Abstract
Introduction
Calcium/calmodulin-dependent (Ca2+/CaM-dependent) regulation of protein signaling has long been recognized for its importance in a number of physiological contexts. Found in almost all eukaryotic cells, Ca2+/CaM-dependent signaling participates in muscle development, immune responses, cardiac myocyte function and regulation of neuronal connectivity. In excitatory neurons, dynamic changes in the strength of synaptic connections, known as synaptic plasticity, occur when calcium ions (Ca2+) flux through NMDA receptors and bind the Ca2+-sensor calmodulin (CaM). Ca2+/CaM, in turn, regulates downstream protein signaling in actin polymerization, receptor trafficking, and transcription factor activation. The activation of downstream Ca2+/CaM-dependent binding proteins (CBPs) is a function of the frequency of Ca2+ flux, such that each CBP is preferentially “tuned” to different Ca2+ input signals. We have recently reported that competition among CBPs for CaM binding is alone sufficient to recreate in silico the observed in vivo frequency-dependence of several CBPs. However, CBP activation may strongly depend on the identity and concentration of proteins that constitute the competitive pool; with important implications in the regulation of CBPs in both normal and disease states.
Methods
Here, we extend our previous deterministic model of competition among CBPs to include phosphodiesterases, AMPAR receptors that are important in synaptic plasticity, and enzymatic function of CBPs: cAMP regulation, kinase activity, and phosphatase activity. After rigorous parameterization and validation by global sensitivity analysis using Latin Hypercube Sampling (LHS) and Partial Rank Correlation Coefficients (PRCC), we explore how perturbing the competitive pool of CBPs influences downstream signaling events. In particular, we hypothesize that although perturbations may decrease activation of one CBP, increased activation of a separate, but enzymatically-related CBP could compensate for this loss, providing a homeostatic effect.
Results and Conclusions
First we compare dynamic model output of two models: a two-state model of Ca2+/CaM binding and a four-state model of Ca2+/CaM binding. We find that a four-state model of Ca2+/CaM binding best captures the dynamic nature of the rapid response of CaM and CBPs to Ca2+ flux in the system. Using global sensitivity analysis, we find that model output is robust to parameter variability. Indeed, although variations in the expression of the CaM buffer neurogranin (Ng) may cause a decrease in Ca2+/CaM-dependent kinase II (CaMKII) activation, overall AMPA receptor phosphorylation is preserved; ostensibly by a concomitant increase in adenylyl cyclase 8 (AC8)-mediated activation of protein kinase A (PKA). Indeed phosphorylation of AMPAR receptors by CaMKII and PKA is robust across a wide range of Ng concentrations, though increases in AMPAR phosphorylation is seen at low Ng levels approaching zero. Our results may explain recent counter-intuitive results in neurogranin knockout mice and provide further evidence that competitive tuning is an important mechanism in synaptic plasticity. These results may be readily translated to other Ca2+/CaM-dependent signaling systems in other cell types and can be used to suggest targeted experimental investigation to explain counter-intuitive or unexpected downstream signaling outcomes.
Electronic supplementary material
The online version of this article (10.1007/s12195-018-0549-4) contains supplementary material, which is available to authorized users.
Keywords: Signal transduction, Dendritic spine, Calcium signaling, Adenylyl cyclase, Ca2+/calmodulin-dependent kinase II (CaMKII), Neurogranin
Introduction
Worldwide, as many as 1 billion people suffer from neurological disorders.52 In the US alone, neurological disorders affect more than 1 in 7 households.4 At the most basic level, these neurological disorders arise from perturbations in protein signaling networks within neuronal synapses. Normal synaptic function requires dynamic, short-timescale regulation of the connective strength of the synapse. This regulation is initiated within the post-synapse by the influx of calcium ions (Ca2+) through NMDA receptors.19 Intracellular Ca2+ binds the Ca2+ sensor protein calmodulin (CaM), which subsequently activates a variety of Ca2+/CaM-dependent protein signaling pathways. Ca2+/CaM-dependent pathways may either potentiate synaptic connective strength via AMPA receptor (AMPAR) phosphorylation and trafficking to the synapse, or they may depress synaptic strength by regulating AMPAR de-phosphorylation and removal from the synapse (recently reviewed in Huganir and Nicoll21). Although many of the proteins that are involved in Ca2+/CaM-dependent AMPAR regulation are well known, the dynamics of the pathway(s) are far from understood. With computer-guided studies, we begin to characterize the dynamics and cross-talk inherent to the protein interactions in these pathways, possibly enabling the identification of new therapeutic targets for treating neurological disorders.
Calmodulin (CaM) regulates synaptic plasticity by selectively activating a number of downstream proteins, termed calmodulin binding proteins (CBPs), within the signaling networks responsible for either the dynamic strengthening or weakening of synaptic connections. The binding of CaM to its many downstream binding partners9,15,27,53 depends on the kinetic rates with which different CaM species bind CBPs, such that each CBP is preferentially activated by, or “tuned” to, different input Ca2+ signals. Aside from binding dynamics, other mechanisms that regulate this tuning include feedback loops, spatial localization, and a recently described phenomenon called competitive tuning.22,45,46 We have recently reported that competitive tuning is sufficient to recreate, in silico, the in vivo Ca2+ frequency-dependence of several CBPs.45 One prediction from competitive tuning is that, in the absence of other mechanisms, signaling outcomes may strongly depend on the abundance and binding dynamics of individual CBPs; parameters susceptible to perturbation either by genetic regulation or by post-translational modification. It follows then that signaling outcomes may lack robustness if competitive tuning occurs in isolation of other regulatory mechanisms.
Indeed, modulating just one parameter (e.g., a protein’s concentration) could cause a shift in competition that influences the signaling outcomes of other proteins in the system, perhaps leading to non-intuitive effects. We have previously shown that simulated knockout of the CaM buffer neurogranin (Ng) shifts the competition for CaM, non-intuitively decreasing Ca2+/CaM-dependent kinase II (CaMKII) activation and concomitantly increasing adenylyl cyclase (AC) activation.45 Our previous results may explain a surprising experimental observation by Krucker et al.26 in which Ng knockout (Ng KO) mice retain the ability to acquire long-term potentiation (LTP), despite a considerable reduction in CaMKII activity.26 In this present work we further explore how competitive tuning regulates LTP, hypothesizing that although reduced CaMKII activation in the Ng KO should reduce the phosphorylation of AMPA receptor GluA1 subunits at residue S831, a coincident increase in AC activation may cause an increase in GluA1 subunit phosphorylation at residue S845. This would lead to robustness in the overall level of GluA1 phosphorylation. Although there is still some debate about the precise roles of phosphorylation at S831 and S845, it is well accepted that phosphorylation of these sites is involved in the function and location of AMPARs and in synaptic plasticity (recently reviewed in Refs.11 and 21). Thus, competitive tuning alone could provide the mechanism by which overall GluA1 phosphorylation levels are maintained and provide a homeostatic effect on synaptic plasticity.
Here, we compare two models of Ca2 +/CaM-dependent activation of CBPs; a 2-state Ca2 +/CaM binding model and a four-state Ca2 +/CaM-binding model. We include well documented CBPs that are highly expressed in neurons, and also include signaling events downstream of Ca2 +/CaM binding, including CaM-dependent enzymatic activity, PKA kinase activation, phosphatase regulation, and AMPAR receptor phosphorylation. After validating the model, we use a global sensitivity analysis to quantify the effect of parameter perturbations on model outcomes. We also find that at short timescales, competitive tuning provides robustness in overall GluA1 phosphorylation levels via upregulation in the activation of PKA-mediated phosphorylation of GluA1 in conditions that simulate the Ng KO. Our results provide further evidence that competitive tuning could be an important mechanism in the regulation and maintenance of synaptic plasticity.
