Abstract
Deiodinase enzymes play an essential role in converting thyroid hormones between active and inactive forms by deiodinating the pro-hormone thyroxine (T4) to the active hormone triiodothyronine (T3) and modifying T4 and T3 to inactive forms. Chemical inhibition of deiodinase activity has been identified as an important endpoint to include in screening chemicals for thyroid hormone disruption. To address the lack of data regarding chemicals that inhibit the deiodinase enzymes, we developed robust in vitro assays that utilized human deiodinase types 1, 2, and 3 and screened over 1,800 unique chemicals from the U.S. EPA’s ToxCast phase 1_v2, phase 2, and e1k libraries. Initial testing at a single concentration identified 411 putative deiodinase inhibitors that produced inhibition of 20% or greater in at least one of the three deiodinase assays, including chemicals that have not previously been shown to inhibit deiodinases. Of these, 228 chemicals produced enzyme inhibition of 50% or greater; these chemicals were further tested in concentration-response to determine relative potency. Comparisons across these deiodinase assays identified 81 chemicals that produced selective inhibition, with 50% inhibition or greater of only one of the deiodinases. This set of three deiodinase inhibition assays provides a significant contribution towards expanding the limited number of in vitro assays used to identify chemicals with the potential to interfere with thyroid hormone homeostasis. Additionally, these results set the groundwork for development and evaluation of structure-activity relationships for deiodinase inhibition, and inform targeted selection of chemicals for further testing to identify adverse outcomes of deiodinase inhibition.
Keywords: thyroid, screening, deiodinase, prioritization, endocrine disruption
Introduction
There is increasing evidence that environmental contaminants have thyroid-disrupting properties (Boas et al., 2012; Brucker-Davis, 1998; Duntas and Stathatos, 2015; Zoeller, 2005). Chemical interference with thyroid hormone (TH) homeostasis is of concern because of the essential role of THs in vertebrate development and metabolic processes (Zoeller et al., 2007). Despite the growing number of studies, information on thyroid disruption is still limited to a small number of the thousands of chemicals that need to be assessed for human and ecological effects (U.S. EPA, 2012, 2014, 2017a). To address this data gap, in vitro and in silico methods must be used to identify potential thyroid-disrupting chemicals (Murk et al., 2013; OECD, 2014).
Thyroid hormone homeostasis is controlled by a complex series of coordinated events dependent on multiple proteins for TH synthesis, transport, and peripheral metabolism and elimination (Brix et al., 2011; Zoeller et al., 2007). Environmental chemicals can perturb this complex system through a variety of mechanisms that result in thyroid disruption (Boas et al., 2012). Until recently, in vitro screening assays had been implemented for only a few of the potential molecular targets for thyroid disruption, and the only thyroid-relevant assays included in the U.S. EPA’s Toxicity Forecaster (ToxCast) program were the receptor transactivation assays and thyrotropin releasing hormone assays (U.S. EPA, 2015). Recent efforts have identified and described priority targets to use in screening assays to detect potential thyroid-disrupting chemicals (Murk et al., 2013; OECD, 2014). Progress in this area includes the development of in vitro screening assays for identifying chemical disruptors of TH synthesis via inhibition of thyroid peroxidase (TPO; Paul et al., 2014) and the sodium-iodide symporter (NIS; Hallinger et al., 2017; Lecat-Guillet et al., 2007), interruption of TH transport (Dong and Wade, 2017; Jayarama-Naidu et al., 2015), and TH activation/inactivation via inhibition of the iodothyronine deiodinases (Hornung et al., 2018; Renko et al., 2012, 2015), with recent screening of chemical libraries for inhibition of TPO (Paul Friedman et al., 2016), NIS (Wang et al., 2018), and one deiodinase isoform (Hornung et al., 2018).
The aim of this study was to screen environmental chemicals for inhibition of the three iodothyronine deiodinase enzymes: deiodinase type 1 (DIO1), deiodinase type 2 (DIO2), and deiodinase type 3 (DIO3). These three distinct deiodinase enzymes are essential in mediating TH action in organs and tissues where they each perform different roles in converting THs between active and inactive forms, with differences in substrate specificities and tissue-specific expression (Gereben et al., 2008; Köhrle, 1999). DIO2 is important for converting the pro-hormone thyroxine (T4) to the more active hormone triiodothyronine (T3) through the removal of the 5’ outer ring iodine. Whereas DIO3 inactivates both T4 and T3 by removing an inner ring iodine, producing reverse T3 (rT3) and diiodotyrosine (T2), respectively. DIO1 targets both the outer and inner rings, and thus can convert T4 to T3 or inactivate either of these THs. These key roles of deiodinases in modulating tissue- and timing-specific levels of T3 and T4 are well-studied, and there is a wealth of knowledge on enzymatic activity and function, tissue and substrate specificity, and relative importance across multiple vertebrates (see reviews: Darras and Van Herck, 2012; Gereben et al., 2008; Köhrle, 1999; Kuiper et al., 2005; Orozco et al., 2012). Although adverse health or developmental effects of chemical inhibition of deiodinases are not well understood, some known thyroid-disrupting compounds may be acting through this pathway (eg, polybrominated diphenyl ethers, Roberts et al., 2015). In addition, altered deiodinase expression has been documented in several types of cancer (Casula and Bianco, 2012) and mammalian knock-out studies demonstrate negative consequences from deiodinase deficiency (Hernandez et al., 2006; Marsili et al., 2011). Chemical inhibition of deiodinase activity has been identified as an important endpoint to include in screening chemicals for TH disruption (Murk et al., 2013; Zoeller et al., 2007). Presently, there is a lack of data regarding the potential of chemicals to inhibit each of the three deiodinase enzymes with few chemicals tested in existing assays (Renko et al., 2015; Schweizer and Steegborn, 2015), apart from recent screening for inhibition of DIO1 (Hornung et al., 2018).
Presented here are development of screening assays for DIO2 and DIO3 and the results for the DIO1, DIO2, and DIO3 inhibition assays from screening over 1,800 chemicals from the ToxCast phase 1_v2 (ph1v2), phase 2 (ph2), and e1k chemical libraries (Richard et al., 2016). First, we screened the ToxCast ph2 and e1k libraries for DIO1 inhibition using the screening assay and approach in Hornung et al. (2018). We then established screening assays for DIO2 and DIO3 through adaptation of the previously reported assays (Hornung et al., 2018; Renko et al., 2015). These assays were used to screen ToxCast ph1v2, ph2, and e1k libraries for DIO2 and DIO3 inhibition using a tiered screening approach with an initial single high concentration (target of 200 μM) of each chemical followed by further concentration-response testing, similar to that used for DIO1 (Hornung et al., 2018) and other thyroid targets (Paul Friedman et al., 2016; Wang et al., 2018). This study significantly expands the current knowledge of chemicals that could disrupt the thyroid axis via inhibition of iodothyronine deiodinase activity and allows comparisons of similarities and differences in chemical inhibitors not previously possible.
Materials and Methods
The development of these deiodinases assays and the chemical screening followed the approach described in Hornung et al. (2018), with adenoviral expression of human deiodinase enzyme, a colorimetric assay that measured the release of iodide from the hormone substrate, and a tiered screening approach with initial testing at a single high concentration followed by a subset of chemicals tested in concentration-response. The Supplementary Data file includes details of the conditions for each assay (Supplementary Table 1) and the assay plate layouts (Supplementary Figs. 1 and 2) as well as results from two test plates used in development of the DIO2 and DIO3 assays (assay performance metrics in Supplementary Table 2 and inhibition produced by each chemical in Supplementary Tables 3 and 4).
Chemicals
A test set of 1,851 unique chemicals from the ToxCast ph1v2, ph2, and e1k chemical libraries (Richard et al. 2016) was obtained through Dr. Ann Richard (U.S. EPA, Research Triangle Park, North Carolina). These test chemicals were supplied with chemical identities masked in a 96-well plate format at a target concentration of 20 mM in dimethyl sulfoxide (DMSO) with one chemical per well. The actual plated concentration of some chemicals differed from the target of 20 mM due to solubility limitations in DMSO or for oils and mixtures with concentrations provided in mg/ml. As described below under Assay quality/performance, 32 chemicals had evidence of assay interference (see Supplementary Table 5). Thus, the final test set was 1,819 unique chemicals, which are listed with maximum concentration tested in Supplementary Table 6, ordered by ToxCast chemical library and Chemical Abstracts Service Registry Numbers (CASRNs).
