Abstract
Rationale: Screening for non–small cell lung cancer is associated with earlier diagnosis and reduced mortality but also increased harm caused by invasive follow-up of benign pulmonary nodules. Lung tumorigenesis activates the immune system, components of which could serve as tumor-specific biomarkers.
Objectives: To profile tumor-derived autoantibodies as peripheral biomarkers of malignant pulmonary nodules.
Methods: High-density protein arrays were used to define the specificity of autoantibodies isolated from B cells of 10 resected lung tumors. These tumor-derived autoantibodies were also examined as free or complexed to antigen in the plasma of the same 10 patients and matched benign nodule control subjects. Promising autoantibodies were further analyzed in an independent cohort of 250 nodule-positive patients.
Measurements and Main Results: Thirteen tumor B-cell–derived autoantibodies isolated ex vivo showed greater than or equal to 50% sensitivity and greater than or equal to 70% specificity for lung cancer. In plasma, 11 of 13 autoantibodies were present both complexed to and free from antigen. In the larger validation cohort, 5 of 13 tumor-derived autoantibodies remained significantly elevated in cancers. A combination of four of these autoantibodies could detect malignant nodules with an area under the curve of 0.74 and had an area under the curve of 0.78 in a subcohort of indeterminate (8–20 mm in the longest diameter) pulmonary nodules.
Conclusions: Our novel pipeline identifies tumor-derived autoantibodies that could effectively serve as blood biomarkers for malignant pulmonary nodule diagnosis. This approach has future implications for both a cost-effective and noninvasive approach to determine nodule malignancy for widespread low-dose computed tomography screening.
Keywords: lung cancer, B cells, computed tomography imaging, indeterminate pulmonary nodules, early detection
At a Glance Commentary
Scientific Knowledge on the Subject
Screening high-risk smokers for non–small cell lung cancer with low-dose computed tomography (CT) imaging reduces lung cancer mortality by 20% and overall mortality by 6%. Lung cancer screening also increases the detection rate of pulmonary nodules, of which up to 96% are determined to be benign with repeat CT scans or invasive procedures. The pathogenesis of lung cancer is intricately involved with the immune system.
What This Study Adds to the Field
We first isolated autoantibodies from 10 resected non–small cell lung cancers and identified the antigenic targets of free autoantibodies found in most resected tumors. We then determined which of these tumor-derived autoantibodies were also present in the plasma of patients with malignant but not benign nodules identified by CT imaging. Consistent with their utility, these free autoantibodies were also found complexed to antigen in plasma and five of these autoantibodies validated in an independent (n = 250) nodule-positive cohort. Moreover, combining four of the individually validated, tumor-specific autoantibodies accurately detected malignant nodules even of indeterminant size. This approach provides a novel path to autoantibody discovery and our combination panel has the potential to influence how indeterminate pulmonary nodules are diagnosed.
The results of the National Lung Screening Trial have ushered in the prospect of low-dose computed tomography (CT) screening for the approximately 7 million individuals in the United States that meet the criteria of high risk for lung cancer (1). If implemented in the eligible population, an estimated 3–4 million pulmonary nodules (PNs) will be detected that require expensive, invasive, and psychologically stressful follow-up to diagnose malignancy (2–4). Because approximately 96% of detected PNs are benign (5), it is clear that for lung cancer screening to be implemented, the ability to identify malignant PN must be improved by supplementing with noninvasive diagnostic tests. Because the lung is highly vascularized and immunologically rich, a viable cancer immunologic marker may be present in blood that could be used for diagnostic purposes. Autoantibodies released from tumor-infiltrating B lymphocytes may be a viable source of such markers because they can be extracted easily from a minimally invasive blood collection and can be detected months or years before clinical symptoms occur (6). A panel of autoantibody biomarkers in blood could be a noninvasive, cost-effective tool to aid CT screening.
The purpose of this study was to demonstrate that autoantibodies produced by tumor-infiltrating B cells are highly specific for cancer antigens and readily identifiable in peripheral blood. To accomplish this, we subjected uncomplexed autoantibodies purified from tumor-infiltrating B cells to a protein array that covers approximately 80% of the human proteome. In a second approach with a larger, independent confirmation set, we tested for autoantibody–antigen complexes using custom antibody arrays (7–14). Using both antibody arrays (to measure antigen-autoantibody complexes) and protein arrays (to measure free autoantibody) in lung tissue and plasma, provides a complete picture of autoantibodies as potential biomarkers of lung cancer. Some of these results have been previously reported in the form of an abstract (14).
Methods
Study Populations
This study used two patient sample sets both collected by the Fred Hutch Lung Cancer Early Detection and Prevention Clinic (LCEDPC) (Table 1). The NSCLC Resection Cohort was comprised of lung tissue and plasma from n = 10 patients with non–small cell lung cancer (NSCLC) undergoing surgical resection of lung cancer for curative intent at the Fred Hutchinson Cancer Research Center/University of Washington Hospital under institutional review board file #6663. This cohort also contained control plasma specimens from n = 10 patients for PN evaluation (nodules were determined to be benign). Plasma was also collected from an independent n = 125 patients with NSCLC and n = 125 patients with benign PNs evaluated in the LCEDPC. Cases and control subjects from both discovery and confirmation cohorts were first matched on sex (exact match), then age (best available match), and lastly pack-years of smoking (best available match). Plasma specimens and CT image data were available for each of these patients. Nonoverlapping aspects of flow cytometry data from these cohorts have been previously published (15, 16).
Table 1.