Results and Discussion
Model Development
Model Structure
Similar to our previous work,45 we constructed models of CaM binding to a number of downstream CBPs and allowed the CBPs to compete for the various states of Ca2+/CaM. CaM has four binding sites for Ca2+ ions, two in EF-hand domains in the amino (N) terminus, and two in EF-hand domains in the carboxy (C) terminus. The binding of Ca2+ within each terminus is highly cooperative, for example upon binding of the first Ca2+ ion at the N-terminus, a second Ca2+ ion binds rapidly to the N-terminus. But binding between the termini is independent (i.e., Ca2+-binding at the N-terminus does not change the binding of Ca2+ at the C-terminus). Many models of Ca2+ signaling cascades in neurons and cardiomyocytes have employed a simplified, yet still relevant model of Ca2+ ions binding to CaM where all four Ca2+ ions are assumed to bind simultaneously (outlined in Fig. 1a). These models have been used extensively and are thought to be quite accurate, at least for scenarios in which the overall magnitude of Ca2+ flux is large, such as in Ca2+-dependent Ca2+ release phenomena or high frequency Ca2+ flux. However, more detailed descriptions of CaM that account for its intermediate, sub-saturated states (outlined in Fig. 1b) may be more appropriate for situations in which Ca2+ flux occurs at moderate to low frequencies, or in conditions of limiting Ca2+.31 It has been shown that several CBPs (MLCK, CaMKII, AC1 and AC8) produce CaM-dependent activity even with small increases in Ca2+ concentration.33,39,48 Additionally, CaMKII has been shown to be enzymatically active with only two Ca2+ ions bound to CaM,13 and AC8 has been shown to bind CaM with only two Ca2+ ions bound to CaM.33 These observations and the fact that binding of Ca2+ to CaM is highly cooperative within each terminus, but not between termini, together have led some to develop models that include more intermediate states of Ca2+ binding to CaM.42,45,46
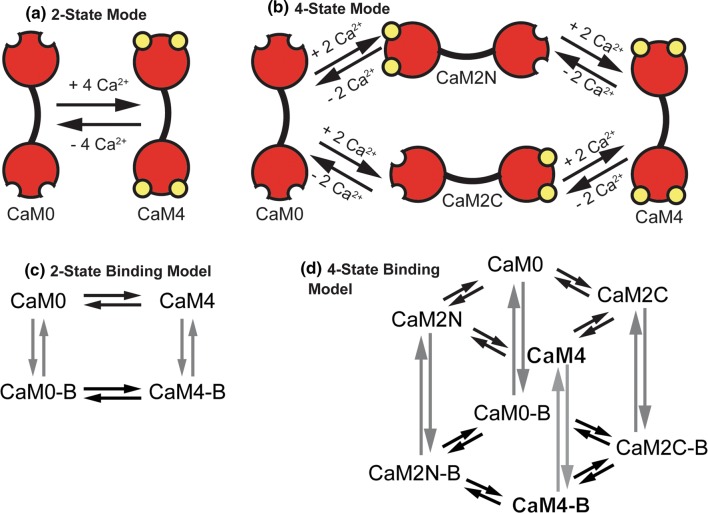
Figure 1.
Schematic representations of binding models. (a, b) Binding models of Ca2+-binding to calmodulin. Four Ca2+ ions (yellow circles) bind to CaM at binding sites in the amino (N) and carboxyl (C) termini. (a) 2-state model of Ca2+ binding to CaM. CaM with no Ca2+ ions bound (CaM0) is assumed to simultaneously bind four Ca2+ ions forming a fully saturated Ca2+ bound CaM (CaM4). (b) 4-state model of Ca2+ binding to CaM where two Ca2+ ions bind simultaneously to either the N- or C- termini of CaM (CaM2 N or CaM2C, respectively). In a subsequent reaction two Ca2+ ions bind to either termini to form fully saturated Ca2+-bound CaM (CaM4). (c, d) Models of Ca2+ binding to CaM and CaM binding to downstream binding partners. (c) 2-state binding model where CaM0 with no Ca2+ ions bound binds to downstream binding partners (represented by CaM0-B). CaM4 can also bind to downstream binding partners (CaM4-B). (d) 4-state binding model where all states of CaM (CaM0, CaM2 N, CaM2C, and CaM4) may bind downstream binding partners (CaM0-B, CaM2N-B, CaM2C-B, and CaM4-B).
We compare a 2-state CaM model to a 4-state CaM model (Figs 1a and 1b). For both models we view CaM as a limiting resource. There are many more CBPs than CaM itself. Thus CBPs compete simultaneously for binding to CaM in its different states (overview in Figs. 1c, 1d and 2). Listed in Table 1, the CBPs that are included in this work are highly expressed in neurons and widely reported to interact with CaM (see Ref.53 for a review). The reactions that describe these interactions are listed in Table S1. To study downstream events in Ca2+/CaM-dependent signaling we additionally include enzymatic activation of CBPs and the relevant proteins and nucleotides that are downstream of CaM-binding. These include the generation of cAMP by AC1 and AC8, hydrolysis of cAMP by PDE1 and PDE4, activation of PKA by cAMP, phosphorylation of GluA1 by PKA and CaMKII, and de-phosphorylation of GluA1 by CaN and PP1 (summarized in Table 2; reactions listed in Table S2). The interactions among Ca2+ ions, proteins, and nucleotides described above and shown schematically in Fig. 2 and the corresponding differential equations are found in Supplemental Material section ‘Model Equations’. In total there are 91 equations and 225 parameters for the 2-state model and 151 equations and 447 parameters for the 4-state model.
Figure 2.
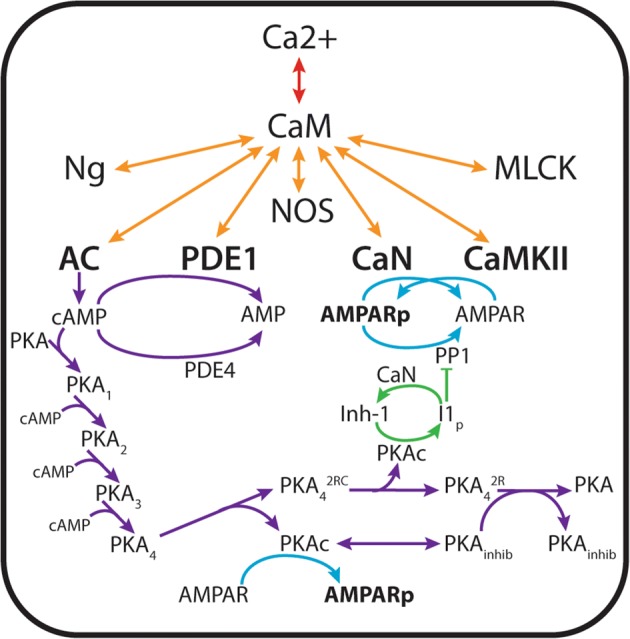
Schematic of the CaM-dependent protein signaling network. Ca2+ ions flux through the NMDA receptor, initiating a cascade of signaling that results in phosphorylation of the AMPA receptors. CaM-binding proteins (neurogranin (Ng), muscle light chain kinase (MLCK), neuronal nitric oxide synthetase (NOS), calcineurin (CaN, also known as protein phosphatase 3), Ca2+/calmodulin-dependent kinase II (CaMKII), phosphodiesterase 1 (PDE1), adenylyl cyclase 1 (AC1) and adenylyl cyclase 8 (AC8)) simultaneously compete for CaM binding. CaM binding activates the catalytic activity of CaMKII, which phosphorylates the AMPA receptor GluA1 subunit at site S831. Generation of cAMP by CaM-bound adenylyl cyclases activates the catalytic activity of protein kinase A (PKA, also known as cAMP-dependent kinase), which phosphorylates Inh-1 as well as GluA1 at site S845. Phosphodiesterases (both CaM-dependent PDE1 and constitutively active PDE4) regulate intracellular levels of cAMP by hydrolysis, thereby regulating PKA activity. PP1 may additionally de-phosphorylate CaMKII or GluA1. PP1 may become sequestered by Inh-1, until Inh-1 is de-phosphorylated by CaN.