Each assay plate included a model inhibitor as a positive control with a complete inhibition curve. For the DIO1 assay, 6-propyl-2-thiouracil (PTU) was used as the model inhibitor, following Hornung et al. (2018) and Renko et al. (2012). For the DIO2 and DIO3 assays, xanthohumol (XTH) was selected for the positive control based on the identification of this chemical as a potent inhibitor of all three deiodinases by Renko et al. (2015). DMSO was the solvent control in all three assays and was considered a negative control that reflected maximum deiodinase activity. The model inhibitors were solubilized in DMSO with graded concentrations to produce an inhibition concentration-response curve (CASRNs and concentrations included in Table 1). To measure intra-assay reproducibility, a small subset of chemicals was replicated across chemical source plates used for the single-point screening. As no standardized set of chemicals exists for deiodinase inhibition, these chemicals were selected based on results in the TPO screening assay (Paul Friedman et al., 2016), with ten ToxCast ph1v2 chemicals in the DIO1 assay and nine of these same chemicals in the DIO2 and DIO3 assays (see Supplementary Figure 3). These chemicals were distributed on the chemical source plates by the supplier with identities masked, with one to six of these chemicals per plate. Chemical plates were sealed and kept at −80 °C when not in use for assays. Chemical reagents, other than the chemical test set, were purchased from Sigma-Aldrich (St. Louis, Missouri).
Table 1.
Summary of the concentration-response curves for the model inhibitors and assay performance across the test chemical plates for both single-point (single high concentration) and concentration-response testing.
| Model Inhibitor |
DMSO |
Z’ factord |
||||||
|---|---|---|---|---|---|---|---|---|
| Enzyme | Chemical Name |
CASRN | Inhibition Curve Concentrations, μM |
IC50a, μM Mean ± SD |
Hill slopea Mean ± SD |
200 μM-MADb Median |
MADc Median |
Mean ± SD |
| DIO1 | 6-Propyl-2-thiouracil | 51-52-5 | 0.033 – 1000 e | 5.4±1.3 | −1.3±0.5 | 3.15 | 4.12 | 0.7±0.1 |
| DIO2 | Xanthohumol | 6754-58-1 | 0.0002 – 200 f | 0.8±0.2 | −0.9±0.3 | 3.06 | 3.85 | 0.7±0.1 |
| DIO3 | Xanthohumol | 6754-58-1 | 0.0002 – 200 f | 0.3±0.1 | −0.9±0.1 | 2.48 | 4.18 | 0.7±0.1 |
IC50 and Hill slope from four-parameter logistic model was calculated from the three replicate runs of each assay plate. Mean and SD across assay plates are reported above for each DIO. Each assay had 26 single-point plates with 16 concentration-response plates for DIO1 and 22 concentration-response plates for DIO2 and DIO3; IC50 and Hillslope not calculated for one DIO2 plate due to errors in the diluted XTH wells.
Model inhibitor-MAD is the platewise positive control median absolute deviation calculated from all the 200 μM wells of the model inhibitor in each of the replicated assay plates.
DMSO-MAD is the platewise median absolution deviation calculated from all the DMSO wells in each of the replicated assay plates.
Z’ factor is a measure of assay quality, with value of >0.5 indicating good separation between the positive and negative controls.
PTU inhibition curve concentrations: 0.033, 0.33, 1, 3.3, 10, 33, and 1000 μM in column A, with 6 wells of 200 μM distributed across each plate.
XTH inhibition curve concentrations: 0.0002, 0.002, 0.02, 0.2, 2, 20, and 200 μM in column A, with 6 additional wells of 200 μM distributed across each plate.
Adenoviral Expression of Human Deiodinases
Adenoviral expression of the human deiodinases followed the methods previously described for DIO1 (Hornung et al., 2018). A summary and specifics for expression of DIO2 and DIO3 are included below. Human deiodinase expression plasmids for DIO2 (hD2D10; Buettner et al., 2000) and DIO3 (hD3CDM; Salvatore et al., 1995) were a gift from Dr. P. Reed Larsen (Harvard Medical School, Boston, Massachusetts). Adenovirus plasmids pDeltaE1sp1A, pBHG10, pCA3 and HEK293 cells were purchased from Microbix Biosystems Inc. (Toronto, Canada). DIO3 expression cassette was liberated from parental plasmids with TaqII (Chimerx, Milwaukee, Wisconsin) and Acc65I (New England Biolabs, Ipswich, Massachuesetts) digests, end filled, gel purified, and blunt end cloned into the EcoRV site of pDeltaE1sp1A. The DIO2 gene was liberated with SacII and Not I, end filled, and blunt end cloned into the EcoRI site of pCA3 which had also been end filled and dephosphorylated. Subclones were isolated and identified by predicted restriction patterns. Adenoviruses expressing deiodinase were constructed by cotransfecting HEK293 cells with the subcloned gene and pBHG10 (Graham, 2000; Graham and Prevec, 1991; Hitt et al., 1994). Isolation of recombinant viruses and subsequent production of DIO2 and DIO3 were identical to DIO1 described earlier (Hornung et al., 2018). Cell lysates were then frozen at −80° C until use in the deiodinase inhibition assays. Protein concentrations were determined using Bradford Reagent with bovine serum albumin as the standard (Sigma-Aldrich). The protein concentration and specific activity varied slightly between the different batches. We previously reported the specific activity of DIO1 to be 9 pmol iodide/h/μg of protein. For DIO2 and DIO3, the specific activities were about 5 and 90 pmol iodide/h/μg of protein, respectively.
Deiodinase Inhibition Assays
Inhibition assay and iodide extraction.
The deiodinase assay methods were first developed for DIO1, closely following Renko et al. (2012, 2015) and are described in Hornung et al. (2018). Briefly, the assay measures deiodinase-liberated iodide with the Sandell-Kolthoff (SK) reaction in a 96-well plate format. The HEK293 cell lysate with expressed DIO1 was thawed, mixed, and diluted in pH 7.0 HEPES buffer. The diluted enzyme (59.4 μl, containing about 10 μg of protein) was then added to the 96-well assay plate (untreated polystyrene, 360 μl well volume, Corning, Corning, New York). Then, 1.2 μl of each test chemical in DMSO was added from the chemical source plate to each well with a Liquidator 96-20 pipettor (Mettler-Toledo Rainin, LLC, Oakland, California) to achieve the final target concentration of 200 μM for each test chemical and 1% DMSO. This maximized the chemical test concentration (given the target concentration on the chemical source plates), and 1% DMSO was found to not inhibit deiodinase enzymes in these assays. The reaction was initiated with the addition of 59.4 μl DIO1 substrate (3,3’,5’-triiodo-L-thyronine, rT3) and dithiothreitol (DTT) in HEPES buffer using a Liquidator 96-200 pipettor (Mettler-Toledo Rainin, LLC). The final assay volume was 120 μl/well with the following conditions: 0.1 M HEPES, 1 mM EDTA, 10 μM rT3, 40 mM DTT, and 1% DMSO. The assay plate was sealed with an adhesive cover sheet (Thermo Fisher Scientific, Waltham, Massachuesetts), mixed in a plate shaker, and incubated for 3 h at 37° C. Following the incubation, 75 μl were transferred to a 96-well, 2 ml polypropylene filtration plate containing Dowex 50WX2 (Biotage USA, Charlotte, North Carolina) and the free iodide was eluted into a 96-well collection plate (Biotage USA) with application of 100 μl of 10% acetic acid.
The DIO2 and DIO3 inhibition assays were adapted from the DIO1 assay described in Hornung et al. (2018) and closely followed methods in Renko et al. (2015), with the following modifications. For DIO2, the HEK293 cell lysate with expressed DIO2 was diluted in pH 7.0 HEPES buffer and then briefly sonicated to obtain a uniform suspension. There were about 20 μg of protein in the 59.4 μl of diluted enzyme added to each well of the assay plate. The substrate used for DIO2 was T4, and final assay conditions were 0.1 M HEPES, pH 7.0, 1 mM EDTA, 5 μM T4, and 40 mM DTT, 1% DMSO and a final assay volume of 120 μl For DIO3, the HEK293 cell lysate with expressed DIO3 was diluted in pH 8.0 HEPES buffer and sonicated as above to obtain about 1 μg protein in the 59.4 μL of added to each well in the assay plate. The substrate used for DIO3 was T3, and final assay conditions were 0.1 M HEPES, pH 8.0, 1 mM EDTA, 5 μM T3, and 40 mM DTT, 1% DMSO and a final assay volume of 120 μl. Final assay conditions of each assay are included in Supplementary Table 1. The incubation period and temperature, and free iodide elution steps in the DIO2 and DIO3 assays were identical to the DIO1 assay. In the development of the DIO2 and DIO3 assays, two test plates including 18 unique chemicals were used in preliminary concentration-response experiments to verify assay performance. The results from these initial test plates are in Supplementary Tables 2-4.
