT. (2009) Characterization of putative GRP- and NMB-receptor antagonist’s interaction with human receptors. Peptides 30:1473–1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González N, Martín-Duce A, Martínez-Arrieta F, Moreno-Villegas Z, Portal-Núñez S, Sanz R, Egido J. (2015a) Effect of bombesin receptor subtype-3 and its synthetic agonist on signaling, glucose transport and metabolism in myocytes from patients with obesity and type 2 diabetes. Int J Mol Med 35:925–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González N, Moreno P, Jensen RT. (2015b) Bombesin receptor subtype 3 as a potential target for obesity and diabetes. Expert Opin Ther Targets 19:1153–1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbulev V, Akhundova A, Büchner H, Fahrenholz F. (1992) Molecular cloning of a new bombesin receptor subtype expressed in uterus during pregnancy. Eur J Biochem 208:405–410. [DOI] [PubMed] [Google Scholar]
- Guan XM, Chen H, Dobbelaar PH, Dong Y, Fong TM, Gagen K, Gorski J, He S, Howard AD, Jian T, et al. (2010) Regulation of energy homeostasis by bombesin receptor subtype-3: selective receptor agonists for the treatment of obesity. Cell Metab 11:101–112. [DOI] [PubMed] [Google Scholar]
- Heinz-Erian P, Coy DH, Tamura M, Jones SW, Gardner JD, Jensen RT. (1987) [D-Phe12]bombesin analogues: a new class of bombesin receptor antagonists. Am J Physiol 252:G439–G442. [DOI] [PubMed] [Google Scholar]
- Ito T, Jensen RT. (2017) Molecular imaging in neuroendocrine tumors: recent advances, controversies, unresolved issues, and roles in management. Curr Opin Endocrinol Diabetes Obes 24:15–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jennings CA, Harrison DC, Maycox PR, Crook B, Smart D, Hervieu GJ. (2003) The distribution of the orphan bombesin receptor subtype-3 in the rat CNS. Neuroscience 120:309–324. [DOI] [PubMed] [Google Scholar]
- Jensen RT, Battey JF, Spindel ER, Benya RV. (2008) International Union of Pharmacology. LXVIII. Mammalian bombesin receptors: nomenclature, distribution, pharmacology, signaling, and functions in normal and disease states. Pharmacol Rev 60:1–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen RT, Charlton CG, Adachi H, Jones SW, O’Donohue TL, Gardner JD. (1983) Use of 125I-secretin to identify and characterize high-affinity secretin receptors on pancreatic acini. Am J Physiol 245:G186–G195. [DOI] [PubMed] [Google Scholar]
- Kiyotsuka Y, Shimada K, Kobayashi S, Suzuki M, Akiu M, Asano M, Sogawa Y, Hara T, Konishi M, Abe-Ohya R, et al. (2016) Synthesis and biological evaluation of novel imidazol-1-ylacetic acid derivatives as non-brain penetrant bombesin receptor subtype-3 (BRS-3) agonists. Bioorg Med Chem Lett 26:4205–4210. [DOI] [PubMed] [Google Scholar]
- Kumagai H, Ikeda Y, Motozawa Y, Fujishiro M, Okamura T, Fujio K, Okazaki H, Nomura S, Takeda N, Harada M, et al. (2015) Quantitative measurement of GPCR endocytosis via pulse-chase covalent labeling. PLoS One 10:e0129394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladenheim EE, Hamilton NL, Behles RR, Bi S, Hampton LL, Battey JF, Moran TH. (2008) Factors contributing to obesity in bombesin receptor subtype-3-deficient mice. Endocrinology 149:971–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lateef DM, Abreu-Vieira G, Xiao C, Reitman ML. (2014) Regulation of body temperature and brown adipose tissue thermogenesis by bombesin receptor subtype-3. Am J Physiol Endocrinol Metab 306:E681–E687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Peng L, Liu C, Tan Y, Xiang Y, Qu X, Weber HC, Dai M, Qin X. (2018) Activation of bombesin receptor subtype-3 promotes antigen-presenting action in human bronchial epithelial cells. Int Arch Allergy Immunol 175:53–60. [DOI] [PubMed] [Google Scholar]
