Abstract
Objectives:
Anti-citrullinated protein antibodies (ACPA) are a hallmark of rheumatoid arthritis (RA). While epitope spreading of the serum ACPA response is believed to contribute to RA pathogenesis, little is understood regarding how this phenomenon occurs. We analyzed the antibody repertoires of individuals with RA to gain insight into the mechanisms leading to epitope spreading of the serum ACPA response in RA.
Methods:
Plasmablasts from the blood of six RA patients were stained with citrullinated peptide tetramers to identify ACPA-producing B cells by flow cytometry. Plasmablasts were single cell sorted and sequenced to obtain antibody repertoires. Sixty-nine antibodies were recombinantly expressed and their anti-citrulline reactivities characterized using CCP ELISA and synovial antigen arrays. Thirty-six mutated antibodies designed either to represent ancestral antibodies or to test paratope residues critical for binding, as determined from molecular modeling studies, were also tested for anti-citrulline reactivities.
Results:
Clonally related monoclonal ACPA and their shared ancestral antibodies each exhibit differential reactivity against citrullinated antigens. Molecular modeling identified residues within the CDR loops and framework regions predicted to be important for citrullinated antigen binding. Affinity maturation resulted in mutations of these key residues, which conferred binding to different citrullinated epitopes and/or increased polyreactivity to citrullinated epitopes.
Conclusions:
These results demonstrate that the different somatic hypermutations accumulated by clonally related B cells during affinity maturation alter the antibody paratope to mediate epitope spreading and polyreactivity of the ACPA response in RA, key properties that likely contribute to the pathogenicity of ACPA.
Keywords: rheumatoid arthritis, ACPA, plasmablast
INTRODUCTION
Rheumatoid arthritis (RA) is a chronic autoimmune disorder of unknown etiology(1, 2). A hallmark of RA is the presence of anti-citrullinated protein antibodies (ACPA) that recognize citrullinated antigens(1, 2). Citrullination is mediated by peptidyl arginine deaminases (PADs) and plays a role in gene regulation, organization of cell structure, apoptosis, and formation of neutrophil extracellular traps(3–5). In RA, however, citrullination of proteins in synovial joint and other tissues, coupled with the production of ACPA, has been hypothesized to contribute to pathogenesis(1–4).
Increasing evidence suggests that ACPA play an important role in the pathogenesis of RA(6). ACPA have been detected in patients as early as ten years prior to diagnosis, with increased titers and epitope spreading of the ACPA response preceding the onset of arthritis(7–9). The presence of ACPA is associated with increased disease severity(10–13) and is a better predictor of erosive disease than rheumatoid factor(14). Finally, in the collagen-induced arthritis mouse model, administration of a monoclonal ACPA exacerbated disease, demonstrating that ACPA can directly promote inflammatory arthritis(15).
Deciphering the mechanisms underlying the development and epitope spreading of the ACPA response could provide new insights into the mechanisms underlying RA. Previous studies using RA patient serum demonstrated that epitope spreading of the ACPA response occurs in the years preceding clinical disease(7–9). Of note, during the two years immediately preceding the onset of clinical arthritis, the number and breadth of ACPA reactivities sharply increases, correlating with an increase in proinflammatory cytokines in the blood(7). Nevertheless, the mechanisms underlying epitope spreading of ACPA and its role in the development of clinical RA remain poorly understood.
Here, we used barcode-enabled single cell sequencing to analyze ACPA plasmablast repertoires in RA. We tested the binding capabilities of 69 recombinant monoclonal antibodies (rMabs) derived from the plasmablast clonal families. We found that plasmablasts within the same clonal family that possess differential somatic hypermutations encode individual ACPA that bind different citrullinated epitopes and/or bind distinct sets of citrullinated epitopes and thus are polyreactive(16). We also found that patient-derived, affinity-matured ACPA frequently exhibit increased polyreactivity, in which they consistently bind a greater number of specific citrullinated epitopes relative to predicted ancestral family members. These findings indicate that somatic hypermutations arising through affinity maturation can result in epitope spreading and increase the polyreactivity of individual ACPA, which in turn increases the ability of the polyclonal serum ACPA repertoire to bind a multitude of citrullinated epitopes in RA. We further used molecular modeling to identify and mutation studies to characterize the key amino acid residues within the CDRs and framework regions forming the paratopes within the antigen-binding sites of ACPA that mediate binding to citrullinated epitopes.
METHODS
Human specimens
Blood samples were collected from individuals recruited at the VA Palo Alto who met the American College of Rheumatology 1987 classification criteria for RA and possessed anti-CCP(17). Clinicopathological characteristics are provided in Supplementary Table 3. All samples were collected with informed consent under human subjects protocols approved by the Stanford University IRB.
Generating fluorescent citrullinated peptide tetramers (cit-tets)
Cit-tets were generated by incubating PE-conjugated streptavidin with biotinylated citrullinated peptides (Supplementary Table 4) previously shown to be targeted by ACPA in RA sera (see Supplementary Materials)(7), (18, 19).
Single cell sorting of ACPA-producing plasmablasts
Cell sorting of CD19+CD20−CD3−CD14−IgD−CD27+CD38hi plasmablasts, which were either IgG+ or IgA+, was performed as described in the Supplementary Methods and our prior work(20, 21). A mix of the PE-conjugated cit-tets was included to identify ACPA producing plasmablasts (details in Supplementary Materials(18, 19)).
Barcode-enabled antibody repertoire sequencing
Antibody repertoire sequencing was performed as previously described(20–23).
Bioinformatic pipeline and repertoire analysis
Paired chain antibody repertoires were generated from sequencing data using a custom bioinformatic pipeline(22). Clonally related antibodies were defined as sharing heavy and light chain VJ genes and having a CDR3 nucleotide Levenshtein distance ≤40% of the total CDR3 length for both chains (i.e. CDR3 sequences are at least 60% identical). The bimodal distribution of the nearest distances for the HC and LC CDR3 sequences was used to set the identity threshold(24). For representing individual clonal family lineages, the heavy chain and light chain sequences of each member were concatenated and analyzed using IgTree(25).
Selection and recombinant expression of monoclonal antibodies
Antibodies sequenced directly from patient blood were produced in-house as described(26). Antibodies sequenced from cit-tet+ clonally expanded plasmablasts were prioritized for expression; however, a handful of cit-tet+ non-clonal and tet- clonal plasmablast antibodies were also expressed. All other antibodies (mutation studies and predicted parent/germline antibody sequences) were commercially produced by LakePharma.
RA planar array
RA antigen microarrays were printed and probed, and datasets analyzed as described(7, 27, 28). Positive hits were confirmed by ELISA.
Molecular modeling
Descriptions of methods used to model the binding of patient-derived ACPA to citrullinated epitopes are available in the Supplementary Materials.
