BCR‐ABL1‐like B‐lymphoblastic leukemia, also termed Philadelphia‐like B‐acute lymphoblastic leukemia (Ph‐like B‐ALL), shares a gene expression profile similar to BCR‐ABL1‐positive B‐ALL1 but lacks BCR‐ABL1 fusion. Because Ph‐like status confers a poor prognosis, identification of Ph‐like B‐ALL cases is important. This brief communication reports 450 cases of B‐ALL evaluated by genomic profiling.
Abstract
BCR‐ABL1‐like B‐Acute Lymphoblastic Leukemia (B‐ALL) is a subset of B‐ALL with a poor prognosis that is found in all age groups. Definitive identification of these patients is difficult in routine clinical practice as gene expression profiling, the gold standard test, is not widely available. Comprehensive genomic profiling performed on 450 patients with extensive fusion profiling revealed a wide range of genomic alterations which were consistent with a classification of BCR‐ABL1‐like B‐ALL in 29% of cases. This manuscript highlights a clinically available alternative method for identifying a large subset of patients with BCR‐ABL1‐like B‐ALL.
Introduction
BCR‐ABL1‐like B‐lymphoblastic leukemia, also termed Philadelphia‐like B‐acute lymphoblastic leukemia (Ph‐like B‐ALL), shares a similar gene expression profile with BCR‐ABL1 positive B‐ALL [1], while lacking the BCR‐ABL1 fusion. Identification of Ph‐like B‐ALL cases is crucial, as Ph‐like status confers a poor prognosis, with increased rates of induction failure and early disease relapse [1], [2], [3]. Ph‐like B‐ALL is found across all age groups, in approximately 10% of pediatric (≤18 years) patients [4], 30% of young adult (>18 to ≤40 years) patients [4], [5], and 20% of adult patients (>40 years) [5] with B‐ALL.
Gene expression profiling is used to definitively determine Ph‐like status [6]; however, as gene expression profiling for B‐ALL is not widely available in routine clinical practice, it is challenging to identify these cases. Previous studies reported that approximately 80% of Ph‐like cases have genomic alterations that activate the JAK‐STAT pathway or other kinases [4], [6]. Comprehensive genomic profiling using parallel DNA and RNA sequencing allows for extensive fusion detection and broad interrogation of the JAK‐STAT pathway as well as other kinases to identify a large majority of patients with Ph‐like B‐ALL and provide potential targeted treatment options.
Discussion
In this study, 450 cases of B‐ALL submitted during routine clinical care were evaluated by comprehensive genomic profiling that interrogated 406 genes via DNA sequencing for all classes of genomic alterations (GAs), including point mutations, insertions/deletions, copy number amplifications, homozygous deletions, and gene rearrangements, and 265 genes via RNA sequencing for rearrangements, using a College of American Pathologists‐accredited, Clinical Laboratory Improvement Amendments (CLIA)‐certified, New York State‐approved, hybrid capture next‐generation sequencing assay [7]. All genomic alterations reported are known or predicted to be pathogenic. Patients for the cohort samples were 46% female and 54% male (median age 26 years, range 1–89 years).
BCR‐ABL rearrangements were detected in 14% (64/450) of cases, a frequency lower than expected because of referral bias against tumors with known BCR‐ABL fusions.
Cases were designated as Ph‐like B‐ALL if activating GAs were detected in the JAK‐STAT pathway or affecting related kinases, specifically GAs in ABL1, ABL2, CRLF2, CSF1R, EPOR, IL7R, NTRK1, NTRK2, NTRK3, JAK1, JAK2, JAK3, PDGFRA, PDGFRB, or SH2B3 [3], [4], [8]. Many of these GAs, especially gene fusions, are not evaluated as part of the routine clinical diagnostic evaluation. The spectrum of alterations detected in the cohort varied by age group, with pediatric (n = 174) and young adult (n = 113) patients more often harboring kinase fusions (77% and 81%, respectively), compared with adult (n = 163) patients (63%).
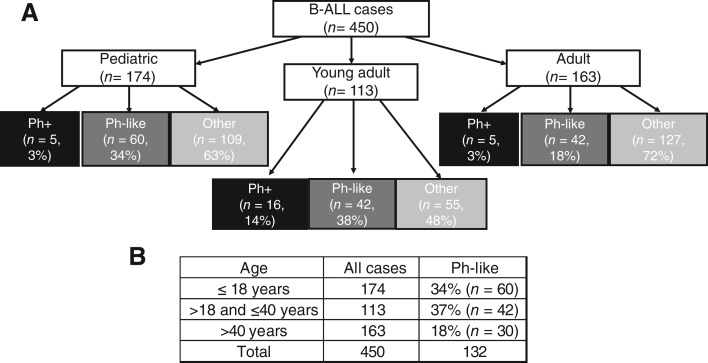
Pediatric (≤18 years) and young adult (>18 to ≤40 years) patients were more often Ph‐like (34% and 38%) compared with adults over 40 years (18%; Fig. 1). This increased incidence compared with the overall B‐ALL patient population is likely secondary to an enrichment for relapsed/refractory cases and a reduced frequency of Ph+ cases because of prior screening.