Table 1.
Calmodulin binding proteins.
| Protein | Description and chemical reactions |
|---|---|
| Adenylyl cyclase 1 (AC1) | A prominent adenylyl cyclase isoform in hippocampal neurons. Binds CaM at its C1b domain and, when activated, converts ATP to cAMP6,7,36,51 |
| Adenylyl cyclase 8 (AC8) | A prominent adenylyl cyclase isoform in hippocampal neurons. Binds Ca2+/CaM at both its N- and C-termini, which are explicitly accounted for in this work.16,33 Ca2+/CaM-AC8ct (C-terminus-bound AC8) may bind ATP for conversion to cAMP36 |
| Ca2+/CaM-dependent kinase II (CaMKII) | Ca2+/CaM-dependent kinase. Highly expressed in brain, and especially hippocampal tissue.34 In this work, CaMKII is modeled as monomers which, when active, may dimerize and subsequently become Ca2+/CaM-independent via autophosphorylation.35 Active CaMKII phosphorylates many downstream proteins such as the GluA1 subunit of AMPA receptors2 |
| Calcineurin (CaN) | A Ca2+/CaM-dependent serine/threonine phosphatase.25,50 CaN dephosphorylates GluA1 at S8453 |
| Myosin light chain kinase (MLCK) | A putatively abundant CBP in dendritic spines, which we model using kinetic parameters derived from studies on smooth muscle.10,28,30 |
| Neurogranin (Ng) | A membrane-associated protein, and one of the few proteins that strongly binds CaM in absence of Ca2+14,20,44,55 |
| Nitric oxide synthetase (NOS) | Typically a membrane-associated protein that binds tightly to CaM and generates Nitric Oxide from citrulline and arginine5,12,40 |
| Phosphodiesterase 1 (PDE1) | A Ca2+/CaM dependent phosphodiesterase that cleaves cAMP into AMP8 |
Table 2.
Non-calmodulin binding proteins included in model.
| GluA1 | One of four subunits of α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptor (AMPA receptor). Is phosphorylated at amino acid residue S831 by CaMKII and residue S845 by PKA18,38 Increase AMPA phosphorylation is implicated in synaptic plasticity, and GluA1-p845 may be necessary for exocytosis of AMPARs to the synaptic membrane18 |
| Inhibitor 1 (Inh-1) | When phosphorylated by active PKAc, Inh-1 may bind PP1, inhibiting the ability of PP1 to de-phosphorylate GluA1. Inh-1 is de-phosphorylated by CaM-activated CaN18 |
| Phosphodiesterase 4 (PDE4) | PDE4 is not Ca2+/CaM dependent but plays a significant role in regulating the levels of cAMP in cells by cleaving cAMP into AMP. Phosphorylation by active PKAc increases the enzymatic activity of PDE417 |
| PKA inhibitor | A generic model species representative of many unspecified off-target binding partners for PKA. These binding partners sequester PKA, preventing its phosphorylation of GluA1, and they additionally participate in the pathway restoring PKAc to the original four-subunit, auto-regulated PKA heteromer17 |
| Protein kinase A (PKA, also known as cAMP-dependent kinase) | Binds up to four cAMP, liberating catalytic subunits that bind and phosphorylate a number of downstream targets such as PDE4, Inh-1, and GluA18,38 |
| Protein phosphatase 1 (PP1) | PP1 de-phosphorylates GluA1 subunits at both S831 and S845, in addition to CaMKII T286.18 In this model, PP1 may only bind CaMKII when CaMKII is un-bound to Ca2+/CaM |
Model Parameterization
Model parameters are either obtained directly from literature or are calculated from published values using the principle of microscopic reversibility and implementing the assumption that Ca2+ binding does not affect the rate of protein dissociation from CaM koff, but rather the association rate (kon) such that the general equation can be used to calculate rate constants. These assumptions have been used regularly in the modeling literature by us and others23,24,42,45 and have been shown to be experimentally validated.41 For the Ca2+/CaM-dependent enzymes AC, PDE1, and CaN the catalytic activity is assumed to scale with the amount of Ca2+ bound to CaM, similar to what has been shown with CaMKII.48 Thus we calculate scaled values for the catalytic activity of sub-saturated Ca2+-bound CaM-CBPs similar to our previous model of CaMKII auto-phosphorylation.45 For example, the catalytic activity of Ca2+/CaM2C-CaN is slower than that of Ca2+/CaM4-CaN. In the absence of experimental measurement of catalytic rate constants for AC, PDE1, and CaN activity when bound to sub-saturated CaM, we calculated these scaled catalytic rate constants such that the rate of activity of the CaM2C-bound state relative to that CaM4-bound state was decreased by a similar proportion to that of the phosphorylation rate CaM2C-CaMKII to CaM4-CaMKII which has been previously measured.48 Catalytic rate constants for CaM2N-bound state of AC, PDE1, and CaN was similarly scaled relative to the ratio of phosphorylation rate constants of CaM2N-CaMKII to CaM4-CaMKII. The values of all parameters are provided in Supplemental Table S5.
Prior to executing Ca2+ flux, each simulation was run for a time course of 30 s at a basal level of 5 nM Ca2+ to allow for steady state binding of CaM to CBPs.32,37 The concentrations of all species at the end of this preliminary simulation were used as the input for simulations in which Ca2+ flux was initiated at varying frequencies.
2-State vs 4-State CaM Binding Models
In previous work we have shown that a thermodynamically complete, 9-state model of Ca2+ binding to CaM did not significantly change model output relative to a 4-state binding model.45 In this work our first task was to compare a 2-state binding model to that of a 4-state binding model (Figs. 1c and 1d) to validate the hypothesis that the dynamics of CBP binding by CaM are optimally represented by a 4-state binding model. For this, we analyzed model output when stimulated at a frequency of 10 Hz (see Methods). In Fig. 3 we used two metrics to show how the 2-state model (Figs. 3a and 3c) and 4-state model (Figs. 3b and 3d) each responded to 10 Hz Ca2+ stimulation. The first metric (Figs. 3a and 3b) monitored CBP binding only to fully-saturated CaM4 over time. The second metric (Figs. 3c and 3d) monitored the total CBP binding to all Ca2+/CaM states. The only difference between metrics for the 2-state model is that the second metric additionally accounts for each CBP bound to apo-CaM (CaM0), if any. Similarly, using the second metric for the 4-state model involves summing the concentrations of each CBP bound to each Ca2+/CaM state: CaM0, CaM2N, CaM2C, and CaM4.
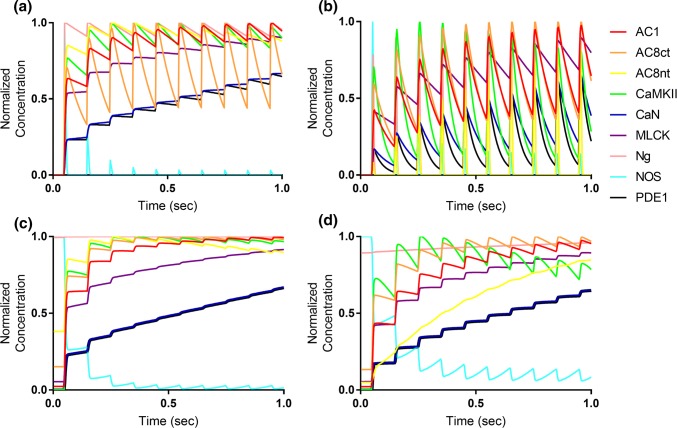
Figure 3.
