Abstract
Immature neurons generated by the subpallial MGE tangentially migrate to the cortex where they become parvalbumin-expressing (PV+) and somatostatin (SST+) interneurons. Here, we show that the Sp9 transcription factor controls the development of MGE-derived cortical interneurons. SP9 is expressed in the MGE subventricular zone and in MGE-derived migrating interneurons. Sp9 null and conditional mutant mice have approximately 50% reduction of MGE-derived cortical interneurons, an ectopic aggregation of MGE-derived neurons in the embryonic ventral telencephalon, and an increased ratio of SST+/PV+ cortical interneurons. RNA-Seq and SP9 ChIP-Seq reveal that SP9 regulates MGE-derived cortical interneuron development through controlling the expression of key transcription factors Arx, Lhx6, Lhx8, Nkx2-1, and Zeb2 involved in interneuron development, as well as genes implicated in regulating interneuron migration Ackr3, Epha3, and St18. Thus, Sp9 has a central transcriptional role in MGE-derived cortical interneuron development.
Keywords: interneurons, Lhx6, Lhx8, medial ganglionic eminence, Nkx2-1, parvalbumin, somatostatin, Sp9, tangential migration
Introduction
The subpallium consists of the lateral, medial, and caudal ganglionic eminences (LGE, MGE, and CGE, respectively) and the preoptic area (POA). The MGE and CGE generate the vast majority of mammalian cortical interneurons. The MGE generates parvalbumin (PV+) and somatostatin (SST+) cortical interneurons, which constitute about 60% of the total (Wonders and Anderson 2006; Gelman and Marin 2010; Rubin et al. 2010; Brown et al. 2011; Rudy et al. 2011; Taniguchi et al. 2013). The MGE also generates striatal GABAergic and cholinergic interneurons and pallidal projection neurons (e.g., the globus pallidus) (Marin et al. 2000; Flandin et al. 2010; Nobrega-Pereira et al. 2010; Wang et al. 2014).
A cascade of transcription factors regulates MGE identity and differentiation. In ventricular zone (VZ) cells (radial glia), Nkx2-1 and Otx2 specify MGE regional identity (Sussel et al. 1999; Butt et al. 2005; Flandin et al. 2010; Hoch et al. 2015; Nord et al. 2015; Silberberg et al. 2016). Otx2 represses ventral identity (POA), whereas Nkx2-1 represses dorsal and caudal identities (LGE and CGE). In addition, Otx2 promotes radial glia maturation into neurons and oligodendrocytes (Hoch et al. 2015) and Nkx2-1 promotes the expression of subventricular zone (SVZ) (secondary progenitors) genes essential for neuronal differentiation (Lhx6 and Lhx8) and patterning/survival (Shh) (Sussel et al. 1999; Du et al. 2008; Silberberg et al. 2016). Nkx2-1 expression needs to be extinguished for interneurons to migrate to the cortex (Nobrega-Pereira et al. 2008); this is controlled in part via Dlx1&2 induction of Zeb2 (Zfhx1b, Sip1) (McKinsey et al. 2013; van den Berghe et al. 2013). Lhx6 is required for the differentiation of GABAergic PV+ and SST+ cortical interneurons (Alifragis et al. 2004; Liodis et al. 2007; Zhao et al. 2008; Neves et al. 2013). Lhx6 drives the expression of Arx and Ackr3 (Cxcr7) to regulate cortical interneuron differentiation and migration, respectively (Vogt et al. 2014). Lhx8 is required for the differentiation of striatal cholinergic neurons (Zhao et al. 2003; Fragkouli et al. 2005, 2009). Lhx6 and Lhx8 are partially redundant, and lacking both causes severe pallidal defects, including loss of Shh expression in pallidal neurons which reduces late-born cortical interneurons, and upregulation of Nkx2-1 expression in a subset of MGE-derived interneurons which induces an ectopic aggregation of them in the ventral telencephalon (Flandin et al. 2011).
Much of the progress in understanding how the MGE generates interneurons and projection neurons has come from the analysis of genes downstream of Dlx1&2 (Anderson et al. 1997; Long, Cobos, et al. 2009; Long, Swan, et al. 2009). The zinc finger transcription factor Sp9 is particularly interesting because its expression is similar to Dlx1&2 in the SVZ of the GEs, and yet its function and targets in the MGE are unknown. In the ventral LGE, Sp9 expression persists in striatopallidal medium-spiny neurons; its function is required for the generation, differentiation and postnatal survival of striatopallidal neurons (Zhang et al. 2016). In the dorsal LGE and postnatal SVZ, Sp9 and Sp8 coordinately regulate the development of olfactory bulb interneurons (Li et al. 2017). Here, we provide evidence that Sp9 regulates MGE-derived interneuron tangential migration in part through promoting Lhx6 expression. Deletion of Sp9 results in ectopic aggregation of MGE-derived interneurons in the embryonic ventral telencephalon, and approximately 50% reduction of MGE-derived cortical interneurons.
Materials and Methods
Mice
Mice were handled in accordance with the institutional guidelines. Nkx2-1-Cre (Xu et al. 2008), Lhx6-Cre (Fogarty et al. 2007), Rosa-YFP (Srinivas et al. 2001), Sp9LacZ/+ (Zhang et al. 2016), and Sp9Flox/+ (Zhang et al. 2016) mice were previously described. The day of vaginal plug detection was defined as embryonic day 0.5 (E0.5).
Immunohistochemistry
Immunohistochemistry was performed on 12 μm (embryonic brains) or 30 μm (postnatal brains) coronal sections according to previous studies (Zhang et al. 2016; Li et al. 2017). Briefly, sections were rinsed in TBS (0.01 M Tris-HCL + 0.9% NaCl, pH = 7.4), blocked with 5% donkey serum/TBS, and then incubated overnight at 4 °C with primary antibodies as followed: rabbit anti-Sp9 (1:500) (Zhang et al. 2016), rabbit anti-calretinin (CR) (1:3000, AB5054, Millipore), chicken anti-GFP (1:2000, GFP-1020, Aves labs), rabbit anti-PV (1:2000, PV25, Swant), rabbit anti-Nkx2-1 (1:500, sc-13 040, Santa Cruz), rabbit anti-Caspase-3 (1:300, 9661, Cell Signaling), goat anti-ChAT (1:400, AB144P, Millipore), mouse anti-Lhx6 (1:500, sc-271 433, Santa Cruz), rabbit anti-NPY (1:500, 22 940, Incstar), and goat anti-SST (1:500, sc-7819, Santa Cruz). Sections were then incubated with secondary antibodies (1:400, Jackson ImmunoResearch) for 2–3 h at room temperature.
In situ Hybridization
RNA in situ hybridization experiments were performed using digoxigenin riboprobes on 20-μm cryostat coronal sections as previously described (Long, Cobos et al. 2009; Long, Swan et al. 2009; McKinsey et al. 2013; Zhang et al. 2016; Li et al. 2017). Riboprobe templates were amplified by PCR with the following primers:
- Lhx6 Fwd: CAGAGAGGGCGCGCATGGTCACCT
- Lhx6 Rev: AATTGGGGGGGGGGTCTTTGGCAC
- Lhx8 Fwd: GCCTTAGTGTGGCTGAGAGA
- Lhx8 Rev: AGGATGGTAGGCTTTGTAAACT
- Shh Fwd: ACAGCTCACAAGTCCTCAGGTT
- Shh Rev: CGTCTCGATCACGTAGAAGACC
- Ackr3 Fwd: ATGGATGTGCACTTGTTTGACTATG
- Ackr3 Rev: GTTCTGTTCCAGGGCAGAGTACTC
- Maf Fwd: GCGCACCTGGAAGACTACTACTGGA
- Maf Rev: CTCCTTGTAGGCGTCCCTTTCG
- Arx Fwd: ATGAGCAATCAGTACCAGGAAGAGG
- Arx Rev: TTAGCACACCTCCTTCCCCGT
- Sp9 Fwd: ACCTGAATCGTGATTCCCAGCAG
- Sp9 Rev: TGCTATGGCTTTTGCAACCCAC
- Sp8 Fwd: TGGGTTGTCTGGTAGAACAGTG
- Sp8 Rev: CGCAAGCAGAGGAAAACTATCT
- NPY Fwd: GTGGATCTCTTCTCTCACAGAGGCA
- NPY Rev: ACAACAACAACAACAAGGGAAATGG
- Rnd2 Fwd: AGAGTGGCCGCTGCAAGATC
- Rnd2 Rev: TCACATGAGGTTACAGCTCTTGGC
- Epha3 Fwd: GTCACCTATGTTCTGGTCGGGAG
- Epha3 Rev: CCTCATGCCGTTAAGCCAATCAC
- Zeb2 Fwd: GCAAATGCTATGACTTAGCTCCCG
- Zeb2 Rev: AAGTAGCGGTGAGTCACATCCACG
- Sox6 Fwd: AAACTCGGCAGGGGCAGTCT
- Sox6: Rev: CGCTGCTATAGTCTGATTTGGGATC
- Id2 Fwd: GGTCTTCCTCCTACGAGCAGCA
- Id2 Rev: CTGGTTGTCTGAAATAAAGCAAGCA
- Dlx2 Fwd: TTTCCTGTCCCGGGTCAGGAT
- Dlx2 Rev: AAGTCTCAGACGCTGTCCACTCGA
- Ccnd2 Fwd: CCCAAGCTTGGGCTAGCAGATGACGAACACG
- Ccnd2 Rev: GCTCTAGATCCCATTCAGCCAAAGGAAGGA
RNA-Sequencing (RNA-Seq)
The MGEs from E12.5 Sp9LacZ/LacZ null mutant and wild-type brains (n = 3 per group) were dissected, and RNA was isolated with the Direct-zol RNA Miniprep kit (Zymo, catalog #R2050) following the manufacturer’s instructions (Li et al. 2017). The values of gene expression level were reported in fragments per kilobase of exon model per million mapped reads (Trapnell et al. 2012). For a gene to be called as differentially expressed, it required a P-value <0.05. RNA data from this experiment has been deposited in the GEO database (GSE99049).