Demographic and Clinical Characteristics of Samples
| Discovery |
Validation |
|||||
|---|---|---|---|---|---|---|
| Patient Characteristics | Malignant Nodule | Benign Nodule | P Value | Malignant Nodule | Benign Nodule | P Value |
| Age, yr | 73.2 | 60.2 | 0.07 | 64.6 | 62.7 | 0.059 |
| Sex | ||||||
| Male | 4 | 4 | 56 | 56 | ||
| Female | 6 | 6 | 69 | 69 | ||
| Race | ||||||
| White | 9 | 8 | 111 | 114 | ||
| African American | 0 | 1 | 2 | 2 | ||
| Asian | 1 | 1 | 8 | 6 | ||
| Other | 0 | 0 | 4 | 3 | ||
| Histology | ||||||
| NSCLC | 10 | — | 125 | — | ||
| AD | 7 | — | 104 | — | ||
| SCC | 3 | — | 18 | — | ||
| Smoking status | ||||||
| Current | 1 | 2 | 46 | 30 | ||
| Former | 7 | 7 | 62 | 68 | ||
| Never | 2 | 1 | 17 | 27 | ||
| Pack-years | 29.8 | 25.4 | 0.12 | 32.1 | 25.3 | 0.00018 |
| BMI | ||||||
| <24.9 kg/m2 | 5 | 4 | 49 | 43 | ||
| 25–29.9 kg/m2 | 1 | 3 | 41 | 42 | ||
| >30 kg/m2 | 4 | 3 | 35 | 39 | ||
| Unknown | 0 | 0 | 0 | 1 | ||
Definition of abbreviations: AD = adenocarcinoma; BMI = body mass index; NSCLC = non–small cell lung cancer; SCC = squamous cell carcinoma.
Tumor B-cell extracts were generated from malignant nodule patients. Plasma was analyzed from both nodule-positive cohorts. Age and pack-years are population means.
B-Cell Isolation and Culture
Tumor and normal adjacent lung (NAL) tissue was received within 2 hours after resection and immediately processed for B-cell quantification as previously described (15). To generate B-cell extracts (BCE), B cells were isolated from fresh NSCLC or NAL cell suspensions using human CD19+ selection kit (EasySep, StemCell) following manufacturer’s instructions. After B-cell selection, 98% of selected cells were CD19+ by flow cytometry. B cells were cultured in IMDM, 10% fetal bovine serum, and 0.5% Pen/Strep for 48 hours. Supernatant and B-cell lysates (Cell Signaling lysis buffer) were harvested and stored at −80°C.
ELISA
IgG and IgM levels were determined using ELISA kits (Abcam: IgG, ab100547; IgM, ab137982) on matched plasma and BCE following manufacturer’s instructions.
Free Autoantibody Detection
Free autoantibodies were detected using the Human Proteome Array (HuProt) from CDI Laboratories, Inc., according to manufacturer’s instructions. These arrays have approximately 17,000 unique full-length human recombinant proteins and controls printed in duplicate (17). Briefly, BCE were diluted to IgG concentrations of approximately 150.00 ng/ml and plasma samples were diluted 1:1000. After blocking, samples were incubated on the array in a humidified chamber for 1 hour. After washing, free autoantibodies bound to proteins on the array were detected simultaneously with Alexa Fluor 647-goat antihuman IgG (Jackson) and DyLight 550 antihuman IgM (Pierce) (both highly cross absorbed). The slides were scanned on a GenePix 4200A microarray scanner (Axon Instruments) and the raw Genepix Array List file was aligned. To identify positive hits on HuProt arrays, both spots of protein printed in duplicate had to be present in samples but not in a blank or secondary antibody-only control. Confirmation of free autoantibody candidates using the second cohort as performed by printing commercially available proteins in triplicate on nitrocellulose PATH slides (Grace Bio) with 14 identical 112-spot arrays printed on a single slide.
After blocking and washing per manufacturer’s protocol, slides were put into Chip Clips (Maine Manufacturing), which creates 16 individual wells (only 14 wells were assayed) per slide, thus separating each array into distinct wells. One microliter of human plasma was diluted 1:80, pipetted into a well containing a single array, and incubated for 60 minutes. Autoantibodies were detected as described previously and the median value of the triplicate was selected for the intensity calculation to reduce the effect of outliers.
Autoantibody–Antigen Complex Detection
The detection of autoantibody–antigen complexes using antibody arrays has been previously described (10).
Immunohistochemistry
Sections were cut from formalin-fixed and paraffin-embedded human lung NSCLC (n = 10; same tumors as BCE). Antigen retrieval was performed in Trilogy (Cell Marque) solution at 100°C for 20 minutes in a steamer. All subsequent steps were at room temperature. After blocking with 3.0% H2O2 followed by 0.25% Casein, sections were incubated with primary antibodies, CD20 (Dako #M0755), FCGR2A (OriGene #UM500051), EPB41L3 (Aviva Sysbio #OAGA01718), CK1/3 (Dako #M3515), or isotype controls for 60 minutes. Subsequently, sections were incubated with Opal antimouse or antirabbit horseradish peroxidase polymer (Perkin-Elmer) for 10 minutes, followed by incubation with DAB+ (Dako) for 5 minutes. After washing the sections were counter stained with Tacha’s Hematoxylin (BioCare Medical). Images were analyzed using the Aperio Positive Pixel Count Algorithm on Imagescope.
CT Imaging Analysis
A radiologist who was blinded to clinical and histologic findings evaluated nodule size.