Statistics
For two group comparisons, the Welch’s t-test was used. For multiple comparisons, the nonparametric Kruskal-Wallis test was used, followed by Dunn’s multiple comparisons test. P values < 0.05 were considered significant.
ELISA
CCP3 and peptide ELISAs are described in the Supplementary Materials.
RESULTS
Sequencing the blood plasmablast antibody repertoire in RA
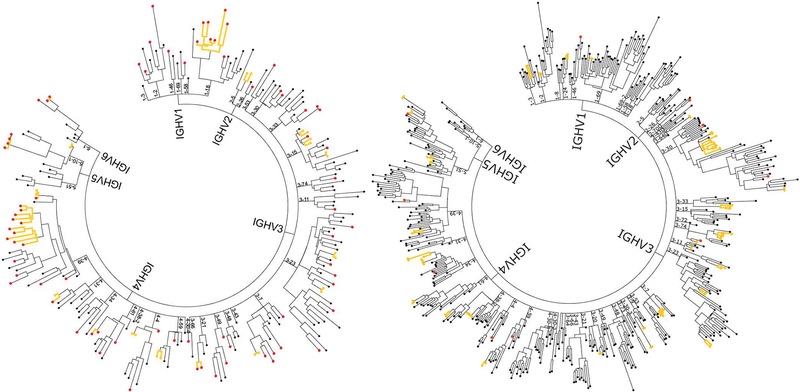
To study ACPA repertoires from activated, disease-relevant B cells in RA, we sequenced the antibodies expressed by plasmablasts from six anti-CCP+ RA patients and generated phylogenetic trees representing the plasmablast antibody repertoires of each individual RA patient (Figure 1). A total of 2,541 antibodies were sequenced (Supplementary Table 1). 182 clonal families consisting of a total of 510 antibodies were identified, with the remaining 2,031 antibodies representing non-clonal family “singletons”. These singletons may in fact represent small clonal families which are not captured at the current sequencing depth; therefore our estimates of clonality are likely underestimates. Sequenced antibodies were split nearly evenly between IgA and IgG isotypes (1,200 for IgA and 1,341 for IgG), although the ratios of IgA to IgG varied between patients. 231 of the sequenced antibodies were derived from tetramer+ (cit-tet+) plasmablasts. While this represents 9% of the total number of plasmablast antibody sequences, actual percentages of cit-tet+ plasmablasts in individual patients ranged from 1.4% to 34.7%. Of the cit-tet+ plasmablasts sequenced, 28.6% belonged to clonal families; of tetramer- (tet-) plasmablasts, 23.8% were clonal. HC and LC V gene usage was comparable between the cit-tet+ plasmablasts and the entire plasmablast repertoires (Supplementary Figure 1). Mutation rates between cit-tet+ and tet- plasmablasts were non-significant, with the exception of subject RA3 in which the cit-tet+ plasmablasts were significantly more mutated (Supplementary Figure 2).
Figure 1.
Representative plasmablast antibody repertoires of CCP+ RA patients. The plasmablast antibody repertoires of CCP+ RA patients were sequenced using a single cell, barcode-based antibody repertoire capture method. Bioinformatic analyses of sequencing data sets were performed to obtain consensus sequences, pair cognate HC and LC sequences for individual plasmablasts, and generate phylogenetic trees. Two representative trees are presented (RA3 on left, RA11 on right; see also Supplementary Table 1). The tip of each branch denotes the paired HC and LC genes of an antibody expressed by a single plasmablast. Antibodies deriving from clonal families of plasmablasts are highlighted by yellow branches. Antibodies from cit-tet+ plasmablasts are colored red.
Recombinant monoclonal ACPA exhibit differential binding and unique polyreactive signatures against citrullinated antigens
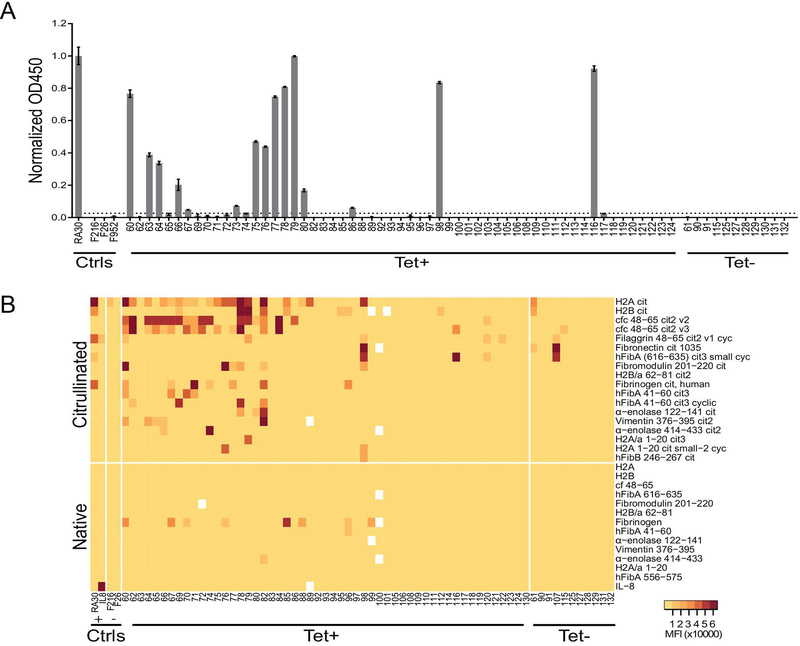
Sixty-nine antibodies spanning samples from the six RA patients were selected for recombinant expression. Fifty-eight of the recombinant monoclonal antibodies (rMabs) were derived from cit-tet+ plasmablasts, the majority of which were clonally expanded. The remaining 11 were derived from tet- plasmablasts. rMabs were tested for citrullinated antigen specificity using a cyclic citrullinated peptide (CCP) assay and synovial antigen planar arrays(27). Of the 69 rMabs expressed, 15 rMabs showed positive reactivity on the CCP ELISA (Figure 2A), while 34 showed reactivity against citrullinated antigens contained on the synovial antigen arrays (Figure 2B). Several rMabs exhibited low-level reactivity against native fibrinogen on the planar array. In comparing the cit-tet+ and cit-tet- rMabs, 33 of 58 cit-tet+ rMabs (56.9%) showed positive reactivity to citrullinated antigens in at least one of the two assays, as compared to only three of 11 tet- rMabs (27.3%). Intriguingly, of the 33 cit-tet+ rMabs that exhibited anti-citrullinated protein reactivity, 24 (72.7%) bound more than one citrullinated antigen based on synovial antigen planar array analysis. Together, these data indicate that we were able to successfully enrich for, express, and characterize recombinant ACPA from RA patient plasmablast repertoires.
Figure 2.