Time-course comparison of Ca2+/CaM binding models. (a, b) Response of binding models to 10 Hz Ca2+ frequency stimulation, monitoring each CBP bound to Ca2+-saturated CaM4. (c, d) Response of binding models to 10 Hz Ca2+ frequency stimulation, monitoring the cumulative concentration of each CBP bound to any Ca2+/CaM state. Similar data for models stimulated at 50 Hz Ca2+ is provided in Fig. S1.
By monitoring either CaM4-bound or all CaM-bound CBP, we assessed the added value of a relatively detailed 4-state model to the simpler 2-state model of Ca2+/CaM. In Fig. 3a, each CBP-CaM4 trace exhibited wavelets implying a frequency detection inherent to CBP activation. For most CBPs such as CaN (blue) and AC8ct (black), relative activation by CaM4 generally increased over time, which was consistent with expectation for CBPs exposed to a continuous, frequency-based stimulus. In contrast, Ng (pink) rapidly decreased in relative CaM4 binding over time though this was also expected due to the high affinity of Ng for CaM0 compared to CaM4. In Fig. 3c, the 2-state model elicited similar time-course trends for each CBP compared to Fig. 3a, and because the traces lack frequency-associated wavelets, it appears that CaM0 was significantly bound to CBPs, particularly during inter-spike intervals of Ca2+. The differences between monitoring solely CaM4- vs. all CaM-bound CBPs were further evidenced by Supplemental Fig. S1, which shows equivalent output but for a model stimulated at 50 Hz.
For the 4-state model, the Ca2+ frequency-associated waveforms were much more prominent than in the 2-state model (compare Figs. 3b and 3d to Figs. 3a and 3c), indicating that the 4-state model is much more responsive to Ca2+ than the 2-state model (some data obscured by the highly dynamic traces). See also results from 50 Hz stimulation in Supplemental Fig. S1. Of note, there is a significant difference in the dynamics of binding of CaM to AC8nt and NOS between the 2-state and 4-state models (compare Figs. 3a and 3c to Figs. 3b and 3c) at 10 Hz. Additionally, the 4-state model is altogether more responsive to rapid changes in Ca2+ concentration, even when all the CaM-bound CBPs are summed (Fig. 3d). Taken together this data provides a strong motivation to move forward with the 4-state model as the best model to simulate the frequency-dependent response of CBPs.
Frequency Analysis
Because our current 4-state model dramatically expands on previous work, we wanted to assess the frequency-dependence of CBP activation. We define two metrics in order to compare the frequency response of all CBPs. In the first metric, Normalized Cb1, the time-averaged integration of all CaM4-bound CBPs (Equation S1) is normalized to the highest Cb1 value of each CBP, respectively. In the second metric, Normalized Cb2, the time-averaged integration of all CaM species-bound CBPs (Equation S2) is normalized to the highest Cb2 value of each CBP, respectively. Normalized Cb1 and Cb2 values for each CBP are plotted at various Ca2+ flux frequencies (Fig. 4).
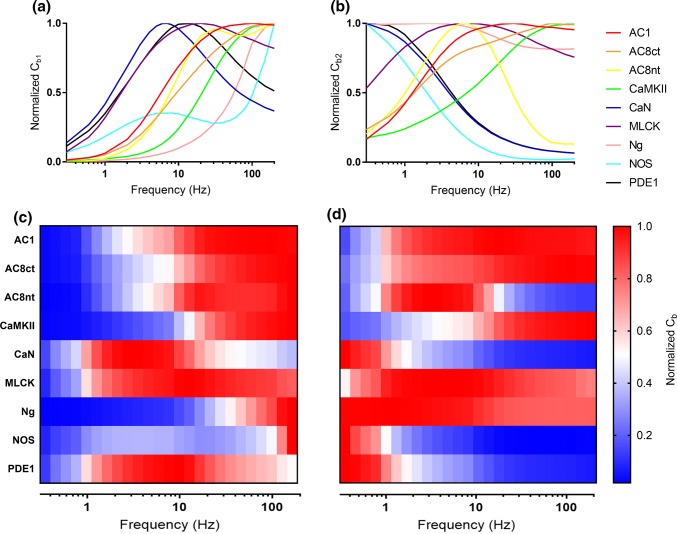
Figure 4.
Frequency-dependence of CBPs. (a, b) Relative time-averaged concentrations of CaM4-bound CBPs (Cb1) and all CaM-bound CBPs (Cb2), respectively, for the 4-state model. (c, d) Heatmap representations of data shown in a and b, where each row denotes the CaM4- and total CaM-binding to each CBP. Blue denotes minimal relative activation of CBP, and red denotes maximal relative activation of CBP. Note the shifts in the frequencies of peak CBP activation when accounting only for CaM4-binding (c) and all CaM-CBP binding (d).
The frequency responses of the various CBPs, when accounting for all CaM-binding, are in good agreement with previously published results. For example, CaMKII (green trace) exhibited its highest CaM4-binding (and thus, activation) at high frequencies, peaking around 100 Hz (Figs. 4a and 4b). High frequency activation of CaMKII coincides with the protein’s putative role in long-term potentiation, which is initiated under high-frequency (~ 100 Hz) Ca2+ flux. Interestingly, Ng, which has a higher affinity for CaM0 than CaM4, exhibited relatively little CaM4-binding across most frequencies, before spiking at about 80 Hz (Figs. 4a and 4c). This spike in average CaM4-bound Ng is explained by the full saturation of CaM4 at high frequencies. Indeed, when accounting for all CaM-binding (Fig. 4b), Ng exhibits a peak activation at lower frequencies, which is more consistent with expectation.54 Most notably, when analyzing Normalized Cb1 (Figs. 4a and 4c) many CBPs such as AC1, AC8, Ng, and NOS have highly similar peak activation frequencies. If distinct Ca2+ signals/frequencies elicit activation of distinct Ca2+/CaM-dependent pathways and were in-part dependent on competitive tuning, then we would have expected each CBP to have a more distinguishable frequency of peak activation.
The frequency dependence of the CBPs is clearer when we analyze the data using the second metric, Normalized Cb2. As seen in Figs. 4b and 4d, this analysis revealed how CaM0 and the intermediate Ca2+/CaM states can, in conjunction with saturated CaM4, contribute to the frequency-dependence of CBP activation. This is additional evidence that suggests that a 4-state model of Ca2+/CaM binding is able to capture the frequency dependent changes in CBP activation. These results also compare favorably with our previous work.45 However, in our previous work the adenylyl cyclase CBPs (AC1, AC8nt, and AC8ct) generally exhibits activation that peaked at higher-frequencies (greater than 50 Hz). In Figs. 4b and 4d, it appears that incorporating the additional CBP (PDE1) caused a downward shift and sharpening in peak activation frequency for AC8nt.
Sensitivity of GluA1 Phosphorylation to Variation in Key Input Parameters
Global sensitivity analysis was performed to assess how variation in input parameters contributed to variation in the overall phosphorylation of GluA1. Latin Hypercube Sampling (LHS) was used to efficiently sample the input parameters over fourfold range (2-fold increased and decreased). Partial Rank Correlation Coefficients (PRCC) was used to quantify how the variation in each input parameter contributed to overall phosphorylation of GluA1 (sum of all phosphorylated GluA1 species). To test the hypothesis that CaMKII significantly contributes to GluA1 phosphorylation under normal Ng concentrations, and PKA (via AC1/8 activation of cAMP) significantly contributes to GluA1 phosphorylation when Ng is knocked down, we performed LHS/PRCC analysis under a both wild-type Ng condition and a total Ng KO condition. For both conditions, the parameters with absolute PRCC’s greater than our threshold of 0.5 are shown in Table 3.
Table 3.