ChIP-Seq
SP9 ChIP-seq was performed using dissected E13.5 lateral and medial ganglionic eminences (MGE) (including VZ, SVZ, and marginal zone, MZ) of wild-type CD1 mice (n = 2 replicas, 2 litters for each independent ChIP experiment) (Sandberg et al. 2016; Pla et al. 2017). Briefly, the E13.5 ganglionic eminences were fixed in 10 ml 1% formaldehyde for 10 min. Sonication was performed using Bioruptor® Pico (15–20 cycles) to obtain optimal fragment size in the range of 200–500 bp. The sheared chromatin (SDS concentration diluted to 0.1%) was incubated with 8 μg anti-SP9 rabbit polyclonal antibody at 4 °C overnight. ChIP-seq sequencing libraries were prepared from both the input DNA and the precipitated DNA using the NEBNext DNA Library Prep Kit and Illumina standard adaptors and were sequenced on the Illumina Hiseq 4000 with 150 bp pair-end reads. Reads from the ChIP and input libraries were mapped to the mouse genome (mm9) using Bowtie with default parameters. Peaks were called considering input as the control sample with filtering to remove peaks in repeat regions. ChIP-seq peaks were visualized using the UCSC genome browser.
Luciferase Assays
The DNA fragments of Lhx6 promoter, Lhx6 and Lhx8 putative enhancers (region 1-3) were amplified by PCR and subsequently cloned into pGL4.10 and pGL4.23 firefly luciferase vector (Promega) upstream of Luc2 gene, respectively (McKinsey et al. 2013).
The primers used for amplifying the Lhx6 promoter were:
Fwd: 5′-CCTGTTATATAAACCGGCGCTGAAC-3′
-
Rev: 5′-AGCGATTTAGCAAAGACATAGGCGA-3′
The primers used for amplifying the Lhx6 putative enhancers (Region 1 and Region 2) and the Lhx8 putative enhancer (Region 3) were the following:
- Region 1 Fwd: 5′-TCTAGACCTGGGAGAGACACACACG-3′
- Rev: 5′-AGCCATTTTCACTCTCTCCGAAGAA-3′
- Region 2 Fwd: 5′-GGAAACCTCAAGCTGCTCTTCGTAG-3′
- Rev: 5′-GAGACCCACCAGCTAGTTGCTAAAA-3′
- Region 3 Fwd: 5′-AGCGGGCGACTCGAGGACT-3′
- Rev: 5′-ACTGAGCACTAAGTGGCAATTCACA-3′
The coding sequence (CDS) of Sp9 was amplified by PCR from mouse genomic DNA and cloned into pCS2 (+) expression vector, using the following primers:
- Sp9 CDS Fwd: 5′-GACCTGAATCGTGATTCCCAGC-3′
- Rev: 5′-TGCAACCCACATAAACTTCATTGC-3′
Mouse embryonal carcinoma cell line P19 were grown in MEMα (Gibco 12571-063) supplemented with 10% fetal bovine serum (FBS, Gibco 10099-141). For luciferase assay, P19 cells transfections were performed in triplicate in 24-well plates by using Fugene HD transfection regent (Promega, E2311) according to the manufacturer’s protocol. The amounts of plasmids for each transfection were: 40 ng pGL4.73 (Promega, Renilla luciferase vector), 240 ng pCS2-empty or pCS2-Sp9, 240 ng pGL4.23-empty (pGL4.10-empty) (Promega), or pGL4.23-Element (pGL4.10-Promoter). Dual-luciferase assay was performed with a GloMax 20/20 Luminometer (Promega). The significance was determined via t-test.
Microscopy
Bright field images (in situ hybridization results) and some fluorescent images were imaged with Olympus BX 51 microscope using a 4× or 10× objective. Other fluorescent images were taken with the Olympus FV1000 confocal microscope system using 10×, 20×, 40×, or 60× objectives. Z-stack confocal images were reconstructed using the FV10-ASW software. All images were merged, cropped, and optimized in Photoshop CS5 without distortion of the original information.
Cell Counting
For embryonic mice, the numbers of GFP+ cells in E15.5 and E17.5 motor cortex were counted in 300-μm-wide bins that spanned from the MZ to VZ. The DAPI staining was used to demarcate MZ, cortical plate (CP), intermediate zone (IZ), SVZ, and VZ. The percentage of GFP+ cells in each layer was calculated. We counted cells from 5 sections of each mouse and analyzed 3 mice of each genotype (Nkx2-1-Cre; Sp9F/+; Rosa-YFP controls and Nkx2-1-Cre; Sp9F/LacZ; Rosa-YFP conditional mutants, Sp9-CKOs). We also counted the numbers of Caspased-3+ cells in the E17.5 ventral telencephalon (n = 3 mice for each genotype, 3 sections per mice).
For P30 mice, the numbers of GFP+, GFP+/PV+, GFP+/SST+, GFP+/NPY+, and GFP+/CR+ cells in primary motor cortex were analyzed in 500-μm-wide bins. The numbers of GFP+, GFP+/PV+, and GFP+/ChAT+ cells in the striatum were analyzed in a 2-mm2 area (n = 3 mice for each genotype, 3–5 sections per mice).
Statistics
Results were calculated as mean ± SEM (standard error of mean), and presented as fold change relative to control. Statistical significance was assessed using unpaired Student’s t-test or χ2 test. P-values less than 0.05 were considered significant.
Results
SP9 is Expressed in MGE Progenitors and Postmitotic Interneurons
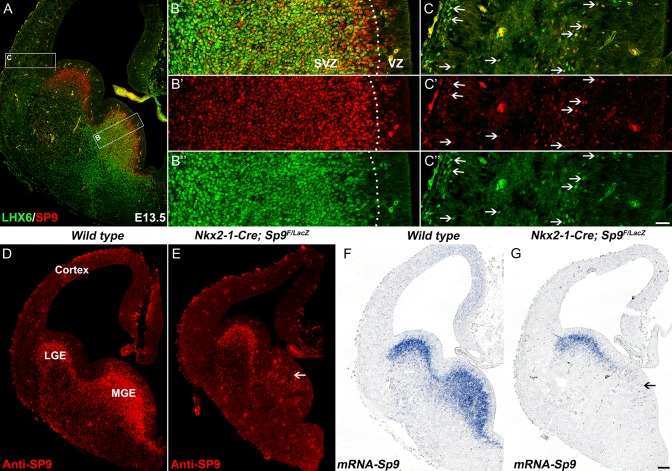
We recently showed that most, if not all, cortical interneurons are derived from Sp9-Cre expressing cells (Zhang et al. 2016). Indeed, in the E13.5 MGE, SP9 is broadly expressed in the MGE (Fig. 1A–B′″). SP9 expression begins in scattered cells in the VZ (Fig. 1B′, D), and then is expressed in virtually all SVZ cells (Fig. 1A–B″′). At the VZ-SVZ boundary (also known as SVZ1) (Petryniak et al. 2007), SP9 expression begins before LHX6 (Fig. 1B). Immature MGE-derived cortical interneurons migrate into the neocortex and continue to coexpress LHX6 and SP9 (Fig. 1C–C″′).
Figure 1.
SP9 is expressed in LHX6+ MGE progenitors and derived interneurons. (A) SP9/LHX6 double-immunostained coronal hemisection at E13.5. (B, C) Higher magnification images of boxed areas in (A) showing that the vast majority of LHX6+ cells expressed SP9 in the MGE SVZ and cortex (arrows). Note that some SP9+ cells were in the MGE VZ (B′). (D–F) SP9 expression was lost in most MGE cells in Nkx2-1-Cre; Sp9F/LacZ conditional knockout mice (Sp9-CKOs) at E13.5 (arrows). Scale bars: 100 μm in G for A, D–G; 20 μm in C″ for B–C″.
To investigate Sp9 function in the development of MGE-derived cells, we mainly used Sp9LacZ/LacZ constitutive null mutants and Nkx2-1-Cre; Sp9F/LacZ conditional (Sp9-CKO) mutants with or without the Rosa-YFP Cre reporter allele. The constitutive and conditional mutants had greatly reduced SP9 protein and Sp9 mRNA in the MGE (Fig. 1D–G) (Zhang et al. 2016).