Statistical Analyses
Array data contain a format identical to two-channel gene expression arrays and analysis proceeds analogously as described previously (11, 18). Autoantibodies discovered in the 20-nodule positive cohort (10 tumor BCE and 20 plasma samples) and validated in the 250-nodule positive cohort were analyzed using logistic regression and a Least Absolute Shrinkage and Selection Operator analysis to reduce overfitting. The identified four-marker combination that best distinguished cases from control subjects was then used to generate a receiver operating characteristic curve, with area under the curve (AUC) computed to evaluate the classification accuracy.
Results
B Cells Isolated from Lung Tumors Produce Antibodies Ex Vivo
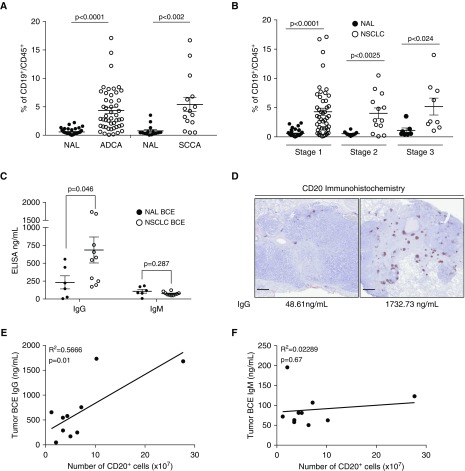
We isolated CD45+ leukocytes from 63 primary NSCLC and NAL (at least 3 cm away from tumor) tissues by creating single cell suspensions within hours of resection. Of the 63 tumors, 48 were adenocarcinomas (ADCA) and 15 were squamous cell carcinomas (SCCA). By flow cytometry, CD19+ B cells account for approximately 4% of total leukocytes in ADCA and 5% in SCCA (Figure 1A). The number of B cells in both ADCA and SCCA was approximately sevenfold higher than in NAL tissue. Tumors staged 1, 2, or 3 all had a 5% B-cell fraction (Figure 1B), suggesting that B-cell infiltration is an early event in lung tumorigenesis and stage independent.
Figure 1.
B-cell number and IgG concentration are significantly higher in tumors compared with normal adjacent lung. (A) Flow cytometry quantification of the proportion of CD19+ B cells in CD45+ leukocytes isolated from adenocarcinoma (ADCA), squamous cell carcinoma (SCCA), and normal adjacent lung (NAL). NAL n = 44 and ADCA n = 48; NAL n = 15 and SCCA n = 15. Data are shown as mean ± SEM. P values were determined via Welch’s t test. (B) Flow cytometry quantification of CD19+ B cells in CD45+ leukocytes by stage of non–small cell lung cancer (NSCLC). Tumors have significantly more CD19+ B cells in all stages of NSCLC than in NAL. Stage 1 n = 47, stage 2 n = 13, and stage 3 n = 9. Data are shown as mean ± SEM. P values were determined via Welch’s t test. (C) ELISA quantification of IgG and IgM antibodies isolated from tumor B-cell extracts (n = 10) and NAL (n = 6) (mean ± SEM). P values were determined via Welch’s t test. (D) Representative images of a tumor with low IgG quantification by ELISA and low CD20 staining by immunohistochemistry compared with a tumor with high CD20 staining and high IgG concentration. Scale bars = 1 mm. (E) Significant correlation of matched ELISA IgG and CD20 immunohistochemistry staining in 10 NSCLC tumors (n = 10, Pearson correlation). (F) ELISA IgM is not correlated with the number of B cells quantified by CD20+ immunohistochemical staining (n = 10, Pearson correlation). BCE = B-cell extract.
We next purified autoantibodies from 10 NSCLC (7seven ADCA, three SCCA) and six NAL samples. We subjected cell suspensions from each case to a CD19+ isolation column and confirmed the isolated cells to be a greater than 96% pure B cells (CD20+ and/or CD19+) by flow cytometry. The isolated B cells were cultured for 48 hours, after which the immunoglobulin-rich supernatant was removed; the B cells were lysed; and the combination of supernatant and lysate, termed BCE, was analyzed via ELISA for its abundance of IgG and IgM protein (Figure 1C). IgG expression was higher in NSCLC BCE with an average of 670.88 ng/ml compared with an average of 233.1 ng/ml in NAL. IgM concentration between NSCLC and NAL BCE were not statistically different (average, 89.4 ng/ml and 107.8 ng/ml, respectively).
In these same tumors, we examined the presence and location of B cells within the tumor microenvironment by CD20+ immunohistochemistry. We observed B cells in all tumors either infiltrating or restricted to the tumor edges. Within each section B cells were predominantly found in tertiary lymphoid structures. Tumors with more CD20+ staining correlated with higher concentrations of IgG by ELISA, whereas tumors with fewer B cells had a lower amount of IgG (Figures 1D and 1E). IgM concentration did not have a significant relationship with the number of B cells (Figure 1F).