Figure 1.
(A): Distribution of cases into pediatric (≤18 years), young adult (>18 to ≤40 years), and adult (>40 years) groups, with a further delineation of the proportion of each group that comprises Ph+ cases, Ph‐like cases, and all other cases; (B): Distribution of cases into pediatric (≤18 years), young adult (>18 to ≤40 years), and adult (>40 years) groups with the percentage of cases that are Ph‐like in each group.Abbreviations: B‐ALL, B‐acute lymphoblastic leukemia; Ph, Philadelphia.
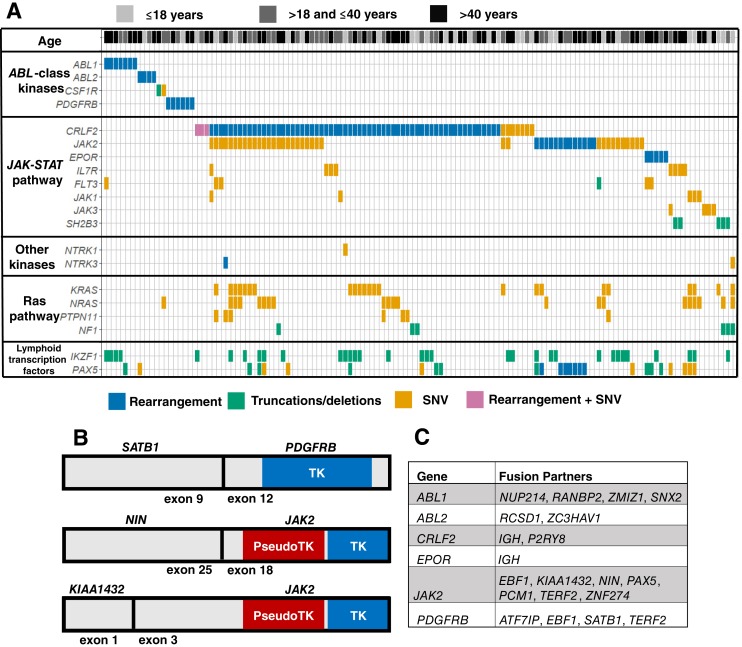
In total, 29% (132/450) of all cases were identified as Ph‐like B‐ALL. As illustrated in Figure 2A, 14% (n = 19) of Ph‐like cases had GAs in an ABL class kinase, whereas 86% (n = 113) of cases harbored a GA in the JAK‐STAT pathway. Of this JAK‐STAT activated group, 41% (n = 46) had an alteration in CRLF2 only, 23% (n = 26) had an alteration in CRLF2 and JAK2, and 20% (n = 23) had an alteration in JAK2 only (Fig. 2A). The JAK2 point mutations were activating alterations at positions other than V617F, consistent with previous reports in Ph‐like B‐ALL [3], [4]. The precise GAs in each gene are outlined in supplemental online Table 1. Of the 132 Ph‐like B‐ALL cases, 42 (32%) harbored concomitant RAS pathway alterations (KRAS, NRAS, PTPN11, or NF1). Three novel fusions were identified in this cohort, one involving PDGFRB and two involving JAK2 (Fig. 2B), all of which preserved the tyrosine kinase and tyrosine kinase‐like domains. Overall, a wide variety of kinase fusion partners was seen (Fig. 2C), which is typical for Ph‐like B‐ALL [4].
Figure 2.
Tile plot, gene schematics, and fusion partners list. (A): Tile plot for 132 cases defined as Ph‐like B‐ALL based on detected genomic alterations in ABL1, ABL2, CRLF2, CSF1R, EPOR, IL7R, NTRK1, NTRK2, NTRK3, JAK1, JAK2, JAK3, PDGFRA, PDGFRB, or SH2B3. Genomic alteration class is indicated by color. Patient age for each case is represented in the top line with light grey for pediatric cases, dark grey for young adult cases, and black for adult cases. (B): Gene schematics for novel kinase fusions identified. Blue represents the tyrosine kinase domains, and red represents pseudo‐tyrosine kinase domains, both of which are intact and in frame for all three fusions. (C): Complete list of fusion partners identified for ABL class kinases or members of the JAK‐STAT pathway. All kinases have intact kinase domains in the fusions reported.Abbreviations: SNV, single nucleotide variant; TK, tyrosine kinase.