Sp9 is Required for the Tangential Migration of MGE-Derived Interneurons
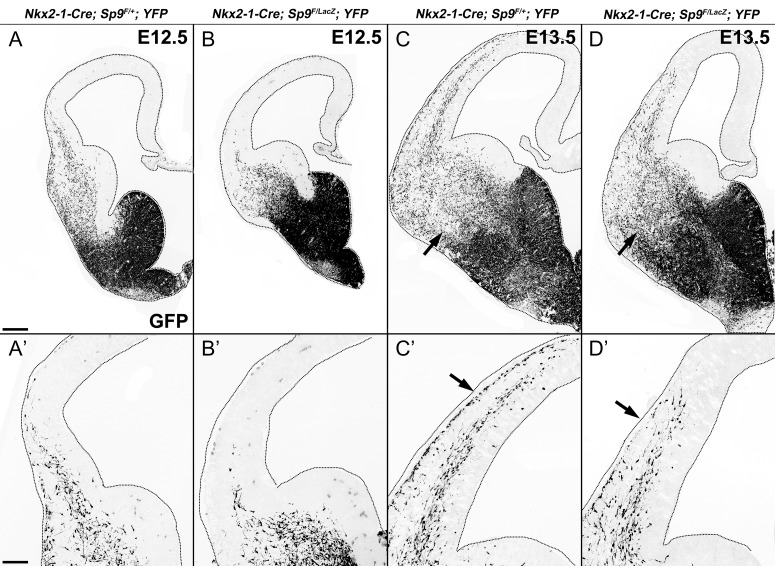
We next examined the role of Sp9 in the tangential migration of MGE-derived cortical inhibitory GABAergic interneurons in Sp9-CKOs. In E12.5 Nkx2-1-Cre; Sp9F/+; Rosa-YFP littermate controls, many GFP+ cells have entered the lateral cortex (Fig. 2A, A′), whereas, in the Sp9-CKOs very few GFP+ cells were detected in this region (Fig. 2B, B′). By E13.5, a large number of GFP+ cells were observed in the 2 cortical migration streams in the MZ and the IZ/SVZ (Fig. 2C, C′); the Sp9-CKOs showed a retardation in the progression of both migration streams, and had many fewer cells in the MZ (Fig. 2D, D′). At E15.5, the Sp9-CKOs continued to show a retardation into the migration into the dorsomedial cortex (Fig. 3A, B). Thus, Sp9 promotes the tangential migration of MGE-derived cortical interneurons.
Figure 2.
Tangential migration delay of MGE-derived cortical interneurons in Sp9-CKOs. (A–D′) GFP immunostaining on E12.5 and E13.5 coronal hemisections showed that MGE-derived Sp9 mutant interneurons migrate into the cortex less efficiently. Note the reduction of GFP+ cells in the marginal zone of Sp9-CKOs compared with controls at E13.5 (arrows in C′,D′). More GFP+ cells were observed in the ventral telencephalon of Sp9-CKOs (arrows in C, D). Scale bars: 200 μm in A for A–D; 100 μm A′ for A–D′.
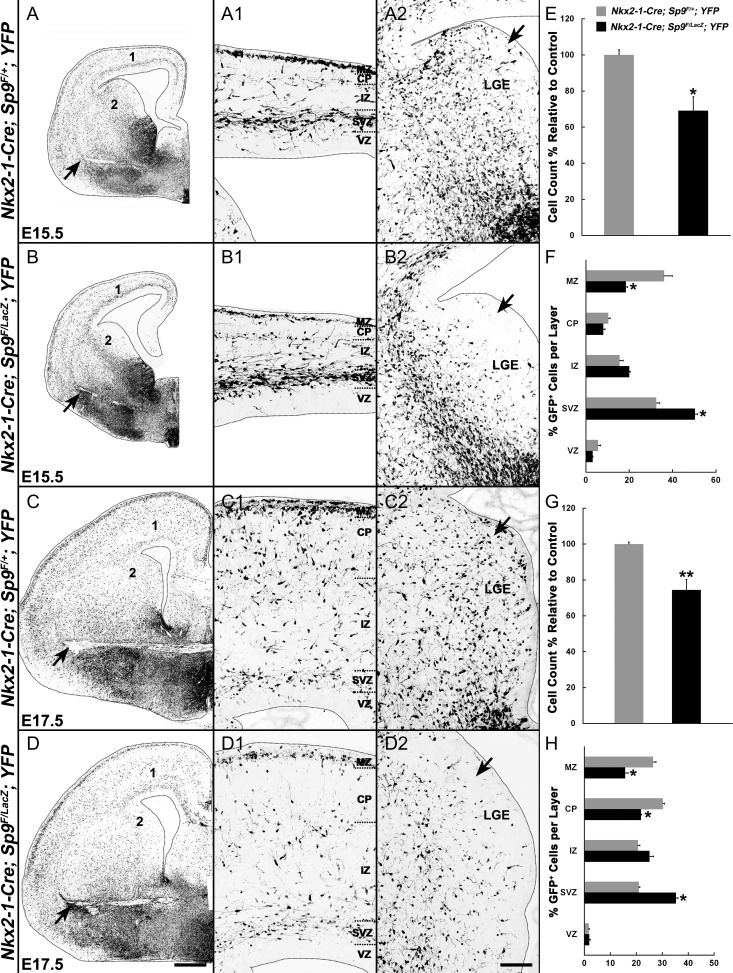
Figure 3.
MGE-derived interneurons are reduced and distributed abnormally in the cortex of Sp9-CKOs. (A–D2) GFP immunostaining on coronal hemisections showing the distribution of MGE-derived cells (GFP+) in telencephalic hemispheres at E15.5 and E17.5. Note that GFP+ cells in the LGE VZ/SVZ of mutants were absent or greatly reduced at E15.5 and at E17.5 (arrows in A2–D2). More GFP+ cells were observed in the ventral telencephalon of Sp9-CKOs (arrows in A–D). (E–H) Quantification showing that Sp9-CKO mice had reduced numbers of GFP+ cells in the cortex at E15.5 (E) and E17.5 (G). The percentage of GFP+ cells was reduced in the marginal zone (MZ) and cortical plate (CP) (F, H), and increased in the SVZ (F, H). IZ, intermediate zone. Student’s t-test (E, G) and χ2 test (F, H), *P < 0.05; **P < 0.01, n = 3. Scale bars: 200 μm in D for A–D; 100 μm D2 for A1–D2.
Sp9 Mutants Have an Altered Allocation of MGE-Derived Interneurons in the Deep and Superficial Tangential Migration Pathways
We then analyzed the numbers and distribution of MGE-derived (GFP+) cells in the cortex of Sp9-CKOs at E15.5 and E17.5. The total numbers of GFP+ cells in E15.5 and E17.5 mutant cortex were reduced by about 30% (Fig. 3A–D, E, G), as were the percentage of GFP+ migrating cells in the MZ and CP (Fig. 3A1–D1, F, H). In contrast, the percentage of mutant GFP+ migrating cells in the cortical SVZ was increased (Fig. 3A1–D1, F, H). Notably, in control embryos, a subpopulation of GFP+ cells tangentially migrates through the LGE VZ/SVZ (arrows, Fig. 3A2, C2). However, very few GFP+ cells were detected in the LGE VZ/SVZ in Sp9 conditional mutants at E15.5 and E17.5 (arrows, Fig. 3B2, D2, cell density was reduced roughly 2- to 3.5-fold). By contrast, within the Sp9-CKO LGE, there is an obvious corridor of migrating MGE-derived cells that are probably immature interneurons (Fig. 3B, B2). Thus, overall, while the loss of Sp9 function reduced E15.5 and E17.5 cortical interneurons in the superficial migration zone, they were increased in the deep migration zone, suggesting that Sp9 controls the allocation of interneurons that follow these distinct pathways.
MGE-Derived Interneurons in the Cortex are Greatly Reduced in Adult Sp9-CKOs
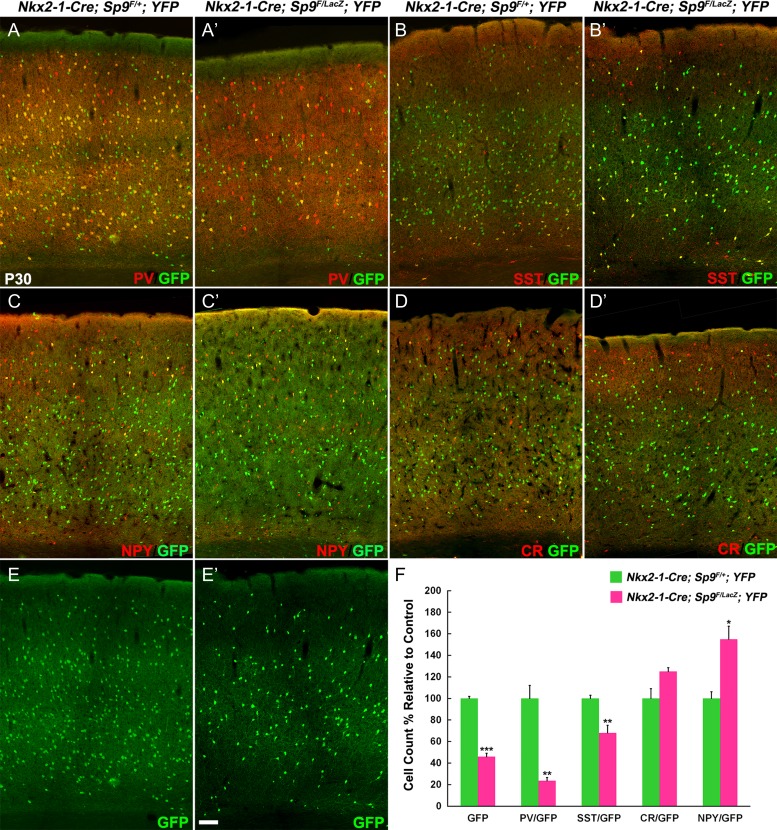
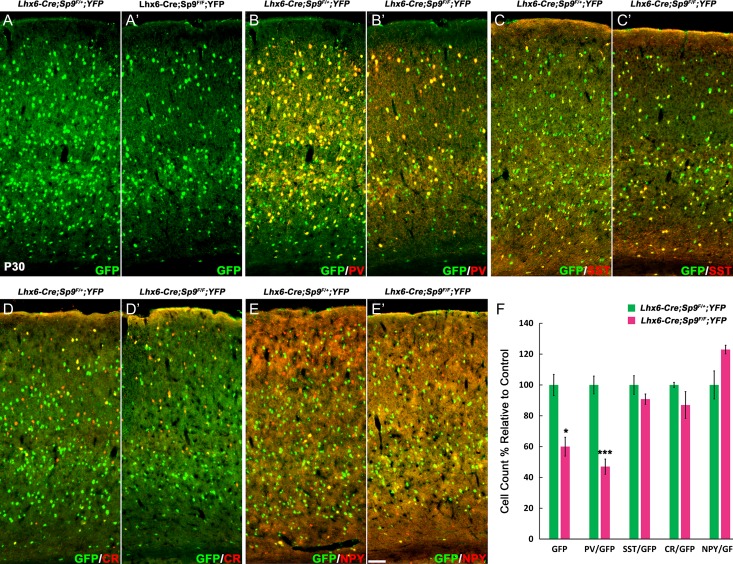
At P30, we found approximately 50% reduction of GFP+ neocortical cells in the Sp9-CKO (Fig. 4E, E′, F). We next quantified the numbers of GFP+/PV+, GFP+/SST+, GFP+/NPY+, and GFP+/CR+ interneurons in the primary motor cortex (Fig. 4A–D′). The Sp9-CKO mutants showed a > 70% reduction in GFP+/PV+ cells, and approximately 30% reduction in GFP+/SST+ cells (Fig. 4F). By contrast, GFP+/NPY+ cells were increased (Fig. 4F); GFP+/CR+ interneurons trended towards an increase (nonsignificant trend; Fig. 4F).