- Liu J, Lao ZJ, Zhang J, Schaeffer MT, Jiang MM, Guan XM, Van der Ploeg LH, Fong TM. (2002) Molecular basis of the pharmacological difference between rat and human bombesin receptor subtype-3 (BRS-3). Biochemistry 41:8954–8960. [DOI] [PubMed] [Google Scholar]
- Maina T, Nock BA, Kulkarni H, Singh A, Baum RP. (2017) Theranostic prospects of gastrin-releasing peptide receptor-radioantagonists in oncology. PET Clin 12:297–309. [DOI] [PubMed] [Google Scholar]
- Majumdar ID, Weber HC. (2012a) Appetite-modifying effects of bombesin receptor subtype-3 agonists. Handb Exp Pharmacol (209):405–432. [DOI] [PubMed] [Google Scholar]
- Majumdar ID, Weber HC. (2012b) Biology and pharmacology of bombesin receptor subtype-3. Curr Opin Endocrinol Diabetes Obes 19:3–7. [DOI] [PubMed] [Google Scholar]
- Mantey S, Frucht H, Coy DH, Jensen RT. (1993) Characterization of bombesin receptors using a novel, potent, radiolabeled antagonist that distinguishes bombesin receptor subtypes. Mol Pharmacol 43:762–774. [PubMed] [Google Scholar]
- Mantey SA, Coy DH, Entsuah LK, Jensen RT. (2004) Development of bombesin analogs with conformationally restricted amino acid substitutions with enhanced selectivity for the orphan receptor human bombesin receptor subtype 3. J Pharmacol Exp Ther 310:1161–1170. [DOI] [PubMed] [Google Scholar]
- Mantey SA, Coy DH, Pradhan TK, Igarashi H, Rizo IM, Shen L, Hou W, Hocart SJ, Jensen RT. (2001) Rational design of a peptide agonist that interacts selectively with the orphan receptor, bombesin receptor subtype 3. J Biol Chem 276:9219–9229. [DOI] [PubMed] [Google Scholar]
- Mantey SA, Weber HC, Sainz E, Akeson M, Ryan RR, Pradhan TK, Searles RP, Spindel ER, Battey JF, Coy DH, et al. (1997) Discovery of a high affinity radioligand for the human orphan receptor, bombesin receptor subtype 3, which demonstrates that it has a unique pharmacology compared with other mammalian bombesin receptors. J Biol Chem 272:26062–26071. [DOI] [PubMed] [Google Scholar]
- Maruyama M, Hotta N, Nio Y, Hamagami K, Nagi T, Funata M, Sakamoto J, Nakakariya M, Amano N, Nishida M, et al. (2017) Bombesin receptor subtype-3-expressing neurons regulate energy homeostasis through a novel neuronal pathway in the hypothalamus. Brain Behav 8:e00881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsufuji T, Shimada K, Kobayashi S, Ichikawa M, Kawamura A, Fujimoto T, Arita T, Hara T, Konishi M, Abe-Ohya R, et al. (2015) Synthesis and biological evaluation of novel chiral diazepine derivatives as bombesin receptor subtype-3 (BRS-3) agonists incorporating an antedrug approach. Bioorg Med Chem 23:89–104. [DOI] [PubMed] [Google Scholar]
- Matsufuji T, Shimada K, Kobayashi S, Kawamura A, Fujimoto T, Arita T, Hara T, Konishi M, Abe-Ohya R, Izumi M, et al. (2014) Discovery of novel chiral diazepines as bombesin receptor subtype-3 (BRS-3) agonists with low brain penetration. Bioorg Med Chem Lett 24:750–755. [DOI] [PubMed] [Google Scholar]
- Moody TW, Chan D, Fahrenkrug J, Jensen RT. (2003) Neuropeptides as autocrine growth factors in cancer cells. Curr Pharm Des 9:495–509. [DOI] [PubMed] [Google Scholar]