Recombinant monoclonal antibodies (rMabs) sequenced from patient plasmablasts exhibit distinct polyreactive binding of citrullinated antigens. (A) Representative antibodies derived from patient plasmablasts were recombinantly expressed and tested for anti-CCP reactivity using a commercial CCP ELISA kit. Results are normalized against a monoclonal ACPA control. Cut-off for positivity was set at three standard deviations above the average of the negative controls (anti-hemagglutinin antibodies). Data is presented as mean +/− SEM of duplicates and represent three independent experiments. (B) Antigen array analysis of recombinant antibody reactivities against RA-associated autoantigens. Arrays were printed with approximately 300 citrullinated and native synovial peptides and proteins, including cfc, a citrullinated filaggrin peptide used in first-generation CCP assays. Arrays were probed with patient-derived recombinant antibodies followed by fluorescent anti-human IgG detection antibody prior to scanning. Medium MFI of 4 replicate spots for each antigen is displayed. Tet+, cit-tetramer-positive (cit-tet+); Tet-, cit-tetramer-negative (cit-tet-); MFI, mean fluorescent intensity; cit, citrullinated; cyc, cyclic; ctrls, controls; H2A, histone 2A; cfc, citrullinated filaggrin cyclic; hFibA, human fibrinogen alpha; hFibB, human fibrinogen beta; cf, citrullinated filaggrin.
Plasmablast clonal families encode differentially mutated antibodies that exhibit differential binding to citrullinated antigens
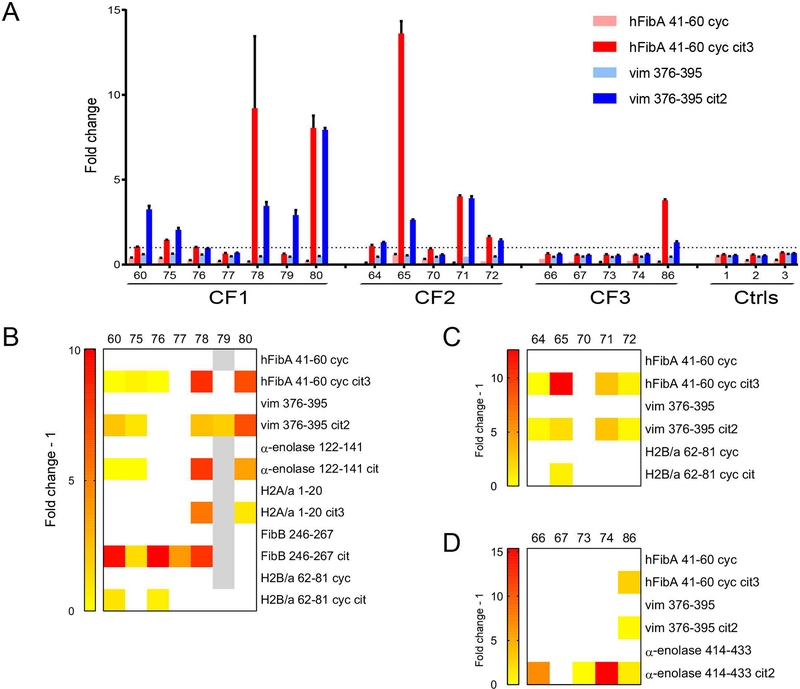
To investigate if epitope spreading arises from affinity maturation, we characterized by synovial antigen array and confirmed by ELISA the specificities of clonally related yet differentially mutated antibodies derived from three clonal families of ACPA-producing plasmablasts. Clonally related rMabs exhibited distinct but overlapping binding specificities to citrullinated antigens (Figure 3). For example, five of the seven rMabs derived from clonal family 1 bound the hFibA 41–60 cyc cit3 peptide (Figure 3B). Of these five, both rMabs 78 and 80 bound H2a/1–20 cit3, but not H2B/a 62–81 cyc cit. In contrast, the reverse is exhibited by rMabs 60 and 76. For both clonal families 2 and 3, three of the expressed rMabs from each family exhibited shared binding reactivities and recognized the same set of citrullinated peptides (with distinct sets of citrullinated peptides recognized by each family) (Figure 3, C and D). However, each family also contained individual ACPA that exhibited increased polyreactivity, binding additional citrullinated peptides, and one antibody that did not bind any peptides. None of the rMabs tested bound the non-citrullinated versions of the peptides (data not shown). This provides evidence of overlapping but distinct epitope reactivity, including the generation of differentially binding and/or polyreactive ACPA, of the divergently mutated antibodies encoded within ACPA plasmablast clonal families.
Figure 3.
Identification of ACPA-producing plasmablast clonal families containing divergent antibodies that exhibit differential polyreactive binding to citrullinated epitopes. (A) rMabs deriving from the three ACPA clonal families were expressed and tested by ELISA for reactivity against citrullinated peptides and their native counterparts. Cut-off values for each peptide were set at three standard deviations above the average of the negative controls. The dotted line represents the normalized cut-off for positive hits (fold change >1). Data are presented as mean +/− SEM and representative of at least three replicated experiments. (B-D) Heatmaps summarizing ELISA data for mAbs deriving from clonal families 1 (B), 2 (C), and 3 (D) binding to various citrullinated peptides. Data are representative of three replicated experiments. Mean reactivity of positive hits (fold change - 1 > 0) are displayed in colored gradient from yellow to red. White indicates no detectable reactivity (fold change - 1 ≤ 0). Gray indicates data not available. CF, clonal family; cyc, cyclic; vim, vimentin
Predicted ancestral antibodies exhibit restricted binding to citrullinated antigens and less polyreactivity
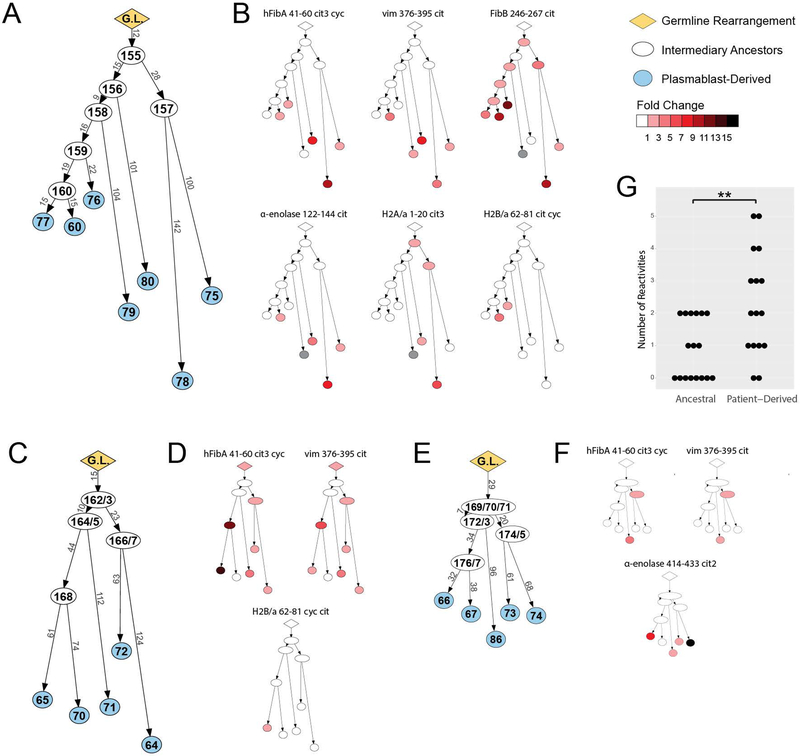