Figure 4.
Cortical interneurons are reduced in Nkx2-1-Cre; Sp9F/LacZ; Rosa-YFP conditional mutants (Sp9-CKOs). (A–E′) GFP and interneuron markers (PV, SST, CR, and NPY) double-immunostaining on P30 cortex sections. (F) Quantification showing that Sp9-CKOs had reduced GFP+, GFP+/PV+, and GFP+/SST+ cells, and had increased GFP+/NPY+ cells in the primary motor cortex. Student’s t-test, *P < 0.05; **P < 0.01, ***P < 0.001, n = 3. Scale bar: 50 μm.
SP9 is mainly expressed in the MGE SVZ and migrating MGE-derived interneurons, but not in the MGE VZ, therefore we hypothesized that loss of PV+ and SST+ cortical interneurons in Sp9-CKOs is mainly due to migration defects, not due to reduced cell proliferation. To test this hypothesis, we bred Sp9 floxed mice with Lhx6-Cre transgenic line in which Cre recombination occurs only in postmitotic MGE-derived neurons (Fogarty et al. 2007; Nobrega-Pereira et al. 2008). We again quantified the numbers of GFP+, GFP+/PV+, GFP+/SST+, GFP+/NPY+, and GFP+/CR+ interneurons in the primary motor cortex in control (Lhx6-Cre; Sp9F/+; Rosa-YFP) and Lhx6-Cre; Sp9F/F; Rosa-YFP mice at P30 (Fig. 5A–E′). Consistent with results from Sp9-CKOs (Fig. 4A–F), Lhx6-Cre; Sp9F/F; Rosa-YFP mice showed approximately 40% reduction in GFP+ cells and approximately 65% reduction in GFP+/PV+ cells (Fig. 5A–F). This result suggests that SP9 only has a minor role in regulating cell proliferation in the MGE, whereas, it has a major role in promoting MGE-derived interneuron migration.
Figure 5.
Cortical interneurons are reduced in Lhx6-Cre; Sp9F/F; Rosa-YFP mice. (A–E′) GFP and interneuron markers double-immunostaining on P30 cortex coronal sections. (F) Quantification of above experiments. GFP+ and GFP+/PV+ cells were significantly reduced. Student’s t-test, *P < 0.05; **P < 0.01, n = 3. Scale bar: 100 μm.
MGE-Derived Interneurons in the Striatum are Reduced in Adult Sp9-CKOs
MGE also generates striatal interneurons. Sp9-CKOs had approximately 70% reduction of GFP+/PV+ and GFP+/SST+ interneurons, whereas the number of GFP+/ChAT+ cells were normal (Supplementary Fig. S1A, A′, C–F′). Once again, Lhx6-Cre; Sp9F/F; Rosa-YFP mice had a reduction of PV+ or SST+ interneurons in the striatum (Supplementary Fig. S1B, B′, C, G–I′). This indicates that SP9 also promotes MGE-derived interneurons migrating into the striatum.
RNA-Seq Reveals Molecular Defects of the Sp9 Mutant MGE
Towards elucidating the mechanisms underlying the Sp9 mutant phenotypes, we used RNA-Seq to compare wild-type with Sp9 null mutant transcriptomes from the E12.5 MGE. We identified 392 dysregulated genes in the mutants; 188 with increased RNA expression and 204 with reduced RNA expression. Some of these genes were listed in the Supplementary Table S1. We have deposited the RNA-Seq data to NCBI’s GEO (GEO database GSE99049).
Differential expression was validated for several genes via in situ hybridization in E12.5–E15.5 embryos (Supplementary Figs S2, S3; Figs 6–8). For example, we observed that expression of Zeb2, Maf (c-maf), Rnd2 and St18 were greatly reduced (Supplementary Fig. S2A–L′), whereas expression of Sp8, Epha3 (Eph receptor A3) and Id2 were increased, and expression of Dlx2 and Ccnd2 (cyclin D2) were at the same level (Supplementary Fig. S3A–O′). Moreover, the expression of Lhx6, Lhx8, Shh, Ackr3, and Arx were also significantly reduced (Figs 6, 7), whereas expression of Sst, Npy, and Sox6 were increased (Fig. 8A–O′). Overall, the altered expression patterns of these genes mainly occurred in the MGE SVZ (Zeb2, Maf, Rnd2, St18, Sp8, Epha3, Id2, Lhx6, Lhx8, Ackr3, and Arx) and MGE-derived migrating interneurons (Epha3, Ackr3, Arx, Sst, Npy, Sox6, and NKX2-1), consistent with the Sp9 expression pattern during embryonic development, and these genes are also known for controlling cortical interneuron development (see below).
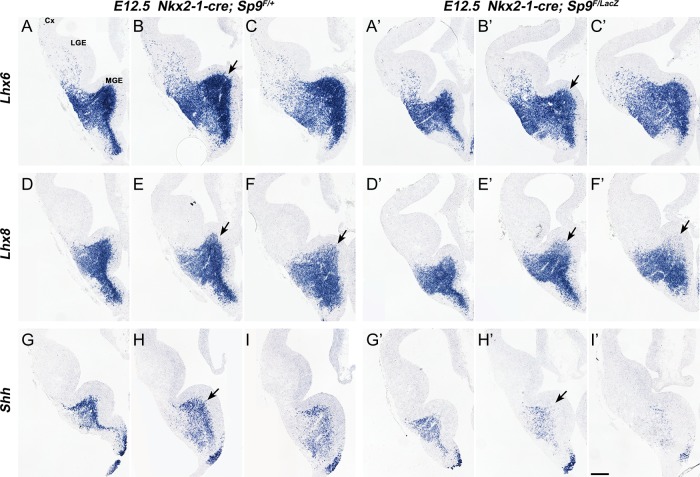
Figure 6.
Lhx6, Lhx8, and Shh expression were downregulated in the Sp9-CKO MGE. (A–I′) In situ RNA hybridization staining for Lhx6, Lhx8, and Shh on E12.5 3 serial coronal hemisections (rostral-most on the left) showed reduced Lhx6 expression in the MGE SVZ (arrows in B, B′), reduced Lhx8 expression in the dorsal MGE SVZ (arrows in E–F′) and reduced Shh expression in the MGE mantle zone (MZ) of Sp9-CKOs (arrows in H–H′). Cx, cortex. Scale bar: 200 μm.