Sandell-Kolthoff (SK) reaction to detect iodide
In the SK reaction, free iodide catalyzes the reduction of cerium IV (Ce+4) which is yellow-colored to the colorless cerium III (Ce+3) in presence of arsenic (As+3), with the rate of this reaction dependent on the concentration of free iodide (Sandell and Kolthoff, 1937). In all three assays, free iodide was detected using the SK reaction following methods previously described in Hornung et al. (2018). Eluent (75 μl) from the wells of the collection plate was transferred to a new untreated polystyrene 96-well plate. With the Liquidator 96-200 pipet, 75 μl of arsenic reagent [25 mM NaAsO2, 0.8 M NaCl, 0.5 M H2SO4] was added and mixed well, followed by the addition of 75 μl of Ce+4 reagent [20 mM (NH4)4Ce(SO4)42H20, 0.44 M H2SO4]. Immediately following the addition of the Ce+4 reagent, the plate was placed in a Synergy 4 plate reader (BioTek Instruments, Inc., Winooski, Vermont), where it was mixed on the fast setting of the plate reader for three seconds, and then absorbance at 420 nm was read every minute under room temperature. Reaction rate was calculated from the change in absorbance between the 1 and 10 min reading in each well.
Chemical Screening
Screening followed the tiered strategy described in Hornung et al. (2018), with all chemicals tested initially at a single concentration followed by further testing in concentration-response mode for those chemicals that showed greater than 50% inhibition. For DIO1, 26 chemical sources plates were used for single-point screening, with further testing of 142 of these chemicals on 16 concentration-response plates. DIO2 and DIO3 were run in parallel, with 26 plates used in single-point screening and 22 concentration-response plates for further testing of 214 of these chemicals. Before use in the single-point screening, chemical source plates were thawed, and test chemicals were mixed by pipetting action. In addition to the concentration-response curve for the model inhibitors, DMSO and 200 μM of the model inhibitor were each plated into six randomly assigned wells per plate, with the DMSO wells used for maximal deiodinase activity and the model inhibitor wells used for maximal inhibition of activity (see eg, plate map in Supplementary Figure 1). In each of the DIO assays, each chemical source plate was tested on three separate assay plates for n = 3 data points for each chemical.
Chemicals that produced less than 20% inhibition were considered ‘inactive’. Those chemicals that produced 20% inhibition or greater were considered putative deiodinase inhibitors. This 20% threshold was based on the background variability of the maximal activity calculated by three times the DMSO median absolute deviation (DMSO-MAD), which was 12.8, 13.4, and 14.0 across all replicates of all plates for DIO1, DIO2, and DIO3, respectively. Chemicals that produced 50% inhibition or greater were further tested in concentration-response; this level of inhibition had greater separation from the background variability (DMSO control) and is more likely to be biologically-relevant. For concentration-response testing, these chemicals were removed from the original chemical plate and added to new 96-well polypropylene plates (Corning). Dilutions in DMSO were made in this new plate so that they could be tested at final target concentrations of 200, 100, 20, 4.0, 0.8, 0.16, and 0.032 μM (see eg, plate map in Supplementary Figure 2). These concentration-response plates were tested on three separate assay plates for n=3 data points for each concentration of each chemical. Test chemical plates were sealed after use in assays with encapsulated pressure sensitive film (Phenix Research Products, Candler, North Carolina).
Data Processing and Analysis
Data were processed and analyzed using R (version 3.3.1; R Core Team 2016). Data from each plate were processed through an automated pipeline for data normalization, calculation of plate diagnostics, and assay-specific flags. Plate-wise normalization was based on the high concentration of the model inhibitor (200 μM PTU or XTH) and the solvent control (DMSO). Data were processed by the following steps: (1) determine the change in absorbance between the 1- and 10-min readings for each well; (2) calculate the net change in absorbance by then subtracting the mean background change in absorbance defined by the completely inhibited reaction (in six wells containing 200 μM model inhibitor); and (3) normalize to % of control by calculating as a % of the mean net change of the uninhibited reactions (in seven DMSO control wells). Percent inhibition was calculated as 100 minus the percent of the DMSO control reaction. For the single-point screening, median of the three replicates was calculated and results are reported as percent inhibition. The median % inhibition produced by each chemical was then compared across the three deiodinase assays. Chemicals were categorized as producing 50% inhibition or greater in one, two, or three of the deiodinase assays. Hierarchical clustering and heat mapping were used to visualize the similarities and differences in the inhibition produced in this single-point screening, with divisive analysis clustering and plotting with the R packages ‘cluster’ version 2.0.5 (Maechler et al., 2016) and ‘gplots’ version 3.0.1 (Warnes et al., 2016).
Concentration-response data were analyzed with the ToxCast Analysis Pipeline (tcpl) package version 1.0 (Filer et al., 2017) using percent inhibition as the response value and 20% inhibition as the threshold cutoff. This method uses all replicates for each concentration of a chemical to fit dose-response curves based on three models (constant, constrained Hill, and constrained gain-loss model) with the best model selected based on lowest Akaike Information Criterion (AIC) value. For chemicals that fit the robust Hill model, absolute IC20, absolute IC50, and Hill slope were calculated from the model fit parameters provided by the tcpl package. Here we display the concentration-response curves in the negative direction (inhibition from maximum response).
Assay quality/performance
To ensure quality and consistency over more than 100 assay plates run for each DIO assay, quality control measures were calculated for each assay plate. The DMSO control and 200 μM model inhibitor wells were used to evaluate variability of and separation between the positive (model inhibitor) and DMSO (solvent/negative) controls. Both a plate-wise DMSO-MAD and a plate-wise positive control median absolute deviation (200 μM-MAD) were calculated and reported in Table 1. The Z’ factor was calculated for each replicate of each assay plate. This metric reflects the dynamic range of the assay and the variability around the maximal and minimal response levels of the assay (Zhang et al., 1999). A Z’ factor of 0.5 or greater was used as a guideline for acceptable plate runs. If quality criteria were not met (eg, low Z’ factor, poor control data), assay plates were typically rerun to replace the data from the poor plate runs. In several instances, individual replicates of single concentration plates with Z’ factor less than 0.5 were not re-run because the other two replicates for those plates met the acceptable quality criteria. For the chemicals that were replicated across multiple plates in single-point screening, the median percent inhibition was compared across all the plates, with the median percent inhibition from testing on chemical source plates for ToxCast ph1v2 used in summaries and analyses.
Test chemical wells or single data points that fell outside of acceptable parameters were automatically flagged for manual review. Flags included high variability or outliers across replicates, no change in absorbance (typically due to a well not receiving one or more reagent), and potential assay interference indicated by absorbance outside of normal ranges. Wells were flagged for high variability when the absolute difference between the mean and median of the three replicate runs was greater than 10%. Potential issues with reagents or assay interference were flagged based on absorbance changing faster than the DMSO control (uninhibited reaction) or slower than the positive control (fully inhibited reaction), with wells flagged when: (a) change in absorbance was less than 0.1 absorbance units; (b) absorbance at one min was less than 85% of the DMSO control; or (c) absorbance at ten min was either less than 50% of the DMSO control or greater than 115% of the positive control. In addition, chemicals were flagged when the median response was greater than 190% or less than −20% of the DMSO control. Wells with observed problems (eg, low volume in a well for one replicate) were excluded from analyses with only two replicates used for these wells as long as variability between the two was less than 20%. Chemicals with high variability across replicates or other anomalous absorbance data were further evaluated with concentration-response testing. Chemicals with evidence of assay interference were excluded from summaries and analyses; such evidence included immediate change to near zero absorbance (typically an indication of the presence of high free iodide), no change in absorbance, and abnormal absorbance time course with values outside the expected range based on the high concentration of the model inhibitor and DMSO control; the 32 chemicals with evidence of assay interference are listed in Supplementary Table 5.