Sensitivity analysis of kinetic parameters at 100 Hz Ca2+.
| Parameter | Description | PRCC |
|---|---|---|
| Catalytic rate of cAMP production by AC1 bound to CaM4 | 0.808 | |
| Association rate of AC1 to CaM4 | 0.758 | |
| Catalytic rate of GluA1 phosphorylation by PKAc | 0.597 | |
| Catalytic rate of GluA1 by CaMKII activated by CaM4 | 0.584 | |
| Association rate of CaMKII to GluA1 | 0.527 | |
| Association rate of CaMKII to CaM4 | − 0.732 | |
| Catalytic rate of cAMP degradation by PDE4 | − 0.805 |
| [Ng] = 0 | ||
|---|---|---|
| Parameter | Description | PRCC |
| Catalytic rate of GluA1 phosphorylation by PKAc | 0.785 | |
| Catalytic rate of cAMP production by AC1 bound to CaM4 | 0.763 | |
| Association rate of GluA1 to PKAc | 0.636 | |
| Catalytic rate of cAMP production by AC8-Ct bound to CaM4 | 0.618 | |
| Catalytic rate of cAMP degradation by PDE | − 0.864 |
In the WT Ng condition at 100 Hz Ca2+ flux, all association and catalytic rate parameters ( or , where B represents the different CBPs) were varied simultaneously. As expected, we find that the phosphorylation rate constant of CaMKII-mediated phosphorylation of GluA1 is highly correlated with total GluA1 phosphorylation (Table 3). Additionally, the catalytic rate constant for AC1-CaM4 production of cAMP () and that of the downstream output of AC1 activation (PKA, ) were also highly correlated with phosphorylation of GluA1. This indicates that at 100 Hz and WT Ng conditions, increases in the rate of CaM4 binding to AC1 leads to increases in phosphorylation of GluA1.
For the second set of conditions the concentration of Ng was set to zero to simulate Ng KO. In comparison to the WT Ng case, it is first interesting to note that the phosphorylation rate constant is no longer highly correlated with GluA1 phosphorylation (PRCC value = 0.467, below our threshold of 0.5). Though admittedly this PRCC value for is still somewhat high, its reduction in the Ng KO analysis suggests a diminished importance for CaMKII-mediated phosphorylation of GluA1 subunits. In accordance with our over-arching hypothesis that competitive tuning mediates a shift in pathway activation, we refer again to Table 3 for clues as to the alternative proteins contributing to GluA1 phosphorylation. Indeed, both the association and catalytic rates of PKAc for GluA1 ( and , respectively) have significantly greater PRCC values in the Ng KO compared to the WT case. Furthermore, the catalytic rate of cAMP production by AC8ct-CaM4 () also exhibits a significant PRCC in the Ng KO case. Note that the short timescale over which we simulate competitive tuning in this sensitivity analysis is biophysically relevant; LTP is classically elicited by a 100 Hz stimulus over one second.19,21 Additionally, previous models indicate that the predominant Ca2+/CaM state may change in fractions of seconds, which is important considering that each Ca2+/CaM state exhibits significant binding affinities for various CBPs.42,50 Altogether, the results of our sensitivity analysis indicate that, at least for 100 Hz Ca2+ and short timescales (< 2 s), competition for Ca2+/CaM-binding may support a shift in signaling in which Ng perturbation causes AC to overtake CaMKII as the primary activator of pathways leading to GluA1 subunit phosphorylation.
Potential Robustness Provided by Competitive Tuning
Having seen in the preceding sections that our expanded model exhibits similar frequency dependence to a previous result, and that perturbing Ng causes a shift in the parameters most strongly associated with GluA1 phosphorylation, we now explore the functional consequences of competitive tuning in this simplified model of synaptic plasticity. One hallmark of synaptic plasticity is the phosphorylation of AMPAR GluA1 subunits at two residues: S831 and S845. S831 is phosphorylated by CaM-bound CaMKII or pCaMKII, and S845 is phosphorylated by catalytic PKA (PKAc).
Previously we have shown that competitive tuning was able to explain counter-intuitive experimental results showing that Ng genetic knock-out (Ng KO) results in decreased CaMKII activation/autophosphorylation.45 This same work also predicted that knocking out Ng concentration would cause a shift in CaM binding away from CaMKII in favor of AC8. Noting that activated AC1 and AC8ct generate cAMP, which in turn activates PKA, it seemed that competitive tuning could provide a mechanism for the system to account for perturbations in Ng expression. That is, because Ng KO reduces CaMKII-facilitated phosphorylation of GluA1 S831, competitive tuning could drive the alternative activation of more AC8ct, increasing PKA activation and in turn, phosphorylation of GluA1 S845.
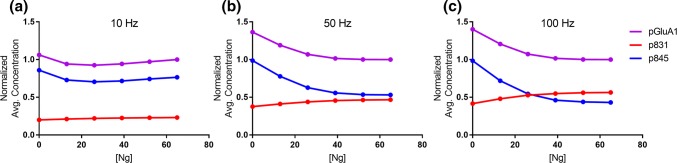
In Fig. 5, we explore this potential mechanism by simulating Ng knockdown, while monitoring the time-averaged concentration of phosphorylated AMPARs (pGluA1) under three different Ca2+ flux frequencies. Note that in the simulations for Fig. 5, the number of Ca2+ pulses is conserved with varying Ca2+ frequency. In order to easily compare across Ca2+ frequencies, each panel in Fig. 5 is additionally normalized against the time-averaged pGluA1 concentration observed for the WT Ng case. For all three stimulation frequencies in Fig. 5, GluA1 subunits phosphorylated by CaMKII at S831 (p831, red) decrease with decreasing Ng concentration. As expected, GluA1 phosphorylation by PKAc at S845 (p845, blue) increases with decreasing Ng concentration. However, only for 10 Hz stimulation (Fig. 5a) does the cumulative GluA1 phosphorylation remain relatively constant for all Ng concentrations. Surprisingly, Ng KO caused phosphorylation at S845 to significantly increase at higher frequencies, whereas the decrease in S831 phosphorylation was relatively modest. It would appear that while competitive tuning could facilitate robustness to perturbations in Ng expression, the system tends to over-correct at early timescales in the absence of additional regulatory mechanisms.
Figure 5.
Phosphorylation of AMPAR GluA1 Subunits at S831 and S845. Time-averaged concentrations of total phosphorylation (pGluA1, purple), CaMKII-mediated phosphorylation at S831 (p831, red), and PKA-mediated phosphorylation at S845 (p845, blue) against Ng concentration. For each panel, the 4-state Ca2+/CaM binding model was stimulated at either (a) 10 Hz (b) 50 Hz, or (c) 100 Hz Ca2+ stimulation. In order to compare Ca2+ frequencies, each panel is additionally normalized against the [pGluA1] observed for WT Ng (65 μM).
These results imply that high frequency stimulation such as those that elicit LTP (100 Hz) should be more easily attainable upon Ng KO. This result is consistent with Krucker et al.,26 who showed that even though CaMKII activation/phosphorylation is reduced upon Ng KO, long-term potentiation can be achieved following stimulation by only a single 100 Hz tetanus. Further, it is worth noting that for all Ca2+ frequencies presenting in Fig. 5, shifts in overall pGluA1 levels with decreasing Ng concentration were largely negligible until Ng expression was reduced by more than half. Thus, it appears that the system is robust to modest perturbations to Ng expression, such as what might be seen in Ng heterozygous animals.