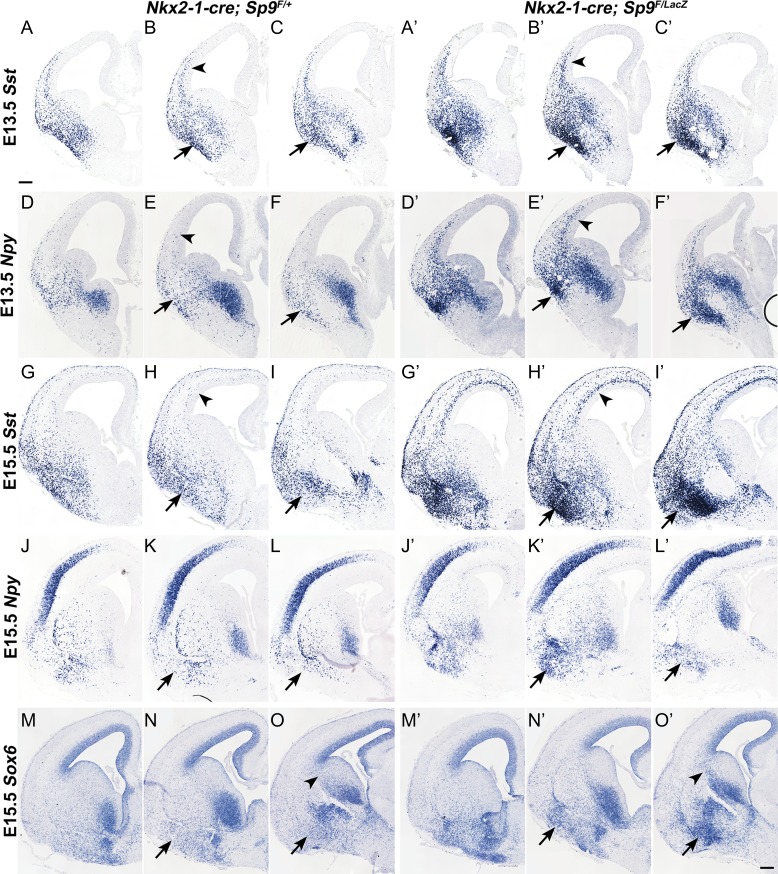
Figure 8.
Ectopic accumulation of MGE-derived interneurons in the Sp9-CKO ventral telencephalon. (A–O′) In situ RNA hybridization showing that MGE-derived Sst+, Npy+, and Sox6+ cells ectopically accumulated in the ventral telencephalon of mutants at E13.5 and E15.5 (arrows). Arrowheads in (B, B′, E, E′, H, H′) indicate upregulation of Sst and Npy expression in Sp9 mutant cortical interneurons. Scale bar: 200 μm.
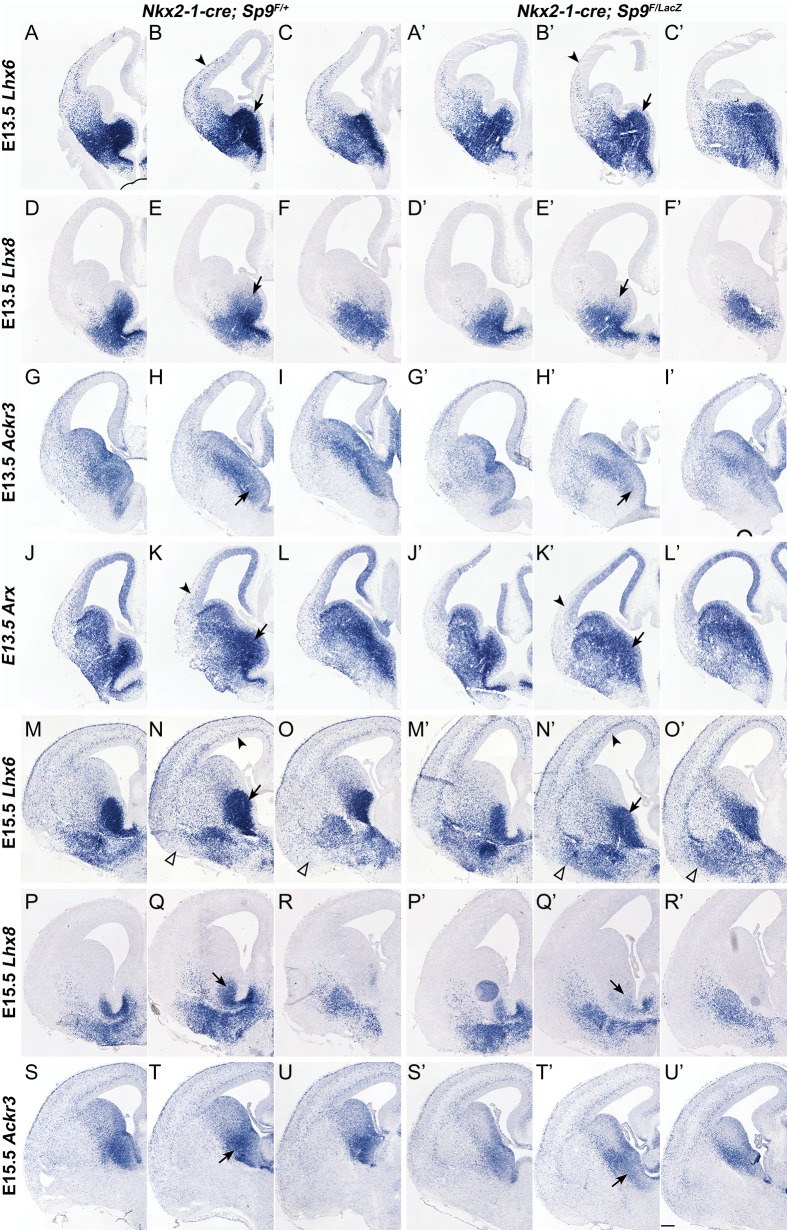
Figure 7.
Sp9-CKOs have reduced Lhx6, Lhx8, Ackr3, and Arx expression in the MGE. (A–U′) In situ RNA hybridization showing that expression of Lhx6, Lhx8, and LHX6 direct target genes, Ackr3 and Arx, were reduced in the mutant MGE at E13.5 and E15.5 (arrows). Arrowheads in (B, B′) indicate less efficient tangential migration of Lhx6+ cortical interneurons. Arrowheads in (K, K′) indicate reduced expression of Arx in cortical interneurons. Arrowheads in (N, N′) indicate increased Lhx6+ interneurons in the Sp9 mutant cortical SVZ. Open arrowheads in (N, N′, O, O′) indicate ectopic accumulation of Lhx6+ interneurons in the Sp9 mutant ventral telencephalon. Scale bar: 200 μm.
Lhx6, Lhx8, Shh, Ackr3, and Arx expression are Reduced in the Sp9 Mutant MGE
Lhx6 and Lhx8 have redundant functions in promoting cortical interneuron generation, in part through driving Shh expression and repressing Nkx2-1 expression in MGE neurons (Flandin et al. 2011). The Sp9 mutant MGE at E12.5 showed reduced Lhx6 expression in the SVZ (Fig. 6A–C′), reduced Lhx8 expression in the dorsal SVZ (arrows, Fig. 6D–F′) and reduced Shh neuronal expression in the MZ (Fig. 6G–I′). Reduced Lhx6 and Lhx8 SVZ expression in the E13.5 and E15.5 MGE was also observed (Fig. 7A–F′, M–R′). We suggest that the combined reduction of Lhx6, Lhx8, and Shh in the Sp9 mutant contributes to the reduced numbers and abnormal migration of the cortical interneurons. Furthermore, we provided evidence that LHX6 directly binds to and promotes the activity of Ackr3 and Arx enhancers (Vogt et al. 2014). Thus, reduced Lhx6 expression in the Sp9 mutant MGE probably contribute to the reduced Ackr3 and Arx expression in the MGE SVZ (in conjunction with the reduced Lhx8) and in the MGE-derived migrating cortical interneurons (Fig. 7G–L′ and S–U′).
MGE-Derived Interneurons Ectopically Accumulate in the Ventral Telencephalon of Sp9-CKO Embryos
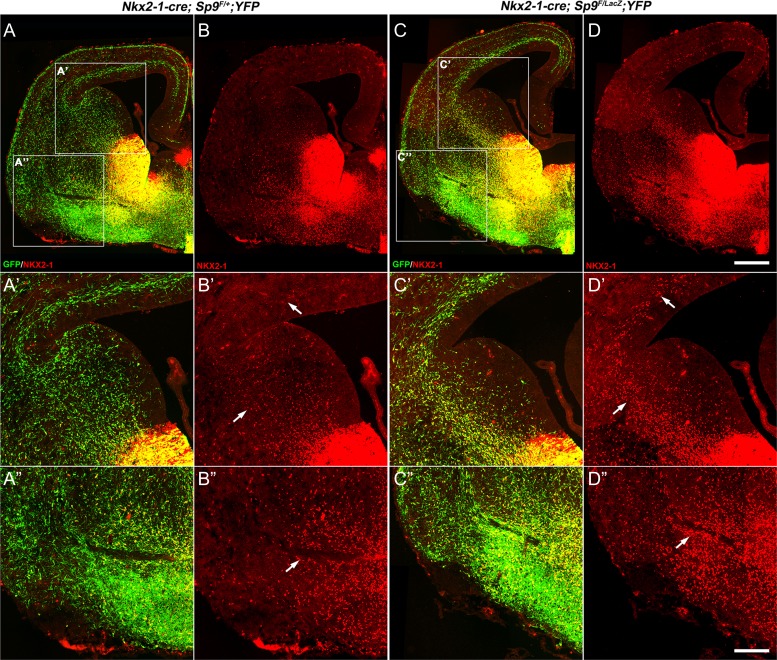
A mechanism that appears to contribute to the reduction in cortical and striatal interneurons in the Sp9-CKOs is the ectopic accumulation of MGE-derived cells in the ventral telencephalon appearing at E13.5, E15.5, and E17.5 (arrows, Figs 2C, D, and 3A–D). Cells in the ectopia are labeled by expression of cortical interneuron and MGE markers, including Lhx6 (arrowheads, Fig. 7M–O′), Npy, Sst, and Sox6 (arrows, Fig. 8A–O′). These cells also express NKX2-1 (arrows, Fig. 9D, D″), whose expression may contribute to their abnormal migration (Nobrega-Pereira et al. 2008). Upregulation of NKX2-1 expression was also observed in the LGE SVZ/MZ and cortical SVZ in Sp9 mutants (arrows, Fig. 9D′), probably contributed by reduced Lhx6 and Lhx8 expression (Flandin et al. 2011). When using the Lhx6-Cre line to conditionally delete Sp9, we also observed the ectopic accumulation of MGE-derived cells (NKX2-1+ and/or GFP+) in the ventral telencephalon (arrows, Supplementary Fig. S4A–F) and upregulation of NKX2-1 expression in the cortical SVZ (Supplementary Fig. S4C′). We found that MGE-derived interneurons expressed much higher levels of Sst, Npy, and Sox6 in Sp9-CKO embryos than those in controls (arrowheads, Fig. 8A–O′). Notably, although the number of MGE-derived cortical interneurons was reduced in the cortical MZ of Sp9 mutants (Figs 2 and 3), the remained cortical MZ interneurons also upregulated SST expression (arrows, Supplementary Fig. S5B′).