Results
Single-Point, High Concentration Screening
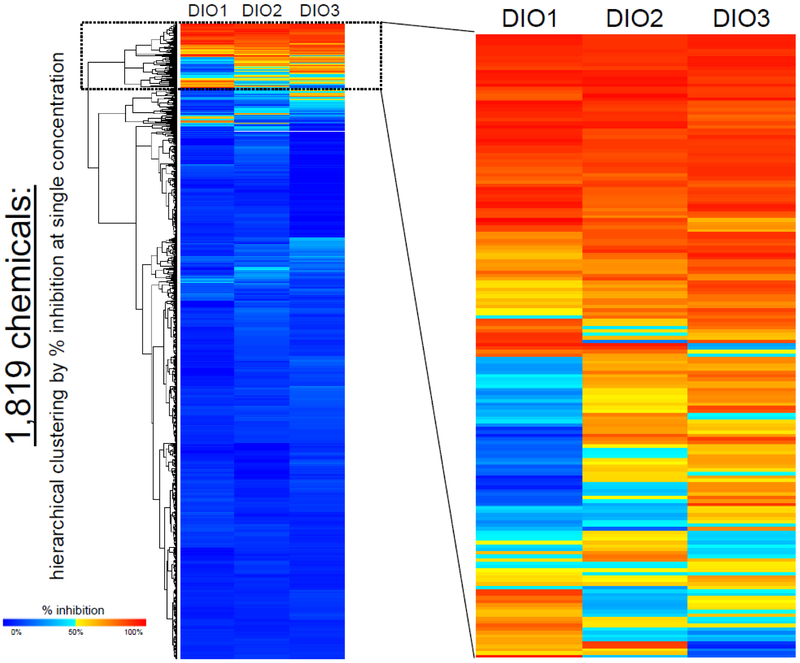
The three deiodinase assays (DIO1, DIO2, and DIO3) were used to assess the ToxCast ph1v2, ph2, and e1k chemical libraries first at a single target concentration of 200 μM (as permissible with solubility). Results and analysis reported here are for 1,819 chemicals due to evidence of assay interference by 32 of the 1,851 chemicals provided (described above in Assay quality/performance and listed in Supplementary Table 5). For each chemical, CASRN, the maximum concentration tested, and the median % inhibition produced in single-point screening are included in the Supplementary Table 6. Most of the chemicals tested did not inhibit deiodinase activity (Table 2, Figure 1). In this single concentration initial screen, 1,407 of the chemicals produced less than 20% inhibition in all three DIO assays compared with activity of the DMSO controls. There were 411 chemicals that produced greater than 20% inhibition, of which 228 chemicals produced greater than 50% inhibition in at least one of the DIO assays, which represent 22.5% and 12.5%, respectively, of the chemicals tested.
Table 2.
Summary of the single-point screening at an initial target concentration of 200 μM: percent and number of chemicals in each library that produced 20% and 50% inhibition.
| Deiodinase Type 1 (DIO1) |
Deiodinase Type 2 (DIO2) |
Deiodinase Type 3 (DIO3) |
|||||
|---|---|---|---|---|---|---|---|
| Chemical Library |
No. Chemicals Testeda |
% With ≥ 20% Inhibition (#) |
% With ≥ 50% Inhibition (#) |
% With ≥ 20% Inhibition (#) |
% With ≥ 50% Inhibition (#) |
% With ≥ 20% Inhibition (#) |
% With ≥ 50% Inhibition (#) |
|
ToxCast ph1_v2 |
290 | 16.9 % (49) |
6.5 % (19) |
18.6 % (54) |
8.6 % (25) |
19.7 % (57) |
9.7 % (28) |
|
ToxCast ph2 |
749b | 12.7 % (95) |
8.0 % (60) |
16.8 % (126) |
9.0 % (67) |
15.6 % (117) |
9.2 % (69) |
|
ToxCast e1k |
780 | 9.9 % (77) |
7.2 % (56) |
15.8 % (123) |
8.5 % (66) |
17.1 % (133) |
10.0 % (78) |
| Total | 1,819b |
12.1 % (221) |
7.4 % (135) |
16.7 % (303) |
8.7 % (158) |
16.9 % (307) |
9.6 % (175) |
The chemicals names, CAS Registry Number (CASRN), maximum tested concentrations, and median % inhibition produced in each of the three deiodinase assays can be found in Supplementary Table 6.
Chemical plates received from ToxCast included 1,851 unique chemicals, however 32 compounds had evidence of interfering with one or more of the deiodinase assays and thus were excluded from all summaries and analyses.
One chemical from ToxCast ph2 was only tested in DIO1 because it was depleted from the chemical inventory, thus a total of 1,818 chemicals were tested in the DIO2 and DIO3 assays.
Figure 1.
Deiodinase inhibition produced by the 1,819 chemicals tested in single-point screening (target concentration of 200 μM), ordered by hierarchical clustering (divisive analysis method), with expanded view of 180 chemicals. Median of n = 3 replicates. The tested concentration and median % inhibition values for each chemical are in Supplementary Table 6.
Concentration-Response Screening
A total of 240 chemicals were tested in concentration-response mode, with 142 chemicals tested in the DIO1 assay and 213 chemicals tested in the DIO2 and DIO3 assays. This concentration-response testing included the chemicals that produced 50% or greater inhibition in the single-point screening in each assay (see summary in Table 2) as well as a subset of those chemicals that produced less than 50% inhibition. These subsets were included as an additional quality control for intra-assay consistency and consisted of 7, 55, and 38 chemicals, respectively, for DIO1, DIO2, and DIO3. For the DIO1 assay, this subset included chemicals with high variability or other issues that were encountered in single-point screening. Because the DIO2 and DIO3 assays were run in parallel, these subsets included the chemicals that produced inhibition of 50% or greater in only one of these two assays. Supplementary Table 7 includes all chemicals that were tested in concentration-response, with sources, chemical name, CASRN, the maximum concentration tested, median % inhibition produced at the maximum tested concentration, and Hill model fit parameters, where appropriate.
The concentration-response results confirmed the single-point results for over 97% of these chemicals. Results were considered consistent when there was less than 25% difference between the median inhibition produced in the single-point screening and that produced at the maximum concentration tested in concentration-response, and the chemical was categorized as ‘inactive’ or a putative deiodinase inhibitor in both tests. In the DIO1 assay, the concentration-response results were consistent with single-point results for 139 of the 142 chemicals tested and only one chemical (chlorophene) with a difference in categorization. Chlorophene had high variability across the multiple single-point plates on which it was tested, producing a range of inhibition in from 11% to 47% (Supplementary Figure 3), but produced less than 20% inhibition in concentration-response testing. In the DIO2 assay, 209 of the 213 chemicals had consistent single-point and concentration-response results, with three chemicals (1-dodecanol, 1-tridecanol, and 1-dodecanamine) that produced 35% inhibition or greater in single-point screening and less than 7% inhibition in concentration-response testing. In the DIO3 assay, concentration-response results for 207 of the 213 chemicals were consistent with the single-point screening results. The six chemicals with differences in the DIO3 assay all consistently produced inhibition greater than 20% and the single-point and concentration-response results differed by 26%-40% (eg, 29% vs 55% inhibition or 67% vs 97% inhibition). Several chemicals were identified as potential false negatives in the DIO2 and DIO3 assays, where the chemical produced less than 20% inhibition in the single-point screening but 20% or greater in the concentration-response testing. However, the maximum % inhibition produced by these chemicals was just above the 20% cut-off and, therefore, these chemicals were considered only borderline active.
The inhibition curves for all chemicals tested in concentration-response mode are available in Supplementary Figure 5 with percent inhibition at maximum tested concentration and Hill model results (Hill slope, absolute IC20, and absolute IC50, when applicable) included in Supplementary Table 7. Hill model parameters are reported for all chemicals that produced inhibition of 20% or greater at the maximum concentration tested in concentration-response; this model was identified in the ToxCast pipeline as the best fit model for most of these chemicals (135 of the 136 chemicals in DIO1, all 192 chemicals in DIO2, and 196 of the 199 chemicals in DIO3) and an acceptable model fit for the rest of these chemicals
For each assay, the top 25 ranked chemicals, based on absolute IC50, are included in Tables 3–5. In the DIO1 assay, nine chemicals had IC50 lower or comparable with the mean IC50 for the control PTU (5.4 μM), including the ToxCast plated PTU and the known DIO1 inhibitor genistein. In the DIO2 assay, no chemical produced an IC50 near that of the mean IC50 for the control chemical XTH (0.8 μM), with the closest chemicals at 2.2 μM (fluazinam), 2.9 μM (triflumizole), and 3.7 μM (bisphenol A diglycidyl ether). In the DIO3 assay, there were two chemicals (nordihyroguaiaretic acid and chlorothalonil) with an IC50 lower or comparable with that of the control XTH (0.3 μM).
Table 3.