Discovery of Candidate Lung Tumor-derived Autoantibodies for Malignant Nodules
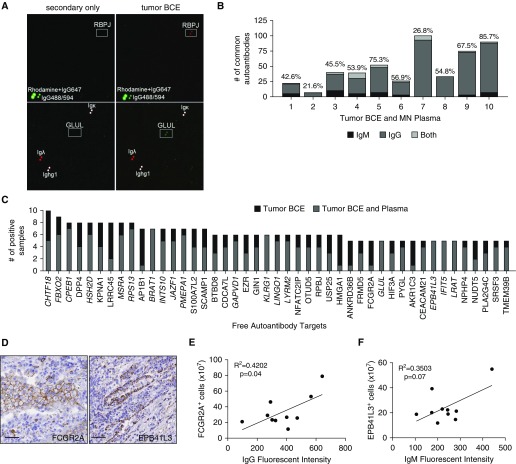
To identify the antigenic targets of the IgG and IgM autoantibodies, tumor BCEs were incubated on the Human Proteome (HuProt) array containing approximately 17,000 yeast-produced human proteins printed in duplicate (11) along with Rhodamine, IgG647, IgG488/594, and immunoglobulin fragments for control subjects (Figure 2A) (17). After incubation, autoantibodies present in BCE bound to their target proteins. Bound IgG and IgM were simultaneously detected using two different fluorescently labeled secondary antibodies. A representative image shows IgG autoantibodies to Recombination Signal Binding Protein for Immunoglobulin Kappa J Region (RBPJ) and IgM autoantibodies to Glutamate-Ammonia Ligase (GLUL) present in tumor BCE but no signal in a secondary-only control (white boxes in Figure 2A). The number of autoantibody targets identified in tumor samples ranged from 29 to 100 IgG and 2 to 265 IgM (see Figure E1A in the online supplement). Most BCEs had more distinct IgG autoantibodies identified than IgM autoantibodies. Signal for both IgG and IgM autoantibodies to the same antigen was relatively uncommon, with an average of only five observed per tumor.
Figure 2.
Common autoantibody targets can be identified in most lung tumors and plasma. (A) Representative images of IgG autoantibody target identification of RPBJ and IgM of GLUL (white boxes, proteins printed in duplicate) in tumor B-cell extracts (BCE) but not in a secondary-only control. Positive controls are rhodamine + IgG647, IgG-Alexa Fluor 488/594, and antibody fragments. (B) The number of autoantibodies found in tumor BCE and corresponding malignant nodule (MN) plasma. The percentage of identity is noted above the bar graph for each sample pair. (C) Targets of autoantibodies isolated from ≥50% tumor BCE identified using the HuProt array. Italicized autoantibodies were also present in ≥50% MN plasmas. (D) Representative images from tumor serial sections of immunohistochemical staining of EPB41L3 and FCGR2A (antigenic targets of lung-derived autoantibodies). Scale bars = 50 μm. (E) The number of FCGR2A-positive cells quantified by immunohistochemistry significantly correlates with the intensity of IgG autoantibody signal on the HuProt array (n = 10, Pearson correlation). (F) The number of EPB41L3-positive cells quantified by immunohistochemistry shows a trend toward correlation with the intensity of IgM autoantibody signal on the HuProt array (n = 10, Pearson correlation).
Autoantibodies Identified in Lung Tumors Can Be Found in Peripheral Plasma
To determine the identity of autoantibodies in peripheral blood, we profiled the plasma (termed malignant nodule [MN] plasma) from the same subject from which the tumor BCE was generated. An average of 105 and 305 autoantibodies were identified in tumor BCE and the corresponding MN plasma, respectively (see Figures E1A and E1B). We found a range of autoantibodies present in tumor BCE that were also present in the corresponding MN plasma from 8 to 101 autoantibodies (Figure 2B) with an average of 56% ± 6.36% SEM in common. Additionally, 8 of 10 subjects had more than 40% of autoantibodies identified in tumor BCE that were also in the corresponding plasma.
After identifying common autoantibodies between tumor BCE and plasma, we sought to determine which autoantibodies were present in most subjects. Forty-five autoantibodies were identified in more than half of the tumor BCE (Figure 2C). A total of 40 out of 45 of these autoantibodies were IgG isotype. Of the remainders, four were IgM (BRAT1, GLUL, TMEM39B, EBP41L3) and one antigen (FBXO2) had both IgG and IgM autoantibodies. All the most common tumor BCE autoantibodies identified were also found in at least one case of MN plasma. A total of 18 out of the 45 most common tumor-specific autoantibodies were also present in most (≥50%) MN plasmas.
Based on a literature search of cancer relevance and presence of well-characterized immunohistochemical antibodies, we chose to further explore two autoantibody targets: Fc Fragment of IgG Receptor IIa (FCGR2A) and Erythrocyte Membrane Protein Band 4.1 Like 3 (EPB41L3). In all 10 tumors we detected a range of expression for both FCGR2A and EPB41L3 in the tumor microenvironment (Figure 2D, representative images). The number of positive cells by immunohistochemistry for FCGR2A and EPB41L3 correlated with the fluorescence intensity of IgG or IgM, respectively, on the HuProt array, with FCGR2A reaching statistical significance (Figures 2E and 2F). This suggests the prevalence of the antigen may be linked to autoantibody production.
Lung Tumor-derived Autoantibodies Also Found in Plasma Are Specific for MN
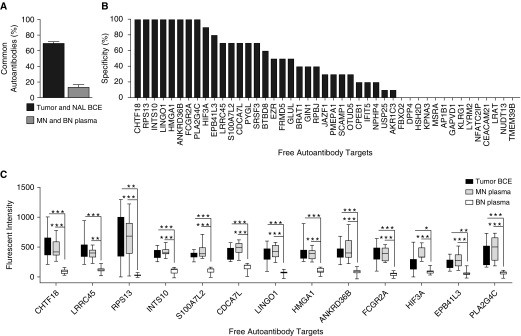
We next sought to determine the specificity of our tumor-derived autoantibodies. Comparing lung tumor and NAL BCE from the same subject showed a high concordance (∼70% ± 1.25 SEM) of IgG and IgM autoantibodies in both tissues suggesting, at least at the autoantibody level, that NAL tissue may not represent true “normal” or healthy tissue. Because our goal was to identify tumor-derived autoantibodies also present in peripheral plasma, we compared plasma from subjects with benign nodules (BN) matched to MN, which showed only 14% ± 2.4% SEM of autoantibodies in common.