Figure 9.
Upregulation of NKX2-1 in MGE-derived cells in Sp9-CKOs. (A–D) NKX2-1/GFP double-immunostaining on coronal hemisections at E15.5. (A′–D′) Higher magnification view of GFP+ and/or NKX2-1+ cells in control (Nkx2-1-Cre; Sp9F/+; Rosa-YFP) and Sp9-CKOs. Note NKX2-1 upregulation in many MGE-derived migrating cortical interneurons in Sp9-CKOs (arrows in B′, D′). (A″–D″) A large ectopic aggregation of NKX2-1+ cells in the ventral telencephalon of Sp9-CKOs (arrows in B″, D″). Scale bars: 200 μm in (D) for (A–D); 100 μm in (D″) for (A′–D″).
Finally, we used Lhx6-GFP BAC transgenic mouse lines to examine Lhx6-GFP expression in the Sp9LacZ/LacZ null mutants at E14.5. We performed GFP/LacZ double-immunostaining. In the Lhx6-GFP; Sp9LacZ/+ control MGE and cortex, virtually all Lhx6-GFP+ cells expressed LacZ (Supplementary Fig. S6A). Lhx6-GFP was strongly expressed in the globus pallidus (Supplementary Fig. S6A, C), whereas LacZ expression was downregulated in this nucleus (Supplementary Fig. S6B) (Zhang et al. 2016). In the Lhx6-GFP; Sp9LacZ/LacZ mutant mice, LacZ expression was stronger relative to the Lhx6-GFP; Sp9LacZ/+ mice (Supplementary Fig. S6B, E), reflecting LacZ dosage. In control mice, more Lhx6-GFP+ cells were found in the MGE SVZ (Supplementary Fig. S6G), than in the mutant MGE SVZ (Supplementary Fig. S6H). Lhx6-GFP expression was also reduced in cells migrating through the mutant LGE VZ/SVZ (Supplementary Fig. S6G, H). Furthermore, the number of Lhx6-GFP+ interneurons in the Sp9 mutant E14.5 cortex was reduced by 60% relative to control (Supplementary Fig. S6I).
The reduction in cortical interneurons does not appear to be caused by an increased cell death, (as marked by cleaved Caspase-3+ cells) as we did not observe increased cell death in the MGE, the migration streams or in the cortex of Sp9 mutants (from E13.5 to P11). On the other hand, increased apoptosis was observed in the mutant’s ventral telencephalon at E17.5 (Supplementary Fig. S7A, B, E) which may reflect the ectopic accumulation of MGE-derived interneurons. Increased apoptosis was also seen in the dorsal striatum at P3 (Supplementary Fig. S7C, D, F).
SP9 Directly Binds to a Putative Enhancer of Lhx6
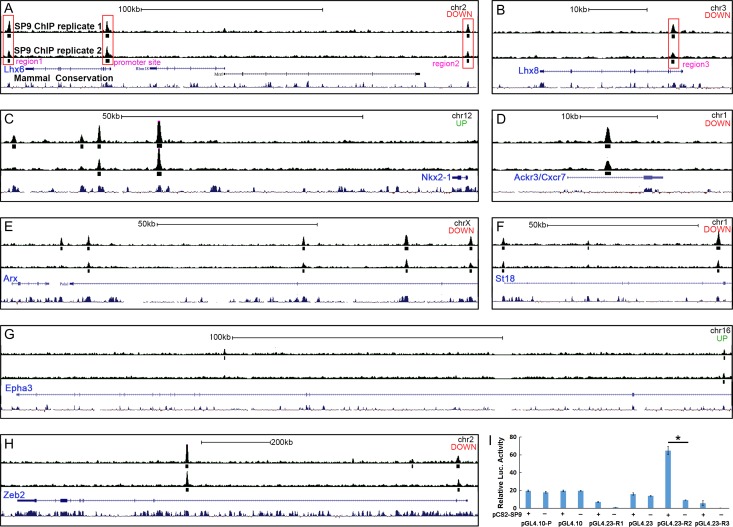
Coexpression of SP9 and LHX6 in MGE-derived migrating interneurons from the MGE SVZ to cortex indicates that SP9 may directly regulate Lhx6 expression. We thus performed chromatin coimmunoprecipitation followed by high-throughput DNA sequencing (ChIP-Seq) (n = 2 replicas) using E13.5 wild-type ganglionic eminences (LGE + MGE) (Sandberg et al. 2016; Pla et al. 2017). There was strong concordance in SP9 binding across replicates (Fig. 10A–H). We derived a set of 2 962 binding regions based on the presence of significant enrichment in SP9 ChIP libraries and no enrichment in the input control datasets. Furthermore, most of those genes that were deregulated in the Sp9 mutant MGE (Supplementary Table 1) had SP9 binding sites (Fig. 10A–H). We found one SP9 ChIP-Seq peak at the Lhx6 promoter region and 3 peaks at Lhx6 and Lhx8 putative enhancers (region 1-3), as each of these loci are evolutionarily (see mammal conservation track in Fig. 10A, B).
Figure 10.
Anti-SP9 ChIP-Seq Results. (A–H) “Called” peak (small rectangles) locations relative to genomic loci are shown for genes that are deregulated in Sp9-CKOs and for which ChIP-seq peaks were identified within 1 Mb of the gene body. Note the different scale bars for individual panels. For each panel, “up” or “down” indicates whether the gene was upregulated or downregulated in the Sp9 mutant MGE. chr, chromosome; kb, kilobase. Large red rectangles highlight where SP9 binds to the promoter and several downstream or upstream regions (regions 1-3) of Lhx6 and Lhx8 genes (A, B). (I) SP9 activated transcription from a putative Lhx6 enhancer (R2, region 2) in a dual-luciferase assay within P19 cells. (Student’s t-test, *P < 0.05, n = 3 individuals, mean ± SEM).
Next, to test the ability of SP9 to regulate these candidate regulatory elements of Lhx6 and Lhx8, we performed a Dual-luciferase transcription activation assay using P19 embryonal carcinoma cells. SP9 robustly activated transcription from an Lhx6′s putative enhancer (Region 2, Fig. 10A, I). In contrast, SP9 did not activate Lhx6 promoter and other Lhx6 and Lhx8 enhancers in P19 embryonal carcinoma cells (Region 1 and 3) (Fig. 10I). Together, these results provide evidence that SP9 directly, at least in part, promotes Lhx6 expression in MGE-derived migrating interneurons through its binding to an Lhx6 enhancer.
Discussion
Here, we show that Sp9 null and CKO mice have approximately 50% reduction in MGE-derived cortical interneurons, which we propose is mainly due to abnormal tangential migration. In the absence of Sp9, a subset of MGE-derived interneurons did not migrate into the cortex or striatum, resulting in their ectopic accumulation in superficial parts of the LGE and CGE and in the external capsule. In the Sp9 mutant embryonic cortex, fewer MGE-derived interneurons migrated in the cortical MZ, whereas, more migrated in the cortical SVZ. Finally, in the Sp9 mutant adult cortex, the SST+/PV+ ratio is increased in cortical interneurons. Lhx6 and Lhx8 expression are reduced in the MGE; we propose that this defect underlies, in part, the cortical interneuron defects.
Sp9 Regulates Tangential Migration of MGE-Derived Cortical Interneurons
Tangential migration to the cortex in the Sp9 mutants appeared reduced at E13.5 and E15.5 using MGE-lineage analysis (Figs 2 and 3). The ectopic accumulation of interneuron-like cells in superficial parts of the LGE and CGE, and in the external capsule (arrows Figs 2, 3, and 7–9), appears to be a major contributor to this phenotype. The persistent expression of NKX2-1 (Fig. 9) and EphA3 (Supplementary Fig. S3D–F′), in these cells, may contribute to their abnormal migration.
Prenatally, Sp9 mutants had a reduction of cortical interneurons in the superficial migration zone, whereas they had more cortical interneurons in the deep migration zone. Thus, Sp9 regulates the relative allocation of interneurons that follow these distinct pathways. It is possible that interneurons migrating along these paths have different fates, in terms of their cell types and laminar positioning. This may account for why the P30 cortex had more interneurons in the superficial layers and a fewer in the deep layers (data not shown). The abnormal ratio of superficial and deep tangentially migrating interneurons may also be related to the altered ratio of PV+ and SST+ interneurons (Figs 4 and 5). Further studies are needed to establish whether Sp9 expression in the MGE SVZ, and in the migrating interneurons, has distinct roles in regulating interneuron migration.