- Moody TW, Mantey SA, Moreno P, Nakamura T, Lacivita E, Leopoldo M, Jensen RT. (2015) ML-18 is a non-peptide bombesin receptor subtype-3 antagonist which inhibits lung cancer growth. Peptides 64:55–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno P, Mantey SA, Lee SH, Ramos-Álvarez I, Moody TW, Jensen RT. (2018) A possible new target in lung-cancer cells: the orphan receptor, bombesin receptor subtype-3. Peptides 101:213–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno P, Mantey SA, Nuche-Berenguer B, Reitman ML, González N, Coy DH, Jensen RT. (2013) Comparative pharmacology of bombesin receptor subtype-3, nonpeptide agonist MK-5046, a universal peptide agonist, and peptide antagonist Bantag-1 for human bombesin receptors. J Pharmacol Exp Ther 347:100–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno P, Ramos-Álvarez I, Moody TW, Jensen RT. (2016) Bombesin related peptides/receptors and their promising therapeutic roles in cancer imaging, targeting and treatment. Expert Opin Ther Targets 20:1055–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamichi Y, Wada E, Aoki K, Ohara-Imaizumi M, Kikuta T, Nishiwaki C, Matsushima S, Watanabe T, Wada K, Nagamatsu S. (2004) Functions of pancreatic beta cells and adipocytes in bombesin receptor subtype-3-deficient mice. Biochem Biophys Res Commun 318:698–703. [DOI] [PubMed] [Google Scholar]
- Nakamura T, Ramos-Álvarez I, Iordanskaia T, Moreno P, Mantey SA, Jensen RT. (2016) Molecular basis for high affinity and selectivity of peptide antagonist, Bantag-1, for the orphan BB3 receptor. Biochem Pharmacol 115:64–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nio Y, Hotta N, Maruyama M, Hamagami K, Nagi T, Funata M, Sakamoto J, Nakakariya M, Amano N, Okawa T, et al. (2017) A selective bombesin receptor subtype 3 agonist promotes weight loss in male diet-induced-obese rats with circadian rhythm change. Endocrinology 158:1298–1313. [DOI] [PubMed] [Google Scholar]
- Nock BA, Kaloudi A, Lymperis E, Giarika A, Kulkarni HR, Klette I, Singh A, Krenning EP, de Jong M, Maina T, et al. (2017) Theranostic perspectives in prostate cancer with the gastrin-releasing peptide receptor antagonist NeoBOMB1: preclinical and first clinical results. J Nucl Med 58:75–80. [DOI] [PubMed] [Google Scholar]
- Ohki-Hamazaki H, Watase K, Yamamoto K, Ogura H, Yamano M, Yamada K, Maeno H, Imaki J, Kikuyama S, Wada E, et al. (1997) Mice lacking bombesin receptor subtype-3 develop metabolic defects and obesity. Nature 390:165–169. [DOI] [PubMed] [Google Scholar]
- Piñol RA, Zahler SH, Li C, Saha A, Tan BK, Škop V, Gavrilova O, Xiao C, Krashes MJ, Reitman ML. (2018) Brs3 neurons in the mouse dorsomedial hypothalamus regulate body temperature, energy expenditure, and heart rate, but not food intake. Nat Neurosci 21:1530–1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popp I, Del Pozzo L, Waser B, Reubi JC, Meyer PT, Maecke HR, Gourni E. (2017) Approaches to improve metabolic stability of a statine-based GRP receptor antagonist. Nucl Med Biol 45:22–29. [DOI] [PubMed] [Google Scholar]
- Porcher C, Juhem A, Peinnequin A, Bonaz B. (2005) Bombesin receptor subtype-3 is expressed by the enteric nervous system and by interstitial cells of Cajal in the rat gastrointestinal tract. Cell Tissue Res 320:21–31. [DOI] [PubMed] [Google Scholar]
- Pradhan TK, Katsuno T, Taylor JE, Kim SH, Ryan RR, Mantey SA, Donohue PJ, Weber HC, Sainz E, Battey JF, et al. (1998) Identification of a unique ligand which has high affinity for all four bombesin receptor subtypes. Eur J Pharmacol 343:275–287. [DOI] [PubMed] [Google Scholar]
- Ramos-Álvarez I, Moreno P, Mantey SA, Nakamura T, Nuche-Berenguer B, Moody TW, Coy DH, Jensen RT. (2015) Insights into bombesin receptors and ligands: highlighting recent advances. Peptides 72:128–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos-Álvarez I, Moreno-Villegas Z, Martín-Duce A, Sanz R, Aparicio C, Portal-Núñez S, Mantey SA, Jensen RT, González N. (2014) Human BRS-3 receptor: functions/role in cell signaling pathways and glucose metabolism in obese or diabetic myocytes. Peptides 51:91–99. [DOI] [PubMed] [Google Scholar]