To directly assess the effects of somatic hypermutation on antigen specificity, we reverted clonally related plasmablast ACPAs back towards their germline sequence. We used IgTree to predict the shared parent antibodies for each of the three clonal families (Figure 4, A, C, E)(25). All ancestral antibodies, including the inferred germline sequences, were expressed and tested by ELISA to determine their binding specificity. In cases where the germline sequence was difficult to discern, particularly for the highly variable CDR3 regions, multiple antibodies were produced. Several of the parent sequences bound citrullinated antigens; however, in general the ancestral rMabs bound fewer antigens than the affinity-matured rMabs. For example, in clonal family 1, rMabs encoded by parent sequences only bound FibB 246–267 cit and H2A/a 1–20 cit3, which represent only two of the six peptides targeted by the corresponding affinity-matured antibodies (Figure 4B). Parent sequences in both clonal families 2 and 3 bound hFib 41–60 cit3 cyc and vim 376–395 cit peptides but showed no reactivity against additional peptides bound by the affinity-matured antibodies, namely H2B/a 62–81 cyc cit or α-enolase 414–433 cit2 (Figure 4, D, F). Of note, the inferred germline antibodies of two of the three clonal families did not bind to any citrullinated antigens contained on the synovial antigen arrays (Figure 4, B, F). These results demonstrate that the somatic hypermutation and affinity maturation of ACPA during clonal expansion can lead to epitope spreading of the ACPA response in individuals with RA.
Figure 4.
Ancestral antibodies of ACPA show divergent anti-citrulline reactivities and less polyreactivity. (A, C, E) Lineage tree analysis of the ACPA clonal families. Sequences of clonally related antibodies were analyzed with IgTree to generate lineage trees representing clonal families 1 (A), 2 (C), and 3 (E). The length of each branch has been adjusted to proportionally match the number of mutations present between two antibody nodes. The sequenced antibodies are highlighted in blue. Inferred germline sequences and intermediary antibodies are highlighted in yellow and white respectively. (B, D, F) ACPA reactivity of predicted ancestral B cell antibodies. Lineage trees for clonal families 1 (B), 2 (D), and 3 (F) are colored based on antigen-specificity as tested by ELISA, representative of at least two independent experiments. Color intensity indicates mean strength of ELISA signal, white indicates no binding, and gray indicates data not available. (G) Polyreactivity graph. The degree of polyreactivity was compared for all ancestral antibodies versus all affinity-matured, patient-derived antibodies across the three clonal family groups, and the degree of polyreactivity between the groups compared by Welch’s t-test. ** P≤0.01.
There were a few cases in which predicted ancestral antibodies bound more citrullinated antigens than affinity-matured descendants. For instance, ancestral antibodies RA168, RA174, and RA175 bind hFibA 41–60 cit3 cyc and vim 376–395 cit, but several of their descendants, RA70, RA73, and RA74, do not (Figure 4, C-F). This suggests that certain somatic hypermutations lead to loss of a specific epitope recognition for some antibodies. Despite this, individual affinity matured RA plasmablast-encoded ACPA exhibit statistically increased polyreactivity as compared to the ancestral antibodies (Figure 4G). These results demonstrate that the affinity maturation of ACPA B cells can lead to generation of individual B cell clones that encode ACPA that bind distinct citrullinated epitopes and/or possess increased polyreactivity, which in turn contributes to the increased number of reactivities against citrullinated antigens by the polyclonal ACPA response in the sera of RA patients.
Molecular modeling predicts regions within the ACPA paratope involved in epitope specificity
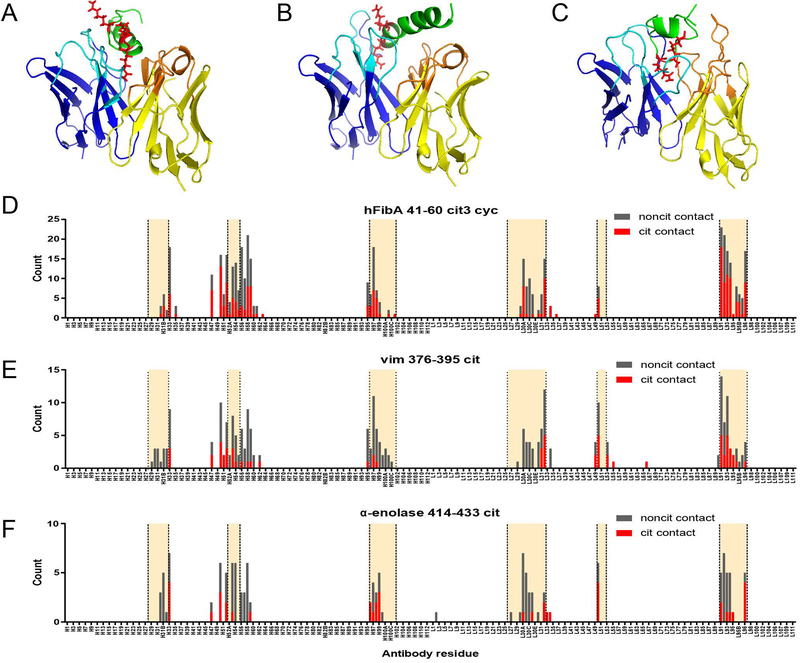
An antibody’s paratope is formed by the specific amino acid residues within the Fv domain that recognize and physically bind to its target antigen. To determine regions within the ACPA paratope responsible for citrullinated antigen binding, we performed molecular modeling to simulate ACPA:citrullinated-antigen interactions. We ran docking experiments for 28 antibody-antigen combinations and generated two top models for each complex, yielding a total of 56 models. A representative set of three docked models is presented in Figure 5, A-C. Using PyMOL, we identified for each model the antibody residues predicted to be in contact with the cit-antigens; the cumulative results across all antibody-antigen models are shown in Figure 5, D-F(29).
Figure 5.