Nodule-positive plasma samples were matched on sex (exact match), age (best available match), and pack-years of smoking (best available match) (Table 1). Out of 45 autoantibodies discovered in most tumor BCE, 10 autoantibodies had greater than or equal to 80% specificity (i.e., were found in two or fewer BN plasmas) (Figure 3B). We chose a top list of 12 IgG and one IgM (EBP41L3) tumor-derived autoantibody candidates with greater than or equal to 50% sensitivity and greater than or equal to 70% specificity to explore further (Figure 3C).
Figure 3.
Autoantibodies specific for malignant nodules. (A) The percentage of common antibodies between tumor B-cell extracts (BCE) and normal adjacent lung BCE or malignant nodule plasma and benign nodules (BN) plasma (n = 2/group). (B) The specificity of free autoantibodies isolated from ≥50% tumor BCE (i.e., if none of the 10 BN plasmas had a given autoantibody, the specificity is indicated as 100%). (C) Quantification of fluorescent intensity of top autoantibodies with ≥50% sensitivity and ≥70% specificity (n = 10/group) (mean ± SEM; unpaired Student’s t test; *P < 0.02, **P < 0.0002, and ***P < 0.00001). MN = malignant nodule; NAL = normal adjacent lung.
Free Autoantibodies Discovered in Lung Tumors Are Also Complexed to Antigen in Peripheral Plasma
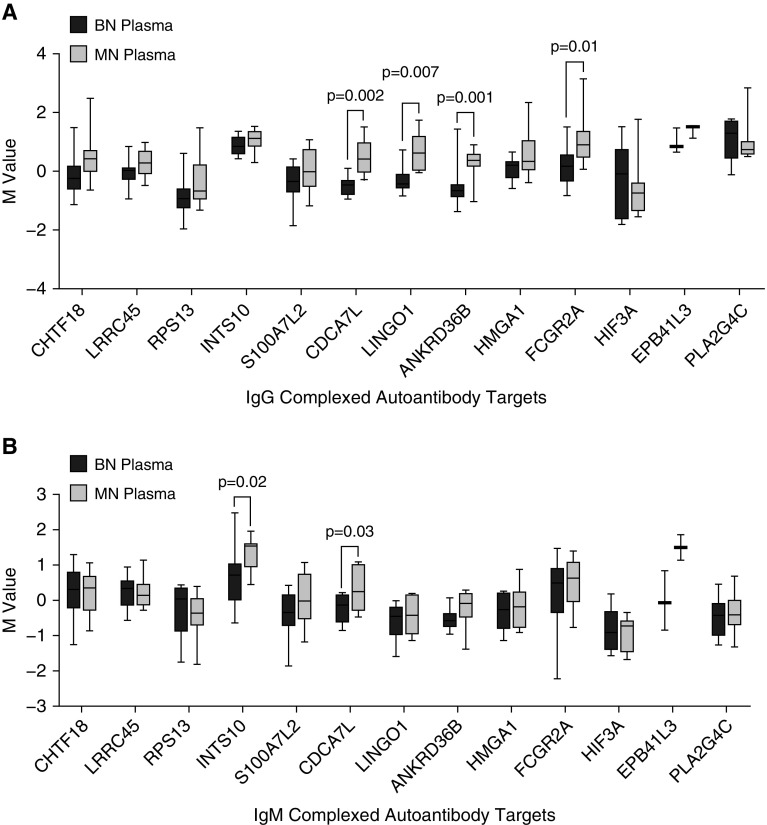
Antibodies can exist as free or bound to antigen and the latter would have been missed in the HuProt array experiments. To test if the free tumor-derived autoantibodies were complexed with antigen in plasma, we created a custom antibody array using commercially available antibodies to the 13 antigens of interest. When possible, we included two to three antibodies per autoantibody target that spanned the length of the protein (i.e., an antibody targeting the c-terminus, middle, and n-terminus) to increase the possibility that at least one arrayed antibody would bind independent of the antigenic site bound by the autoantibody. We discovered that in 11 of the 13 candidates, more IgG autoantibodies were complexed to antigen in MN plasma compared with BN plasma, four of which reached statistical significance (Figure 4A). Five of the 13 candidates also had higher levels of complexed IgM autoantibodies in MN plasma, two of which reached statistical significance (Figure 4B). Thus, most of the tumor-derived autoantibodies are present in plasma as both free and complexed to antigen.
Figure 4.
Autoantibodies identified in lung tumors can be found in peripheral plasma complexed with antigen. Targeted antibody arrays were created by covalently linking human protein–specific antibodies to the top free autoantibody targets onto the slide surface. After hybridization with whole plasma, autoantibody–antigen complexes are detected via fluorescently labeled antihuman (A) IgG and (B) IgM secondary antibodies and quantified to compare complexed autoantibodies in plasma from malignant (n = 10) and benign (n = 10) nodule subjects (mean ± SEM; unpaired Student’s t test). BN = benign nodule; M value = log2(red channel or green channel signal) or the expression on the log2 scale after background correction; MN = malignant nodule.