- Ramos-Álvarez I, Nakamura T, Mantey SA, Moreno P, Nuche-Berenguer B, Jensen RT. (2016) Novel chiral-diazepines function as specific, selective receptor agonists with variable coupling and species variability in human, mouse and rat BRS-3 receptor cells. Peptides 75:8–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reubi JC. (2013) Old and new peptide receptor targets in cancer: future directions. Recent Results Cancer Res 194:567–576. [DOI] [PubMed] [Google Scholar]
- Reubi JC, Wenger S, Schmuckli-Maurer J, Schaer JC, Gugger M. (2002) Bombesin receptor subtypes in human cancers: detection with the universal radioligand (125)I-[D-TYR(6), beta-ALA(11), PHE(13), NLE(14)] bombesin(6-14). Clin Cancer Res 8:1139–1146. [PubMed] [Google Scholar]
- Reynolds TS, Bandari RP, Jiang Z, Smith CJ. (2016) Lutetium-177 labeled bombesin peptides for radionuclide therapy. Curr Radiopharm 9:33–43. [DOI] [PubMed] [Google Scholar]
- Rowley WH, Sato S, Huang SC, Collado-Escobar DM, Beaven MA, Wang LH, Martinez J, Gardner JD, Jensen RT. (1990) Cholecystokinin-induced formation of inositol phosphates in pancreatic acini. Am J Physiol 259:G655–G665. [DOI] [PubMed] [Google Scholar]
- Ryan RR, Katsuno T, Mantey SA, Pradhan TK, Weber HC, Coy DH, Battey JF, Jensen RT. (1999) Comparative pharmacology of the nonpeptide neuromedin B receptor antagonist PD 168368. J Pharmacol Exp Ther 290:1202–1211. [PubMed] [Google Scholar]
- Ryan RR, Weber HC, Hou W, Sainz E, Mantey SA, Battey JF, Coy DH, Jensen RT. (1998a) Ability of various bombesin receptor agonists and antagonists to alter intracellular signaling of the human orphan receptor BRS-3. J Biol Chem 273:13613–13624. [DOI] [PubMed] [Google Scholar]
- Ryan RR, Weber HC, Mantey SA, Hou W, Hilburger ME, Pradhan TK, Coy DH, Jensen RT. (1998b) Pharmacology and intracellular signaling mechanisms of the native human orphan receptor BRS-3 in lung cancer cells. J Pharmacol Exp Ther 287:366–380. [PubMed] [Google Scholar]
- Sancho V, Di Florio A, Moody TW, Jensen RT. (2011) Bombesin receptor-mediated imaging and cytotoxicity: review and current status. Curr Drug Deliv 8:79–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancho V, Moody TW, Mantey SA, Di Florio A, Uehara H, Coy DH, Jensen RT. (2010) Pharmacology of putative selective hBRS-3 receptor agonists for human bombesin receptors (BnR): affinities, potencies and selectivity in multiple native and BnR transfected cells. Peptides 31:1569–1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sano H, Feighner SD, Hreniuk DL, Iwaasa H, Sailer AW, Pan J, Reitman ML, Kanatani A, Howard AD, Tan CP. (2004) Characterization of the bombesin-like peptide receptor family in primates. Genomics 84:139–146. [DOI] [PubMed] [Google Scholar]
- Sebhat IK, Franklin C, Lo MM, Chen D, Jewell JP, Miller R, Pang J, Palyha O, Kan Y, Kelly TM, et al. (2010) Discovery of MK-5046, a potent, selective bombesin recptor subtype-3 agonist for the treatment of obesity. ACS Med Chem Lett 2:43–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegfried JM, Krishnamachary N, Gaither Davis A, Gubish C, Hunt JD, Shriver SP. (1999) Evidence for autocrine actions of neuromedin B and gastrin-releasing peptide in non-small cell lung cancer. Pulm Pharmacol Ther 12:291–302. [DOI] [PubMed] [Google Scholar]
- Strosberg JR, Wolin EM, Chasen B, Kulke M, Bushnell DL, Caplin ME, Baum RP, Kunz PL, Hobday TJ, Hendifar AE, et al. (2016) NETTER-1 phase III: progression-free survival, radiographic response, and preliminary overall survival results in patients with midgut neuroeneodcrine tumors treated with 177-Lu-Dotatate (Abstract). J Clin Oncol 34 (4 suppl):194.