Predicted contact residues of ACPA clonal family antibodies based on molecular modeling. Molecular models of antibodies from clonal families 1, 2, and 3 from Fig. 4 were producing using the Rosetta software package. Three peptides, hFibA 41–60 cit3 cyc (A, D), vim 376–395 cit (B, E), and α-enolase cit2 (C, F), were modelled using PEP-FOLD 2.0 and converted to citrullinated forms using PyTM. Antibody-antigen docking was performed using Rosetta (see Methods). Three representative molecular models of patient-derived mAbs docked with citrullinated peptides are presented: rMab 80 docked with hFibA 41–60 cit3 cyc peptide (A), rMab 78 with vim 376–395 cit peptide (B), and rMab 66 with α-enolase 414–433 cit2 peptide (C). The predicted contact residues for each docked model were counted and the cumulative results of all antibody-antigen combinations is presented (D-F). Antibody sequences are presented using the Chothia (post-1997) numbering scheme with the heavy and light chains concatenated along the X-axis. CDR regions are highlighted between the vertical dotted lines.
As expected, the majority of the contact residues were found in the CDR regions of the antibody sequences (Figure 5, D-F). However, while contact residues are predicted in the HCDR3 region, which is the predominant region conferring the binding specificity of most antibodies, these data indicate a stronger likelihood of involvement of the LCDR3 (189 counts) than the HCDR3 (85 counts), at least with regards to hFib41–60 cit3 cyc binding. We also observed that a substantial number of contact residues were predicted to occur in the framework regions flanking the HCDR2 (Figure 5, D-F). Taken together, our molecular modeling studies identified multiple regions within the ACPA paratopes predicted to be important for citrullinated antigen binding and that confer polyreactivity against specific sets of citrullinated epitopes.
Mutagenesis of critical ACPA paratope regions alters specificity and recapitulates epitope spreading
To confirm the relevant ACPA paratope regions identified from molecular modeling, we used the predicted contact residue information from the docked models to guide mutation studies on five pairs of clonally related ACPA. In each pair, we compared one antibody with a particular anti-citrullinated antigen reactivity to a clonally related antibody without that reactivity. Residues predicted to be relevant to binding were transferred from the reactive antibody to the non-reactive antibody (Supplementary Table 2). Mutated antibodies were tested alongside the subject-derived recombinant plasmablast antibodies using citrullinated peptide ELISAs to determine their antigen specificities (Figure 6).
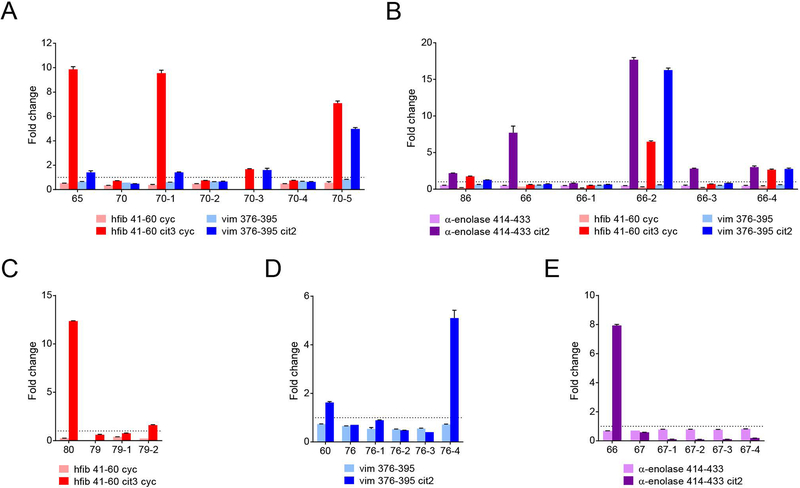
Figure 6.
Paratope mutagenesis studies of clonally-related ACPA alter epitope specificity and polyreactivity. Detection of altered cit-antigen reactivity of mutated recombinant ACPA. Pairs of antibodies from within the ACPA-producing clonal families were selected based on having divergent reactivity against one or more citrullinated peptides. Specifically, (A) rMabs 65 and 70 were selected from the clonal family presented in Figure 4C. rMab pairs (B) 66 and 86 and (E) 66 and 67 were selected from the clonal family in Figure 4E, and rMab pairs (C) 79 and 80 and (D) 60 and 76 were selected from Figure 4A. Mutated versions of the ACPA were designed based on the predicted residues identified in the modeling studies that were predicted to be critical for antigen binding (Figure 5, Supplementary Table 2). The original pair of recombinant antibodies and their corresponding mutated versions were then analyzed by peptide ELISAs for reactivity against the same antigens used in the modeling studies. Antibodies above the dotted line are considered to be positive hits (fold change > 1). Data are presented as mean +/− SEM and representative of at least two replicated experiments.
We were able to generate mutated antibodies that gained novel specificity(ies) against the citrullinated antigens and/or epitopes of interest. For instance, from clonal family 2, rMab 65 bound both hFib 41–60 cit3 cyc and vim 376–395 cit2, whereas rMab 70 bound neither. Transferring heavy chain residues 56–59 from rMab 65 to rMab 70 induced binding by modified rMab 70 against both peptides (Figure 6A, rMab 70–1). Importantly, these four residues lie in the framework region flanking the HCDR2 and were predicted by the modeling studies to be involved in binding. Mutating another region adjacent to the LCDR1 also induced binding to both antigens, although the reactivity against hFibA 41–60 was largely abrogated (rMab 70–3). Mutating the L31 residue or LCDR3 (rMabs 70–2 and 70–4) did not alter binding. Interestingly, combining all the mutations together (rMab 70–5) induced not only binding to hFibA 41–60 cit3 cyc but also greater reactivity to vim 376–395 cit2, when compared to either of the original, subject-derived, native rMabs.
Similar results were found for the other pairs of antibodies. Sequences from rMab 86 were transferred to rMab 66, resulting in binding activity to hFib 41–60 cit3 cyc and vim 376–395 cit2 (Figure 6B). In particular, transferring residues from LCDR1 region of rMab 86 induced strong binding reactivity against hFib, vim, and α-enolase peptides (rMab 66–2). The HCDR3 from rMab 80, when grafted into rMab 79, resulted in a modest amount of binding to hFib 41–60 cit3 cyc (rMab 79–2) (Figure 6C). Finally, transferring a combination of HCDR2, HCDR3, and LCDR3 from rMab 60 to rMab 76 induced strong reactivity against vim 376–395 cit2 peptide (Figure 6D, rMab 76–4). Only one set of mutated antibodies failed to exhibit new differential reactivities –transferring sequences from rMab 66 to rMab 67 resulted in no change in binding against α-enolase 414–433 cit (Figure 6E).
Overall, using the predicted ACPA paratope contact residues identified from the molecular models enabled mutation of clonally related antibody pairs to convey new and/or polyreactive binding to citrullinated epitopes in four of the five antibodies which previously showed no reactivity. These mutagenesis studies mimic the somatic hypermutation process and thus provide further evidence that affinity maturation of ACPA during clonal expansion can lead to both binding to distinct citrullinated epitopes and/or increased polyreactivity of individual ACPA in RA.
