ABSTRACT
Chronic caloric restriction (CR) without malnutrition is known to affect different cellular processes such as stem cell function, cell senescence, inflammation, and metabolism. Despite the differences in the implementation of CR, the reduction of calories produces a widespread beneficial effect in noncommunicable chronic diseases, which can be explained by improvements in immuno-metabolic adaptation. Cellular adaptation that occurs in response to dietary patterns can be explained by alterations in epigenetic mechanisms such as DNA methylation, histone modifications, and microRNA. In this review, we define these modifications and systematically summarize the current evidence related to CR and the epigenome. We then explain the significance of genome-wide epigenetic modifications in the context of disease development. Although substantial evidence exists for the widespread effect of CR on longevity, there is no consensus regarding the epigenetic regulations of the underlying cellular mechanisms that lead to improved health. We provide compelling evidence that CR produces long-lasting epigenetic effects that mediate expression of genes related to immuno-metabolic processes. Epigenetic reprogramming of the underlying chronic low-grade inflammation by CR can lead to immuno-metabolic adaptations that enhance quality of life, extend lifespan, and delay chronic disease onset.
Keywords: energy intake, DNA methylation, histone acetylation, sirtuin, microRNA, dietary restriction
Introduction
Caloric restriction (CR) without malnutrition constitutes a safe and effective way to promote weight loss, decrease metabolic complications, increase lifespan, and improve quality of life. CR refers to a reduction in the intake of net calories, while meeting the necessary micronutrient requirements. Caloric intake can be restricted in a variety of ways: percentage CR, macronutrient limitation, or exercise-induced restriction. Percentage of restriction of total calories spans from mild (15% energy restriction) to severe (60% restriction) (1–4) and can be supplemented with physical activity (1, 4, 5). Alternatives for CR include changes in the proportion of macronutrients, such as hyperproteic compared with hypoproteic diets (6), or ketogenic diets with a high lipid, low carbohydrate content (7–9). In animal studies, CR can also be achieved by increasing the litter size or access to food, but, although this approach provides a natural method of energy limitation, often the amount of food cannot be measured (10, 11).
CR has been used to treat and prevent chronic disease development. In obese patients, CR provides additional health benefits to weight loss, such as decreased visceral adipose tissue (12) and inflammation (13), while improving kidney function (14, 15) and cellular quality control (16). Moreover, improved insulin sensitivity has also been documented for type 2 diabetic animals (17) and patients (18–22). In addition, a 12-wk CR treatment (30% restriction) reduced the circulating concentration of fetuin-A (a biomarker for multiple metabolic diseases), improved blood pressure in patients, and hepatic steatosis in rats (23). Lastly in preclinical and preliminary clinical studies, CR or fasting can effectively prevent malignancies through a variety of cellular responses and can improve the efficacy of therapeutic agents (24). It has been shown that various forms of CR reduce the progression of cancers with high morbidity rates, including colorectal, pancreas, breast, liver, prostate, esophagus, and kidney malignancies. In monkeys from the Wisconsin National Primate Research Center (WNPRC), 30% lifelong restriction was sufficient to cause a 50% reduction in neoplastic events, gastrointestinal adenocarcinoma being the most common (25). Similarly, CR at a young age (30% restriction) reduced the incidence of cancer in monkeys from the National Institute of Aging (NIA), despite great genetic differences between them and the WNPRC monkeys. Overall, chronic CR has been demonstrated to be an effective dietary intervention for the treatment of many noncommunicable chronic diseases.
Given the impact that CR has on the prevention, development, and treatment of chronic diseases, we sought to review 3 epigenetic mechanisms that might be related to the long-term programming of cellular functions. Epigenetic mechanisms are at the forefront of cellular changes that can be modulated by CR and lead to long-lasting cellular adaptations and improved health outcomes. Modulation of important growth mechanisms that lead to increased longevity [phosphatidylinositol-3-kinase/protein kinase B/mammalian target of rapamycin (PI3K/Akt/mTOR)] among others is known to be regulated by epigenetic factors, but the wide range and complexity of these pathways are not well understood (26). We will provide an overview of different epigenetic mechanisms that can be targeted. We will then explore the effect of CR on such mechanisms. Lastly, we will delineate the epigenetic regulation of immuno-metabolic processes that leads to improvements for many noncommunicable chronic diseases.
CR and DNA Methylation
CR has been found to affect the methylation pattern of certain genes involved in biological processes such as metabolism, oxidative stress, senescence, and aging (27–29) (Figure 1). CR protocols that compensate for malnutrition can combat chronic disease development by shaping the epigenome during early developmental stages. The timing of the restriction onset as well as the diet formulation (30) are the most important factors that can predict the positive outcomes related to CR (Figure 1A). Moreover, important caveats exist in CR research, where sex-specific and strain-specific effects are observed, and higher restrictions result in improvements of neither lifespan nor health span (31). Thus, a clear consensus is necessary to address the methodological differences between studies regarding percentage restriction and health outcomes.
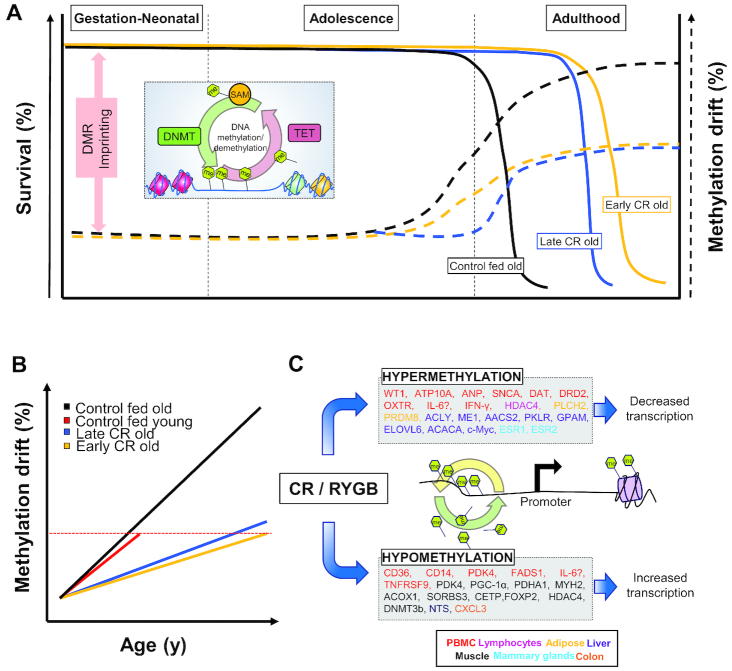
FIGURE 1.
Age-related changes in DNA methylation drift and the effect of CR. (A) DNA methylation is a dynamic process that is regulated during development and throughout life. Early-life DNA methylation patterns are established through genetic and epigenetic imprinting of DMRs. Both early (yellow solid line) and late onset of CR (blue solid line) are able to extend lifespan with differences in health span compared with control without CR (black solid line). Both early (yellow dotted line) and late onsets (blue dotted line) are able to ameliorate age-related methylation drifts. (B) DNA methylation dysregulation with age. (C) Weight loss strategies such as CR and RYGB are able to produce distinct patterns of either hyper- or hypomethylation of many genes (listed in the text boxes) in many metabolic tissues (see color code by tissue), compared with their obese counterparts. AACS2, acetoacetyl-CoA synthetase; ACACA, acetyl-CoA carboxylase α; ACLY, ATP citrate lyase; ACOX1, acyl-CoA oxidase 1; ANP, natriuretic peptide A; ATP10A, ATPase phospholipid transporting 10A; CD, cluster determinant; CETP, cholesteryl ester transfer protein; c-MYC, MYC proto-oncogene; CR, caloric restriction; CXCL3, C-X-C motif chemokine ligand 3; DAT, dopamine transporter; DMR, differentially methylated region; DNMT, DNA methyltransferase; DNMT3b, DNMT 3 β; DRD2, dopamine receptor D2; ELOVL6, fatty acid elongase 6; ESR, estrogen receptor; FADS1, fatty acid desaturase 1; FOXP2, forkhead box P2; GPAM, glycerol-3-phosphate acyltransferase, mitochondrial; HDAC4, histone deacetylase 4; IFN-γ, interferon-γ; Me, methyl (CH3); ME1, malic enzyme 1; MYH2, myosin heavy chain 2; NTS, neurotensin; OXTR, oxytocin receptor; PBMC, peripheral blood mononuclear cell; PDHA1, pyruvate dehydrogenase E1 α 1 subunit; PDK4, pyruvate dehydrogenase kinase 4; PGC-1α, peroxisome proliferator–activated receptor γ coactivator 1 α; PKLR, pyruvate kinase L/R; PLCH2, phospholipase C η 2; PRDM8, PR/SET domain 8; RYGB, Roux-en-Y gastric bypass; SAM, S-adenosyl methionine; SNCA, synuclein α; SORBS3, sorbin and SH3 domain containing 3; TET, Ten Eleven Translocation protein; TNFRSF9, TNF receptor superfamily member 9; WT1, Wilms tumor 1.
Dynamic regulation of DNA methylation
DNA methylation, or the modification of a cytosine nucleotide to 5-methyl cytosine (5mC), is an epigenetic process intricately associated with the regulation of gene expression. DNA methylation has been associated with the control of gene expression at all stages of development (32, 33). DNA methyltransferases (DNMTs) are the enzymes involved in de novo and maintenance methylation of cytosine residues (33, 34). DNMT1 is responsible for the establishment of methylation patterns that define different tissues, whereas DNMT3A and 3B oversee the dynamic turnover of novel methylation marks, or de novo methylation. Other DNMTs include DNMT2, which is involved in the methylation of transfer RNA (tRNA), and DNMT3L, which activates DNMT3A and 3B. Transcriptionally related DNMTs are required for the silencing and transcription of target genes.
The mechanisms and enzymes involved in the process of DNA demethylation are poorly understood. In eukaryotes, the recently described Ten Eleven Translocation family of proteins has been found to play an important role in DNA demethylation (35). The proposed mechanism for DNA demethylation involves Ten Eleven Translocation–mediated oxidation, starting with the conversion of 5mC to 5-hydroxymethyl cytosine, followed by oxidation to 5-formyl cytosine, and finally to 5-carboxyl cytosine (36). Both 5-carboxyl cytosine and 5-formyl cytosine DNA residues are recognized by other proteins that restore the nucleotide to the demethylated cytosine (37).
CR induces changes in DNA methylation
CR without malnutrition provides protection against chronic illness, produces weight loss, and helps prevent the development of metabolic abnormalities. Comparison of 2 nonhuman primate longitudinal CR studies from the NIA and the WNPRC revealed differences in CR onset and diet formulation (30). In the studies, very early CR onset appears to be linked to lower life expectancy: for instance, in the NIA study, before the control group, CR monkeys reached 80% mortality. Meanwhile, early CR onset did not affect the average body weight as expected: compared with the control group, female CR monkeys in the NIA studies showed no significantly different body weight for any age categories. On the other hand, male CR monkeys in the NIA study weighed significantly less than controls, among the juvenile and adult categories. In addition, early CR onset was also linked to reduced quality of life, whereas mid- to late-onset CR produced significant health benefits (Figure 1B). Hence, we sought to review the effects of mid- and late-onset CR and diet specifications associated with longevity and better quality of life, with an epigenetic focus. In a clinical population, 8 wk of CR-induced weight loss significantly reduced DNA methylation of the inflammatory cytokine tumor necrosis factor (TNF), and provided a striking difference in biomarkers for the prediction of weight reduction (38). Moreover, similar methylation patterns for TNF and leptin were observed for obese women who were prescribed an 8-wk-long low-calorie diet (39), thus lessening the inflammatory burden of obese patients. Other genes that are known to be modified by CR-induced weight loss are ATPase phospholipid transporting 10A (ATP10A) and cluster determinant (CD) 44 molecule (CD44) in overweight and obese men (40). Similarly, in overweight and obese postmenopausal women, CR differentially affected the methylation pattern of genomic loci involved in weight control and insulin secretion, analyzed in adipose tissue biopsies (41). In the group of weight loss maintainers, DNA methylation patterns were also more similar to those of normal-weight individuals, rather than those of the obese counterparts who did not lose weight (42). Interestingly, weight loss methods (CR or bariatric surgery) affect the methylation status differently (43).
CR-induced DNA methylation changes and immuno-metabolism
In human studies, weight loss by CR has been shown to alter DNA methylation in blood, adipose tissue, and skeletal muscle (Figure 1C). In 1 study, obese and overweight men were subjected to 30% energy restriction for 8 wk (40). Greater weight loss was associated with hypermethylation of genes for Wilms tumor 1 (WT1) and ATP10A in peripheral blood mononuclear cells (PBMCs). In a similar study involving overweight women, an 8-wk intervention involving 30% CR resulted in decreased methylation of cluster determinant 36 (CD36), cluster determinant 14 (CD14), pyruvate dehydrogenase kinase 4 (PDK4), and fatty acid desaturase 1 (FADS1) in PBMCs (44). Examination of subcutaneous adipose tissue from these women revealed no change in DNA methylation within the leptin (LEP) promoter (39). Another study focused on overweight and obese postmenopausal women who underwent a 6-mo CR weight loss program. Subcutaneous adipose tissue biopsies were performed after an additional 4-wk weight stability period. Those participants that had >3% reduction in body fat percentage were found to have hypermethylated loci associated with phospholipase C η 2 (PLCH2) and PR/SET domain 8 (PRDM8) (41). Thus, it appears that weight loss strategies, regardless of the form or severity, are able, to some extent, to alter DNA methylation patterns of metabolism- or immune-related genes, but significant variability in the response is expected.