The remaining samples (n = 291), designated BCR‐ABL‐negative and Ph‐like alteration‐negative cases, harbored genomic alterations in CDKN2A and/or CDKN2B (31%), TP53 (31%), KRAS (19%), NRAS (17%), FLT3 (10%), or ETV6‐RUNX1 fusions (14%). A subset of cases with RAS pathway alterations and FLT3 alterations likely represent Ph‐like B‐ALL cases that cannot be definitively identified by genomics alone [4], [8].
Conclusion
Approximately 80% of Ph‐like B‐ALL cases can be identified by the detection of genomic alterations affecting the JAK‐STAT pathway or other kinases, including a diverse array of gene rearrangements [4], [6]. Thus, a significant subset of Ph‐like B‐ALL cases can be identified through comprehensive genomic profiling that includes extensive fusion detection. In our cohort, 31% of patients with B‐ALL had genomic alterations consistent with Ph‐like B‐ALL. Ph‐like B‐ALL is a critically important subgroup of B‐ALL to identify, as many of these patients either fail standard of care induction therapy or have early relapse. Improved therapy is a critical need for this population of patients. Clinical trials treating patients with Ph‐like B‐ALL by targeting the driver alteration with tyrosine kinases inhibitors are in progress. Ideally, Ph‐like status would be determined by gene expression profiling for prognostication, and comprehensive genomic profiling would be used to direct patients toward targeted therapies. Currently gene expression profiling is not available in routine clinical practice, leaving many clinicians with few options to identify and appropriately treat these patients. This article highlights an alternate method for identifying a majority of Ph‐like B‐ALL patients using a clinically available CLIA‐certified assay for comprehensive genomic profiling with extensive fusion detection, with data from a large cohort showing the spectrum of genomic alterations detected in this manner.
See http://www.TheOncologist.com for supplemental material available online.
Disclosures
Eric A. Severson: Foundation Medicine (E, OI); Jo‐Anne Vergilio: Foundation Medicine (E, OI); Laurie M. Gay: Foundation Medicine (E, OI); Sugganth Daniel: Foundation Medicine (E, OI); Amanda C. Hemmerich: Foundation Medicine (E, OI); Julia A. Elvin: Foundation Medicine (E, OI); Nicholas Britt: Foundation Medicine (E); Michelle Nahas: Foundation Medicine (E, OI); Charlotte Brown: Foundation Medicine (E, OI); Pratheesh Sathyan: Foundation Medicine (E, OI); Andrew Rankin: Foundation Medicine (E, OI); Vincent Miller: Foundation Medicine (E, OI), Revolution Medicines (H, OI); Jeffrey S. Ross: Foundation Medicine (E, OI); Shakti H. Ramkissoon: Foundation Medicine (E, OI). The other author indicated no financial relationships.
(C/A) Consulting/advisory relationship; (RF) Research funding; (E) Employment; (ET) Expert testimony; (H) Honoraria received; (OI) Ownership interests; (IP) Intellectual property rights/inventor/patent holder; (SAB) Scientific advisory board
References
- 1.Den Boer ML, van Slegtenhorst M, De Menezes RX et al. A subtype of childhood acute lymphoblastic leukaemia with poor treatment outcome: A genome‐wide classification study. Lancet Oncol 2009;10:125–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.van der Veer A, Waanders E, Pieters R et al. Independent prognostic value of BCR‐ABL1‐like signature and IKZF1 deletion, but not high CRLF2 expression, in children with B‐cell precursor ALL. Blood 2013;122:2622–2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jain N, Roberts KG, Jabbour E et al. Ph‐like acute lymphoblastic leukemia: A high‐risk subtype in adults. Blood 2017;129:572–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Roberts KG, Li Y, Payne‐Turner D et al. Targetable kinase‐activating lesions in Ph‐like acute lymphoblastic leukemia. N Engl J Med 2014;371:1005–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roberts KG, Gu Z, Payne‐Turner D et al. High frequency and poor outcome of Philadelphia chromosome‐like acute lymphoblastic leukemia in adults. J Clin Oncol 2017;35:394–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reshmi SC, Harvey RC, Roberts KG et al. Targetable kinase gene fusions in high‐risk B‐ALL: A study from the Children's Oncology Group. Blood 2017;129:3352–3361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.He J, Abdel‐Wahab O, Nahas MK et al. Integrated genomic DNA/RNA profiling of hematologic malignancies in the clinical setting. Blood 2016;127:3004–3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Roberts KG, Morin RD, Zhang J et al. Genetic alterations activating kinase and cytokine receptor signaling in high‐risk acute lymphoblastic leukemia. Cancer Cell 2012;22:153–166. [DOI] [PMC free article] [PubMed] [Google Scholar]