DISCUSSION
The phenomenon of epitope spreading of the ACPA response against citrullinated antigens is a well-established feature of RA, but little is understood regarding how and why it occurs. Here, by sequencing the plasmablast antibody repertoire, performing molecular modeling, and characterizing the encoded antibodies, we demonstrate a greater level of ACPA functional diversity at the single cell level than has been previously reported. We additionally report data suggesting that somatic hypermutations occurring during affinity maturation confer antibody paratopes that mediate both binding to distinct citrullinated epitopes and/or increased polyreactivity, which results in epitope spreading of the serum ACPA response.
To guide our analysis of the ACPA repertoire, we developed cit-peptide tetramers as a sort reagent. This sort reagent enabled identification of ACPA-producing B cells in blood from individuals with RA, as confirmed by positive reactivities of the encoded rMabs in CCP ELISAs and against citrullinated antigens on synovial antigen arrays. Several of the expressed cit-tet+ plasmablast rMabs did not bind citrullinated antigens in either ELISA or on planar arrays. There are several potential explanations for this, including potential differences in the conformation of the peptides, which is known to be important for ACPA recognition, which may differ in solution during flow cytometry as compared to when printed on epoxy slides. Additionally, some cit-tet- rMabs showed positive reactivity to citrullinated antigens. Since we used a cocktail of 14 citrullinated peptides to generate our tetramers—a relatively small representation of the spectrum of citrullinated antigens known to be present in RA joints—which would be expected to only enrich for a subset of ACPA-producing B cells, we likely missed some ACPA-producing B cells.
From the ELISAs and antigen array analysis, we determined that many of the rMab ACPA were polyreactive. This is in line with previous reports where ACPA have been shown to cross-react against multiple citrullinated epitopes(21, 30–32). Of note, while most studies have been limited to analysis of the polyclonal antibodies present in RA blood, only a few studies have characterized monoclonal ACPA. Of these, one group identified 36 patient-derived ACPA, 66.7% of which cross-reacted against multiple epitopes(33). This is similar to our own data in which 75.8% of our rMabs bound citrullinated epitopes, suggesting that the majority of ACPA display some degree of polyreactivity. Additionally, through testing for reactivity against a greater number of citrullinated antigens via synovial antigen arrays, we demonstrated distinct polyreactivity patterns between individual ACPA and that affinity maturation can increase the polyreactivity of individual ACPA.
While the heterogeneity of ACPA reactivity is well established, it is unclear if the epitope spreading and overall breadth of the ACPA response arises from de novo activation of ACPA B cells targeting distinct citrullinated epitopes versus diversification of the citrullinated epitope binding specificity within individual clonal families. An important finding of this study is the divergence of reactivities observed amongst clonally related ACPA, along with the expanded polyreactivity of individual affinity matured ACPA against distinct sets of citrullinated antigens.
By definition, clonal families of antibodies derive from a single activated parent B cell. They share the same germline genes and junctional rearrangements, differing only in the somatic hypermutations that each antibody has individually accumulated over time. According to this dogma, B cells would be selected for continued clonal expansion if their somatic hypermutations increase the affinity and specificity against the original antigen. However, our data suggest that this is not the case for ACPA, at least not in terms of their antigen specificity. We demonstrate multiple cases in which clonally related ACPA exhibit differential binding to citrullinated epitopes. Additionally, we demonstrate differences in the ability of ancestral antibodies to bind citrullinated epitopes, often recognizing fewer targets than the affinity-matured antibodies. Finally, we utilized molecular modeling and mutagenesis to recreate somatic hypermutations, proving that they can change the antigen specificities of ACPA. Together, our results reveal that somatic hypermutation alters the specific citrullinated epitopes bound by clonally related ACPA. We propose that somatic hypermutation during clonal expansion directly causes epitope spreading of the ACPA response.
In a healthy immune response, the majority of B cells encoding autoreactive antibodies arising from affinity maturation are tolerized(34). In autoimmune diseases, these checkpoints can be disrupted resulting in the escape of B cells encoding autoreactive antibodies. This phenomenon is not unique to RA; autoantibodies against double-stranded DNA can arise from non-autoreactive B cells undergoing somatic hypermutation in mouse models(35, 36) and SLE patients(37). Similarly, autoantibodies against DSG3 in pemphigus vulgaris can be generated through somatic hypermutation(38). Our study is the first to reveal the role of affinity maturation-mediated somatic hypermutation in the generation of ACPA. Somatic hypermutations induced novel reactivities and/or polyreactivities against citrullinated antigens, resulting in epitope spreading within clonal families of ACPA B cells. Additionally, ACPA paratopes that were artificially reverted to the inferred germline sequences lost their anti-citrulline reactivity and polyreactivity, suggesting that ACPA paratopes may arise from non-autoreactive B cells.
It is important to recognize the role of citrullinated epitopes in driving the evolution of ACPA. Based on our studies, we believe that citrulline acts as a “quasi-hapten”, increasing the antigenicity of an otherwise heterogeneous set of antigens, allowing ACPA to mutate in ways that increase their polyreactivity, so long as they retain a strong affinity for the citrulline residue itself. In this way, antibodies switching from protective reactivity against foreign citrullinated antigens, such as those arising from P. gingivalis infection or smoking, to pathogenic autoreactivity against citrullinated self-antigens are easily conceivable(33, 39). We hypothesize that citrulline recognition increases the likelihood of an ACPA-producing B cell receiving an activation signal because its B cell receptor is less restricted in which antigens it can bind. Increased B cell activation coupled with polyreactive ACPA leads to the continued expansion of B cell populations targeting multiple citrullinated antigens. Accordingly, our findings suggest that affinity maturation during clonal expansion drives epitope spreading of the ACPA response.
If citrulline is indeed acting as a quasi-hapten, elucidating the mechanisms by which ACPA B cells are generated could give rise to next-generation diagnostics and therapeutics. Conceivably, a common feature responsible for targeting citrullinated epitopes may exist amongst ACPA. Identification of a conserved binding paratope could lead to treatments that selectively target ACPA-producing B cells. We characterized the ACPA paratope through molecular modeling and mutagenesis. We were able to predict and verify key contact residues important for binding to citrullinated antigens, including the framework region next to HCDR2. This is an interesting finding given that framework regions are believed to primarily maintain loop structures and not direct antigen binding. The role of the framework regions in modulating the specificity of the ACPA paratope should be further investigated. While our studies identified regions within the ACPA paratope important for citrullinated antigen-specific binding, crystallization studies will be necessary to confirm the critical citrulline-binding residues.