In extreme cases of obesity, bariatric surgery may be used as a weight loss strategy to reduce the capacity of the stomach and reduce food intake, thus providing some form of artificial CR. One study found that gastric bypass patients had higher methylation of PDK4 in whole blood at 12 mo after surgery (45). In skeletal muscle biopsies 6 mo after surgery, gastric bypass patients had normalized DNA methylation in 11 metabolic gene promoters, including PDK4, peroxisome proliferator–activated receptor γ coactivator 1 α (PPARGC1A), pyruvate dehydrogenase E1 α 1 subunit (PDHA1), myosin heavy chain 2 (MYH2), acyl-CoA oxidase 1 (ACOX1), and others (46). Another study found reduced methylation in skeletal muscle at 30 CpGs associated with sorbin and SH3 domain containing 3 (SORBS3) after Roux-en-Y gastric bypass surgery (47). In addition to muscle, DNA methylation profiles of omentum and subcutaneous adipose tissue were altered by gastric bypass surgery (48). Results showed 3601 differentially methylated CpGs in subcutaneous adipose tissue and 15 in omentum. CpGs were associated with genes involved in obesity and epigenetic regulation, such as cholesteryl ester transfer protein (CETP), forkhead box P2 (FOXP2), histone deacetylase (HDAC) 4, and DNMT3B. Overall, CR via bariatric surgery mediates DNA methylation of metabolic genes from metabolic tissues such as muscle and adipose tissue in humans.
Whereas human studies have primarily focused on CR as a means of weight loss, animal models have examined CR in the context of longevity and cancer. Previous studies have discussed the role of CR on lifespan through modification of DNA methylation changes (49, 50). Studies in Drosophila found dietary restriction to extend the lifespan without changing methylation patterns, whereas several others have shown that CR mitigates age-associated DNA methylation in a range of tissues (28, 51–53). A study in mice used 60% energy restriction starting at 12 wk of age and examined genome-wide methylation in the liver (54). Not only did restricted animals have longer lifespans, but dietary restriction was also shown to ameliorate age-related hepatic DNA methylation changes. In a set of 1,167,959 bins covering 29 million CpGs, 3176 bins showed a significant methylation difference. Additional analysis revealed hypermethylation of gene bodies. CR-induced methylation was enriched for fatty acid, TG, and ketone body metabolism-related genes, including ATP-citrate lyase (Acly), malic enzyme 1 (Me1), acetoacetyl-CoA synthetase (Aacs2), pyruvate kinase (Pklr), glycerol-3-phosphate acyltransferase (Gpam), fatty acid elongase 6 (Elovl6), and acetyl-CoA carboxylase 1 (Acaca). In hippocampal tissue of mice exposed to 40% CR, >30% of age-related differentially methylated CpGs were prevented by CR (55). Genes affected by CR were enriched for pathways related to energy regulation, inflammation, and phagocytosis. DNA methylation in other brain regions also depends on age and diet. In mouse cerebellum, aging induced a significant increase in 5mC immunoreactivity (56). A 15% calorie reduction had no effect on Purkinje cell methylation in 12-mo-old mice. However, the same CR regimen decreased global DNA methylation in Purkinje cells of 24-mo-old mice. In normal WI-38 lung fibroblasts, glucose restriction extended the lifespan and downregulated expression of the senescence gene p16 (57). This effect might be due to elevated DNMT activity and hypermethylation of the promoter of p16 in the restricted cells. In addition to metabolic and aging pathways, CR also affects methylation of cancer-related genes. One study showed that the livers of aging mice steadily decreased methylation in the promoter and increased methylation in the gene body of proto-oncogene (58). After both 11 mo and 21 mo of 42% CR, age-related methylation changes surrounding MYC proto-oncogene (Myc) were significantly diminished. Thus, CR improves health span by altering DNA methylation in a variety of tissues.
The duration of CR appears to be critical in producing methylation changes. Female mice were 30% calorie restricted starting at 6–8 wk of age, and genome-wide DNA methylation in mammary tissue was measured at 5 mo and 22 mo (59). After 5 mo of dietary treatment, there were 511 significantly more methylated along with 248 significantly less methylated CpGs in CR compared with control mice. After prolonged treatment, the number of differentially methylated loci substantially increased to 7552 with the majority (6901) being hypermethylated in the CR group. Closer investigation of Estrogen receptor 1 and 2 (Esr1 and Esr2) uncovered minimal methylation differences at 5 mo. However, in aged animals, CR resulted in greater methylation at 3 loci within the first intron. Aged CR mice also showed hypermethylation upstream and downstream of the Esr2 gene body. Another study in mice found that 40% CR for 4 mo was sufficient to observe an increase in neurotensin (Nts) expression in the colon as well as lower promoter DNA methylation in restricted mice. Interestingly, this DNA methylation change persisted even when mice were switched back to an ad libitum diet for 5 mo (27).
Although CR is known to regulate systemic inflammation, it remains unclear how DNA methylation contributes to altered cytokine concentrations. In humans, weight loss studies have revealed various changes across adipose tissue and blood. One report found that after an 8-wk 30% energy-restricted intervention there was no change in promoter DNA methylation of TNF subcutaneous adipose tissue of obese women (39). However, results in whole blood were contradictory, as gastric bypass patients had elevated methylation of IL-1B, IL-6, and TNF 12 mo after surgery (45). Findings regarding IL-6 were reproduced in white blood cells, because women subjected to 6 mo of 30% energy restriction had increased IL-6 methylation (43). However, IL-6 methylation in bariatric surgery patients decreased. Interestingly, these changes were not correlated with circulating concentrations of the cytokines. In another study, obese and overweight men were subjected to 30% energy restriction for 8 wk. Analysis revealed hypomethylation of TNF receptor superfamily member 9 (TNFRSF9) and hypermethylation of interferon-γ (IFNG) in PBMCs (40). Overall, investigation of CR-mediated DNA methylation of inflammatory genes has yielded inconsistent results in humans.
In animal models, CR and DNA methylation in inflammatory pathways has been examined in a limited number of studies. One study showed that age-related DNA methylation drift is accelerated under conditions of chronic inflammation (60). In ulcerative colitis patients there was hypermethylation in age-related CpG islands in colon epithelial cells (this observation was found in 12 patients out of 18 cases, with 5 controls). These results suggest an association between CR, DNA methylation, and inflammation. Indeed, in mouse hippocampus, CR resulted in hypermethylation of CpGs that fell within genes that were enriched for inflammatory pathways, including FC ε receptor signaling, signaling by the B cell receptor (BCR), antigen activation of B cell receptor leading to generation of second messengers, FC γ receptor–dependent phagocytosis, and IL-2 signaling (55). Similarly, in monkey liver, CR reduced age-related DNA methylation drift associated with several genes including the neutrophil chemoattractant C-X-C motif chemokine ligand 3 (CXCL3) (28). Collectively, evidence suggests an association between CR and inflammation as well as CR and DNA methylation; however, more work is necessary to uncover the role of DNA methylation in mediating inflammatory outcomes in CR.
Histone Modifications and CR
The basis of histone remodeling
The chromatin landscape determines the availability of the genome, whether it is found in an open and accessible conformation (euchromatin) or in a tightly packed, closed arrangement (heterochromatin). Chromatin changes are mainly driven by covalent modifications to nucleosomal histones, producing a change in the availability of genes for gene expression; specific amino acid residues within histones can be modified covalently, thus altering the interaction with the DNA that winds around the histone octamer (61). Modifications to the histones, such as acetylation, methylation, phosphorylation, or addition of small ubiquitin-like modifers (SUMOylation), can interact with each other and other factors (61) to control the rate of transcription, which in turn dictates tissue-specific gene expression. One such modification, histone acetylation, is a posttranscriptional addition of an acetyl group facilitated by histone acetyltransferases, and removed by HDACs. Histone acetyltransferases transfer acetyl groups from acetyl CoA onto the histone tails (lysine residues), neutralizing their positive charge and causing DNA to decondense to allow transcription, mainly driven by an increased frequency of transcription factor binding within the target genes. For instance, the addition of acetyl groups to histone H4 lysine (H4K) 16 increases transcription in vivo and in vitro (62, 63). Conversely, HDACs remove acetyl groups from the histones and restore the positive charge on the histone, restoring the heterochromatic state (64, 65). Other histone modifications have been described and their activating or repressive mechanisms have been reviewed previously (61, 66). Furthermore, the dynamic regulation of histone modifications and their modifiers has been established previously (67–69) and, thus, will not be the focus of this review. Important aging pathways regulated by sirtuins (SIRTs) include the PI3K/Akt/mTOR axis (70, 71), which highlights the great therapeutic potential of CR and SIRTs. Studies that investigate the relation between nutrient sensing and chromatin modifications are reviewed here to illustrate the potential effects of CR on the health span (72).
CR as regulator of protein modifications
During CR, energy depletion in the cell is evidenced by the increased catabolic state, alteration of nutrient-sensing pathways (73), and increased NAD+ production (74, 75). Alteration of the nutrient pool and its sensors will in turn influence nuclear gene transcription, which can be mediated by protein, and specifically histone modifiers susceptible to CR (67, 76). CR is known to affect SIRTs, a family of nutrient-sensing HDACs (77) (Figure 2). SIRTs are NAD+-dependent protein deacetylases (78) and exert their function on lysine residues found mostly on the histone tails. The first identified SIRT in Saccharomyces cerevisiae, Silent mating type information regulation 2 (Sir2), is indicated in lifespan extension through silencing ribosomal DNA, decreasing the frequency of ribosomal DNA circles that cause aging in yeast (79). Deletion of Sir2 results in a shorter lifespan (80). In addition, telomeres, the genomic regions that protect the ends of each chromosome from deterioration, are shortened when Sir2 expression is low (81). When Sir2 is overexpressed, the longevity phenotype is restored. Increased cellular stress coming from increased ribosomal circles, shorter telomeres, or epigenomic insults could lead to cell senescence and long-term exposure could affect aging, and Sir2 appears to be a strong regulator of this process.
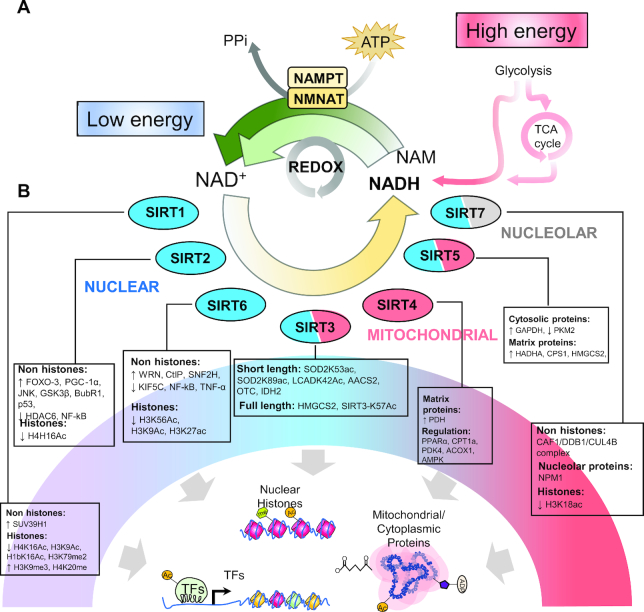
FIGURE 2.