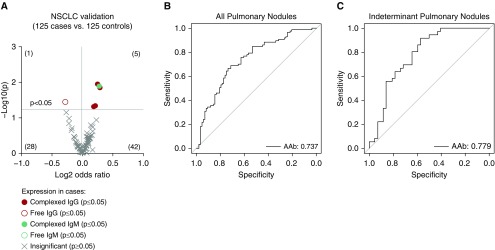
We next performed a validation study with a sample size of 250 independent plasma samples from the LCEDPC. We printed a protein array with commercially available proteins and a targeted antibody array containing the top 13 autoantibody targets. With these arrays we confirmed 5 (IgG: EPB41L3, ANKRD36B, FGCR2A, and LINGO1; IgM: S100A7L2) of our 13 autoantibody targets as significantly higher in cancers than control subjects (Figure 5A; see Table E1). We could also test marker performance in the different histologic subtypes and the candidates performed better with 6 and 17 autoantibodies significantly upregulated in ADCA or SCCA, respectively (see Figures E2A and E2B). Because IgM autoantibodies would switch to the IgG isotype during an ongoing immune response, we reasoned that IgM and IgG autoantibodies to the same target represent a collective immune response against the tumor and their detection largely depends on the stage of the antitumor immune response in each patient.
Figure 5.
Validated tumor-derived autoantibodies in plasma can detect malignant nodules. (A) Five autoantibodies are significantly higher in plasma from non–small cell lung cancer (NSCLC) nodule (n = 125) compared with benign nodule (n = 125) patients (unpaired Student’s t test). (B) Performance of a four-autoantibody panel by receiver operating characteristic curves in NSCLC cohort (n = 125 cases, 125 control subjects). (C) Performance of the same four-autoantibody panel in only indeterminant pulmonary nodules with the largest diameter 8–20 mm (n = 37 cases; n = 41 control subjects). AAb = autoantibody.
To find validated autoantibodies that could complement each other’s performance, we used logistic regression to determine the four-marker panel that would maximize the prediction AUC. We found a panel of FCGR2A, EPB41L3, and LINGO1 IgG-complexed autoantibodies combined with a S100A7L2 IgM-complexed autoantibody had an AUC of 0.737 (33.3% sensitivity at 90% specificity) in PNs (Figure 5B). Outside of our reasoning for isotype variation, if we only included autoantibodies discovered and validated with the same immunoglobulin class, the optimal three-marker panel had a comparable AUC of 0.727. Indeterminate PNs are clinically challenging with a risk of malignancy between 5% and 60% and account for the largest number of invasive biopsies for benign disease (19). The performance of our four-autoantibody panel had an AUC of 0.78 (91.7% sensitivity at 57.1% specificity) in indeterminate PNs, defined here as nodules greater than or equal to 8 mm but less than or equal to 20 mm in the largest diameter, suggesting our panel could still accurately classify MN (Figure 5C). Our autoantibodies also provide novel information about indeterminate PNs because they are not correlated (highest R2 value = 0.15) with any of the currently used independent risk factors for malignancy including smoking status, stage, age, histology, or nodule volume (see Table E2).
Discussion
Here, we demonstrate for the first time that truly tumor-derived autoantibodies can serve as excellent plasma biomarkers of lung cancer. We first identified the antigen targets of tumor-derived autoantibodies and demonstrated their increased presence in plasma from subjects with malignant verses BN. Many of our top candidates discovered as free autoantibodies were also confirmed to be complexed with antigen in plasma at higher levels in malignancy. Five autoantibodies validated in an additional plasma set of 250 low-dose CT-identified nodules. When four of the confirmed autoantibodies were combined the combination could accurately detect MN.
We note that our top four autoantibodies potentially reflect multiple mechanisms of autoantibody generation. For example, LINGO1 is an extracellular receptor typically only expressed in the central nervous system during development and in the adult brain (20), and therefore presumably would not be seen by B cells under homeostatic conditions. Computational analysis using The Cancer Genome Atlas Firebrowse portal of RNA expression data showed an approximate threefold increase in LINGO1 expression in NSCLC compared with normal lung (21). Whether this translates to increases in protein expression that could stimulate autoantibody production (22) is undetermined but LINGO1 protein is overexpressed in other cancer types, such as Ewing sarcoma (23). Although not much is known about S100A7L2, its putative receptor, S100A7, showed a 50-fold increase in mRNA in SCCA TGCA samples compared with normal lung (24) and S100A7 protein is upregulated in breast cancer and melanoma (25). FCGR2A is expressed on phagocytic cells in normal lung tissue but can be further upregulated after exposure to cigarette smoke (26). Because FCGR2A promotes phagocytosis of antibody-coated antigens (27), autoantibodies targeting it may be an example of loss of self-tolerance and immune escape. EPB41L3 is expressed intracellularly in lung epithelial cells and is thought to act as a tumor suppressor in lung cancer transformation (24). Autoantibody-mediated targeting, as observed with other tumor suppressors, such as p53 (28), could play a contributing role to its reduced expression during lung tumorigenesis.
There have been many attempts to identify plasma biomarkers for the early detection of lung cancer including autoantibodies, miRNA, mRNA, tumor cell DNA methylation, or cell-free circulating tumor DNA (29). Currently there are five commercially available biomarker tests for PN diagnosis with panels ranging from 4 to 23 biomarkers (30). Two of these tests incorporate autoantibodies to prevalent cancer antigens including p53 and NY-ESO1. Comparisons between panel performances are difficult because of variations in cohort populations and statistical reporting but overall the AUCs reported here are comparable with published panels ranging from AUCs of 0.634 to 0.84. Because none have yet to achieve Food and Drug Administration approval, it may be necessary to expand or combine autoantibody panels with other biomarker types. As is best practice for biomarker research, both cohorts were matched on sex, age, and pack-years to identify true biomarkers of disease. This matching unfortunately limited us from directly comparing our autoantibody panel with existing CT-based risk models by reducing the contributions of smoking and age to lung tumorigenesis. Studies are currently underway to validate our four-autoantibody panel alone and in combination with existing CT-based risk models in unmatched cohorts.