Conclusions
Competitive tuning is a recently-described phenomenon in which signaling molecules compete for binding of a common activator. Although others have either alluded to or invoked competition as being important in signal transduction,1,29,49,54 our work shows that competition may play a larger role in informing signaling outcomes than previously thought. In this work we first compare a 2-state and 4-state model of competitive binding for Ca2+/CaM and determined that a 4-state model is best-suited to accurately detect the frequency dependence of a competitive CaM-CBP reaction network under dynamic stimulation. We also verify that expanding our 4-state model network to include an additional CBPs and downstream species preserves the overall competitive tuning phenomenon. We explore the functional implications of competitive tuning by using sensitivity analysis to identify the parameters most strongly correlated with a hallmark of synaptic plasticity, AMPAR GluA1 subunit phosphorylation. We further explore the implications of competitive tuning by building on a previous result showing that Ng KO counter-intuitively decreases CaMKII activation and autophosphorylation. Because competitive models indicate that Ng KO causes an increase in Ca2+/CaM binding to AC8, we hypothesize that competitive tuning could provide a mechanism that compensates for a loss in CaMKII-mediated phosphorylation of GluA1 by promoting (via AC) PKA-mediated phosphorylation of GluA1. Indeed, in Fig. 5 we observe that for high Ca2+ flux frequencies, Ng knockdown results in a switch between CaMKII and PKA in cumulative kinase activity.
Interestingly, stimulating at an LTP-inducing Ca2+ frequency of 100 Hz (Fig. 5c) further causes an upregulation of GluA1 phosphorylation by PKA in the Ng KO case. This upregulation could explain results by Krucker et al. who observed that LTP could be elicited by just a single tetanus of stimulation at 100 Hz in brain slices of Ng KOs. Note that the current model includes both PP1- and CaN-dependent dephosphorylation of GluA1. Contrary to expectation, the combined action of PP1 and Ca2+/CaM-dependent dephosphorylation by CaN seemingly did not dramatically decrease GluA1 phosphorylation levels in the presence of compensation facilitated by competitive tuning. Of course, other regulatory mechanisms could additionally be affecting GluA1 phosphorylation levels. For example, spatial localizations of proteins such as Ng and PP1, or perhaps A-kinase-anchoring proteins (AKAPs), could be required to increase, through avidity effects, the local concentrations of CaMKII, AC, and/or CaN in the dendritic spine. Alternatively, it may be required to analyze the reaction network presented here on longer timescales in order to discern the complete downstream effects of competitive tuning. As it currently stands, our current results are focused on short-term effects of Ca2+-dependent signaling. Future work is needed to analyze longer term processes (e.g., receptor localization, other feedback mechanisms, receptor trafficking). It remains unclear whether increased GluA1 phosphorylation in Ng KO is an expected consequence of Ng perturbation and represents an example how competitive tuning can impart robustness to model outcomes across a wide range of Ng concentrations, or whether the maintenance of GluA1 phosphorylation is a result of missing layers of model regulation. Future work will include these additional mechanisms of regulation and will explore other protein activation dynamics in GluA1 phosphorylation. A key prediction from our model that could be readily experimentally tested is that the relative level of phosphorylation of GluA1 at p845 would increase in Ng KO mice and phosphorylation of GluA1 at p831 would decrease.
Methods
Simulation Methods
Chemical reactions between species were written and converted to ordinary differential equations according to laws of mass action using the XCellerator package.47 Mathematica was used to solve the differential equations using the NDSolve command. All other data analysis (including integration of species concentration over time and the LHS/PRCC sensitivity analysis) were performed in Mathematica (Wolfram Research, Mathematical version 11). Model output is plotted in the 2D graphing program GraphPad Prism (GraphPad, version 7).
Ca2+ stimulation was implemented as a boundary condition forcing function for the system of differential equations solved in Mathematica. To control for the magnitude of Ca2+ exposure from a single Ca2+ pulse, Ca2+ spike half widths are set to 5 ms regardless of frequency. To reduce computational complexity and control for the cumulative magnitude of Ca2+ exposure across all simulations, total magnitude of calcium flux is conserved at 100 pulses regardless of Ca2+ frequency.
All the equations for this model can be found in S1 Appendix. Mathematica files for the complete models can be found on the Purdue PURR database.43
Sensitivity Analysis
A global sensitivity analysis was used to investigate how variation in input parameter values contributed to variation in model output. LHS was used to efficiently sample input parameters over a range of values that were determined by experimental measurement or calculated by thermodynamic equilibrium (see Model Parameterization for a more in-depth discussion of model parameterization). In all variations of our sensitivity analysis, each parameter was sampled at least 250 times using values within 2-fold of each parameter’s listed value (See S1 Appendix). PRCC were used correlate the rank contribution of parameter variation on the variation in desired model output as described in our previous work.24,42,45 We have previously shown that allowing the Kd of an interaction to vary by either varying the kon or koff (but not both simultaneously) produced similar results in the sensitivity analysis.24 Therefore only association rate constants (kon) and catalytic rate constants (kp) were varied. PRCC analysis produces correlation factors over a range of 1 to − 1, where 1 indicates perfect positive correlation and − 1 indicates perfect negative correlation. In order to show the parameters whose variation most greatly impacted model output (total phosphorylation of GluA1 AMPAR subunits), a threshold PRCC value of absolute magnitude of 0.5 was used to generate the parameter list in Table 3.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
The authors thank members of the Kinzer-Ursem Lab for helpful discussion and comment on the manuscript. We gratefully acknowledge support from the National Institute of Neurological Disorders And Stroke (NINDS) of The National Institutes of Health (NIH) under Award Number R21NS095218. The content is solely the responsibility of the authors and does not necessarily represent the official view of the NIH. Also, this material is based on upon work supported by the National Science Foundation CAREER Award Grant No. 1752366. Any opinions, findings, and conclusions or recommendations expressed in this material are those of the authors and do not necessarily reflect the views of the National Science Foundation.
Conflict of interest
Authors Matthew Pharris, Neal Patel, and Tamara Kinzer-Ursem declare that they have no conflicts of interest.
Ethical Standards
No animal studies were carried out by the authors for this article. Additionally, no human subjects research was conducted in this study.
Abbreviations
- AC1
Adenylyl cyclase 1
- AC8nt, AC8ct
Adenylyl cyclase 8N/C-terminus
- AMPAR
α-Amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptor
- Ca2+
Calcium ion
- CaM
Calmodulin
- CBP
Calmodulin binding protein
- CaMKII
Calcium/calmodulin-dependent protein kinase II
- CaN
Calcineurin
- Inh-1
Inhibitor 1
- LTP
Long-term potentiation
- MLCK
Myosin light chain kinase
- NOS
Nitric oxide synthetase
- Ng
Neurogranin
- NMDAR
N-methyl-d-aspartic acid receptor
- PDE1, PDE4
Phosphodiesterase 1/4
- PKA
Protein kinase A
- PP1
Protein phosphatase 1
Footnotes
Tamara L. Kinzer-Ursem is an Assistant Professor in the Weldon School of Biomedical Engineering. She received her B.S. in Bioengineering from the University of Toledo, M.S. and Ph.D. degrees in Chemical Engineering from the University of Michigan, and her post-doctoral training in Molecular Neuroscience at the California Institute of Technology. Prior to joining Purdue she was the Head of R&D in Biochemistry at Maven Biotechnologies and Visiting Associate in Chemical Engineering at the California Institute of Technology. Research in the Kinzer-Ursem lab focuses on developing tools to advance quantitative descriptions of cellular processes and disease within three areas of expertise: (1) Computational modeling of signal transduction mechanisms to understand cellular processes. Using computational techniques, we have recently described “competitive tuning” as a mechanism that might be used to regulate information transfer through protein networks, with implications in cell behavior and drug target analysis; (2) Development of novel protein tagging technologies that are used to label proteins in vivo to enable quantitative description of protein function and elucidate disease mechanisms; and (3) Using particle diffusivity measurements to quantify biomolecular processes. Particle diffusometry is being used as a sensitive biosensor to detect the presence of pathogens in environmental and patient samples.