Epigenetic and genetic regulation of sirtuins by caloric restriction. (A) High energy levels after a feeding period contribute directly to the elevated concentrations of NAM and NADH originated from catabolic pathways. Diverse cells and cellular processes deplete the concentration of NADH, and together with the biosynthetic transformation by NAMPT and NMNAT, high intracellular concentrations of NAD+ are produced. (B) Intracellular NAD+ is sensed by NAD+-dependent enzymes, such as sirtuins that add or remove posttranslational protein modifications from nuclear (blue), cytosolic, nucleolar (grey), and mitochondrial (red) proteins. Seven sirtuins, or SIRTs, have been defined in mammals, and participate in deacetylation, mono ADP-ribosylation, and defatty-acylation [demyristoylation, desuccinylation, demalonylation, deglutarylation, demethylglutarylation, and de-3-hydroxy-3-methylglutaryl(HMG)-ation] of nuclear transcription factors, nucleosomal histones, and various nuclear, nucleolar, and mitochondrial proteins. AACS2, acetoacetyl-CoA synthetase; ACOX1, acyl-CoA oxidase 1; AMPK, AMP-activated protein kinase; BubR1, BUB1 mitotic checkpoint serine/threonine kinase B; CAF1/DDB1/CUL4B, ubiquitin complex CAF1/DDB1/CUL4B; CPS1, carbamoyl-phosphate synthase 1; CPT1a, carnitine palmitoyltransferase 1A; CtIP, C-terminal-binding protein interacting protein; FOXO-3, forkhead box O3; GSK3β, glycogen synthase kinase 3 β; HADHA, hydroxyacyl-CoA dehydrogenase subunit α; HDAC6, histone deacetylase 6; HMGCS2, 3-hydroxy-3-methylglutaryl-CoA synthase 2; H1bK16Ac, histone 1b lysine 16 acetylation; H3K9Ac, histone 3 lysine 9 acetylation; H3K9me3, histone 3 lysine 9 trimethylation; H3K18ac, histone 3 lysine 18 acetylation; H3K56Ac, histone H3 lysine 56 acetylation; H3K79me2, histone 3 lysine 79 dimethylation; H4H16Ac, histone 4 histidine 16 acetylation; H4K16Ac, histone 4 lysine 16 acetylation; H4K20me, histone 4 lysine 20 monomethylation; IDH2, isocitrate dehydrogenase 2, mitochondrial; JNK, JUN N-terminal kinase; KIF5C, kinesin family member 5C; LCADK42Ac, acyl-CoA dehydrogenase long chain lysine 42 acetylation; NAM, nicotinamide; NAMPT, nicotinamide phosphoribosyltransferase; NF-kB, nuclear factor κB; NMNAT, nicotinamide mononucleotide adenylyltransferase 1; NPM1, nucleophosmin 1; OTC, ornithine carbamoyltransferase; PDH, pyruvate dehydrogenase; PDK4, pyruvate dehydrogenase kinase 4; PGC-1α, peroxisome proliferator–activated receptor γ coactivator 1 α; PKM2, pyruvate kinase M1/2; PPARα, peroxisome proliferator–activated receptor α; p53, tumor protein P53; SIRT, sirtuin; SIRT3-K57Ac, sirtuin 3 lysine 57 acetylation; SNF2H, sucrose nonfermenting protein 2 homolog; SOD2K53ac, superoxide dismutase lysine 53 acetylation; SOD2K89ac, superoxide dismutase lysine 89 acetylation; SUV39H1, suppressor of variegation 3-9 homolog 1; TCA cycle, tricarboxylic acid cycle; TF, transcription factor; TNF, tumor necrosis factor; WRN, Werner Syndrome RecQ-like helicase; H3K27ac, histone H3 lysine 27 acetylation; Ac, acetylation; Me, metylation; PPi, pyrophosphate.
The SIRT family is comprised of 7 mammalian isoforms, which localize in the nucleus (SIRT1, SIRT2, SIRT6) where they influence histones and other trans-activating factors, in the mitochondria (SIRT3–5) where they can participate in metabolic-related processes and modulate oxidative stress (72), or in the nucleolus (SIRT7) where they aid in cell division (72). SIRTs function as intracellular nutrient sensors allowing them to detect the energy status of the cell, which then is coupled with deacetylation of target proteins (such as histones within target genes or target proteins), as well as ADP-ribosylation in the cytosol (82) or deacylation of mitochondrial matrix proteins (83).
Nutrient and energy sensing within the cells can be achieved through the monitoring of cellular energetics during feeding and fasting periods. Among the determinants of cellular energetics, the ratio of NAD+:NADH is an adequate indicator of the energy status. The NAD+:NADH ratio is high when energy levels are low, whereas the NAD+:NADH ratio decreases when nutrient and energy availability is high. High concentrations of NADH provide reducing agents for oxidized substrates (redox reactions), and together with biosynthetic pathways that use a nicotinamide phosphoribosyl transferase and nicotinamide nucleotide adenylyl transferase to convert nicotinamide, both can yield high amounts of oxidized NAD+ (Figure 2A). During fasting or CR, the concentrations of NAD+ increase and NAD+ catalyzes reactions mediated by all SIRTs (Figure 2B), given that during each round of deacetylation, SIRTs consume 1 NAD+ molecule (84). After activation of the SIRTs, a variety of posttranslational modifications are removed (acetyl, acyl, myristoyl, succinyl, etc.) or added (ADP-ribosylation) to proteins and histones within the nucleus, mitochondria, and nucleolus. We have subdivided the functions of SIRTs according to their subcellular localization, and we will give a brief overview of their effects on metabolism and immunity through epigenetic modifications.
SIRT1
SIRT1 is the mammalian homolog of yeast Sir2 (78) and it is thought to have a major role in extending lifespan with CR in mammals. It localizes within the nucleus and cytosol and is involved in deacetylation reactions of different substrates such as cytosolic proteins, and nuclear histones and transcription factors. Although SIRT1 has many targets within the cytosol, the long-lasting effects of CR that are known to be mediated by SIRT1 are thought to be linked to its HDAC role in the nucleus. In general, SIRT1 maintains a heterochromatic environment by deacetylation of histone H1, H3, and H4 at specific lysine residues. The combined reactions result in the recruitment of histone H1 and the loss of the active transcription mark histone H3 lysine (H3K) 79 methylation (85). In addition, SIRT1 interacts with histone methyltransferase (HMT) suppressor of variegation 3-9 homolog 1 (SUV39H1) to allow the methylation of H3K9 (H3K9me3) and H4K20 (H4K20me), both hallmark histone modifications of heterochromatin (86, 87). Interestingly, the state of constitutive and facultative chromatin is responsive to both micronutrient and macronutrient availability, as we have previously reviewed (88). SIRT1 provides stability to facultative heterochromatin, as it deacetylates H3K9Ac and H4K16Ac marks, and is able to interact with linker histone modification H1bK26Ac and recruit it to promote higher-order organization (85), which in turn is modified by Enhancer of zeste 2 (EZH2) to generate H1bK26me (67). Thus, it seems that SIRT1 can respond to nutrient availability through the increased NAD+ concentration in CR and induce several changes within the acetylation landscape of cytosolic and nuclear proteins and histones to mount an adaptive response to the low energy status.
SIRT2
SIRT2 is regarded as the most conserved SIRT across species and, owing to this feature, it is thought to regulate important cellular processes that are shared by multiple organisms. SIRT2 localizes within the nucleus and cytosol where it can act on cell cycle control and cell division as well as metabolism of fatty acids, which is thought to be mediated through its control on acetylation and myristoylation. This SIRT deacetylates and activates the transcription factors forkhead box o3 (FOXO-3), PPARG coactivator 1 α (PGC-1α), p53, and NF-κB, key regulatory kinases Jun kinase (JNK) and glycogen synthase kinase 3b (GSK3β), and HDAC6. The main function of SIRT2 is to localize to microtubules where it deacetylates the α-tubulin, thus promoting their polymerization/depolymerization cycle. This process is sensitive to the concentrations of NAD+ (89), which is of particular importance given that during CR the concentrations of NAD+ rise and can directly regulate the SIRT2-mediated deacetylation of α-tubulin. Moreover, the deletion of SIRT2 in mouse oocytes results in higher rates of spindle defects, chromosome disorganization, and impaired kinetochore interaction with the centromere (90). This function appears to be intricately related to the direct acetylation of a component of the spindle assembly checkpoint complex BubR1 (BubR1-K243) (91) and deacetylation of histone H4 at K16 (90). CR may prevent genomic instability by regulating mitotic and spindle control, inspecting cell division checkpoints, and promoting the fidelity of the chromosomal distribution.
SIRT3
The response of mitochondrial protein deacetylase SIRT3 to the cell's energy status is primordial for the coupling of metabolic reactions and preservation of mitochondrial integrity (92, 93). SIRT3 exists in 2 forms, the full-length form (FL, 40 kD) and the mitochondrial-exclusive short-length form (SL, 28 kD). FL SIRT3 is known to act as an HDAC. Although the regulation of stress-response genes is important in the context of disease, limited information is available on the genome-wide binding targets of the FL SIRT3 form and its regulation during CR. The SL, mitochondrial form of SIRT3 has been well characterized in relation to the mitochondrial acetylome (mitochondria-wide acetylation/deacetylation) and its regulation. In the mitochondrial matrix, the SL SIRT3 is able to interact with a myriad of factors that are involved in energy metabolism, such as the electron transport chain, tricarboxylic acid cycle, β-oxidation, and ketogenesis, as well as stress resilience and reactive oxygen species quenching (94–99). It appears that the effects of SL SIRT3 revolve around mitochondrial-performance enhancement and increasing resistance to stress. Interestingly, higher NAD+ availability in CR is able to activate SL SIRT3 that in turn directs the deacetylation of 2 lysine residues within superoxide dismutase 2 (SOD2) (K53 and K89), thus improving the response to oxidative stress (97). Similarly, fasting or CR could increase fatty acid β-oxidation by deacetylating long-chain acyl CoA dehydrogenase (LCAD) at lysine 42 (K42) (98). Moreover, in addition to LCAD, SL SIRT3 is known to directly deacetylate acetyl-CoA synthetase 2 (ACSS2), ornithine carbamoyltransferase (OTC), and isocitrate dehydrogenase 2 (IDH2) (99, 100), to promote acetate and urea metabolism and antioxidant defenses.
SIRT3 is highly responsive to CR through various regulation pathways. SIRT3 is able to direct the deacetylation of 3-hydroxy-3-methylglutaryl CoA synthase 2 (HMGCS2), thus increasing its activity and enabling the generation of ketone bodies during fasting and CR (101). Moreover, SIRT3 modifies mitochondrial enzymes that participate in the hallmark processes of CR (102), which makes it an attractive target for therapeutic strategies. SIRT3 is also able to reshape the entire acetylome of the liver in response to CR with 3 different types of acetyl residues: those that are responsive to CR and are acted on by SIRT3, those that are responsive to CR and are unresponsive to SIRT3 (class II), and those that are unresponsive to both CR and SIRT3 (class III) (101). Furthermore, evidence also suggests that SIRT3 activity may be inactivated by other SIRTs. In obese and aged mice, SIRT1 concentrations are reduced and SIRT3 becomes hyperacetylated owing to the inability of SIRT1 to remove SIRT3-K57Ac (103).
SIRT4
Unlike the other mitochondrial SIRTs, SIRT4 is strictly localized within the matrix and its primary function does not involve the deacetylation of target proteins, but rather entails ADP-ribosylation and deacylation, mainly removing glutaryl, methylglutaryl, and de-3-hydroxy-3-methylglutaryl (HMGyl) residues from mitochondrial proteins. Unlike other SIRTs, SIRT4 is negatively regulated by CR and its crystal structure indicates a higher inhibitory potential of NADH due to its higher preference over NAD+ (104).
The effects of SIRT4 oppose that of CR. Studies indicate that it participates in crucial steps of glycolysis by controlling the pyruvate dehydrogenase complex by removing lipoyl and biotinyl residues from the complex (105). Moreover, SIRT4 is known to regulate fatty acid β-oxidation in the liver by controlling PPAR-α, the master regulator of fatty acid oxidation, which controls carnitine palmitoyl transferase 1a (CPT1a), PDK4, and ACOX1 (106), as well as AMP-activated protein kinase (AMPK) (107). During fasting conditions or prolonged nutrient limitation, SIRT4 is suppressed and SIRT1 is upregulated to initiate the oxidative program of the mitochondria.
SIRT5
The final “mitochondrial” SIRT actor is SIRT5, a particularly interesting SIRT whose function spans not only mitochondrial protein modifications, but nuclear and cytosolic ones as well. SIRT5 does not possess deacetylase activity, which sets it apart from other SIRTs. Rather it removes glutaryl, malonyl, and succinyl residues from its target proteins (83). SIRT5 (and SIRT4) is involved in deglutarylation of proteins belonging to oxidation/reduction, generation of precursor metabolites and energy, fatty acid and coenzyme metabolism, as well as aerobic respiration (108). SIRT5 targets carbamoyl phosphate synthase 1 (CPS1), and possibly other glutarylation targets such as hydroxyacyl-CoA dehydrogenase trifunctional multienzyme complex subunit α (HADHA). Regarding energy metabolism, SIRT5 is known to affect the lysine (K) malonylation of several glycolytic enzymes such as GAPDH and pyruvate kinase (PK), thus activating glycolytic flux, as well as urea cycle and other mitochondrial enzymes (109). Likewise, SIRT5 is also able to regulate several mitochondrial proteins related to β-oxidation and ketogenesis; for instance, SIRT5 removes succinyl residues from 3-hydroxy-3-methylglutaryl-CoA synthase 2 (HMGCS2), thus regulating the critical step in ketogenesis (110). SIRT5 not only regulates the mitochondrial acylome, but also protects against mitochondrial fragmentation and mitophagy (mitochondrial degradation). Thus, it is vital for starvation-induced (CR) mitochondrial elongation (111). Finally, although the role of SIRT5 in metabolism has been defined, several avenues of research indicate the great potential of this SIRT in health and age-related diseases (112–115).
SIRT6
SIRT6 primarily remains in the nuclear compartment where it is involved in deacetylation, ADP-ribosylation, and defatty-acylation (myristoyl residues) of numerous proteins related to cell cycle and metabolism. Interestingly, unlike other SIRTs, SIRT6 is capable of binding NAD+ in the absence of acetylated substrate, implying that this SIRT might act as an NAD+ metabolite sensor (116). NAD+ sensing constitutes a key function of SIRT6 in health and disease (117). SIRT6 deacetylates and activates the C-terminal-binding protein interacting protein (CtIP) and recruits sucrose nonfermenting protein 2 homolog (SNF2H) to promote double-strand break and resection, which in turn facilitates homologous recombination of chromosomes in collaboration with breast cancer type 1 susceptibility protein (BRCA1) (118, 119).