The top 25 ranked chemicals for inhibition of deiodinase type 1 (DIO1), based on absolute IC50, with activity across the deiodinases (DIOs) indicating which chemicals produced 50% inhibition or greater in each of the deiodinase assays in single-point screening.
| DIO1 |
Across DIOs |
||||||
|---|---|---|---|---|---|---|---|
| Rank | Chemical | CASRN | Max Tested Conc., μM |
% Inhibition at Max Conc.a |
IC20 (μM) |
IC50 (μM) |
≥ 50% Inhibition |
| 1 | 4,5-Dichloro-3H-1,2-dithiol-3-one | 1192-52-5 | 200 | 100 | 0.1 | 0.2 | DIO 1, 2, 3 |
| 2 | 4,5-Dichloro-2-octyl-3(2H)-isothiazolone | 64359-81-5 | 200 | 99 | 0.2 | 0.4 | DIO 1, 2, 3 |
| 3 | C.I. Direct Yellow 12 | 2870-32-8 | 200 | 103 | 0.2 | 0.8 | DIO 1, 2, 3 |
| 4 | Bisphenol A diglycidyl ether | 1675-54-3 | 200 | 104 | 0.4 | 1.3 | DIO 1, 2, 3 |
| 5 | Temephos | 3383-96-8 | 200 | 93 | 0.1 | 1.3 | DIO 1 |
| 6 | Genisteinb | 446-72-0 | 200 | 95 | 0.8 | 2.6 | DIO 1 |
| 7 | 6-Hydroxy-2-naphthyl disulfide | 6088-51-3 | 200 | 93 | 0.5 | 2.7 | DIO 1, 2, 3 |
| 8 | 6-Propyl-2-thiouracilb | 51-52-5 | 200 | 101 | 1.1 | 3.8 | DIO 1 |
| 9 | C12-14-Alkyl glycidyl ether | 68609-97-2 | 119 | 65 | 0.8 | 4.1 | DIO 1, 2 |
| 10 | Darbufelone mesylate | 139340-56-0 | 100 | 74 | 1.4 | 4.7 | DIO 1, 2, 3 |
| 11 | Morin hydrate | 654055-01-3 | 200 | 94 | 0.6 | 7.4 | DIO 1, 3 |
| 12 | Linoleic acid | 60-33-3 | 200 | 96 | 0.7 | 10.4 | DIO 1, 2, 3 |
| 13 | Nordihydroguaiaretic acid | 500-38-9 | 200 | 80 | 1.6 | 11.0 | DIO 1, 2, 3 |
| 14 | Tributyltetradecylphosphonium chloride | 81741-28-8 | 200 | 94 | 5.5 | 12.4 | DIO 1, 2, 3 |
| 15 | Captafol | 2425-06-1 | 200 | 94 | 4.8 | 12.9 | DIO 1, 2, 3 |
| 16 | Hexadecyltrimethylammonium bromide | 57-09-0 | 200 | 96 | 5.9 | 12.9 | DIO 1, 2, 3 |
| 17 | Chlorhexidine diacetate | 56-95-1 | 200 | 87 | 1.2 | 13.0 | DIO 1, 2, 3 |
| 18 | Zoxamide | 156052-68-5 | 200 | 98 | 8.2 | 17.0 | DIO 1, 2, 3 |
| 19 | Didecyldimethylammonium chloride | 7173-51-5 | 200 | 106 | 4.9 | 17.1 | DIO 1, 2, 3 |
| 20 | Tannic acid | 1401-55-4 | 60 | 96 | 9.7 | 17.4 | DIO 1, 2, 3 |
| 21 | 6-Methyl-2-thiouracilb | 56-04-2 | 200 | 88 | 3.9 | 18.4 | DIO 1 |
| 22 | Triflumizole | 68694-11-1 | 200 | 74 | 13.4 | 18.8 | DIO 1, 2, 3 |
| 23 | 2-Chloro-N-phenylacetamide | 587-65-5 | 200 | 105 | 7.0 | 18.8 | DIO 1 |
| 24 | 2-Ethylhexyl acrylate | 103-11-7 | 200 | 88 | 3.9 | 19.4 | DIO 1, 2, 3 |
| 25 | Linolenic acid | 463-40-1 | 200 | 94 | 5.5 | 20.0 | DIO 1, 2, 3 |
Median of three replicates at maximum concentration of chemical in concentration-response testing; inhibition in single-point results differed by 15% or less.
Previously reported to inhibit DIO1 (Renko et al., 2015; Visser et al., 1992; Wassen et al., 2004).
Table 5.
The top 25 ranked chemicals for inhibition of deiodinase type 3 (DIO3), based on absolute IC50, with activity across the deiodinases (DIOs) indicating which chemicals produced 50% inhibition or greater in each of the deiodinase assays in single-point screening.
| DIO3 |
Across DIOs |
||||||
|---|---|---|---|---|---|---|---|
| Rank | Chemical | CASRN | Max Tested Conc., μM |
% Inhibition at Max Conc.a |
IC20 (μM) |
IC50 (μM) |
≥ 50% Inhibition |
| 1 | Nordihydroguaiaretic acid | 500-38-9 | 200 | 97 | 0.01 | 0.1 | DIO 1, 2, 3 |
| 2 | Chlorothalonil | 1897-45-6 | 200 | 76 | 0.1 | 0.5 | DIO 1, 2, 3 |
| 3 | Pentachloropyridine | 2176-62-7 | 200 | 98 | 0.2 | 1.0 | DIO 2, 3 |
| 4 | Fipronil | 120068-37-3 | 200 | 101 | 0.3 | 1.2 | DIO 1, 2, 3 |
| 5 | Quinoxyfen | 124495-18-7 | 200 | 88 | 0.2 | 1.3 | DIO 2, 3 |
| 6 | Hexadecyltrimethylammonium bromide | 57-09-0 | 200 | 97 | 1.0 | 1.6 | DIO 1, 2, 3 |
| 7 | AVE6324 | NOCAS_47377 | 200 | 86 | 0.4 | 1.8 | DIO 1, 2, 3 |
| 8 | Fluazinam | 79622-59-6 | 180 | 101 | 0.4 | 2.0 | DIO 1, 2, 3 |
| 9 | Triflumizole | 68694-11-1 | 200 | 102 | 0.5 | 2.3 | DIO 1, 2, 3 |
| 10 | Ergocalciferol | 50-14-6 | 200 | 95 | 0.5 | 2.6 | DIO 1, 2, 3 |
| 11 | Chlorhexidine diacetate | 56-95-1 | 200 | 103 | 1.3 | 2.6 | DIO 1, 2, 3 |
| 12 | 6-Hydroxy-2-naphthyl disulfide | 6088-51-3 | 200 | 102 | 0.6 | 3.1 | DIO 1, 2, 3 |
| 13 | Octadecyl sulfate sodium salt | 1120-04-3 | 200 | 93 | 1.7 | 3.3 | DIO 1, 2, 3 |
| 14 | Hexachlorocyclopentadiene | 77-47-4 | 200 | 99 | 0.7 | 3.3 | DIO 1, 2, 3 |
| 15 | Didecyldimethylammonium chloride | 7173-51-5 | 200 | 100 | 2.8 | 3.3 | DIO 1, 2, 3 |
| 16 | 4,5-Dichloro-3H-1,2-dithiol-3-one | 1192-52-5 | 200 | 103 | 0.7 | 3.3 | DIO 1, 2, 3 |
| 17 | Tributyltetradecylphosphonium chloride | 81741-28-8 | 200 | 81 | 2.8 | 3.3 | DIO 1, 2, 3 |
| 18 | 1,2-Benzenedicarboxaldehyde | 643-79-8 | 200 | 93 | 0.7 | 3.5 | DIO 1, 2, 3 |
| 19 | Methyltrioctylammonium chloride | 5137-55-3 | 200 | 89 | 3.2 | 3.8 | DIO 1, 2, 3 |
| 20 | 2-(8-Heptadecenyl)-2-imidazoline-1-ethanol | 95-38-5 | 200 | 101 | 3.3 | 3.9 | DIO 1, 2, 3 |
| 21 | Bisphenol A diglycidyl ether | 1675-54-3 | 200 | 94 | 0.9 | 4.1 | DIO 1, 2, 3 |
| 22 | Ascorbyl palmitate | 137-66-6 | 200 | 98 | 1.2 | 4.2 | DIO 1, 2, 3 |
| 23 | Kepone | 143-50-0 | 200 | 100 | 2.2 | 4.4 | DIO 1, 2, 3 |
| 24 | 2-Benzylideneoctanal | 101-86-0 | 200 | 84 | 1.1 | 4.7 | DIO 2, 3 |
| 25 | SSR69071 | 344930-95-6 | 50 | 90 | 1.6 | 5.1 | DIO 2, 3 |
Median of three replicates at maximum concentration of chemical in concentration-response testing; inhibition in single-point results differed by 11% or less except quinoxyfen, which produced 49.4% inhibition, and 2-benzylideneoctanal which produced 65.0% inhibition.