Normally, a subset of MGE-derived interneurons tangentially migrate into the cortex via the LGE VZ/SVZ, however, in the Sp9-CKOs very few GFP+ cells were found in this region (arrows in Fig. 3A2–D2). This could be explained by the upregulation of Epha3 expression in the Sp9 mutant MGE-derived migrating interneurons (Supplementary Fig. S3D–F′), as enhanced Eph/ephrin signaling in the LGE VZ/SVZ increases the repulsive effect on migrating interneurons (Zimmer et al. 2008; Rudolph et al. 2010; Villar-Cervino et al. 2015). Accordingly, in the postnatal brain, fewer GFP+ cells were observed in the medial part of the Sp9 mutant striatum (Supplementary Fig. S1A, A′). Notably, Ephrin-A3 is strongly expressed in the CP (Rudolph et al. 2010). Thus, upregulation of Epha3 expression also prevents migrating Sp9 mutant MGE-derived interneurons from entering the CP and pushes these interneurons into the SVZ (Fig. 3).
Sp9 Mutants Have an Increased Ratio of SST+ to PV+ Cortical Interneurons
Multiple transcriptional and signaling mechanisms control the specification and ratio of SST+ to PV+ cortical interneurons (Hu et al. 2017). Sp9 mutants have an increased ratio of SST+ to PV+ cortical interneurons (Figs 4 and 5). This could be due to differential cell death, although increases in apoptosis were only observed in the area of the ectopic interneuron accumulation and in the dorsal striatum (Supplementary Fig. S7). Alternatively, Sp9 may differentially regulate the generation of SST+ and PV+ interneurons. This possibility seems plausible given the robust increase in SST expression in cortical immature interneurons present at E13.5–15.5 (Fig 8, Supplementary Fig. S5).
SST interneurons are generally generated before PV interneurons (Xu et al. 2004; Butt et al. 2005; Miyoshi et al. 2007), and thus, Sp9 could alter their balance by controlling the timing of SST and PV specification. There is also evidence that SST+ interneurons arise via direct neurogenesis from VZ progenitors, whereas PV+ interneurons originate from SVZ progenitors of the MGE (Petros et al. 2015). Thus, perhaps Sp9, by virtue of its SVZ expression (Fig. 1), is particularly important in generating PV interneurons. Note that there is a selective deficit in cortical PV+ interneurons in mice lacking Ccnd2 in the SVZ of the MGE (Glickstein et al. 2007); however, Ccnd2 expression was not affected in the Sp9 mutant MGE, according to our RNA-Seq and in situ hybridization analysis (Supplementary Fig. S3M–O′).
Sp9 Regulation of Multiple Transcription Factors Controls Cortical Interneuron Development
There is evidence dorsal MGE has a bias for generating SST cortical interneurons whereas the ventral MGE has a bias for generating PV interneurons and the globus pallidus (Flandin et al. 2010; Nobrega-Pereira et al. 2010; Inan et al. 2012). Although Sp9 expression appears uniform across the MGE, development of pallidal projection neurons (e.g., globus pallidus) appears grossly normal, perhaps because Sp9 is not expressed in these neurons (Zhang et al. 2016). On the other hand, the Sp9 mutants have reduced numbers of both striatal and cortical interneurons (Figs 4 and 5; Supplementary Fig. S1). The specificity for MGE-derived interneurons, and the increase in the cortical interneuron SST/PV ratio, may reflect the combination of transcription factors that are altered in the Sp9 mutants.
Sp9 mutants have reduced expression of the Lhx6, Lhx8, Zeb2, and Maf transcription factors. Reductions in both Lhx6 and Lhx8 could explain why both cortical and striatal interneurons are affected (Flandin et al. 2011). The lack of a defect in striatal cholinergic neurons (Supplementary Fig. S1), which require Lhx8 function LGE (Zhao et al. 2003), implies further specificity of Sp9′s function. Dlx1&2 and Zeb2 mutants fail to express the Maf and Mafb transcription factors, whose expression is highly enriched in developing cortical interneurons (McKinsey et al. 2013). Downregulation of Zeb2 expression could explain why NKX2-1 expression was upregulated in several MGE derivatives: the ectopic interneurons in the superficial LGE, and interneurons migrating through the LGE SVZ and cortical SVZ (Fig. 9). Dlx1&2 control the generation of cortical interneurons through increasing expression of the Zeb2 transcription factor in the SVZ of the MGE, which in turn represses Nkx2-1 expression (McKinsey et al. 2013; van den Berghe et al. 2013). Thus, we propose that reduced expression of Lhx6, Lhx8, Zeb2, and Maf in the Sp9 mutant together contribute to the defects in cortical interneuron development.
To investigate whether these genes are directly regulated by SP9, we performed SP9 ChIP-seq using E13.5 ganglionic eminences. We found that SP9 binds to loci in the region of genes dysregulated in the Sp9 mutant: AckR3, Arx, EphA3, Lhx6, Lhx8, Nkx2-1, St18, and Zeb2 (Fig. 10A–H). Thus, Sp9, through direct activation (AckR3, Arx, Lhx6, Lhx8, St18, and Zeb2) and repression (Nkx2-1 and Epha3), has multiple molecular targets, and modes of function, in regulating development of MGE-derived cortical interneurons. As discussed above, this function appears to be particularly important of interneuron tangential migration. In this regard we recently demonstrated that Sp9 and Sp8 in the dLGE and postnatal SVZ also have critical roles in promoting migration of olfactory bulb interneurons (Li et al. 2017).
Most Lhx6 mutant MGE-derived cortical interneurons fail to differentiate to mature PV+ and SST+ interneurons, while this defect was not observed in Sp9 mutants. However, Sp9 mutants partially phenocopy the migration defects of MGE-derived interneurons in Lhx6 and Lhx6/Lhx8 mutants (Liodis et al. 2007; Zhao et al. 2008; Flandin et al. 2011; Vogt et al. 2014). A putative Lhx6 enhancer (Region 2 in Fig. 10A) was strongly activated by SP9 in luciferase reporter assays. Given that enhancers are required for normal development (Dickel et al. 2018), our results suggest that the Lhx6 enhancer may have a crucial role in promoting the tangential migration of MGE-derived interneurons.
Supplementary Material
Notes
The GEO accession number for the RNA-Seq data reported in this article is GSE 99 049. Conflict of Interest: J.L.R. is cofounder, stockholder, and currently serve on the scientific board of Neurona, a company studying the potential therapeutic use of interneuron transplantation. Conflict of interest: none declared.
Authors’ Contributions
Z.L. and Z.Z. performed experiments and analysis. S.L. performed SP9 ChIP-Seq. Z.L., Z.X., S.W., Q.L., Y.W., G.T., and Y.Y. helped conduct experiments and analyze the data. B.C., Y.W., and J.L.R. helped guide the project and discussed some of the results. Z.Y. designed the experiments and analyzed the results. Z.Y. and J.L.R. wrote the article.
Funding
Research grants to Z.Y. from the National Natural Science Foundation of China (NSFC 31630032, 31425011, and 31429002), research grant to Y.Y. (NSFC 31 700 889), research grants to B.C. from NIH (MH094589 and NS089777), and J.L. Rubenstein from NIH (MH049428).