Limitations to our study include the patient population of the VA hospital which is predominantly male, and thus the subjects utilized do not fully reflect the general RA patient population. Further, at the time of our study, our technology was only optimized to sequence plasmablasts expressing IgG or IgA isotype antibodies, thus only B cells expressing these two antibody subclasses were analyzed. Additionally, given the limited number of patients analyzed, the repertoire sequencing depth, and the number of recombinant antibodies expressed, it is difficult to estimate the extent to which affinity maturation within clonal families results in epitope spreading within individual patients and across the entire RA population. In our study, we selected the largest cit-tet+ clonal families with polyreactivity against citrullinated antigens for in depth characterization; however, these clonal families all derived from a single donor (RA3). Therefore, while we anticipate that multiple clonal families with differentially mutated members will exhibit epitope spreading in other patient repertoires, it is also possible that there will be families with differentially mutated members that exhibit a shared specificity without evidence of epitope expansion. The mechanisms driving the evolution of the antibody repertoire may vary between clonal families within a patient as well as between patients, based on the antigen(s) targeted, or over the course of disease. Nevertheless, our study establishes that affinity maturation is a mechanism by which epitope spreading of ACPA can occur. Although it is possible that our observations regarding RA3 may represent an extreme case, given the ease with which a few mutations to the variable region of an ACPA can alter its epitope specificity we suspect that epitope spreading via somatic hypermutation is a common occurrence. We anticipate that affinity maturation-driven epitope spreading has the potential to result in diversification of the specificity of the autoantibody response in multiple directions, depending on the specific mutations conferred in the process. Additional studies utilizing deeper sequencing in a larger cohort would help further define the extent to which this mechanism mediates epitope spreading in RA.
Together, our findings shed light on a mechanism mediating the pathogenesis of RA, in which somatic hypermutation and affinity maturation result in epitope spreading within lineages of ACPA-encoding B cells as well as generation of individual ACPA with increased polyreactivity that contribute to the reactivity of the overall ACPA response in RA. Further structural analysis of ACPA:citrullinated antigen interactions may help identify citrulline-specific and epitope-specific paratope regions that could inform the development of next-generation diagnostics and therapeutics. Finally, defining and characterizing the germline versions of ACPA could reveal key antigens involved in breaking tolerance to initiate the development of RA and thereby open avenues for disease prevention.
Supplementary Material
ACKNOWLEDGMENTS
We thank Jeremy Sokolove for assisting in acquiring samples and for his scientific insights, and Michelle Bloom for editing and proofreading. We also thank the Stanford Functional Genomics Facility and Stanford Shared FACS Facility for advice and technical assistance.
Funding: This research was supported by NIH \ NIAMS R01 AR063676, U19 AI11049103 and U01 AI101981.
Footnotes
Competing interests: W.H.R. is a consultant to, director of, and owns equity in Atreca, Inc. All other authors declare no conflict of interest.
DATA AND MATERIALS AVAILABILITY: Raw sequencing data have been deposited in NCBI dbGaP and are accessible through accession number <<submission in process>>.
References:
- 1.McInnes IB, Schett G. The pathogenesis of rheumatoid arthritis. N Engl J Med. 2011;365(23):2205–19. [DOI] [PubMed] [Google Scholar]
- 2.Firestein GS. Evolving concepts of rheumatoid arthritis. Nature. 2003;423(6937):356–61. [DOI] [PubMed] [Google Scholar]
- 3.Schellekens GA, de Jong BA, van den Hoogen FH, van de Putte LB, van Venrooij WJ. Citrulline is an essential constituent of antigenic determinants recognized by rheumatoid arthritis-specific autoantibodies. J Clin Invest. 1998;101(1):273–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van Venrooij WJ, van Beers JJBC, Pruijn GJM. Anti-CCP antibodies: the past, the present and the future. Nat Rev Rheumatol. 2011;7(7):391–8. [DOI] [PubMed] [Google Scholar]
- 5.Muller S, Radic M. Citrullinated Autoantigens: From Diagnostic Markers to Pathogenetic Mechanisms. Clin Rev Allergy Immunol. 2015;49(2):232–9. [DOI] [PubMed] [Google Scholar]
- 6.Whiting PF, Smidt N, Sterne JAC, Harbord R, Burton A, Burke M, et al. Systematic review: accuracy of anti-citrullinated Peptide antibodies for diagnosing rheumatoid arthritis. Ann Intern Med. 2010;152(7):456–64; W155–66. [DOI] [PubMed] [Google Scholar]
- 7.Sokolove J, Bromberg R, Deane KD, Lahey LJ, Derber LA, Chandra PE, et al. Autoantibody epitope spreading in the pre-clinical phase predicts progression to rheumatoid arthritis. PLoS ONE. 2012;7(5):e35296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van de Stadt LA, de Koning MH, van de Stadt RJ, Wolbink G, Dijkmans BA, Hamann D, et al. Development of the anti-citrullinated protein antibody repertoire prior to the onset of rheumatoid arthritis. Arthritis Rheum. 2011;63(11):3226–33. [DOI] [PubMed] [Google Scholar]
- 9.van der Woude D, Rantapaa-Dahlqvist S, Ioan-Facsinay A, Onnekink C, Schwarte CM, Verpoort KN, et al. Epitope spreading of the anti-citrullinated protein antibody response occurs before disease onset and is associated with the disease course of early arthritis. Ann Rheum Dis. 2010;69(8):1554–61. [DOI] [PubMed] [Google Scholar]
- 10.de Vries-Bouwstra JK, Goekoop-Ruiterman YPM, Verpoort KN, Schreuder GMT, Ewals JaPM, Terwiel JP, et al. Progression of joint damage in early rheumatoid arthritis: association with HLA-DRB1, rheumatoid factor, and anti-citrullinated protein antibodies in relation to different treatment strategies. Arthritis Rheum. 2008;58(5):1293–8. [DOI] [PubMed] [Google Scholar]
- 11.Lindqvist E, Eberhardt K, Bendtzen K, Heinegård D, Saxne T. Prognostic laboratory markers of joint damage in rheumatoid arthritis. Ann Rheum Dis. 2005;64(2):196–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meyer O, Labarre C, Dougados M, Goupille P, Cantagrel A, Dubois A, et al. Anticitrullinated protein/peptide antibody assays in early rheumatoid arthritis for predicting five year radiographic damage. Ann Rheum Dis. 2003;62(2):120–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rönnelid J, Wick MC, Lampa J, Lindblad S, Nordmark B, Klareskog L, et al. Longitudinal analysis of citrullinated protein/peptide antibodies (anti-CP) during 5 year follow up in early rheumatoid arthritis: anti-CP status predicts worse disease activity and greater radiological progression. Ann Rheum Dis. 2005;64(12):1744–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nishimura K, Sugiyama D, Kogata Y, Tsuji G, Nakazawa T, Kawano S, et al. Meta-analysis: diagnostic accuracy of anti-cyclic citrullinated peptide antibody and rheumatoid factor for rheumatoid arthritis. Ann Intern Med. 2007;146(11):797–808. [DOI] [PubMed] [Google Scholar]