Comparison across deiodinases
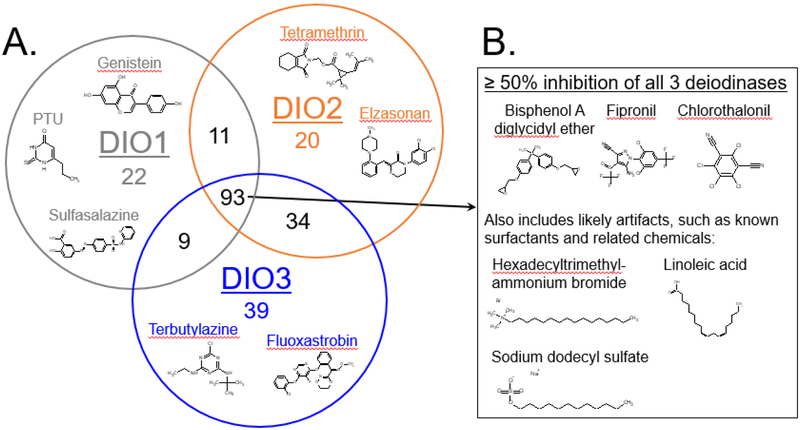
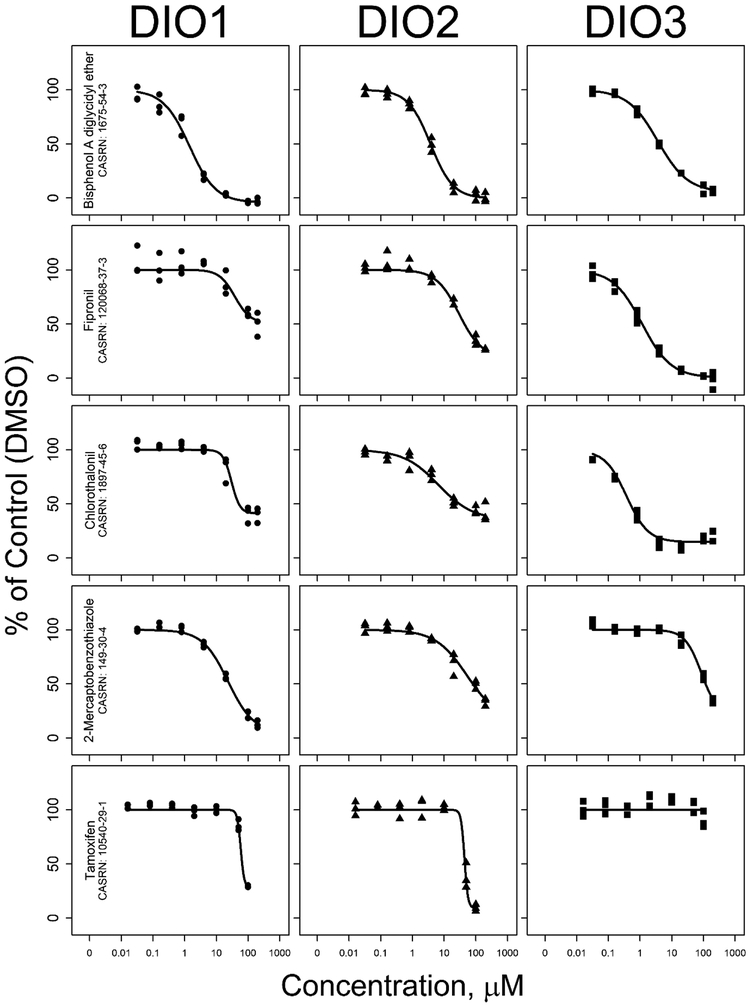
The inhibition results were similar across the three deiodinases for over 90% of the tested chemicals. In single-point screening, 1,590 chemicals produced less than 50% inhibition in all three assays (Supplementary Table 6). Of the 228 chemicals that produced inhibition of 50% or greater in one or more of the assays, 93 chemicals produced this result in all three assays (Figure 2). This comparison also identified 54 chemicals that inhibited only two of the three deiodinases (DIO1/DIO2 = 11, DIO2/DIO3 = 34, and DIO1/DIO3 = 9 chemicals) and isoform-specific inhibition by 81 chemicals that produced inhibition of 50% or greater in only one of the three deiodinase assays (DIO1=22, DIO2=20, and DIO3=39 chemicals). Concentration-response testing also showed similarities and differences in chemical inhibition of the three deiodinases, with inhibition curve shapes nearly identical for some and differing greatly for other chemicals, as shown with five example chemicals in Figure 3.
Figure 2.
A, The number of chemicals that produced 50% inhibition or greater in one or more deiodinase assay (Venn diagram) with examples that produced ≥ 50% inhibition specific to a single enzyme; and B, examples that produced ≥ 50% inhibition in all three deiodinases.
Figure 3.
Concentration-response curves for deiodinase inhibition by five example compounds, tested at seven concentrations.
Assay performance and quality control
Performance of the assay was monitored in each assay plate through Z’ factor, variability of the positive and solvent controls, and IC50 for the model inhibitor (Table 1). All three assays performed reproducibly with excellent dynamic range, low variability of the control chemicals, and consistent responses of the model inhibitor concentration-response curves on each plate. The Z’ factor was generally near 0.7 for individual replicates of each assay plate and mean Z’ factor was above 0.5 for all single-point and concentration-response plates, with the exception of one single-point DIO2 plate (mean Z’ factor = 0.4). The plate-wise MADs for the DMSO control and the 200 μM model inhibitor wells were 10% or less for nearly all of assay plates (93% of DIO1 and 95% of DIO2 and DIO3 assay plates). The concentration-response inhibition curves for the model inhibitors were consistent across plates within each assay (see eg, in Supplementary Figure 4). The IC50 values ranges were 3.1–8.9 μM PTU for DIO1, 0.4–1.1 μM XTH for DIO2, and 0.1–0.9 μM XTH for DIO3.
The ten chemicals used for intra-assay reproducibility had very consistent results across multiple plates (Supplementary Figure 3). In each assay, there were four to six chemicals that consistently produced less than 20% inhibition and two to five chemicals that produced inhibition of 20% or greater. These chemicals had highly reproducible results with the exceptions of chlorophene and PFOS in DIO1, which could possibly be explained by chemical degradation or different lots of a chemical provided on the chemical source plates.
Chemicals with potential assay interference
The 32 chemicals that were flagged as potentially interfering in these DIO assays are listed in the Supplementary Table 5. The assay results for these chemicals fell outside of acceptable parameters as described above (in the Materials and Methods section). Several of these chemicals are from chemical classes with known interferences with the SK reaction (eg, thiocyanate, Sandell and Kolthoff, 1937). Iodine-containing chemicals may act as substrates for deiodinases (eg, iopanoic acid, Renko et al., 2012) or may have free iodide that causes false results in these assay (eg, sodium iodide). These chemicals were excluded from summaries and analyses; further evaluation of the source of interference in each of these chemicals was outside of the scope of this study.
Discussion
The extensive screening effort presented here identified 411 putative deiodinase inhibitors through testing 1,819 chemicals at a single, high concentration in recently developed 96-well plate assays described in Hornung et al. (2018) for DIO1 and here for DIO2 and DIO3. Of these, 228 chemicals produced inhibition of 50% or greater in one or more of the deiodinases including chemicals that, to our knowledge, have not previously been known to inhibit deiodinases. In addition, the single-point and concentration-response results suggest differential inhibition with varied maximum inhibition and potency for some chemicals when compared across DIO1, DIO2, and DIO3.
The consistent performance in these assays demonstrated their reproducibility and fitness for screening and prioritizing chemicals for potential thyroid disruption. Across multiple months of screening, each assay maintained low variability in controls, reproducible inhibition curves for model inhibitors, high Z’ factors, and reliable results across replicated chemicals. Although there are currently no established reference chemicals for deiodinase inhibition, the results from these assays are consistent with earlier studies. PTU, methylthiouracil, and genistein produced DIO1-specific inhibition, aurothioglucose and xanthohumol inhibited all three DIO isoforms (data not shown for DIO1), and previously confirmed non-inhibitors such as methimazole and bisphenol A were also identified as inactive in these assays (Renko et al., 2015; Tuarog et al., 1994; Visser et al., 1992; Wassen, et al., 2004). In addition, these assay results match studies that identified some halogenated phenolic compounds (Butt et al., 2011) and flavonoids (Auf’mkolk et al., 1986; Ferreira et al., 2002; Spanka et al., 1990) as inhibitors of DIO1 enzyme activity.
Results from in vitro screening assays have limitations and uncertainties inherent to them, including nonspecific chemical activity and assay interference (Judson et al., 2013, 2016; Thorne et al., 2010). Chemicals producing disruption by these means could be identified through a comparison across multiple in vitro assays. Of the 93 chemicals with greater than 50% inhibition in all three deiodinases assays, over 20 chemicals were identified as surfactants (eg, sodium dodecyl sulfate, hexadecyltrimethyl-ammonium bromide) or related chemicals (eg, linoleic acid), which may disrupt membranes, the test system, or be related to nonspecific enzyme inhibition. However, this set of chemicals that produced greater than 50% inhibition in all three assays included a range of chemical types and further analysis is required to differentiate false positives from specific deiodinase inhibition. The screening strategy of initial testing with a single concentration has a potential risk of false negatives, as active compounds can only be defined based on highest concentration tested. Ideally, the tested concentration would be consistent across all chemicals; however chemical solubility and purity, and protein sequestration of chemical could alter actual tested concentrations. In these deiodinase assays, 96% of the chemicals were tested in single-point at or near the target concentration of 200 μM (150 – 210 μM) and efforts were taken to reduce the possibility of chemical degradation (eg, limiting number of freeze/thaw cycles); however, 19 chemicals were initially tested at less than 100 μM (see Supplementary Table 6 for tested concentrations of all chemicals). We also employed a set of interference flags specific to these deiodinase assays to identify chemicals that were producing false positives/negatives. With this data-based approach, we identified 32 compounds with evidence of interfering with one or more of the assays, as described in the Results section above and listed in Supplementary Table 5. This set of interfering compounds included both potential false negatives and false positives, all of which were excluded from assay results and summaries. However, further testing with other methods (eg, monitoring for changes in substrate with liquid chromatography-tandem mass spectrometry) is warranted for these chemicals, as several are suspected thyroid disruptors or have been previously documented to produce inhibition in deiodinases (eg, Tetrac, FD&C Red 3, amiodarone hydrochloride, iopanoic acid) (Bartalena et al., 2018; Braga and Cooper, 2001; Capen and Martin, 1989; Koehrle et al., 1986; Renko et al. 2012, 2015; Rosene et al., 2010).