In addition to chromosomal stability, SIRT6 also cooperates with different telomeric maintenance systems to protect these important regions. In the nucleus, SIRT6 is able to deacetylate H3K9Ac and H3K56Ac within telomeric regions and facilitates the recruitment of Werner Syndrome RecQ-like helicase to aid in telomere capping during cell division (67, 120). Moreover, upon oxidative damage, SIRT6 promotes directional telomeric movement, which grants protection and is related to telomeric length conservation (121). In a similar way, SIRT6 can act on equally important genomic regions such as enhancers; SIRT6 deacetylates H3K27ac and thus activates cis-regulatory loci (122, 123). Another function that has been described for SIRT6 is related to energy and nutrient metabolism in liver, muscle, and brain (124). As an NAD+-dependent deacetylase, SIRT6 can affect insulin secretion in response to glucose by deacetylating H3K56Ac within the thioredoxin interacting protein (Txnip) promoter in pancreatic β-cells to enhance insulin secretion (125). SIRT6 downregulates microRNA (miR)-122 in the liver by deacetylating H3K56Ac within the promoter, prevents the inhibitory effect from miR-122, and thus increases fatty acid β-oxidation (126). Lastly, SIRT6 inhibits adipogenesis by blocking mitotic clonal expansion through the inactivation of kinesin family member 5C (KIF5C), thus preventing hyperplasia in adipose tissue (127).
When NAD+ is present, SIRT6 deacetylates myristoyl residues, and this process mediates the secretion of various proteins (128). SIRT6 regulates secretion of TNF-α, a potent inflammatory and signaling cytokine, through removal of fatty acyl modifications on K19 and K20, thus stimulating its cellular export in macrophages (129), other immune cells (130), and pancreatic cancer cells (131). SIRT6 deacetylates H3K9Ac tails within p65 (NF-κB)-responsive regions, thus impeding binding of this transcription factor and inhibiting aging-associated inflammation (132). Finally, SIRT6-mediated inactivation of cytokines might be related to the potent inhibition of inflammation by CR in aged animals (133) and humans (16), which is sensitive to cellular NAD+ concentrations.
SIRT7
The last member of the SIRT family is one of the least understood and most understudied NAD+-dependent deacetylases. Nucleolar SIRT7 is depleted in senescent cells, which indicates that SIRT7 is related to replicative senescence (134). Disorganized spindles and disruption of chromosomal syzygy are observed in SIRT7-depleted cells and obese mice (135), but overexpression of SIRT7 is associated with cancer growth (136, 137). The latter is thought to be regulated through miR-125b-5p and miR-340 (136, 138), or CCAAT enhancer binding protein (C/EBPα)-mediated HDAC3 recruitment (139), which negatively regulates SIRT7 expression. Deacetylation of histone H3K18 by SIRT7 decreases mRNA transcription mediated by the Pol II machinery, which demonstrates its effect on chromatin and ability to promote cell transformation and tumorigenesis (137). Further, SIRT7 interacts with and represses the RNA pol I and other nucleolar chromatin remodeling complexes, which emphasizes the role of SIRT7 in transcription (140).
SIRT7 is known to be affected differently by aging and CR in a tissue-specific manner (141). SIRT7 and SIRT6 appear to have a shared proportion of protein targets that are related to DNA repair, chromatin assembly, and aging (142). Furthermore, aging-dependent nucleophosmin (NPM1) acetylation is dependent on SIRT6–SIRT7, thus shedding light on the antiaging effects of SIRT7. Finally, SIRT7 is known to play a role in fat uptake, opposing the effects of SIRT1, SIRT3, and SIRT6 in fat utilization (143). SIRT7 enhances fat uptake by upregulating hepatic CD36 and promotes TG synthesis and storage through the elevation of Mogat, Monoacylglycerol O-Acyltransferase; Cidea, death-inducing DFFA-like effector A; Cidec, cell death-inducing DFFA-like effector C (144). Moreover, SIRT7 was identified as a direct inhibitor of the E3-ubiquitin complex CAF1/DDB1/CUL4B, which is known to target the nuclear receptor testicular receptor 4 (TR4) (145). In turn, TR4 activates genes involved in fat uptake and lipid storage in the liver (144). SIRT7 regulation therefore might constitute an attractive target to counteract the effects of a high-fat diet to prevent fatty liver disease.
Small Noncoding RNA
Epigenetic basis of small noncoding RNA
When discussing the epigenetic regulation of genes, one must consider the ubiquitous and silencing/activating nature of small noncoding RNAs. miRs are small noncoding RNA molecules that regulate posttranscriptional stability of genes through base pair recognition within the 3’-UTR in the target gene. Technologies such as microarray, q-PCR–based, or sequencing approaches have allowed for the identification of numerous miRs present in vivo (146). The binding of miRs to the target gene recruits the multiprotein complex RNA-induced silencing complex (RISC) that cleaves the target gene through one of its components, Argonaute (147–149). This highly orchestrated process is responsible for the posttranscriptional stability of mRNAs in the cytosol. Therefore, environmental stimuli that affect a particular set of miRs will determine the gene expression pattern. During early eukaryotic mRNA translation, cytoplasmic poly(A)-binding protein (PABPC) and the poly(A) tail of the mature mRNA interact to form a complex that can then associate with the eukaryotic translation-initiation factor (eIF) 4G to protect the mRNA, but miRs hinder the interaction of PABPC and eIF4G at the early stages of translation. Binding of miRs to 3’-UTR recruits the RISC complex, which activates one of its components, trinucleotide repeat containing 6A (GW182) (150). GW182 activation recruits the CAF1-CCR4-NOT deadenylase complex, causing deadenylation of the mRNA, leading to mRNA degradation (151). RISC-mediated degradation is a fine-tuned degradation machinery, but its specificity is ultimately dependent on the presence of the miRs.
Owing to the ubiquitous nature of miRs (151), research in this area can provide valuable knowledge in the exploration of the etiology of human diseases (152). In time, miRs could be used as a predictive biomarker for different tissues given their adaptability to different environmental stimuli (153–155). The mechanisms by which CR promotes health benefits are thought to be mediated through alteration of miR patterns in different tissues.
CR and miRs
The effects of CR on miR expression patterns appear to be conserved in different species, from Caenorhabditis elegans and Drosophila, to rodents (Rattus norvegicus and Mus musculus), Rhesus macaques (Macaca mulatta), and humans (Homo sapiens). Different CR concentrations and protocols have been investigated for their miR-modulating effect in many tissue types (Figure 3). In this section, we will review all the evidence regarding CR using a systematic review algorithm in PubMed (Supplemental Table 1).
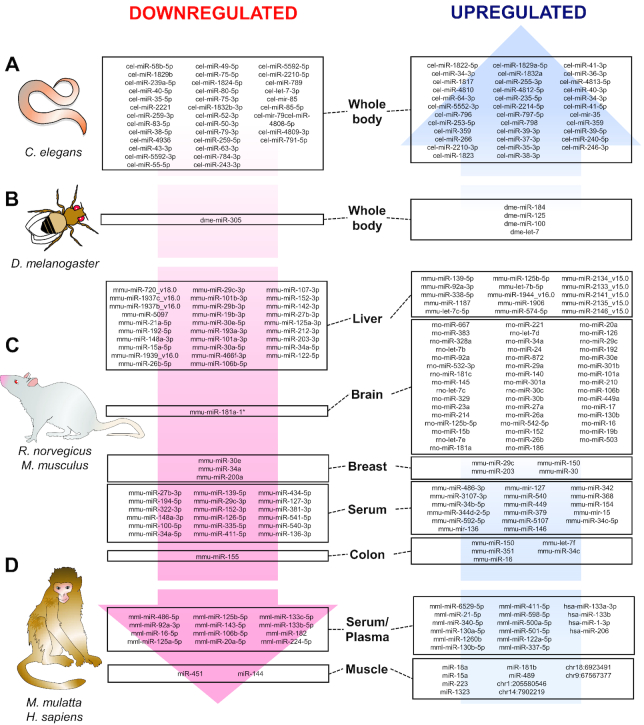
FIGURE 3.
CR conservation of miR changes across species and tissues. CR and starvation are able to upregulate or downregulate miR signatures in (A) Caenorhabditis elegans, (B) Drosophila melanogaster, (C) rodents (Rattus norvegicus and Mus musculus), and (D) Rhesus macaques (Macaca mulatta) and humans (Homo sapiens). miR signatures associated with CR are involved in antiaging pathways such as immuno-metabolic regulation in different peripheral and central tissues. CR, caloric restriction; miR, the mature for of the miRNA; mir, the pre-miRNA and the pri-miRNA; *, asterisk following the name indicate the mature species found at low levels from the opposite arm of a hairpin.
In C. elegans, 12-h starvation is sufficient to produce significant changes in miRs whose target genes are related to metabolism, development, and oogenesis processes (156) (Figure 3A; Supplemental Table 1). Further, 2-d deprivation of food produced whole-body miR changes that were related to longevity, and such changes were dependent on Drosha ortholog (DRSH-1) (157). In addition, 12-h deprivation of food induced physiological changes seen in higher organisms such as decreased lipid accumulation, reduced reproductive function, and increased lifespan (156) (Figure 3A; Supplemental Table 1). Limited evidence exists for miR regulation by CR in fruit flies (Drosophila melanogaster), pointing to metabolic improvements and an increased lifespan. Comparing high- with low-nutrient diets (CR mimic), CR flies had altered expression of miR-184, let-7, miR-125, and miR-100 (out of a total of 18 miRs examined), but it was let-7 overexpression in female nervous tissue that was responsible for an ∼22% increase in lifespan (158). Finally, starvation-induced downregulation of miR-305 activates Dp53 (p53) in the fat body and thus produces metabolic adaptations through nutrient-sensing pathways (159). Altogether, CR in lower organisms highlights the great contribution of nutrient-sensing pathways to lifespan and health span.
In rodents, different tissue types with varied restriction protocols have demonstrated the effectiveness of CR at controlling miR production and exocytosis (Figure 3C; Supplemental Table 1). In long-lived B6C3F1 mice, 40% CR for 27 mo produced an miR pattern in serum that is related to longevity (160). Certain miRs appear to be genotype-specific and age-specific in long-lived Ames dwarf mice. In addition, the pathways these miRs affect are related to tumor suppression, inflammation, WNT, wingless/integrated-, insulin-, mTOR-, and MAPK-signaling pathways (161). Similarly, miRs that are commonly observed in serum (160) were upregulated in liver in C57B6J mice after increasing CR from 10% to 30% for 2 y, and miR-125a-5p was identified as a direct contributor to age-related CR effects (162). When compared with metformin, another life-extending intervention, CR was able to produce a distinguishable signature in liver of mice leading to the alteration of miR-20a, miR-34a, miR-130a, miR-106b, miR-125, and let-7 expression (out of a total of 64 miRs examined) (163) (Figure 3C; Supplemental Table 1).
In colon and colon mucosa, CR seems to be related to anti-inflammatory and anticarcinogenic effects. In a murine colon cancer model, 10 miRs were found to be significantly differentially expressed between the 3 treatment groups: CR, diet-induced obese, and control (164). In particular, miR-150 was upregulated, whereas miR-155 was downregulated by CR. Interestingly, another study showed that one of the functions of miR-150 is increasing cell susceptibility to apoptosis and reducing cell proliferation through decreasing cell cycle progression via initiator of the eukaryotic translation protein, eIF5A (165). A previous study indicated a potential mechanism by which eIF5A is used to regulate apoptosis by upregulating p53 protein expression, which in turn increases Bax, Bcl-2 associated X expression [a preapoptotic member from the B-cell lymphoma 2 (Bcl-2) family] while decreasing expression of Bcl2 (166). Consequently, p53-dependent apoptosis is promoted by the activation of Bax, while cell survival signals are repressed (167). A balance between these BCL2/BAX and BAX/BAX homodimer formations in mammalian cells is necessary to regulate survival and death signals (168). Thus, the upregulation of miR-150 by CR might be linked to the modulation of apoptosis and alteration of cell proliferation. CR significantly downregulates the expression of miR-155 in colon, which is linked to cell apoptosis (169) and proliferation (170). Interestingly, miR-155 targets tumor protein P53 inducible nuclear protein 1 (TP53INP1), which is a proapoptotic stress-induced gene that activates p53 (Figure 3C; Supplemental Table 1).
In breast tissue, CR is sufficient to produce miR patterns that are associated with longevity and aging (149). This study demonstrated several miRs that were altered after CR treatment, among which the most significantly increased were miR-29c, miR-203, miR-150, and miR-30 (out of an analyzed data set containing >100 miRs generated from 7.0 MiRNA microarray analysis by LC Sciences on 5 mg of total RNA). Co-transfection assays suggested that miR-203 can downregulate the translation of Caveolin-1 (Cav-1). Cav-1 is a scaffolding protein that functionally interacts with and regulates signaling molecules such as protein kinase A (PKA), protein kinase C (PKC), H-Ras, epidermal growth factor receptor (EGFR), and G-protein α subunit, and its interaction with such proteins is related to Ku70-mediated apoptosis regulation (171). In a similar way, other miRs such as miR-10a, miR-10b, miR-21, miR-124, miR-125b, miR-126, miR-145, and miR-200a were also identified (out of 101 miRs examined by an Affymetrix GeneChip miRNA 2.0 array) to be associated with mammary tumorigenesis (172). Specifically, miR-200a concentration is elevated throughout cancer progression and it has a pro-proliferative function in mammary cancer cells. Interestingly, miR-200a was found to be downregulated significantly in mammary tumor of CR rats. Considering the function of miR-200a, CR may reduce mammary tumor burden by suppressing miR-200a expression, which consequently inhibits cellular proliferation. Importantly, miR-200a may be useful for the detection of stage-specific breast cancer and may constitute a beneficial CR target for breast cancer patients.