References
- Alifragis P, Liapi A, Parnavelas JG. 2004. Lhx6 regulates the migration of cortical interneurons from the ventral telencephalon but does not specify their GABA phenotype. J Neurosci. 24:5643–5648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson SA, Eisenstat DD, Shi L, Rubenstein JL. 1997. Interneuron migration from basal forebrain to neocortex: dependence on Dlx genes. Science. 278:474–476. [DOI] [PubMed] [Google Scholar]
- Brown KN, Chen S, Han Z, Lu CH, Tan X, Zhang XJ, Ding L, Lopez-Cruz A, Saur D, Anderson SA, et al. 2011. Clonal production and organization of inhibitory interneurons in the neocortex. Science. 334:480–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butt SJ, Fuccillo M, Nery S, Noctor S, Kriegstein A, Corbin JG, Fishell G. 2005. The temporal and spatial origins of cortical interneurons predict their physiological subtype. Neuron. 48:591–604. [DOI] [PubMed] [Google Scholar]
- Dickel DE, Ypsilanti AR, Pla R, Zhu Y, Barozzi I, Mannion BJ, Khin YS, Fukuda-Yuzawa Y, Plajzer-Frick I, Pickle CS, et al. 2018. Ultraconserved enhancers are required for normal development. Cell. 172:491–499 e415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du T, Xu Q, Ocbina PJ, Anderson SA. 2008. NKX2.1 specifies cortical interneuron fate by activating Lhx6. Development. 135:1559–1567. [DOI] [PubMed] [Google Scholar]
- Flandin P, Kimura S, Rubenstein JL. 2010. The progenitor zone of the ventral medial ganglionic eminence requires Nkx2-1 to generate most of the globus pallidus but few neocortical interneurons. J Neurosci. 30:2812–2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flandin P, Zhao Y, Vogt D, Jeong J, Long J, Potter G, Westphal H, Rubenstein JL. 2011. Lhx6 and Lhx8 coordinately induce neuronal expression of Shh that controls the generation of interneuron progenitors. Neuron. 70:939–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogarty M, Grist M, Gelman D, Marin O, Pachnis V, Kessaris N. 2007. Spatial genetic patterning of the embryonic neuroepithelium generates GABAergic interneuron diversity in the adult cortex. J Neurosci. 27:10935–10946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fragkouli A, Hearn C, Errington M, Cooke S, Grigoriou M, Bliss T, Stylianopoulou F, Pachnis V. 2005. Loss of forebrain cholinergic neurons and impairment in spatial learning and memory in LHX7-deficient mice. Eur J Neurosci. 21:2923–2938. [DOI] [PubMed] [Google Scholar]
- Fragkouli A, van Wijk NV, Lopes R, Kessaris N, Pachnis V. 2009. LIM homeodomain transcription factor-dependent specification of bipotential MGE progenitors into cholinergic and GABAergic striatal interneurons. Development. 136:3841–3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelman DM, Marin O. 2010. Generation of interneuron diversity in the mouse cerebral cortex. Eur J Neurosci. 31:2136–2141. [DOI] [PubMed] [Google Scholar]
- Glickstein SB, Moore H, Slowinska B, Racchumi J, Suh M, Chuhma N, Ross ME. 2007. Selective cortical interneuron and GABA deficits in cyclin D2-null mice. Development. 134:4083–4093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoch RV, Lindtner S, Price JD, Rubenstein JL. 2015. OTX2 transcription factor controls regional patterning within the medial ganglionic eminence and regional identity of the septum. Cell Rep. 12:482–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu JS, Vogt D, Sandberg M, Rubenstein JL. 2017. Cortical interneuron development: a tale of time and space. Development. 144:3867–3878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inan M, Welagen J, Anderson SA. 2012. Spatial and temporal bias in the mitotic origins of somatostatin- and parvalbumin-expressing interneuron subgroups and the chandelier subtype in the medial ganglionic eminence. Cereb Cortex. 22:820–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Wang C, Zhang Z, Wen Y, An L, Liang Q, Xu Z, Wei S, Li W, Guo T, et al. 2017. Transcription factors Sp8 and Sp9 coordinately regulate olfactory bulb interneuron development. Cereb Cortex. 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liodis P, Denaxa M, Grigoriou M, Akufo-Addo C, Yanagawa Y, Pachnis V. 2007. Lhx6 activity is required for the normal migration and specification of cortical interneuron subtypes. J Neurosci. 27:3078–3089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long JE, Cobos I, Potter GB, Rubenstein JL. 2009. Dlx1&2 and Mash1 transcription factors control MGE and CGE patterning and differentiation through parallel and overlapping pathways. Cereb Cortex. 19(Suppl 1):i96–i106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long JE, Swan C, Liang WS, Cobos I, Potter GB, Rubenstein JL. 2009. Dlx1&2 and Mash1 transcription factors control striatal patterning and differentiation through parallel and overlapping pathways. J Comp Neurol. 512:556–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin O, Anderson SA, Rubenstein JL. 2000. Origin and molecular specification of striatal interneurons. J Neurosci. 20:6063–6076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinsey GL, Lindtner S, Trzcinski B, Visel A, Pennacchio LA, Huylebroeck D, Higashi Y, Rubenstein JL. 2013. Dlx1&2-dependent expression of Zfhx1b (Sip1, Zeb2) regulates the fate switch between cortical and striatal interneurons. Neuron. 77:83–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi G, Butt SJ, Takebayashi H, Fishell G. 2007. Physiologically distinct temporal cohorts of cortical interneurons arise from telencephalic Olig2-expressing precursors. J Neurosci. 27:7786–7798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neves G, Shah MM, Liodis P, Achimastou A, Denaxa M, Roalfe G, Sesay A, Walker MC, Pachnis V. 2013. The LIM homeodomain protein Lhx6 regulates maturation of interneurons and network excitability in the mammalian cortex. Cereb Cortex. 23:1811–1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobrega-Pereira S, Gelman D, Bartolini G, Pla R, Pierani A, Marin O. 2010. Origin and molecular specification of globus pallidus neurons. J Neurosci. 30:2824–2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobrega-Pereira S, Kessaris N, Du T, Kimura S, Anderson SA, Marin O. 2008. Postmitotic Nkx2-1 controls the migration of telencephalic interneurons by direct repression of guidance receptors. Neuron. 59:733–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nord AS, Pattabiraman K, Visel A, Rubenstein JL. 2015. Genomic perspectives of transcriptional regulation in forebrain development. Neuron. 85:27–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petros TJ, Bultje RS, Ross ME, Fishell G, Anderson SA. 2015. Apical versus basal neurogenesis directs cortical interneuron subclass fate. Cell Rep. 13:1090–1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petryniak MA, Potter GB, Rowitch DH, Rubenstein JL. 2007. Dlx1 and Dlx2 control neuronal versus oligodendroglial cell fate acquisition in the developing forebrain. Neuron. 55:417–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pla R, Stanco A, Howard MA, Rubin AN, Vogt D, Mortimer N, Cobos I, Potter GB, Lindtner S, Price JD, et al. 2017. Dlx1 and Dlx2 promote interneuron GABA synthesis, synaptogenesis, and dendritogenesis. Cereb Cortex. doi: 10.1093/cercor/bhx241. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin AN, Alfonsi F, Humphreys MP, Choi CK, Rocha SF, Kessaris N. 2010. The germinal zones of the basal ganglia but not the septum generate GABAergic interneurons for the cortex. J Neurosci. 30:12050–12062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudolph J, Zimmer G, Steinecke A, Barchmann S, Bolz J. 2010. Ephrins guide migrating cortical interneurons in the basal telencephalon. Cell Adh Migr. 4:400–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudy B, Fishell G, Lee S, Hjerling-Leffler J. 2011. Three groups of interneurons account for nearly 100% of neocortical GABAergic neurons. Dev Neurobiol. 71:45–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandberg M, Flandin P, Silberberg S, Su-Feher L, Price JD, Hu JS, Kim C, Visel A, Nord AS, Rubenstein JL. 2016. Transcriptional networks controlled by NKX2-1 in the development of forebrain GABAergic neurons. Neuron. 91:1260–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silberberg SN, Taher L, Lindtner S, Sandberg M, Nord AS, Vogt D, McKinsey GL, Hoch R, Pattabiraman K, Zhang D, et al. 2016. Subpallial enhancer transgenic lines: a data and tool resource to study transcriptional regulation of GABAergic cell fate. Neuron. 92:59–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivas S, Watanabe T, Lin CS, William CM, Tanabe Y, Jessell TM, Costantini F. 2001. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev Biol. 1:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sussel L, Marin O, Kimura S, Rubenstein JL. 1999. Loss of Nkx2.1 homeobox gene function results in a ventral to dorsal molecular respecification within the basal telencephalon: evidence for a transformation of the pallidum into the striatum. Development. 126:3359–3370. [DOI] [PubMed] [Google Scholar]
- Taniguchi H, Lu J, Huang ZJ. 2013. The spatial and temporal origin of chandelier cells in mouse neocortex. Science. 339:70–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. 2012. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 7:562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Berghe V, Stappers E, Vandesande B, Dimidschstein J, Kroes R, Francis A, Conidi A, Lesage F, Dries R, Cazzola S, et al. 2013. Directed migration of cortical interneurons depends on the cell-autonomous action of Sip1. Neuron. 77:70–82. [DOI] [PubMed] [Google Scholar]
- Villar-Cervino V, Kappeler C, Nobrega-Pereira S, Henkemeyer M, Rago L, Nieto MA, Marin O. 2015. Molecular mechanisms controlling the migration of striatal interneurons. J Neurosci. 35:8718–8729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt D, Hunt RF, Mandal S, Sandberg M, Silberberg SN, Nagasawa T, Yang Z, Baraban SC, Rubenstein JL. 2014. Lhx6 directly regulates Arx and CXCR7 to determine cortical interneuron fate and laminar position. Neuron. 82:350–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, You Y, Qi D, Zhou X, Wang L, Wei S, Zhang Z, Huang W, Liu Z, Liu F, et al. 2014. Human and monkey striatal interneurons are derived from the medial ganglionic eminence but not from the adult subventricular zone. J Neurosci. 34:10906–10923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wonders CP, Anderson SA. 2006. The origin and specification of cortical interneurons. Nat Rev Neurosci. 7:687–696. [DOI] [PubMed] [Google Scholar]
- Xu Q, Cobos I, De La Cruz E, Rubenstein JL, Anderson SA. 2004. Origins of cortical interneuron subtypes. J Neurosci. 24:2612–2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Q, Tam M, Anderson SA. 2008. Fate mapping Nkx2.1-lineage cells in the mouse telencephalon. J Comp Neurol. 506:16–29. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Zhang Y, Wang C, Xu Z, Liang Q, An L, Li J, Liu Z, You Y, He M, et al. 2016. The zinc finger transcription factor Sp9 is required for the development of striatopallidal projection neurons. Cell Rep. 16:1431–1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Flandin P, Long JE, Cuesta MD, Westphal H, Rubenstein JL. 2008. Distinct molecular pathways for development of telencephalic interneuron subtypes revealed through analysis of Lhx6 mutants. J Comp Neurol. 510:79–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Marin O, Hermesz E, Powell A, Flames N, Palkovits M, Rubenstein JL, Westphal H. 2003. The LIM-homeobox gene Lhx8 is required for the development of many cholinergic neurons in the mouse forebrain. Proc Natl Acad Sci USA. 100:9005–9010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmer G, Garcez P, Rudolph J, Niehage R, Weth F, Lent R, Bolz J. 2008. Ephrin-A5 acts as a repulsive cue for migrating cortical interneurons. Eur J Neurosci. 28:62–73. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.