These data provide the most comprehensive examination of chemical inhibition of deiodinase activity currently available, greatly expanding on previous studies that included ten to fifteen compounds for one or more deiodinases (eg, Butt et al., 2011; Ferreira et al., 2002; Renko et al., 2015), as well as our recent report on approximately 300 chemicals for inhibition of DIO1 (Hornung et al. 2018). The chemicals that produced maximum inhibition and potency in each assay are of most interest. In the top 25 chemicals in each assay, we identified 12 chemicals for DIO1 and 2 chemicals each for DIO2 and DIO3 that exhibited potency similar or greater than the model inhibitors based on IC50. Few of these most potent chemicals, however, have been reported in the literature to inhibit deiodinases. The few chemicals previously reported to produce deiodinase inhibition were only on the list of potent chemicals in the DIO1 assay. These included the well-documented inhibition of DIO1 produced by genistein, PTU, and 6-methyl-2-thiouracil (Kaplan and Utiger, 1978; Renko et al., 2015; Schweizer and Steegborn, 2015; Visser et al., 1992; Wassen et al., 2004). In addition, Chopra et al. (1985) reported inhibition of the conversion of T4 to T3 by fatty acids, including linoleic acid, and Ferreira et al. (2002) demonstrated that morin reduced thyroid DIO1 activity. Previous reports of deiodinase inhibition are limited for these most potent chemicals; however, several of the potent deiodinase inhibitors in this study have been documented to affect thyroid axis-endpoints. Two examples are fipronil, which is suspected to enhance hepatic metabolism and excretion of TH (Hurley et al., 1998; Leghait et al., 2009), and nordihydroguaiaretic acid, which has been shown to inhibit growth of thyroid cells (Gartner et al. 1985). While a comprehensive review of the literature for potential thyroid disruption by the chemicals producing deiodinase inhibition in these assays is warranted, it is beyond the scope of this study. In addition, targeted in vivo testing is needed to expand the limited understanding of how in vitro inhibition assay results relate to effects of inhibition of deiodinase activity in vivo.
Results from this set of three assays provide a unique opportunity to compare chemical inhibition across the deiodinase isoforms for many chemicals. In the set of 93 chemicals that produced inhibition of 50% or greater in all three assays, we recognized the potential for nonspecific enzyme inhibition. However, activity across multiple deiodinase assays could also be used as a method to prioritize chemicals for further investigation as chemicals disrupting multiple molecular targets on the thyroid-axis could result in more extreme adverse organismal outcomes. This will be especially informative when it is possible to conduct a comprehensive comparison across a suite of thyroid-relevant in vitro assays. This could include the deiodinase results reported here, the recently completed screening for NIS inhibition (Wang et al., 2018), and the TPO, TH receptor transactivation, and thyrotropin releasing hormone results previously completed for these chemical libraries (Martin et al., 2010; Paul Friedman et al., 2016; Sipes et al., 2013). Chemicals with differences across the deiodinases are also of great interest, especially the 81 chemicals that produced inhibition of 50% or greater in only one of the three deiodinases. This set of assays identified 22 chemicals that produced inhibition of 50% or greater in DIO1 but not in DIO2 or DIO3, including the known DIO1-specific inhibitors PTU, 6-methyl-2-thiouracil, and genistein (Renko et al., 2015; Schweizer and Steegborn, 2015; Wassen et al., 2004). In addition, 20 chemicals were identified as possible DIO2-specific inhibitors, and 39 chemicals were identified as possible DIO3-specific inhibitors. An initial examination of classes of the chemicals with differential inhibition showed potentially interesting patterns. For example, 11 of the 23 chemicals classified as triazines (aromatic or aliphatic) in ECOSAR version 2.0 (U.S. EPA, 2017b) produced inhibition greater than 20% in DIO3 but not DIO1 or DIO2, and four of these chemicals produced DIO3-specific inhibition greater than 50%. As another example, the three closely related tamoxifen compounds (tamoxifen, tamoxifen citrate, and 4-hydroxytamoxifen) each produced almost complete inhibition in DIO1 and DIO2, but not DIO3 (Figure 3). To our knowledge, these chemical class-specific differences in deiodinase inhibition have not previously been reported and provide the impetus for the development of structure-activity relationship models.
In summary, this set of three deiodinase inhibition assays is a significant contribution towards expanding the limited number of in vitro assays used to identify chemicals having the potential to interfere with TH homeostasis. Of the 1,819 chemicals tested, 22% were identified as putative deiodinase inhibitors, with between 12% and 17% in each assay. This study sets the groundwork for development and evaluation of structure-activity relationships for deiodinase inhibition, and informs targeted selection of chemicals for further testing to evaluate effects on THs and identify adverse outcomes of deiodinase inhibition.
Supplementary Material
Table 4.
The top 25 ranked chemicals for inhibition of deiodinase type 2 (DIO2), based on absolute IC50, with activity across the deiodinases (DIOs) indicating which chemicals produced 50% inhibition or greater in each of the deiodinase assays in single-point testing.
| DIO2 |
Across DIOs |
||||||
|---|---|---|---|---|---|---|---|
| Rank | Chemical | CASRN | Max Tested Conc., μM |
% Inhibition at Max Conc.a |
IC20 (μM) |
IC50 (μM) |
≥ 50% Inhibition |
| 1 | Fluazinam | 79622-59-6 | 180 | 100 | 0.5 | 2.2 | DIO 1, 2, 3 |
| 2 | Triflumizole | 68694-11-1 | 200 | 98 | 0.7 | 2.9 | DIO 1, 2, 3 |
| 3 | Bisphenol A diglycidyl ether | 1675-54-3 | 200 | 101 | 1.2 | 3.7 | DIO 1, 2, 3 |
| 4 | Octhilinone | 26530-20-1 | 200 | 79 | 3.4 | 12.2 | DIO 1, 2, 3 |
| 5 | Zoxamide | 156052-68-5 | 200 | 95 | 3.1 | 12.9 | DIO 1, 2, 3 |
| 6 | Chlorhexidine diacetate | 56-95-1 | 200 | 99 | 5.7 | 13.1 | DIO 1, 2, 3 |
| 7 | SSR69071 | 344930-95-6 | 50 | 77 | 4.5 | 13.8 | DIO 2, 3 |
| 8 | 4,5-Dichloro-3H-1,2-dithiol-3-one | 1192-52-5 | 200 | 92 | 3.0 | 15.6 | DIO 1, 2, 3 |
| 9 | 4,5-Dichloro-2-octyl-3(2H)-isothiazolone | 64359-81-5 | 200 | 95 | 2.4 | 16.9 | DIO 1, 2, 3 |
| 10 | Tributyltetradecylphosphonium chloride | 81741-28-8 | 200 | 97 | 9.0 | 17.8 | DIO 1, 2, 3 |
| 11 | Dinocap | 39300-45-3 | 200 | 93 | 3.2 | 18.2 | DIO 1, 2, 3 |
| 12 | (Dicyclopentadienyloxy)ethyl methacrylate | 68169-03-9 | 200 | 83 | 4.4 | 18.4 | DIO 2, 3 |
| 13 | Isooctyl acrylate | 29590-42-9 | 200 | 96 | 4.6 | 18.7 | DIO 1, 2, 3 |
| 14 | Quinoxyfen | 124495-18-7 | 200 | 63 | 4.9 | 18.7 | DIO 2, 3 |
| 15 | Hydramethylnon | 67485-29-4 | 200 | 86 | 16.3 | 19.8 | DIO 2 |
| 16 | Hexadecyltrimethylammonium bromide | 57-09-0 | 200 | 101 | 16.6 | 19.9 | DIO 1, 2, 3 |
| 17 | 2-Ethylhexyl acrylate | 103-11-7 | 200 | 88 | 3.0 | 20.1 | DIO 1, 2, 3 |
| 18 | Methyltrioctylammonium chloride | 5137-55-3 | 200 | 99 | 12.4 | 20.8 | DIO 1, 2, 3 |
| 19 | 6-Hydroxy-2-naphthyl disulfide | 6088-51-3 | 200 | 85 | 2.7 | 21.1 | DIO 1, 2, 3 |
| 20 | Benzalkonium chloride | 8001-54-5 | 200 | 100 | 16.7 | 22.5 | DIO 1, 2, 3 |
| 21 | Dichlone | 117-80-6 | 200 | 94 | 4.0 | 22.6 | DIO 1, 2, 3 |
| 22 | AVE5638 | 725228-45-5 | 200 | 96 | 5.0 | 22.8 | DIO 1, 2, 3 |
| 23 | Hexachlorocyclopentadiene | 77-47-4 | 200 | 87 | 6.0 | 25.5 | DIO 1, 2, 3 |
| 24 | Calcium dodecylbenzene sulfonate | 26264-06-2 | 200 | 94 | 16.5 | 25.6 | DIO 1, 2, 3 |
| 25 | Ascorbyl palmitate | 137-66-6 | 200 | 97 | 6.9 | 26.4 | DIO 1, 2, 3 |
Median of three replicates at maximum concentration of chemical in concentration-response testing; inhibition in single-point results differed by 12% or less, except for dinocap, which produced 49.4% inhibition.