Besides the beneficial effects of CR in peripheral tissues, it also plays a neuroprotective role by modulating expression of age-dependent miRs, thus establishing a balance between proapoptotic and survival signals in the brain. Data has shown age-dependent miRs, miR-181a-1, miR-30e, and miR-34a (56 miRs were examined out of 367 antisense mature miR sequences, scanned by an Expression 1680 scanner, and analyzed using Array-Pro Analyzer 4.5 software), to be downregulated by CR in brain tissue, which correspond with the upregulation of Bcl-2 and downregulation of Bax, leading to apoptosome inhibition (173) (Figure 3C; Supplemental Table 1). In mouse breast tissue, miR-30 was discovered to be significantly upregulated by CR (149). miR-30 downregulates its 2 target genes ubiquitin-conjugating enzyme 9 (UBC9) and integrin β3 (ITGB3) by translational repression (174). UBC9 promotes transcription of the Bcl-2 gene and translation of the Bcl-2 protein. ITGB3 is indicated to have a proapoptotic effect resulting from unligated integrin-mediated cell death, a pathway induced by unligated integrins acting as a negative modulator of cell survival. Therefore, CR-induced expression of miR-30 is implicated in cellular stemness and senescence. CR is related to cell development and physiology in primary cerebromicrovascular endothelial cells, where CR reduced oxidative stress, enhanced nuclear factor erythroid 2 (Nrf2) function, and increased miRs related to angiogenic, proliferative, adhesive, antiapoptotic, and anti-inflammatory processes (175). Collectively, evidence from rodent models shows that CR modulates the expression of several immuno-metabolic and oncogenic miRs across tissue types.
Although the evidence in distinct animal models is strong, clinical trials that aim to identify markers relevant to human populations are needed to provide efficacious and sensitive miR biomarkers (Figure 3D; Supplemental Table 1). Relevant studies have been conducted in Rhesus monkeys to assess the CR miR signature, showing a conserved miR pattern related to growth and insulin signaling as well as regulation of ribosomal, mitochondrial, and spliceosomal pathways (176). Similarly, a study analyzing old monkeys revealed an age-dependent decline in muscle-specific miRs (35 significantly regulated miRs out of a total of 451 miRs examined by Ingenuity Pathway Analysis), but CR improved health span and rescued the expression of miR-181b and chr1:205580546, while decreasing miR-451 and miR-144 concentrations (177). Finally, one of the only interventions in humans revealed that whole-body protein synthesis was inversely related with circulating concentrations of muscle-specific miRs (myomiRs) (miR-1-3p, miR-133a-3p, miR-133b, and miR-206) in energy-restricted (35 d) overweight men (178). Altogether, the evidence suggests that CR-based therapies could target muscle miRs and muscle immuno-metabolism to prevent age-related comorbidities.
Through the investigations using several animal models, it is clear that CR treatment affects cellular processes and the cell cycle via regulating miR expression. This knowledge can help build a better understanding of how CR modifies the epigenome and will shed light on future discoveries of treatments and medicines aimed at antiaging and inhibiting tumorigenesis.
Conclusion
Modulation of age-related decline by CR is robust and is related to genetic and epigenetic adaptations to nutrient availability. Short- and long-term CRs produce significant changes in different tissues and across species, in some animal models even with sex-specific effects, thus indicating that CR acts through conserved mechanisms, such as immuno-metabolic pathways. Notably, early CR onset may cause a different and even an opposite effect on physiological outcomes in animal models such as body weight. Furthermore, CR directly affects the DNA methylation/demethylation cycle, histone and protein modifiers like SIRTs, and miRs, to orchestrate the adaptive and long-lasting response, leading to increased lifespan and health span.
Supplementary Material
ACKNOWLEDGEMENTS
The authors’ responsibilities were as follows—DH-S and Y-XP: conceptualized the manuscript; DH-S, LM, GBX, HC, and Y-XP: wrote the manuscript; DH-S, LM, GBX, and HC: edited the manuscript; and all authors: read and approved the final manuscript.
Notes
Supported by USDA HATCH funds ILLU-698391 (to Y-XP) and ILLU-698369 (to HC).
Author disclosures: DH-S, LM, GBX, HC, and Y-XP, no conflicts of interest.
Supplemental Table 1 is available from the “Supplementary data” link in the online posting of the article and from the same link in the online table of contents at https://academic.oup.com/advances/.
Address correspondence to Y-XP (e-mail: yxpan@illinois.edu).
Abbreviations used: 5mC, 5-methyl cytosine; CD, cluster determinant; CR, caloric restriction; DNMT, DNA methyltransferase; eIF, eukaryotic translation-initiation factor; FL, full length; HDAC, histone deacetylase; H3K, histone H3 lysine; H4K, histone H4 lysine; miR, microRNA; mTOR, mammalian target of rapamycin; NIA, National Institute of Aging; PBMC, peripheral blood mononuclear cell; PDK4, pyruvate dehydrogenase kinase 4; RISC, RNA-induced silencing complex; SIRT, sirtuin; Sir2, silent mating type information regulation 2; SL, short length; WNPRC, Wisconsin National Primate Research Center.
References
- 1. Civitarese AE, Carling S, Heilbronn LK, Hulver MH, Ukropcova B, Deutsch WA, Smith SR, Ravussin E, The CALERIE Pennington Team. Calorie restriction increases muscle mitochondrial biogenesis in healthy humans. PLoS Med. 2007;4(3):e76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Heilbronn LK, Ravussin E. Calorie restriction and aging: review of the literature and implications for studies in humans. Am J Clin Nutr. 2003;78(3):361–9. [DOI] [PubMed] [Google Scholar]
- 3. Parra D, Gonzalez A, Martinez JA, Labayen I, Diez N. In vivo assessment of the mitochondrial response to caloric restriction in obese women by the 2-keto[1-13C]isocaproate breath test. Metabolism. 2003;52(4):463–7. [DOI] [PubMed] [Google Scholar]
- 4. Anton SD, Han H, York E, Martin CK, Ravussin E, Williamson DA. Effect of calorie restriction on subjective ratings of appetite. J Hum Nutr Diet. 2009;22(2):141–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lefevre M, Redman LM, Heilbronn LK, Smith JV, Martin CK, Rood JC, Greenway FL, Williamson DA, Smith SR, Ravussin E et al.. Caloric restriction alone and with exercise improves CVD risk in healthy non-obese individuals. Atherosclerosis. 2009;203(1):206–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Larsen TM, Dalskov SM, van Baak M, Jebb SA, Papadaki A, Pfeiffer AF, Martinez JA, Handjieva-Darlenska T, Kunesova M, Pihlsgard M et al.. Diets with high or low protein content and glycemic index for weight-loss maintenance. N Engl J Med. 2010;363(22):2102–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Goday A, Bellido D, Sajoux I, Crujeiras AB, Burguera B, Garcia-Luna PP, Oleaga A, Moreno B, Casanueva FF. Short-term safety, tolerability and efficacy of a very low-calorie-ketogenic diet interventional weight loss program versus hypocaloric diet in patients with type 2 diabetes mellitus. Nutr Diabetes. 2016;6(9):e230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lv M, Zhu X, Wang H, Wang F, Guan W. Roles of caloric restriction, ketogenic diet and intermittent fasting during initiation, progression and metastasis of cancer in animal models: a systematic review and meta-analysis. PLoS One. 2014;9(12):e115147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rogovik AL, Goldman RD. Ketogenic diet for treatment of epilepsy. Can Fam Physician. 2010;56(6):540–2. [PMC free article] [PubMed] [Google Scholar]
- 10. Li N, Guenancia C, Rigal E, Hachet O, Chollet P, Desmoulins L, Leloup C, Rochette L, Vergely C. Short-term moderate diet restriction in adulthood can reverse oxidative, cardiovascular and metabolic alterations induced by postnatal overfeeding in mice. Sci Rep. 2016;6:30817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Branquinho NTD, Cruz GHP, Borrasca CL, Alves LPS, de Godoy Gomes CR, Ferreira de Godoi VA, Pedrosa MMD. Early-onset obesity and food restriction alter hepatocyte metabolism in adult Wistar rats. Arch Physiol Biochem. 2017;123(5):297–305. [DOI] [PubMed] [Google Scholar]
- 12. Verheggen RJ, Maessen MF, Green DJ, Hermus AR, Hopman MT, Thijssen DH. A systematic review and meta-analysis on the effects of exercise training versus hypocaloric diet: distinct effects on body weight and visceral adipose tissue. Obes Rev. 2016;17(8):664–90. [DOI] [PubMed] [Google Scholar]
- 13. Meydani SN, Das SK, Pieper CF, Lewis MR, Klein S, Dixit VD, Gupta AK, Villareal DT, Bhapkar M, Huang M et al.. Long-term moderate calorie restriction inhibits inflammation without impairing cell-mediated immunity: a randomized controlled trial in non-obese humans. Aging (Albany NY). 2016;8(7):1416–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Giordani I, Malandrucco I, Donno S, Picconi F, Di Giacinto P, Di Flaviani A, Chioma L, Frontoni S. Acute caloric restriction improves glomerular filtration rate in patients with morbid obesity and type 2 diabetes. Diabetes Metab. 2014;40(2):158–60. [DOI] [PubMed] [Google Scholar]
- 15. Ruggenenti P, Abbate M, Ruggiero B, Rota S, Trillini M, Aparicio C, Parvanova A, Petrov Iliev I, Pisanu G, Perna A et al.. Renal and systemic effects of calorie restriction in patients with type 2 diabetes with abdominal obesity: a randomized controlled trial. Diabetes. 2017;66(1):75–86. [DOI] [PubMed] [Google Scholar]
- 16. Yang L, Licastro D, Cava E, Veronese N, Spelta F, Rizza W, Bertozzi B, Villareal DT, Hotamisligil GS, Holloszy JO et al.. Long-term calorie restriction enhances cellular quality-control processes in human skeletal muscle. Cell Rep. 2016;14(3):422–8. [DOI] [PubMed] [Google Scholar]
- 17. Kanda Y, Hashiramoto M, Shimoda M, Hamamoto S, Tawaramoto K, Kimura T, Hirukawa H, Nakashima K, Kaku K. Dietary restriction preserves the mass and function of pancreatic β cells via cell kinetic regulation and suppression of oxidative/ER stress in diabetic mice. J Nutr Biochem. 2015;26(3):219–26. [DOI] [PubMed] [Google Scholar]
- 18. Li C, Sadraie B, Steckhan N, Kessler C, Stange R, Jeitler M, Michalsen A. Effects of a one-week fasting therapy in patients with type-2 diabetes mellitus and metabolic syndrome – a randomized controlled explorative study. Exp Clin Endocrinol Diabetes. 2017;125(9):618–24. [DOI] [PubMed] [Google Scholar]
- 19. Urbanova M, Mraz M, Durovcova V, Trachta P, Klouckova J, Kavalkova P, Haluzikova D, Lacinova Z, Hansikova H, Wenchich L et al.. The effect of very-low-calorie diet on mitochondrial dysfunction in subcutaneous adipose tissue and peripheral monocytes of obese subjects with type 2 diabetes mellitus. Physiol Res. 2017;66(5):811–22. [DOI] [PubMed] [Google Scholar]
- 20. Bhatt AA, Choudhari PK, Mahajan RR, Sayyad MG, Pratyush DD, Hasan I, Javherani RS, Bothale MM, Purandare VB, Unnikrishnan AG. Effect of a low-calorie diet on restoration of normoglycemia in obese subjects with type 2 diabetes. Indian J Endocrinol Metab. 2017;21(5):776–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ghalandari H, Kamalpour M, Alimadadi A, Nasrollahzadeh J. Comparison of two calorie-reduced diets of different carbohydrate and fiber contents and a simple dietary advice aimed to modify carbohydrate intake on glycemic control and inflammatory markers in type 2 diabetes: a randomized trial. Int J Endocrinol Metab. 2018;16(1):e12089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Oshakbayev K, Dukenbayeva B, Togizbayeva G, Durmanova A, Gazaliyeva M, Sabir A, Issa A, Idrisov A. Weight loss technology for people with treated type 2 diabetes: a randomized controlled trial. Nutr Metab (Lond). 2017;14:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Choi KM, Han KA, Ahn HJ, Lee SY, Hwang SY, Kim BH, Hong HC, Choi HY, Yang SJ, Yoo HJ et al.. The effects of caloric restriction on fetuin-A and cardiovascular risk factors in rats and humans: a randomized controlled trial. Clin Endocrinol (Oxf). 2013;79(3):356–63. [DOI] [PubMed] [Google Scholar]