The autoantibody dataset generated here could also potentially be useful for immunotherapeutic target selection. The positive results of clinical trials using immune checkpoint blockade for NSCLC serve as an important proof of concept that tumor neoantigens can be targeted by the immune system (31). Related strategies using adoptive T-cell therapies, such as chimeric antigen receptor T-cell therapy, have demonstrated great success in lymphoma and are being tested in solid tumors (32). Because the targeting aspect of a chimeric antigen receptor T cell is the single-chain variant fragment on an antibody, our data could be used to identify autoantibodies displaying specificity against cell-surface tumor antigens. Mining the autoantibody dataset for this purpose is likely to identify novel immunotherapeutic targets.
In conclusion, this is the first study to provide evidence that plasma autoantibodies distinguishing MN are truly tumor-derived and exist complexed to and free from antigen. To prove clinical utility of these autoantibodies, we are continuing to further validate our model in multiple plasma sets with corresponding CT imaging data. If validated, the autoantibody assay would be an inexpensive and noninvasive test to aid PN diagnosis.
Footnotes
Supported by National Cancer Institute/NIH (U01 185097, U01 186157, R01 188,341, and F32 CA200265). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Author Contributions: K.J.L. designed, performed, and analyzed autoantibody array data, and wrote the manuscript. J.K. designed, performed, and analyzed all human flow-cytometry experiments and ex vivo antibody isolation. Y.Z. performed statistical analysis of microarray. X.Z. performed all immunohistochemistry. E.L. performed free autoantibody validation array. D.S. performed antigen-complexed validation array. J.J.L. wrote R code and performed statistical analyses of marker combinations. W.W. designed and analyzed all computed tomography (CT) images. P.K. supervised design and analysis of CT images. S.N.J.P. analyzed all patient CT images. T.W.R. supervised design and statistical analyses. M.S. managed clinical specimen database. P.D.L. and A.M.H. supervised the design and performance of all experiments, reviewed all data, and wrote and edited the manuscript.
This article has an online supplement, which is accessible from this issue’s table of contents at www.atsjournals.org.
Originally Published in Press as DOI: 10.1164/rccm.201804-0628OC on November 13, 2018
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Humphrey L, Deffebach M, Pappas M, Baumann C, Artis K, Mitchell JP, et al. Screening for lung cancer: systematic review to update the US Preventive Services Task Force recommendation; US Preventive Services Task Force Evidence Syntheses, formerly Systematic Evidence Reviews. Rockville, MD: Agency for Healthcare Research and Quality; 2013. [PubMed]
- 2.Aberle DR, Adams AM, Berg CD, Black WC, Clapp JD, Fagerstrom RM, et al. National Lung Screening Trial Research Team. Reduced lung-cancer mortality with low-dose computed tomographic screening. N Engl J Med. 2011;365:395–409. doi: 10.1056/NEJMoa1102873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Smith RA, Andrews KS, Brooks D, Fedewa SA, Manassaram-Baptiste D, Saslow D, et al. Cancer screening in the United States, 2017: A review of current American Cancer Society guidelines and current issues in cancer screening. CA Cancer J Clin. 2017;67:100–121. doi: 10.3322/caac.21392. [DOI] [PubMed] [Google Scholar]
- 4.Freiman MR, Clark JA, Slatore CG, Gould MK, Woloshin S, Schwartz LM, et al. Patients’ knowledge, beliefs, and distress associated with detection and evaluation of incidental pulmonary nodules for cancer: results from a multicenter survey. J Thorac Oncol. 2016;11:700–708. doi: 10.1016/j.jtho.2016.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kinsinger LS, Anderson C, Kim J, Larson M, Chan SH, King HA, et al. Implementation of lung cancer screening in the Veterans Health Administration. JAMA Intern Med. 2017;177:399–406. doi: 10.1001/jamainternmed.2016.9022. [DOI] [PubMed] [Google Scholar]
- 6.Tan EM. Autoantibodies as reporters identifying aberrant cellular mechanisms in tumorigenesis. J Clin Invest. 2001;108:1411–1415. doi: 10.1172/JCI14451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mirus JE, Zhang Y, Li CI, Lokshin AE, Prentice RL, Hingorani SR, et al. Cross-species antibody microarray interrogation identifies a 3-protein panel of plasma biomarkers for early diagnosis of pancreas cancer. Clin Cancer Res. 2015;21:1764–1771. doi: 10.1158/1078-0432.CCR-13-3474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ramirez AB, Lampe PD. Discovery and validation of ovarian cancer biomarkers utilizing high density antibody microarrays. Cancer Biomark. 2010-2011-2011;8:293–307. doi: 10.3233/CBM-2011-0215. [DOI] [PubMed] [Google Scholar]
- 9.Rho JH, Mead JR, Wright WS, Brenner DE, Stave JW, Gildersleeve JC, et al. Discovery of sialyl Lewis A and Lewis X modified protein cancer biomarkers using high density antibody arrays. J Proteomics. 2014;96:291–299. doi: 10.1016/j.jprot.2013.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rho JH, Lampe PD. High-throughput analysis of plasma hybrid markers for early detection of cancers. Proteomes. 2014;2:1–17. doi: 10.3390/proteomes2010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rho JH, Lampe PD. High-throughput screening for native autoantigen–autoantibody complexes using antibody microarrays. J Proteome Res. 2013;12:2311–2320. doi: 10.1021/pr4001674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rho JH, Ladd JJ, Li CI, Potter JD, Zhang Y, Shelley D, et al. Protein and glycomic plasma markers for early detection of adenoma and colon cancer. Gut. 2018;67:473–484. doi: 10.1136/gutjnl-2016-312794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Loch CM, Ramirez AB, Liu Y, Sather CL, Delrow JJ, Scholler N, et al. Use of high density antibody arrays to validate and discover cancer serum biomarkers. Mol Oncol. 2007;1:313–320. doi: 10.1016/j.molonc.2007.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lastwika K, Kargl J, Zhang Y, Zhu X, Shelley D, Lo E, et al. Non-small cell lung tumor-derived autoantibodies can distinguish benign from malignant pulmonary nodules [abstract]. Presented at the Fifth AACR-IASLC International Joint Conference: Lung Cancer Translational Science from the Bench to the Clinic. January 8-11, 2018, San Diego, CA. Poster A12.