This article is part of the 2018 CMBE Young Innovators special issue.
References
- 1.Antunes G, Roque AC, Souza FMS. Modelling intracellular competition for calcium: kinetic and thermodynamic control of different molecular modes of signal decoding. Sci. Rep. 2015;6:23730. doi: 10.1038/srep23730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barria A, Derkach V, Soderling T. Identification of the Ca2+/calmodulin-dependent protein kinase II regulatory phosphorylation site in the alpha-amino-3-hydroxyl-5-methyl-4-isoxazole-propionate-type glutamate receptor. J. Biol. Chem. 1997;272:32727–32730. doi: 10.1074/jbc.272.52.32727. [DOI] [PubMed] [Google Scholar]
- 3.Beattie EC, Carroll RC, Yu X, Morishita W, Yasuda H, von Zastrow M, et al. Regulation of AMPA receptor endocytosis by a signaling mechanism shared with LTD. Nat. Neurosci. 2000;3(12):1291–1300. doi: 10.1038/81823. [DOI] [PubMed] [Google Scholar]
- 4.Borlongan CV, Burns J, Tajiri N, Stahl CE, Weinbren NL, et al. Epidemiological survey-based formulae to approximate incidence and prevalence of neurological disorders in the United States: a meta-analysis. PLoS ONE. 2013;2:e78490. doi: 10.1371/journal.pone.0078490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bredt DS, Snyder SH. Isolation of nitric-oxide synthetase, a calmodulin-requiring enzyme. Proc. Nat. Acad. Sci. 1990;87(2):682–685. doi: 10.1073/pnas.87.2.682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brostrom CO, Huang YC, Breckenridge BM, Wolff DJ. Identification of a calcium-binding protein as a calcium-dependent regulator of brain adenylate cyclase. Proc. Nat. Acad. Sci. 1975;72(1):64–68. doi: 10.1073/pnas.72.1.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cali JJ, Zwaagstra JC, Mons N, Cooper DMF, Krupinski J. Type-VIII adenylyl-cyclase - a Ca2+/calmodulin-stimulated enzyme expressed in discrete regions of rat-brain. J. Biol. Chem. 1994;269(16):12190–12195. [PubMed] [Google Scholar]
- 8.Chay A, Zamparo I, Koschinski A, Zaccolo M, Blackwell KT. Control of beta AR- and N-methyl-d-aspartate (NMDA) receptor-dependent cAMP dynamics in hippocampal neurons. PLoS Comput. Biol. 2016;12(2):32. doi: 10.1371/journal.pcbi.1004735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chin D, Means AR. Calmodulin: a prototypical calcium sensor. Trends Cell Biol. 2000;10(8):322–328. doi: 10.1016/S0962-8924(00)01800-6. [DOI] [PubMed] [Google Scholar]
- 10.Dabrowska R, Sherry JMF, Aromatorio DK, Hartshorne DJ. Modulator protein as a component of myosin light chain kinase from chicken gizzard. Biochemistry. 1978;17(2):253–258. doi: 10.1021/bi00595a010. [DOI] [PubMed] [Google Scholar]
- 11.Diering GH, Heo S, Hussain NK, Liu B, Huganir RL. Extensive phosphorylation of AMPA receptors in neurons. Proc. Nat. Acad. Sci. 2016;113(33):4920–4927. doi: 10.1073/pnas.1610631113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Doyle CA, Slater P. Localization of neuronal and endothelial nitric oxide synthase isoforms in human hippocampus. Neuroscience. 1997;76(2):387–395. doi: 10.1016/S0306-4522(96)00297-7. [DOI] [PubMed] [Google Scholar]
- 13.Forest A, Swulius MT, Tse JKY, Bradshaw JM, Gaertner T, Waxham MN. Role of the N- and C-lobes of calmodulin in the activation of Ca2+/calmodulin-dependent protein kinase II. Biochemistry. 2008;47(40):10587–10599. doi: 10.1021/bi8007033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gerendasy DD, Herron SR, Watson JB, Sutcliffe JG. Mutational and biophysical studies suggest RC3/neurogranin regulates calmodulin availability. J. Biol. Chem. 1994;269(35):22420–22426. [PubMed] [Google Scholar]
- 15.Ghosh A, Greenberg ME. Calcium signaling in neurons: molecular mechanisms and cellular consequences. Science. 1995;268(5208):239–247. doi: 10.1126/science.7716515. [DOI] [PubMed] [Google Scholar]
- 16.Gu C, Cooper DMF. Calmodulin-binding sites on adenylyl cyclase type VIII. J. Biol. Chem. 1999;274(12):8012–8021. doi: 10.1074/jbc.274.12.8012. [DOI] [PubMed] [Google Scholar]
- 17.Hao HP, Zak DE, Sauter T, Schwaber J, Ogunnaike BA. Modeling the VPAC(2)-activated cAMP/PKA signaling pathway: from receptor to circadian clock gene induction. Biophys. J. 2006;90(5):1560–1571. doi: 10.1529/biophysj.105.065250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hayer A, Bhalla US. Molecular switches at the synapse emerge from receptor and kinase traffic. PLoS Comput. Biol. 2005;1(2):137–154. doi: 10.1371/journal.pcbi.0010020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Herring BE, Nicoll RA. Long-term potentiation: from CaMKII to AMPA receptor trafficking. Ann. Rev. Physiol. 2016;78:351–365. doi: 10.1146/annurev-physiol-021014-071753. [DOI] [PubMed] [Google Scholar]
- 20.Huang KP, Huang FL, Chen HC. Characterization of a 7.5-kDa protein-kinase-C substrate (RC3 protein, neurogranin) from rat-brain. Arch. Biochem. Biophys. 1993;305(2):570–580. doi: 10.1006/abbi.1993.1463. [DOI] [PubMed] [Google Scholar]
- 21.Huganir RL, Nicoll RA. Ampars and synaptic plasticity: the last 25 years. Neuron. 2013;80(3):704–717. doi: 10.1016/j.neuron.2013.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Keller DX, Franks KM, Bartol TM, Sejnowski TJ. Calmodulin activation by calcium transients in the postsynaptic density of dendritic spines. PLoS ONE. 2008;3(4):16. doi: 10.1371/journal.pone.0002045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim M, Park AJ, Havekes R, Chay A, Guercio LA, Oliveira RF, et al. Colocalization of protein kinase A with adenylyl cyclase enhances protein kinase A activity during induction of long-lasting long-term potentiation. PLoS Comput. Biol. 2011;7(6):18. doi: 10.1371/journal.pcbi.1002084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kinzer-Ursem TL, Linderman JJ. Both ligand- and cell-specific parameters control ligand agonism in a kinetic model of G protein-coupled receptor signaling. PLoS Comput. Biol. 2007;3(1):84–94. doi: 10.1371/journal.pcbi.0030006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klee CB, Krinks MH. Purification of cyclic 3′,5′-nucleotide phosphodiesterase inhibitory protein by affinity chromatography on activator protein coupled to sepharose. Biochemistry. 1978;17(1):120–126. doi: 10.1021/bi00594a017. [DOI] [PubMed] [Google Scholar]
- 26.Krucker T, Siggins GR, McNamara RK, Lindsley KA, Dao A, Allison DW, et al. Targeted disruption of RC3 reveals a calmodulin-based mechanism for regulating metaplasticity in the hippocampus. J. Neurosci. 2002;22(13):5525–5535. doi: 10.1523/JNEUROSCI.22-13-05525.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kulkami C, Lo M, Fraseur JG, Tirrell DA, Kinzer-Ursem TL. Bioorthogonal chemoenzymatic functionalization of calmodulin for bioconjugation applications. Bioconj Chem. 2015;26(10):2153–2160. doi: 10.1021/acs.bioconjchem.5b00449. [DOI] [PubMed] [Google Scholar]