- 24. Brandhorst S, Longo VD. Fasting and caloric restriction in cancer prevention and treatment. Recent Results Cancer Res. 2016;207:241–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Simpson SJ, Le Couteur DG, Raubenheimer D, Solon-Biet SM, Cooney GJ, Cogger VC, Fontana L. Dietary protein, aging and nutritional geometry. Ageing Res Rev. 2017;39:78–86. [DOI] [PubMed] [Google Scholar]
- 26. Lamming DW. Diminished mTOR signaling: a common mode of action for endocrine longevity factors. Springerplus. 2014;3:735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Unnikrishnan A, Jackson J, Matyi SA, Hadad N, Wronowski B, Georgescu C, Garrett KP, Wren JD, Freeman WM, Richardson A. Role of DNA methylation in the dietary restriction mediated cellular memory. Geroscience. 2017;39(3):331–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Maegawa S, Lu Y, Tahara T, Lee JT, Madzo J, Liang S, Jelinek J, Colman RJ, Issa JJ. Caloric restriction delays age-related methylation drift. Nat Commun. 2017;8(1):539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mendelsohn AR, Larrick JW. Epigenetic drift is a determinant of mammalian lifespan. Rejuvenation Res. 2017;20(5):430–6. [DOI] [PubMed] [Google Scholar]
- 30. Mattison JA, Colman RJ, Beasley TM, Allison DB, Kemnitz JW, Roth GS, Ingram DK, Weindruch R, de Cabo R, Anderson RM. Caloric restriction improves health and survival of rhesus monkeys. Nat Commun. 2017;8:14063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mitchell SJ, Madrigal-Matute J, Scheibye-Knudsen M, Fang E, Aon M, Gonzalez-Reyes JA, Cortassa S, Kaushik S, Gonzalez-Freire M, Patel B et al.. Effects of sex, strain, and energy intake on hallmarks of aging in mice. Cell Metab. 2016;23(6):1093–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Klose RJ, Bird AP. Genomic DNA methylation: the mark and its mediators. Trends Biochem Sci. 2006;31(2):89–97. [DOI] [PubMed] [Google Scholar]
- 33. Jurkowska RZ, Jurkowski TP, Jeltsch A. Structure and function of mammalian DNA methyltransferases. Chembiochem. 2011;12(2):206–22. [DOI] [PubMed] [Google Scholar]
- 34. Zemach A, McDaniel IE, Silva P, Zilberman D. Genome-wide evolutionary analysis of eukaryotic DNA methylation. Science. 2010;328(5980):916–19. [DOI] [PubMed] [Google Scholar]
- 35. Kinney SM, Chin HG, Vaisvila R, Bitinaite J, Zheng Y, Esteve PO, Feng S, Stroud H, Jacobsen SE, Pradhan S. Tissue-specific distribution and dynamic changes of 5-hydroxymethylcytosine in mammalian genomes. J Biol Chem. 2011;286(28):24685–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wu H, Zhang Y. Mechanisms and functions of Tet protein-mediated 5-methylcytosine oxidation. Genes Dev. 2011;25(23):2436–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kohli RM, Zhang Y. TET enzymes, TDG and the dynamics of DNA demethylation. Nature. 2013;502(7472):472–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Campion J, Milagro FI, Goyenechea E, Martinez JA. TNF-α promoter methylation as a predictive biomarker for weight-loss response. Obesity (Silver Spring). 2009;17(6):1293–7. [DOI] [PubMed] [Google Scholar]
- 39. Cordero P, Campion J, Milagro FI, Goyenechea E, Steemburgo T, Javierre BM, Martinez JA. Leptin and TNF-alpha promoter methylation levels measured by MSP could predict the response to a low-calorie diet. J Physiol Biochem. 2011;67(3):463–70. [DOI] [PubMed] [Google Scholar]
- 40. Milagro FI, Campion J, Cordero P, Goyenechea E, Gomez-Uriz AM, Abete I, Zulet MA, Martinez JA. A dual epigenomic approach for the search of obesity biomarkers: DNA methylation in relation to diet-induced weight loss. FASEB J. 2011;25(4):1378–89. [DOI] [PubMed] [Google Scholar]
- 41. Bouchard L, Rabasa-Lhoret R, Faraj M, Lavoie ME, Mill J, Perusse L, Vohl MC. Differential epigenomic and transcriptomic responses in subcutaneous adipose tissue between low and high responders to caloric restriction. Am J Clin Nutr. 2010;91(2):309–20. [DOI] [PubMed] [Google Scholar]
- 42. Huang YT, Maccani JZ, Hawley NL, Wing RR, Kelsey KT, McCaffery JM. Epigenetic patterns in successful weight loss maintainers: a pilot study. Int J Obes (Lond). 2015;39(5):865–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Nicoletti CF, Nonino CB, de Oliveira BA, Pinhel MA, Mansego ML, Milagro FI, Zulet MA, Martinez JA. DNA methylation and hydroxymethylation levels in relation to two weight loss strategies: energy-restricted diet or bariatric surgery. Obes Surg. 2016;26(3):603–11. [DOI] [PubMed] [Google Scholar]
- 44. do Amaral CL, Milagro FI, Curi R, Martinez JA. DNA methylation pattern in overweight women under an energy-restricted diet supplemented with fish oil. Biomed Res Int. 2014:675021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kirchner H, Nylen C, Laber S, Barres R, Yan J, Krook A, Zierath JR, Naslund E. Altered promoter methylation of PDK4, IL1 B, IL6, and TNF after Roux-en Y gastric bypass. Surg Obes Relat Dis. 2014;10(4):671–8. [DOI] [PubMed] [Google Scholar]
- 46. Barres R, Kirchner H, Rasmussen M, Yan J, Kantor FR, Krook A, Naslund E, Zierath JR. Weight loss after gastric bypass surgery in human obesity remodels promoter methylation. Cell Rep. 2013;3(4):1020–7. [DOI] [PubMed] [Google Scholar]
- 47. Day SE, Garcia LA, Coletta RL, Campbell LE, Benjamin TR, De Filippis EA, Madura JA 2nd, Mandarino LJ, Roust LR, Coletta DK. Alterations of sorbin and SH3 domain containing 3 (SORBS3) in human skeletal muscle following Roux-en-Y gastric bypass surgery. Clin Epigenetics. 2017;9:96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Benton MC, Johnstone A, Eccles D, Harmon B, Hayes MT, Lea RA, Griffiths L, Hoffman EP, Stubbs RS, Macartney-Coxson D. An analysis of DNA methylation in human adipose tissue reveals differential modification of obesity genes before and after gastric bypass and weight loss. Genome Biology. 2015;16:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lian T, Gaur U, Yang D, Li D, Li Y, Yang M. Epigenetic mechanisms of dietary restriction induced aging in Drosophila. Exp Gerontol. 2015;72:38–44. [DOI] [PubMed] [Google Scholar]
- 50. Unnikrishnan A, Freeman WM, Jackson J, Wren JD, Porter H, Richardson A. The role of DNA methylation in epigenetics of aging. Pharmacol Ther. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wang T, Tsui B, Kreisberg JF, Robertson NA, Gross AM, Yu MK, Carter H, Brown-Borg HM, Adams PD, Ideker T. Epigenetic aging signatures in mice livers are slowed by dwarfism, calorie restriction and rapamycin treatment. Genome Biol. 2017;18(1):57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Cole JJ, Robertson NA, Rather MI, Thomson JP, McBryan T, Sproul D, Wang TN, Brock C, Clark W, Ideker T et al.. Diverse interventions that extend mouse lifespan suppress shared age-associated epigenetic changes at critical gene regulatory regions. Genome Biology. 2017;18:58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Kim CH, Lee EK, Choi YJ, An HJ, Jeong HO, Park D, Kim BC, Yu BP, Bhak J, Chung HY. Short-term calorie restriction ameliorates genomewide, age-related alterations in DNA methylation. Aging Cell. 2016;15(6):1074–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Hahn O, Gronke S, Stubbs TM, Ficz G, Hendrich O, Krueger F, Andrews S, Zhang Q, Wakelam MJ, Beyer A et al.. Dietary restriction protects from age-associated DNA methylation and induces epigenetic reprogramming of lipid metabolism. Genome Biol. 2017;18(1):56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Hadad N, Unnikrishnan A, Jackson JA, Masser DR, Otalora L, Stanford DR, Richardson A, Freeman WM. Caloric restriction mitigates age-associated hippocampal differential CG and non-CG methylation. Neurobiol Aging. 2018;67:53–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Lardenoije R, van den Hove DLA, Vaessen TSJ, Iatrou A, Meuwissen KPV, van Hagen BTJ, Kenis G, Steinbusch HWM, Schmitz C, Rutten BPF. Epigenetic modifications in mouse cerebellar Purkinje cells: effects of aging, caloric restriction, and overexpression of superoxide dismutase 1 on 5-methylcytosine and 5-hydroxymethylcytosine. Neurobiol Aging. 2015;36(11):3079–89. [DOI] [PubMed] [Google Scholar]
- 57. Li YY, Liu L, Tollefsbol TO. Glucose restriction can extend normal cell lifespan and impair precancerous cell growth through epigenetic control of hTERT and p16 expression. FASEB J. 2010;24(5):1442–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Miyamura Y, Tawa R, Koizumi A, Uehara Y, Kurishita A, Sakurai H, Kamiyama S, Ono T. Effects of energy restriction on age-associated changes of DNA methylation in mouse liver. Mutat Res. 1993;295(2):63–9. [DOI] [PubMed] [Google Scholar]
- 59. Rossi EL, Dunlap SM, Bowers LW, Khatib SA, Doerstling SS, Smith LA, Ford NA, Holley D, Brown PH, Estecio MR et al.. Energy balance modulation impacts epigenetic reprogramming, ERα and ERβ expression, and mammary tumor development in MMTV-neu transgenic mice. Cancer Res. 2017;77(9):2500–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Issa JP, Ahuja N, Toyota M, Bronner MP, Brentnall TA. Accelerated age-related CpG island methylation in ulcerative colitis. Cancer Res. 2001;61(9):3573–7. [PubMed] [Google Scholar]
- 61. Lawrence M, Daujat S, Schneider R. Lateral thinking: how histone modifications regulate gene expression. Trends Genet. 2016;32(1):42–56. [DOI] [PubMed] [Google Scholar]
- 62. Shogren-Knaak M, Ishii H, Sun JM, Pazin MJ, Davie JR, Peterson CL. Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science. 2006;311(5762):844–7. [DOI] [PubMed] [Google Scholar]
- 63. Akhtar A, Becker PB. Activation of transcription through histone H4 acetylation by MOF, an acetyltransferase essential for dosage compensation in Drosophila. Mol Cell. 2000;5(2):367–75. [DOI] [PubMed] [Google Scholar]
- 64. Taunton J, Hassig CA, Schreiber SL. A mammalian histone deacetylase related to the yeast transcriptional regulator Rpd3p. Science. 1996;272(5260):408–11. [DOI] [PubMed] [Google Scholar]
- 65. Ng HH, Bird A. Histone deacetylases: silencers for hire. Trends Biochem Sci. 2000;25(3):121–6. [DOI] [PubMed] [Google Scholar]
- 66. Zhang Y, Reinberg D. Transcription regulation by histone methylation: interplay between different covalent modifications of the core histone tails. Genes Dev. 2001;15(18):2343–60. [DOI] [PubMed] [Google Scholar]
- 67. Vaquero A, Reinberg D. Calorie restriction and the exercise of chromatin. Genes Dev. 2009;23(16):1849–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Han S, Brunet A. Histone methylation makes its mark on longevity. Trends Cell Biol. 2012;22(1):42–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Watroba M, Dudek I, Skoda M, Stangret A, Rzodkiewicz P, Szukiewicz D. Sirtuins, epigenetics and longevity. Ageing Res Rev. 2017;40:11–19. [DOI] [PubMed] [Google Scholar]
- 70. Mazucanti CH, Cabral-Costa JV, Vasconcelos AR, Andreotti DZ, Scavone C, Kawamoto EM. Longevity pathways (mTOR, SIRT, insulin/IGF-1) as key modulatory targets on aging and neurodegeneration. Curr Top Med Chem. 2015;15(21):2116–38. [DOI] [PubMed] [Google Scholar]
- 71. Sharples AP, Hughes DC, Deane CS, Saini A, Selman C, Stewart CE. Longevity and skeletal muscle mass: the role of IGF signalling, the sirtuins, dietary restriction and protein intake. Aging Cell. 2015;14(4):511–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Chang HC, Guarente L. SIRT1 and other sirtuins in metabolism. Trends Endocrinol Metab. 2014;25(3):138–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Fontana L, Partridge L. Promoting health and longevity through diet: from model organisms to humans. Cell. 2015;161(1):106–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Mouchiroud L, Houtkooper RH, Auwerx J. NAD+ metabolism: a therapeutic target for age-related metabolic disease. Crit Rev Biochem Mol Biol. 2013;48(4):397–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Mitchell SJ, Bernier M, Aon MA, Cortassa S, Kim EY, Fang EF, Palacios HH, Ali A, Navas-Enamorado I, Di Francesco A et al.. Nicotinamide improves aspects of healthspan, but not lifespan, in mice. Cell Metab. 2018;27(3):667–76..e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Wood SH, van Dam S, Craig T, Tacutu R, O'Toole A, Merry BJ, de Magalhaes JP. Transcriptome analysis in calorie-restricted rats implicates epigenetic and post-translational mechanisms in neuroprotection and aging. Genome Biol. 2015;16:285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Guarente L. Sir2 links chromatin silencing, metabolism, and aging. Genes Dev. 2000;14(9):1021–6. [PubMed] [Google Scholar]