- 15.Kargl J, Busch SE, Yang GHY, Kim K-H, Hanke ML, Metz HE, et al. Neutrophils dominate the immune cell composition in non-small cell lung cancer. Nat Commun. 2017;8:14381. doi: 10.1038/ncomms14381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mark NM, Kargl J, Busch SE, Yang GHY, Metz HE, Zhang H, et al. Chronic obstructive pulmonary disease alters immune cell composition and immune checkpoint inhibitor efficacy in non-small cell lung cancer. Am J Respir Crit Care Med. 2018;197:325–336. doi: 10.1164/rccm.201704-0795OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jeong JS, Jiang L, Albino E, Marrero J, Rho HS, Hu J, et al. Rapid identification of monospecific monoclonal antibodies using a human proteome microarray. Mol Cell Proteomics. 2012;11:016253. doi: 10.1074/mcp.O111.016253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smyth GK, Speed T. Normalization of cDNA microarray data. Methods. 2003;31:265–273. doi: 10.1016/s1046-2023(03)00155-5. [DOI] [PubMed] [Google Scholar]
- 19.Massion PP, Walker RC. Indeterminate pulmonary nodules: risk for having or for developing lung cancer? Cancer Prev Res (Phila) 2014;7:1173–1178. doi: 10.1158/1940-6207.CAPR-14-0364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.de Wit J, Hong W, Luo L, Ghosh A. Role of leucine-rich repeat proteins in the development and function of neural circuits. Annu Rev Cell Dev Biol. 2011;27:697–729. doi: 10.1146/annurev-cellbio-092910-154111. [DOI] [PubMed] [Google Scholar]
- 21.Broad Institute TCGA Genome Data Analysis Center. doi: 10.7908/C11G0KM9. Analysis-ready standardized TCGA data from Broad GDAC Firehose 2016_01_28 run. Broad Institute of MIT and Harvard. Dataset. 2016 [accessed 2018 Mar 30]. Available from: [DOI]
- 22.Tsou P, Katayama H, Ostrin EJ, Hanash SM. The emerging role of B cells in tumor immunity. Cancer Res. 2016;76:5597–5601. doi: 10.1158/0008-5472.CAN-16-0431. [DOI] [PubMed] [Google Scholar]
- 23.Town J, Pais H, Harrison S, Stead LF, Bataille C, Bunjobpol W, et al. Exploring the surfaceome of Ewing sarcoma identifies a new and unique therapeutic target. Proc Natl Acad Sci USA. 2016;113:3603–3608. doi: 10.1073/pnas.1521251113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen X, Guan X, Zhang H, Xie X, Wang H, Long J, et al. DAL-1 attenuates epithelial-to mesenchymal transition in lung cancer. J Exp Clin Cancer Res. 2015;34:3. doi: 10.1186/s13046-014-0117-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bresnick AR, Weber DJ, Zimmer DB. S100 proteins in cancer. Nat Rev Cancer. 2015;15:96–109. doi: 10.1038/nrc3893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hoonhorst SJM, Timens W, Koenderman L, Lo Tam Loi AT, Lammers J-WJ, Boezen HM, et al. Increased activation of blood neutrophils after cigarette smoking in young individuals susceptible to COPD. Respir Res. 2014;15:121. doi: 10.1186/s12931-014-0121-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lu J, Marnell LL, Marjon KD, Mold C, Du Clos TW, Sun PD. Structural recognition and functional activation of FcgammaR by innate pentraxins. Nature. 2008;456:989–992. doi: 10.1038/nature07468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pedersen AE, Stryhn A, Justesen S, Harndahl M, Rasmussen S, Donskov F, et al. Wildtype p53-specific antibody and T-cell responses in cancer patients. J Immunother. 2011;34:629–640. doi: 10.1097/CJI.0b013e3182281381. [DOI] [PubMed] [Google Scholar]
- 29.Blandin Knight S, Crosbie PA, Balata H, Chudziak J, Hussell T, Dive C. Progress and prospects of early detection in lung cancer. Open Biol. 2017;7:170070. doi: 10.1098/rsob.170070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Atwater T, Cook CM, Massion PP. The pursuit of noninvasive diagnosis of lung cancer. Semin Respir Crit Care Med. 2016;37:670–680. doi: 10.1055/s-0036-1592314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Remon J, Vilariño N, Reguart N. Immune checkpoint inhibitors in non-small cell lung cancer (NSCLC): Approaches on special subgroups and unresolved burning questions. Cancer Treat Rev. 2018;64:21–29. doi: 10.1016/j.ctrv.2018.02.002. [DOI] [PubMed] [Google Scholar]
- 32.Newick K, O’Brien S, Moon E, Albelda SM. CAR T cell therapy for solid tumors. Annu Rev Med. 2017;68:139–152. doi: 10.1146/annurev-med-062315-120245. [DOI] [PubMed] [Google Scholar]