- 28.Lei SB, Czerwinska E, Czerwinski W, Walsh MP, MacDonald JF. Regulation of NMDA receptor activity by F-actin and myosin light chain kinase. J. Neurosci. 2001;21(21):8464–8472. doi: 10.1523/JNEUROSCI.21-21-08464.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li L, Stefan MI, Le Novere N. Calcium input frequency, duration and amplitude differentially modulate the relative activation of calcineurin and CaMKII. PLoS ONE. 2012;7(9):17. doi: 10.1371/annotation/0e8ec400-2a84-45fe-81d4-bbf20d8124c9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lukas TJ, Burgess WH, Prendergast FG, Lau W, Watterson DM. Calmodulin binding domains: characterization of a phosphorylation and calmodulin binding-site from myosin light chain kinase. Biochemistry. 1986;25(6):1458–1464. doi: 10.1021/bi00354a041. [DOI] [PubMed] [Google Scholar]
- 31.Malenka RC, Lancaster B, Zucker RS. Temporal limits on the rise in postsynaptic calcium required for the induction of long-term potentiation. Neuron. 1992;9(1):121–128. doi: 10.1016/0896-6273(92)90227-5. [DOI] [PubMed] [Google Scholar]
- 32.Maravall M, Mainen ZF, Sabatini BL, Svoboda K. Estimating intracellular calcium concentrations and buffering without wavelength ratioing. Biophys. J. 2000;78(5):2655–2667. doi: 10.1016/S0006-3495(00)76809-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Masada N, Schaks S, Jackson SE, Sinz A, Cooper DMF. Distinct mechanisms of calmodulin binding and regulation of adenylyl cyclases 1 and 8. Biochemistry. 2012;51(40):7917–7929. doi: 10.1021/bi300646y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miller SG, Kennedy MB. Distinct forebrain and cerebellar isozymes of type II Ca2+/calmodulin-dependent protein kinase associate differently with the postsynaptic density fraction. J. Biol. Chem. 1985;260(15):9039–9046. [PubMed] [Google Scholar]
- 35.Miller SG, Kennedy MB. Regulation of brain type-II Ca2+/calmodulin-dependent protein-kinase by autophosphorylation: a Ca2+-triggered molecular switch. Cell. 1986;44(6):861–870. doi: 10.1016/0092-8674(86)90008-5. [DOI] [PubMed] [Google Scholar]
- 36.Mons N, Yoshimura M, Cooper DMF. Discrete expression of Ca2+/calmodulin-sensitive and Ca2+-insensitive adenylyl cyclases in the rat-brain. Synapse. 1993;14(1):51–59. doi: 10.1002/syn.890140108. [DOI] [PubMed] [Google Scholar]
- 37.Nabavi S, Kessels HW, Alfonso S, Aow J, Fox R, Malinow R. Metabotropic NMDA receptor function is required for NMDA receptor-dependent long-term depression. Proc. Nat. Acad. Sci. 2013;110(10):4027–4032. doi: 10.1073/pnas.1219454110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Oliveira RF, Kim M, Blackwell KT. Subcellular location of PKA controls striatal plasticity: stochastic simulations in spiny dendrites. PLoS Comput. Biol. 2012;8(2):19. doi: 10.1371/journal.pcbi.1002383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Olwin BB, Edelman AM, Krebs EG, Storm DR. Quantitation of energy coupling between Ca2+, calmodulin, skeletal-muscle myosin light chain kinase, and kinase substrates. J. Biol. Chem. 1984;259(17):949–955. [PubMed] [Google Scholar]
- 40.Panda K, Ghosh S, Stuehr DJ. Calmodulin activates intersubunit electron transfer in the neuronal nitric-oxide synthase dimer. J. Biol. Chem. 2001;276(26):23349–23356. doi: 10.1074/jbc.M100687200. [DOI] [PubMed] [Google Scholar]
- 41.Peersen OB, Madsen TS, Falke JJ. Intermolecular tuning of calmodulin by target peptides and proteins: differential effects on Ca2+ binding and implications for kinase activation. Protein Sci. 1997;6(4):794–807. doi: 10.1002/pro.5560060406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pepke S, Kinzer-Ursem T, Mihalas S, Kennedy MB. A dynamic model of interactions of Ca2+, calmodulin, and catalytic subunits of Ca2+/Calmodulin-dependent protein kinase II. PLoS Comput. Biol. 2010;6(2):15. doi: 10.1371/journal.pcbi.1000675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pharris, M. C., N. M. Patel, and T. L. Kinzer-Ursem. Competitive tuning of Ca2+/calmodulin-activated proteins provides a compensatory mechanism for AMPA receptor phosphorylation in synaptic plasticity. Purdue University Research Repository. 10.4231/r7st7n11.
- 44.Represa A, Deloulme JC, Sensenbrenner M, Benari Y, Baudier J. Neurogranin: immunocytochemical localization of a brain-specific protein-kinase-c substrate. J. Neurosci. 1990;10(12):3782–3792. doi: 10.1523/JNEUROSCI.10-12-03782.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Romano DR, Pharris MC, Patel NM, Kinzer-Ursem TL. Competitive tuning: competition’s role in setting the frequency-dependence of Ca2+-dependent proteins. PLoS Comput. Biol. 2017;13(11):10–45. doi: 10.1371/journal.pcbi.1005820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Saucerman JJ, Bers DM. Calmodulin mediates differential sensitivity of CaMKII and calcineurin to local Ca2+ in cardiac myocytes. Biophys. J. 2008;95(10):4597–4612. doi: 10.1529/biophysj.108.128728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shapiro BE, Levchenko A, Meyerowitz EM, Wold BJ, Mjolsness ED. Cellerator: extending a computer algebra system to include biochemical arrows for signal transduction simulations. Bioinformatics. 2003;19(5):677–678. doi: 10.1093/bioinformatics/btg042. [DOI] [PubMed] [Google Scholar]
- 48.Shifman JM, Choi MH, Mihalas S, Mayo SL, Kennedy MB. Ca2+/calmodulin-dependent protein kinase II (CaMKII) is activated by calmodulin with two bound calciums. Proc. Nat. Acad. Sci. 2006;103(38):13968–13973. doi: 10.1073/pnas.0606433103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Slavov N, Carey J, Linse S. Calmodulin transduces Ca2+ oscillations into differential regulation of its target proteins. ACS Chem. Neurosci. 2013;4(4):601–612. doi: 10.1021/cn300218d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stemmer PM, Klee CB. Dual calcium ion regulation of calcineurin by calmodulin and calcineurin B. Biochemistry. 1994;33(22):6859–6866. doi: 10.1021/bi00188a015. [DOI] [PubMed] [Google Scholar]
- 51.Swislocki NI, Tierney J. Solubilization, stabilization, and partial-purification of brain adenylate cyclase from rat. Biochemistry. 1973;12(10):1862–1866. doi: 10.1021/bi00734a004. [DOI] [PubMed] [Google Scholar]
- 52.WHO . Neurological Disorders: Public Health Challenges. Geneva: WHO Press; 2006. [Google Scholar]
- 53.Xia ZG, Storm DR. The role of calmodulin as a signal integrator for synaptic plasticity. Nat. Rev. Neurosci. 1995;6(4):267–276. doi: 10.1038/nrn1647. [DOI] [PubMed] [Google Scholar]
- 54.Zhabotinsky AM, Camp RN, Epstein IR, Lisman JE. Role of the neurogranin concentrated in spines in the induction of long-term potentiation. J. Neurosci. 2006;26(28):7337–7347. doi: 10.1523/JNEUROSCI.0729-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhong L, Gerges NZ. Neurogranin targets calmodulin and lowers the threshold for the induction of long-term potentiation. PLoS ONE. 2012;7(7):8. doi: 10.1371/journal.pone.0041275. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.