- 78. Wang F, Nguyen M, Qin FX, Tong Q. SIRT2 deacetylates FOXO3a in response to oxidative stress and caloric restriction. Aging Cell. 2007;6(4):505–14. [DOI] [PubMed] [Google Scholar]
- 79. Sinclair DA, Guarente L. Extrachromosomal rDNA circles—a cause of aging in yeast. Cell. 1997;91(7):1033–42. [DOI] [PubMed] [Google Scholar]
- 80. Kennedy BK, Austriaco NR Jr, Guarente L. Daughter cells of Saccharomyces cerevisiae from old mothers display a reduced life span. J Cell Biol. 1994;127(6 Pt 2):1985–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Dang W, Steffen KK, Perry R, Dorsey JA, Johnson FB, Shilatifard A, Kaeberlein M, Kennedy BK, Berger SL. Histone H4 lysine 16 acetylation regulates cellular lifespan. Nature. 2009;459(7248):802–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Tanny JC, Dowd GJ, Huang J, Hilz H, Moazed D. An enzymatic activity in the yeast Sir2 protein that is essential for gene silencing. Cell. 1999;99(7):735–45. [DOI] [PubMed] [Google Scholar]
- 83. Carrico C, Meyer JG, He W, Gibson BW, Verdin E. The mitochondrial acylome emerges: proteomics, regulation by sirtuins, and metabolic and disease implications. Cell Metab. 2018;27(3):497–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Imai S, Armstrong CM, Kaeberlein M, Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403(6771):795–800. [DOI] [PubMed] [Google Scholar]
- 85. Vaquero A, Scher M, Lee D, Erdjument-Bromage H, Tempst P, Reinberg D. Human SirT1 interacts with histone H1 and promotes formation of facultative heterochromatin. Mol Cell. 2004;16(1):93–105. [DOI] [PubMed] [Google Scholar]
- 86. Trojer P, Reinberg D. Facultative heterochromatin: is there a distinctive molecular signature?. Mol Cell. 2007;28(1):1–13. [DOI] [PubMed] [Google Scholar]
- 87. Peters AH, O'Carroll D, Scherthan H, Mechtler K, Sauer S, Schofer C, Weipoltshammer K, Pagani M, Lachner M, Kohlmaier A et al.. Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell. 2001;107(3):323–37. [DOI] [PubMed] [Google Scholar]
- 88. Hernandez-Saavedra D, Strakovsky RS, Ostrosky-Wegman P, Pan Y-X. Epigenetic regulation of centromere chromatin stability by dietary and environmental factors. Adv Nutr. 2017;8(6):889–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Skoge RH, Dolle C, Ziegler M. Regulation of SIRT2-dependent α-tubulin deacetylation by cellular NAD levels. DNA Repair (Amst). 2014;23:33–8. [DOI] [PubMed] [Google Scholar]
- 90. Zhang L, Hou X, Ma R, Moley K, Schedl T, Wang Q. Sirt2 functions in spindle organization and chromosome alignment in mouse oocyte meiosis. FASEB J. 2014;28(3):1435–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Qiu D, Hou X, Han L, Li X, Ge J, Wang Q. Sirt2-BubR1 acetylation pathway mediates the effects of advanced maternal age on oocyte quality. Aging Cell. 2018;17(1):e12698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Albani D, Ateri E, Mazzuco S, Ghilardi A, Rodilossi S, Biella G, Ongaro F, Antuono P, Boldrini P, Di Giorgi E et al.. Modulation of human longevity by SIRT3 single nucleotide polymorphisms in the prospective study “Treviso Longeva (TRELONG)”. Age (Dordr). 2014;36(1):469–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Dhillon RS, Denu JM. Using comparative biology to understand how aging affects mitochondrial metabolism. Mol Cell Endocrinol. 2017;455:54–61. [DOI] [PubMed] [Google Scholar]
- 94. Giralt A, Villarroya F. SIRT3, a pivotal actor in mitochondrial functions: metabolism, cell death and aging. Biochem J. 2012;444(1):1–10. [DOI] [PubMed] [Google Scholar]
- 95. Jing E, Emanuelli B, Hirschey MD, Boucher J, Lee KY, Lombard D, Verdin EM, Kahn CR. Sirtuin-3 (Sirt3) regulates skeletal muscle metabolism and insulin signaling via altered mitochondrial oxidation and reactive oxygen species production. Proc Natl Acad Sci U S A. 2011;108(35):14608–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Shimazu T, Hirschey MD, Hua L, Dittenhafer-Reed KE, Schwer B, Lombard DB, Li Y, Bunkenborg J, Alt FW, Denu JM et al.. SIRT3 deacetylates mitochondrial 3-hydroxy-3-methylglutaryl CoA synthase 2 and regulates ketone body production. Cell Metab. 2010;12(6):654–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Qiu X, Brown K, Hirschey MD, Verdin E, Chen D. Calorie restriction reduces oxidative stress by SIRT3-mediated SOD2 activation. Cell Metab. 2010;12(6):662–7. [DOI] [PubMed] [Google Scholar]
- 98. Hirschey MD, Shimazu T, Goetzman E, Jing E, Schwer B, Lombard DB, Grueter CA, Harris C, Biddinger S, Ilkayeva OR et al.. SIRT3 regulates mitochondrial fatty-acid oxidation by reversible enzyme deacetylation. Nature. 2010;464(7285):121–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Hallows WC, Yu W, Smith BC, Devries MK, Ellinger JJ, Someya S, Shortreed MR, Prolla T, Markley JL, Smith LM et al.. Sirt3 promotes the urea cycle and fatty acid oxidation during dietary restriction. Mol Cell. 2011;41(2):139–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Yu W, Dittenhafer-Reed KE, Denu JM. SIRT3 protein deacetylates isocitrate dehydrogenase 2 (IDH2) and regulates mitochondrial redox status. J Biol Chem. 2012;287(17):14078–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Hebert AS, Dittenhafer-Reed KE, Yu W, Bailey DJ, Selen ES, Boersma MD, Carson JJ, Tonelli M, Balloon AJ, Higbee AJ et al.. Calorie restriction and SIRT3 trigger global reprogramming of the mitochondrial protein acetylome. Mol Cell. 2013;49(1):186–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Hirschey MD, Shimazu T, Huang JY, Schwer B, Verdin E. SIRT3 regulates mitochondrial protein acetylation and intermediary metabolism. Cold Spring Harb Symp Quant Biol. 2011;76:267–77. [DOI] [PubMed] [Google Scholar]
- 103. Kwon S, Seok S, Yau P, Li X, Kemper B, Kemper JK. Obesity and aging diminish sirtuin 1 (SIRT1)-mediated deacetylation of SIRT3, leading to hyperacetylation and decreased activity and stability of SIRT3. J Biol Chem. 2017;292(42):17312–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Pannek M, Simic Z, Fuszard M, Meleshin M, Rotili D, Mai A, Schutkowski M, Steegborn C. Crystal structures of the mitochondrial deacylase Sirtuin 4 reveal isoform-specific acyl recognition and regulation features. Nat Commun. 2017;8(1):1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Mathias RA, Greco TM, Oberstein A, Budayeva HG, Chakrabarti R, Rowland EA, Kang Y, Shenk T, Cristea IM. Sirtuin 4 is a lipoamidase regulating pyruvate dehydrogenase complex activity. Cell. 2014;159(7):1615–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Laurent G, de Boer VC, Finley LW, Sweeney M, Lu H, Schug TT, Cen Y, Jeong SM, Li X, Sauve AA et al.. SIRT4 represses peroxisome proliferator-activated receptor α activity to suppress hepatic fat oxidation. Mol Cell Biol. 2013;33(22):4552–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Nasrin N, Wu X, Fortier E, Feng Y, Bare OC, Chen S, Ren X, Wu Z, Streeper RS, Bordone L. SIRT4 regulates fatty acid oxidation and mitochondrial gene expression in liver and muscle cells. J Biol Chem. 2010;285(42):31995–2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Tan M, Peng C, Anderson KA, Chhoy P, Xie Z, Dai L, Park J, Chen Y, Huang H, Zhang Y et al.. Lysine glutarylation is a protein posttranslational modification regulated by SIRT5. Cell Metab. 2014;19(4):605–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Nishida Y, Rardin MJ, Carrico C, He W, Sahu AK, Gut P, Najjar R, Fitch M, Hellerstein M, Gibson BW et al.. SIRT5 regulates both cytosolic and mitochondrial protein malonylation with glycolysis as a major target. Mol Cell. 2015;59(2):321–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Rardin MJ, He W, Nishida Y, Newman JC, Carrico C, Danielson SR, Guo A, Gut P, Sahu AK, Li B et al.. SIRT5 regulates the mitochondrial lysine succinylome and metabolic networks. Cell Metab. 2013;18(6):920–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Guedouari H, Daigle T, Scorrano L, Hebert-Chatelain E. Sirtuin 5 protects mitochondria from fragmentation and degradation during starvation. Biochim Biophys Acta. 2017;1864(1):169–76. [DOI] [PubMed] [Google Scholar]
- 112. Kumar S, Lombard DB. Functions of the sirtuin deacylase SIRT5 in normal physiology and pathobiology. Crit Rev Biochem Mol Biol. 2018;53(3):311–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Zou R, Shi W, Tao J, Li H, Lin X, Yang S, Hua P. SIRT5 and post-translational protein modifications: a potential therapeutic target for myocardial ischemia-reperfusion injury with regard to mitochondrial dynamics and oxidative metabolism. Eur J Pharmacol. 2018;818:410–18. [DOI] [PubMed] [Google Scholar]
- 114. Bringman-Rodenbarger LR, Guo AH, Lyssiotis CA, Lombard DB. Emerging roles for SIRT5 in metabolism and cancer. Antioxid Redox Signal. 2018;28(8):677–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. van de Ven RAH, Santos D, Haigis MC. Mitochondrial sirtuins and molecular mechanisms of aging. Trends Mol Med. 2017;23(4):320–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Pan PW, Feldman JL, Devries MK, Dong A, Edwards AM, Denu JM. Structure and biochemical functions of SIRT6. J Biol Chem. 2011;286(16):14575–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Etchegaray JP, Chavez L, Huang Y, Ross KN, Choi J, Martinez-Pastor B, Walsh RM, Sommer CA, Lienhard M, Gladden A et al.. The histone deacetylase SIRT6 controls embryonic stem cell fate via TET-mediated production of 5-hydroxymethylcytosine. Nat Cell Biol. 2015;17(5):545–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Toiber D, Erdel F, Bouazoune K, Silberman DM, Zhong L, Mulligan P, Sebastian C, Cosentino C, Martinez-Pastor B, Giacosa S et al.. SIRT6 recruits SNF2H to DNA break sites, preventing genomic instability through chromatin remodeling. Mol Cell. 2013;51(4):454–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Kugel S, Mostoslavsky R. Chromatin and beyond: the multitasking roles for SIRT6. Trends Biochem Sci. 2014;39(2):72–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Michishita E, McCord RA, Berber E, Kioi M, Padilla-Nash H, Damian M, Cheung P, Kusumoto R, Kawahara TL, Barrett JC et al.. SIRT6 is a histone H3 lysine 9 deacetylase that modulates telomeric chromatin. Nature. 2008;452(7186):492–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Gao Y, Tan J, Jin J, Ma H, Chen X, Leger B, Xu J, Spagnol ST, Dahl KN, Levine AS et al.. SIRT6 facilitates directional telomere movement upon oxidative damage. Sci Rep. 2018;8(1):5407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Wang WW, Zeng Y, Wu B, Deiters A, Liu WR. A chemical biology approach to reveal Sirt6-targeted histone H3 sites in nucleosomes. ACS Chem Biol. 2016;11(7):1973–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Tasselli L, Zheng W, Chua KF. SIRT6: novel mechanisms and links to aging and disease. Trends Endocrinol Metab. 2017;28(3):168–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Cui X, Yao L, Yang X, Gao Y, Fang F, Zhang J, Wang Q, Chang Y. SIRT6 regulates metabolic homeostasis in skeletal muscle through activation of AMPK. Am J Physiol Endocrinol Metab. 2017;313(4):E493–505. [DOI] [PubMed] [Google Scholar]
- 125. Qin K, Zhang N, Zhang Z, Nipper M, Zhu Z, Leighton J, Xu K, Musi N, Wang P. SIRT6-mediated transcriptional suppression of Txnip is critical for pancreatic beta cell function and survival in mice. Diabetologia. 2018;61(4):906–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Elhanati S, Ben-Hamo R, Kanfi Y, Varvak A, Glazz R, Lerrer B, Efroni S, Cohen HY. Reciprocal regulation between SIRT6 and miR-122 controls liver metabolism and predicts hepatocarcinoma prognosis. Cell Rep. 2016;14(2):234–42. [DOI] [PubMed] [Google Scholar]