- Tokita K, Hocart SJ, Katsuno T, Mantey SA, Coy DH, Jensen RT. (2001) Tyrosine 220 in the 5th transmembrane domain of the neuromedin B receptor is critical for the high selectivity of the peptoid antagonist PD168368. J Biol Chem 276:495–504. [DOI] [PubMed] [Google Scholar]
- Tsuda T, Kusui T, Hou W, Benya RV, Akeson MA, Kroog GS, Battey JF, Jensen RT. (1997) Effect of gastrin-releasing peptide receptor number on receptor affinity, coupling, degradation, and modulation. Mol Pharmacol 51:721–732. [DOI] [PubMed] [Google Scholar]
- Uehara H, González N, Sancho V, Mantey SA, Nuche-Berenguer B, Pradhan T, Coy DH, Jensen RT. (2011) Pharmacology and selectivity of various natural and synthetic bombesin related peptide agonists for human and rat bombesin receptors differs. Peptides 32:1685–1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varasteh Z, Aberg O, Velikyan I, Lindeberg G, Sörensen J, Larhed M, Antoni G, Sandström M, Tolmachev V, Orlova A. (2013) In vitro and in vivo evaluation of a (18)F-labeled high affinity NOTA conjugated bombesin antagonist as a PET ligand for GRPR-targeted tumor imaging. PLoS One 8:e81932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Schrenck T, Wang LH, Coy DH, Villanueva ML, Mantey S, Jensen RT. (1990) Potent bombesin receptor antagonists distinguish receptor subtypes. Am J Physiol 259:G468–G473. [DOI] [PubMed] [Google Scholar]
- Wang LH, Coy DH, Taylor JE, Jiang NY, Kim SH, Moreau JP, Huang SC, Mantey SA, Frucht H, Jensen RT. (1990a) Desmethionine alkylamide bombesin analogues: a new class of bombesin receptor antagonists with potent antisecretory activity in pancreatic acini and antimitotic activity in Swiss 3T3 cells. Biochemistry 29:616–622. [DOI] [PubMed] [Google Scholar]
- Wang LH, Coy DH, Taylor JE, Jiang NY, Moreau JP, Huang SC, Frucht H, Haffar BM, Jensen RT. (1990b) des-Met carboxyl-terminally modified analogues of bombesin function as potent bombesin receptor antagonists, partial agonists, or agonists. J Biol Chem 265:15695–15703. [PubMed] [Google Scholar]
- Waser B, Eltschinger V, Linder K, Nunn A, Reubi JC. (2007) Selective in vitro targeting of GRP and NMB receptors in human tumours with the new bombesin tracer 177Lu-AMBA. Eur J Nucl Med Mol Imaging 34:95–100. [DOI] [PubMed] [Google Scholar]
- Weber D, Berger C, Eickelmann P, Antel J, Kessler H. (2003) Design of selective peptidomimetic agonists for the human orphan receptor BRS-3. J Med Chem 46:1918–1930. [DOI] [PubMed] [Google Scholar]
- Weber D, Berger C, Heinrich T, Eickelmann P, Antel J, Kessler H. (2002) Systematic optimization of a lead-structure identities for a selective short peptide agonist for the human orphan receptor BRS-3. J Pept Sci 8:461–475. [DOI] [PubMed] [Google Scholar]
- Weber HC. (2009) Regulation and signaling of human bombesin receptors and their biological effects. Curr Opin Endocrinol Diabetes Obes 16:66–71. [DOI] [PubMed] [Google Scholar]
- Xiao C, Piñol RA, Carlin JL, Li C, Deng C, Gavrilova O, Reitman ML. (2017) Bombesin-like receptor 3 (Brs3) expression in glutamatergic, but not GABAergic, neurons is required for regulation of energy metabolism. Mol Metab 6:1540–1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao C, Reitman ML. (2016) Bombesin-like receptor 3: physiology of a functional orphan. Trends Endocrinol Metab 27:603–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada K, Ohki-Hamazaki H, Wada K. (2000a) Differential effects of social isolation upon body weight, food consumption, and responsiveness to novel and social environment in bombesin receptor subtype-3 (BRS-3) deficient mice. Physiol Behav 68:555–561. [DOI] [PubMed] [Google Scholar]
- Yamada K, Wada E, Wada K. (2000b) Bombesin-like peptides: studies on food intake and social behaviour with receptor knock-out mice. Ann Med 32:519–529. [DOI] [PubMed] [Google Scholar]
- Zang J, Mao F, Wang H, Zhang J, Liu Q, Peng L, Li F, Lang L, Chen X, Zhu Z. (2018) 68Ga-NOTA-RM26 PET/CT in the evaluation of breast cancer: a pilot prospective study. Clin Nucl Med 43:663–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Parks GS, Wang Z, Wang L, Lew M, Civelli O. (2013) Anatomical characterization of bombesin receptor subtype-3 mRNA expression in the rodent central nervous system. J Comp Neurol 521:1020–1039. [DOI] [PubMed] [Google Scholar]