- 15.Kuhn KA, Kulik L, Tomooka B, Braschler KJ, Arend WP, Robinson WH, et al. Antibodies against citrullinated proteins enhance tissue injury in experimental autoimmune arthritis. J Clin Invest. 2006;116(4):961–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wardemann H, Yurasov S, Schaefer A, Young JW, Meffre E, Nussenzweig MC. Predominant autoantibody production by early human B cell precursors. Science. 2003;301(5638):1374–7. [DOI] [PubMed] [Google Scholar]
- 17.Arnett FC, Edworthy SM, Bloch DA, McShane DJ, Fries JF, Cooper NS, et al. The American Rheumatism Association 1987 revised criteria for the classification of rheumatoid arthritis. Arthritis Rheum. 1988;31(3):315–24. [DOI] [PubMed] [Google Scholar]
- 18.Elliott SE, Kongpachith S, Lingampalli N, Adamska JZ, Cannon BJ, Mao R, et al. Affinity Maturation Drives Epitope Spreading and Generation of Pro-inflammatory Anti-Citrullinated Protein Antibodies in Rheumatoid Arthritis. Arthritis Rheumatol. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lu DR, McDavid AN, Kongpachith S, Lingampalli N, Glanville J, Ju CH, et al. T cell-dependent affinity maturation and innate immune pathways differentially drive autoreactive B cell responses in rheumatoid arthritis. Arthritis Rheumatol. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tan Y-C, Blum LK, Kongpachith S, Ju C-H, Cai X, Lindstrom TM, et al. High-throughput sequencing of natively paired antibody chains provides evidence for original antigenic sin shaping the antibody response to influenza vaccination. Clin Immunol. 2014;151(1):55–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tan Y-C, Kongpachith S, Blum LK, Ju C-H, Lahey LJ, Lu DR, et al. Barcode-Enabled Sequencing of Plasmablast Antibody Repertoires in Rheumatoid Arthritis. Arthritis & Rheumatology. 2014;66(10):2706–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kinslow JD, Blum LK, Deane KD, Demoruelle MK, Okamoto Y, Parish MC, et al. Elevated IgA Plasmablast Levels in Subjects at Risk of Developing Rheumatoid Arthritis. Arthritis & Rheumatology (Hoboken, NJ). 2016;68(10):2372–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Blum LK, Cao RRL, Sweatt AJ, Bill M, Lahey LJ, Hsi AC, et al. Circulating plasmablasts are elevated and produce pathogenic anti‐endothelial cell autoantibodies in idiopathic pulmonary arterial hypertension. European Journal of Immunology.0(0). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gupta NT, Vander Heiden JA, Uduman M, Gadala-Maria D, Yaari G, Kleinstein SH. Change-O: a toolkit for analyzing large-scale B cell immunoglobulin repertoire sequencing data. Bioinformatics. 2015;31(20):3356–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barak M, Zuckerman NS, Edelman H, Unger R, Mehr R. IgTree: creating Immunoglobulin variable region gene lineage trees. J Immunol Methods. 2008;338(1–2):67–74. [DOI] [PubMed] [Google Scholar]
- 26.Nair N, Feng N, Blum LK, Sanyal M, Ding S, Jiang B, et al. VP4- and VP7-specific antibodies mediate heterotypic immunity to rotavirus in humans. Science Translational Medicine. 2017;9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hueber W, Kidd BA, Tomooka BH, Lee BJ, Bruce B, Fries JF, et al. Antigen microarray profiling of autoantibodies in rheumatoid arthritis. Arthritis Rheum. 2005;52(9):2645–55. [DOI] [PubMed] [Google Scholar]
- 28.Robinson WH, DiGennaro C, Hueber W, Haab BB, Kamachi M, Dean EJ, et al. Autoantigen microarrays for multiplex characterization of autoantibody responses. Nat Med. 2002;8(3):295–301. [DOI] [PubMed] [Google Scholar]
- 29.Schrodinger LLC. The PyMOL Molecular Graphics System, Version 1.8. 2015. [Google Scholar]
- 30.Ioan-Facsinay A, el-Bannoudi H, Scherer HU, van der Woude D, Ménard HA, Lora M, et al. Anti-cyclic citrullinated peptide antibodies are a collection of anti-citrullinated protein antibodies and contain overlapping and non-overlapping reactivities. Ann Rheum Dis. 2011;70(1):188–93. [DOI] [PubMed] [Google Scholar]
- 31.Goules JD, Goules AV, Tzioufas AG. Fine specificity of anti-citrullinated peptide antibodies discloses a heterogeneous antibody population in rheumatoid arthritis. Clin Exp Immunol. 2013;174(1):10–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Trier NH, Leth ML, Hansen PR, Houen G. Cross-reactivity of a human IgG₁ anticitrullinated fibrinogen monoclonal antibody to a citrullinated profilaggrin peptide. Protein Sci. 2012;21(12):1929–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li S, Yu Y, Yue Y, Liao H, Xie W, Thai J, et al. Autoantibodies From Single Circulating Plasmablasts React With Citrullinated Antigens and Porphyromonas gingivalis in Rheumatoid Arthritis. Arthritis Rheumatol. 2016;68(3):614–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pelanda R, Torres RM. Central B-cell tolerance: where selection begins. Cold Spring Harb Perspect Biol. 2012;4(4):a007146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shlomchik M, Mascelli M, Shan H, Radic MZ, Pisetsky D, Marshak-Rothstein A, et al. Anti-DNA antibodies from autoimmune mice arise by clonal expansion and somatic mutation. J Exp Med. 1990;171(1):265–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guo W, Smith D, Aviszus K, Detanico T, Heiser RA, Wysocki LJ. Somatic hypermutation as a generator of antinuclear antibodies in a murine model of systemic autoimmunity. J Exp Med. 2010;207(10):2225–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wellmann U, Letz M, Herrmann M, Angermüller S, Kalden JR, Winkler TH. The evolution of human anti-double-stranded DNA autoantibodies. Proc Natl Acad Sci USA. 2005;102(26):9258–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Di Zenzo G, Di Lullo G, Corti D, Calabresi V, Sinistro A, Vanzetta F, et al. Pemphigus autoantibodies generated through somatic mutations target the desmoglein-3 cis-interface. J Clin Invest. 2012;122(10):3781–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Klareskog L, Stolt P, Lundberg K, Källberg H, Bengtsson C, Grunewald J, et al. A new model for an etiology of rheumatoid arthritis: Smoking may trigger HLA–DR (shared epitope)–restricted immune reactions to autoantigens modified by citrullination. Arthritis & Rheumatism. 2006;54(1):38–46. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.