- 127. Chen Q, Hao W, Xiao C, Wang R, Xu X, Lu H, Chen W, Deng CX. SIRT6 is essential for adipocyte differentiation by regulating mitotic clonal expansion. Cell Rep. 2017;18(13):3155–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Zhang X, Khan S, Jiang H, Antonyak MA, Chen X, Spiegelman NA, Shrimp JH, Cerione RA, Lin H. Identifying the functional contribution of the defatty-acylase activity of SIRT6. Nat Chem Biol. 2016;12(8):614–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Jiang H, Khan S, Wang Y, Charron G, He B, Sebastian C, Du J, Kim R, Ge E, Mostoslavsky R et al.. SIRT6 regulates TNF-α secretion through hydrolysis of long-chain fatty acyl lysine. Nature. 2013;496(7443):110–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Van Gool F, Galli M, Gueydan C, Kruys V, Prevot PP, Bedalov A, Mostoslavsky R, Alt FW, De Smedt T, Leo O. Intracellular NAD levels regulate tumor necrosis factor protein synthesis in a sirtuin-dependent manner. Nat Med. 2009;15(2):206–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Bauer I, Grozio A, Lasiglie D, Basile G, Sturla L, Magnone M, Sociali G, Soncini D, Caffa I, Poggi A et al.. The NAD+-dependent histone deacetylase SIRT6 promotes cytokine production and migration in pancreatic cancer cells by regulating Ca2+ responses. J Biol Chem. 2012;287(49):40924–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Kawahara TL, Michishita E, Adler AS, Damian M, Berber E, Lin M, McCord RA, Ongaigui KC, Boxer LD, Chang HY et al.. SIRT6 links histone H3 lysine 9 deacetylation to NF-κB-dependent gene expression and organismal life span. Cell. 2009;136(1):62–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Phillips T, Leeuwenburgh C. Muscle fiber specific apoptosis and TNF-α signaling in sarcopenia are attenuated by life-long calorie restriction. FASEB J. 2005;19(6):668–70. [DOI] [PubMed] [Google Scholar]
- 134. Kiran S, Chatterjee N, Singh S, Kaul SC, Wadhwa R, Ramakrishna G. Intracellular distribution of human SIRT7 and mapping of the nuclear/nucleolar localization signal. FEBS J. 2013;280(14):3451–66. [DOI] [PubMed] [Google Scholar]
- 135. Gao M, Li X, He Y, Han L, Qiu D, Ling L, Liu H, Liu J, Gu L. SIRT7 functions in redox homeostasis and cytoskeletal organization during oocyte maturation. FASEB J. 2018;32(11):6228–38. [DOI] [PubMed] [Google Scholar]
- 136. Li W, Sun Z, Chen C, Wang L, Geng Z, Tao J. Sirtuin7 has an oncogenic potential via promoting the growth of cholangiocarcinoma cells. Biomed Pharmacother. 2018;100:257–66. [DOI] [PubMed] [Google Scholar]
- 137. Barber MF, Michishita-Kioi E, Xi Y, Tasselli L, Kioi M, Moqtaderi Z, Tennen RI, Paredes S, Young NL, Chen K et al.. SIRT7 links H3K18 deacetylation to maintenance of oncogenic transformation. Nature. 2012;487(7405):114–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Wang X, Song Y. MicroRNA-340 inhibits the growth and invasion of angiosarcoma cells by targeting SIRT7. Biomed Pharmacother. 2018;103:1061–8. [DOI] [PubMed] [Google Scholar]
- 139. Liu GF, Lu JY, Zhang YJ, Zhang LX, Lu GD, Xie ZJ, Cheng MB, Shen YF, Zhang Y. C/EBPα negatively regulates SIRT7 expression via recruiting HDAC3 to the upstream-promoter of hepatocellular carcinoma cells. Biochim Biophys Acta. 2016;1859(2):348–54. [DOI] [PubMed] [Google Scholar]
- 140. Tsai YC, Greco TM, Boonmee A, Miteva Y, Cristea IM. Functional proteomics establishes the interaction of SIRT7 with chromatin remodeling complexes and expands its role in regulation of RNA polymerase I transcription. Mol Cell Proteomics. 2012;11(5):60–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Wronska A, Lawniczak A, Wierzbicki PM, Kmiec Z. Age-related changes in Sirtuin 7 expression in calorie-restricted and refed rats. Gerontology. 2016;62(3):304–10. [DOI] [PubMed] [Google Scholar]
- 142. Lee N, Kim DK, Kim ES, Park SJ, Kwon JH, Shin J, Park SM, Moon YH, Wang HJ, Gho YS et al.. Comparative interactomes of SIRT6 and SIRT7: implication of functional links to aging. Proteomics. 2014;14(13–14):1610–22. [DOI] [PubMed] [Google Scholar]
- 143. Houtkooper RH, Pirinen E, Auwerx J. Sirtuins as regulators of metabolism and healthspan. Nat Rev Mol Cell Biol. 2012;13(4):225–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Yoshizawa T, Karim MF, Sato Y, Senokuchi T, Miyata K, Fukuda T, Go C, Tasaki M, Uchimura K, Kadomatsu T et al.. SIRT7 controls hepatic lipid metabolism by regulating the ubiquitin-proteasome pathway. Cell Metab. 2014;19(4):712–21. [DOI] [PubMed] [Google Scholar]
- 145. Lee J, Zhou P. DCAFs, the missing link of the CUL4-DDB1 ubiquitin ligase. Mol Cell. 2007;26(6):775–80. [DOI] [PubMed] [Google Scholar]
- 146. Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: microRNAs can up-regulate translation. Science. 2007;318(5858):1931–4. [DOI] [PubMed] [Google Scholar]
- 147. Carthew RW, Sontheimer EJ. Origins and mechanisms of miRNAs and siRNAs. Cell. 2009;136(4):642–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Orom UA, Nielsen FC, Lund AH. MicroRNA-10a binds the 5'UTR of ribosomal protein mRNAs and enhances their translation. Mol Cell. 2008;30(4):460–71. [DOI] [PubMed] [Google Scholar]
- 149. Orom UA, Lim MK, Savage JE, Jin L, Saleh AD, Lisanti MP, Simone NL. MicroRNA-203 regulates caveolin-1 in breast tissue during caloric restriction. Cell Cycle. 2012;11(7):1291–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Braun JE, Huntzinger E, Izaurralde E. The role of GW182 proteins in miRNA-mediated gene silencing. Adv Exp Med Biol. 2013;768:147–63. [DOI] [PubMed] [Google Scholar]
- 151. Calvopina DA, Coleman MA, Lewindon PJ, Ramm GA. Function and regulation of microRNAs and their potential as biomarkers in paediatric liver disease. Int J Mol Sci. 2016;17(11):1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Nezami BG, Mwangi SM, Lee JE, Jeppsson S, Anitha M, Yarandi SS, Farris AB 3rd, Srinivasan S. MicroRNA 375 mediates palmitate-induced enteric neuronal damage and high-fat diet-induced delayed intestinal transit in mice. Gastroenterology. 2014;146(2):473–83..e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S, Shimizu M, Rattan S, Bullrich F, Negrini M et al.. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci U S A. 2004;101(9):2999–3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Negrini M, Nicoloso MS, Calin GA. MicroRNAs and cancer—new paradigms in molecular oncology. Curr Opin Cell Biol. 2009;21(3):470–9. [DOI] [PubMed] [Google Scholar]
- 155. Simone NL, Soule BP, Ly D, Saleh AD, Savage JE, Degraff W, Cook J, Harris CC, Gius D, Mitchell JB. Ionizing radiation-induced oxidative stress alters miRNA expression. PLoS One. 2009;4(7):e6377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Garcia-Segura L, Abreu-Goodger C, Hernandez-Mendoza A, Dimitrova Dinkova TD, Padilla-Noriega L, Perez-Andrade ME, Miranda-Rios J. High-throughput profiling of Caenorhabditis elegans starvation-responsive microRNAs. PLoS One. 2015;10(11):e0142262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Kogure A, Uno M, Ikeda T, Nishida E. The microRNA machinery regulates fasting-induced changes in gene expression and longevity in Caenorhabditis elegans. J Biol Chem. 2017;292(27):11300–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Gendron CM, Pletcher SD. MicroRNAs mir-184 and let-7 alter Drosophila metabolism and longevity. Aging Cell. 2017;16(6):1434–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Barrio L, Dekanty A, Milan M. MicroRNA-mediated regulation of Dp53 in the Drosophila fat body contributes to metabolic adaptation to nutrient deprivation. Cell Rep. 2014;8(2):528–41. [DOI] [PubMed] [Google Scholar]
- 160. Dhahbi JM, Spindler SR, Atamna H, Yamakawa A, Guerrero N, Boffelli D, Mote P, Martin DI. Deep sequencing identifies circulating mouse miRNAs that are functionally implicated in manifestations of aging and responsive to calorie restriction. Aging (Albany NY). 2013;5(2):130–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Victoria B, Dhahbi JM, Nunez Lopez YO, Spinel L, Atamna H, Spindler SR, Masternak MM. Circulating microRNA signature of genotype-by-age interactions in the long-lived Ames dwarf mouse. Aging Cell. 2015;14(6):1055–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Makwana K, Patel SA, Velingkaar N, Ebron JS, Shukla GC, Kondratov R. Aging and calorie restriction regulate the expression of miR-125a-5p and its target genes Stat3, Casp2 and Stard13. Aging (Albany NY). 2017;9(7):1825–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Noren Hooten N, Martin-Montalvo A, Dluzen DF, Zhang Y, Bernier M, Zonderman AB, Becker KG, Gorospe M, de Cabo R, Evans MK. Metformin-mediated increase in DICER1 regulates microRNA expression and cellular senescence. Aging Cell. 2016;15(3):572–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Olivo-Marston SE, Hursting SD, Perkins SN, Schetter A, Khan M, Croce C, Harris CC, Lavigne J. Effects of calorie restriction and diet-induced obesity on murine colon carcinogenesis, growth and inflammatory factors, and microRNA expression. PLoS One. 2014;9(4):e94765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Watanabe A, Tagawa H, Yamashita J, Teshima K, Nara M, Iwamoto K, Kume M, Kameoka Y, Takahashi N, Nakagawa T et al.. The role of microRNA-150 as a tumor suppressor in malignant lymphoma. Leukemia. 2011;25(8):1324–34. [DOI] [PubMed] [Google Scholar]
- 166. Li AL, Li HY, Jin BF, Ye QN, Zhou T, Yu XD, Pan X, Man JH, He K, Yu M et al.. A novel eIF5A complex functions as a regulator of p53 and p53-dependent apoptosis. J Biol Chem. 2004;279(47):49251–8. [DOI] [PubMed] [Google Scholar]
- 167. Basu A, Haldar S. The relationship between BcI2, Bax and p53: consequences for cell cycle progression and cell death. Mol Hum Reprod. 1998;4(12):1099–109. [DOI] [PubMed] [Google Scholar]
- 168. Mo YY, Yu Y, Theodosiou E, Ee PL, Beck WT. A role for Ubc9 in tumorigenesis. Oncogene. 2005;24(16):2677–83. [DOI] [PubMed] [Google Scholar]
- 169. Shibuya H, Iinuma H, Shimada R, Horiuchi A, Watanabe T. Clinicopathological and prognostic value of microRNA-21 and microRNA-155 in colorectal cancer. Oncology. 2010;79(3–4):313–20. [DOI] [PubMed] [Google Scholar]
- 170. He B, Gao SQ, Huang LD, Huang YH, Zhang QY, Zhou MT, Shi HQ, Song QT, Shan YF. MicroRNA-155 promotes the proliferation and invasion abilities of colon cancer cells by targeting quaking. Mol Med Rep. 2015;11(3):2355–9. [DOI] [PubMed] [Google Scholar]
- 171. Martinez-Outschoorn UE, Sotgia F, Lisanti MP. Caveolae and signalling in cancer. Nat Rev Cancer. 2015;15(4):225–37. [DOI] [PubMed] [Google Scholar]
- 172. Devlin KL, Sanford T, Harrison LM, LeBourgeois P, Lashinger LM, Mambo E, Hursting SD. Stage-specific microRNAs and their role in the anticancer effects of calorie restriction in a rat model of ER-positive luminal breast cancer. PLoS One. 2016;11(7):e0159686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. Khanna A, Muthusamy S, Liang R, Sarojini H, Wang E. Gain of survival signaling by down-regulation of three key miRNAs in brain of calorie-restricted mice. Aging (Albany NY). 2011;3(3):223–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174. Yu F, Deng H, Yao H, Liu Q, Su F, Song E. Mir-30 reduction maintains self-renewal and inhibits apoptosis in breast tumor-initiating cells. Oncogene. 2010;29(29):4194–204. [DOI] [PubMed] [Google Scholar]
- 175. Csiszar A, Gautam T, Sosnowska D, Tarantini S, Banki E, Tucsek Z, Toth P, Losonczy G, Koller A, Reglodi D et al.. Caloric restriction confers persistent anti-oxidative, pro-angiogenic, and anti-inflammatory effects and promotes anti-aging miRNA expression profile in cerebromicrovascular endothelial cells of aged rats. Am J Physiol Heart Circ Physiol. 2014;307(3):H292–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176. Schneider A, Dhahbi JM, Atamna H, Clark JP, Colman RJ, Anderson RM. Caloric restriction impacts plasma microRNAs in rhesus monkeys. Aging Cell. 2017;16(5):1200–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Mercken EM, Majounie E, Ding J, Guo R, Kim J, Bernier M, Mattison J, Cookson MR, Gorospe M, de Cabo R et al.. Age-associated miRNA alterations in skeletal muscle from rhesus monkeys reversed by caloric restriction. Aging (Albany NY). 2013;5(9):692–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. Margolis LM, Rivas DA, Pasiakos SM, McClung JP, Ceglia L, Fielding RA. Upregulation of circulating myomiR following short-term energy restriction is inversely associated with whole body protein synthesis. Am J Physiol Regul Integr Comp Physiol. 2017;313(3):R298–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.