Acknowledgements:
The authors thank Dr. P. Reed Larson for sharing plasmids for DIO1, DIO2, and DIO3 as well as the gift of rat DIO enzyme for assay development. We also thank Dr. Kostja Renko for advice and the gift of DIO enzyme that aided the methods optimization in our lab. Additionally, we thank Dr. Ann Richard, Katherine Coutros, and Dr. Chris Grulke (U.S. EPA, National Center for Computational Toxicology) for their valuable role in obtaining the ToxCast chemical libraries for screening and Dr. Katie Paul Friedman (U.S. EPA, National Center for Computational Toxicology) for guidance on ToxCast pipeline data analysis. The authors thank Dr. Susan Laws (U.S. EPA, National Health and Environmental Effects Research Laboratory) for providing comments on an early draft of the manuscript.
Funding Information: This work was supported by the US Environmental Protection Agency.
Footnotes
Disclaimer: The views expressed in this paper are those of the authors and do not necessarily reflect the views or policies of the U.S. Environmental Protection Agency. Mention of trade names or commercial products does not constitute endorsement or recommendation for use.
Conflict of Interest: The authors claim no conflicts of interest.
Supplementary Data: Supplementary data are available at Toxicological Sciences online.
References.
- Auf’mkolk M, Koehrle J, Hesch R-D, and Cody V (1986). Inhibition of rat liver iodothyronine deiodinase. J. Biol. Chem 261, 11623–11630. [PubMed] [Google Scholar]
- Bartalena L, Bogazzi F, Chiovato L, Hubalewska-Dydejczyk A, Links TP, and Vanderpump M (2018). 2018 European Thyroid Association (ETA) guidelines for the management of amiodarone-associated thyroid dysfunction. Eur. Thyroid J 7, 55–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braga M, and Cooper DS (2001). Clinical Review 129. Oral cholecystographic agents and the thyroid. J. Clin. Endocrinol. Metab 86, 1853–1860. [DOI] [PubMed] [Google Scholar]
- Boas M, Feldt-Rasmussen U, and Main KM (2012). Thyroid effects of endocrine disrupting chemicals. Mol. Cell. Endocrinol 355, 240–248. [DOI] [PubMed] [Google Scholar]
- Brix K, Fuhrer D, and Biebermann H (2011). Molecules important for thyroid hormone synthesis and action – known facts and future perspectives. Thyroid Res. 4 (Suppl 1), S9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brucker-Davis F (1998). Effects of environmental synthetic chemicals on thyroid function. Thyroid 8, 827–856. [DOI] [PubMed] [Google Scholar]
- Buettner C, Harney JW, and Larsen PR (2000). The role of selenocysteine 133 in catalysis by the human type 2 iodothyronine deiodinase. Endocrinology 141, 4606–4612 [DOI] [PubMed] [Google Scholar]
- Butt CM, Wang DL, and Stapleton HM (2011). Halogenated phenolic contaminants inhibit the in vitro activity of the thyroid-regulating deiodinases in human liver. Toxicol. Sci 124, 339–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capen CC, and Martin SL (1989). The effects of xenobiotics on the structure and function of thyroid follicular and C-cells. Toxicol. Pathol 17, 266–293. [DOI] [PubMed] [Google Scholar]
- Casula S, and Bianco AC (2012). Thyroid hormone deiodinases and cancer. Front. Endocrinol 3, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chopra IJ, Huang T-S, Beredo A, Solomon DH, Chua Teco GN, and Mead JF (1985). Evidence for an inhibitor of extrathyroidal conversion of thyroxine to 3,5,3’-triiodothyronine in sera of patients with nonthyroidal illness. J. Clin. Endocr. Metab 60, 666–672. [DOI] [PubMed] [Google Scholar]
- Darras VM, and Van Herck SLJ (2012). Iodothyronine deiodinase structure and function: from ascidians to humans. J. Endocrinol 215, 189–206. [DOI] [PubMed] [Google Scholar]
- Dong H, and Wade MG (2017). Application of a nonradioactive assay for high throughput screening for inhibition of thyroid hormone uptake via the transmembrane transporter MCT8. Toxicol. In Vitro 40, 234–242. [DOI] [PubMed] [Google Scholar]
- Duntas LH, and Stathatos N (2015). Toxic chemicals and thyroid function: hard facts and lateral thinking. Rev. Endocr. Metab. Disord 16, 311–318. [DOI] [PubMed] [Google Scholar]
- Ferreira ACF, Lisboa PC, Oliveira KJ, Lima LP, Barros IA, and Carvalho DP (2002). Inhibition of thyroid type 1 deiodinase activity by flavonoids. Food Chem. Toxicol 40, 913–917. [DOI] [PubMed] [Google Scholar]
- Filer DL, Kothiya P, Setzer RW, Judson RS, and Martin MT (2017). tcpl: the ToxCast pipeline for high-throughput screening data. Bioinformatics 33, 618–620. [DOI] [PubMed] [Google Scholar]
- Gartner R, Greil W, Demharter R, and Horn K (1985). Involvement of cyclic AMP, iodide and metabolites of arachidonic acid in regulation of cell proliferation of isolated porcine thyroid follicles. Mol. Cell. Endocrinol 42, 145–155. [DOI] [PubMed] [Google Scholar]
- Gereben B, Zavacki AM, Ribich S, Kim BW, Huang SA, Simonides WS, Zeold A, and Bianco AC (2008). Cellular and molecular basis of deiodinase-regulated thyroid hormone signaling. Endocr. Rev 29, 898–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham FL (2000). Adenovirus vectors for high-efficiency gene transfer into mammalian cells. Immunol. Today 21, 426–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham FL, and Prevec L (1991). Manipulation of adenovirus vectors In Methods in Molecular Biology (Murray EJ, Ed.) pp. 109–128. The Humana Press, Inc: Clifton, NJ, USA. [DOI] [PubMed] [Google Scholar]
- Hallinger DR, Murr AS, Buckalew AR, Simmons SO, Stoker TE, and Laws SE (2017). Development of a screening approach to detect thyroid disrupting chemicals that inhibit the human sodium iodide symporter (NIS). Toxicol. In Vitro 40, 66–78. [DOI] [PubMed] [Google Scholar]
- Hernandez A, Martinez ME, Fiering S, Galton VA, and St. Germain D (2006). Type 3 deiodinase is critical for the maturation and function of the thyroid axis. J. Clin. Invest 116, 476–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitt MM, Ng P, and Graham FL (1994). Construction and Propagation of Human Adenovirus vectors In Cell Biology: A Laboratory Handbook (Celis JE, Ed.) Vol 1, pp 479–490. Academic Press: San Diego, CA, USA. [Google Scholar]
- Hornung MW, Korte JJ, Olker JH, Denny JS, Knutsen C, Hartig PC, Cardon MC, and Degitz SJ (2018). Screening the ToxCast phase 1 chemical library for inhibition of deiodinase type 1 activity. Toxicol. Sci 162, 570–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley PM, Hill RN, and Whiting RJ (1998). Mode of carcinogenic action of pesticides inducing thyroid follicular cell tumors in rats. Environ. Health Persp 106, 437–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayarama-Naidu R, Johannes J, Meyer F, Wirth EK, Schomburg L, Köhrle J, and Renko K (2015). A nonradioactive uptake assay for rapid analysis of thyroid hormone transporter function. Endocrinology 156, 2739–2745. [DOI] [PubMed] [Google Scholar]
- Judson R, Kavlock R, Martin M, Reif D, Houck K, Knudsen T, Richard A, Tice RR, Whelan M, Xia M, et al. (2013). Perspectives on validation of high-throughput assays supporting 21st century toxicity testing. ALTEX 30, 51–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Judson R, Houck K, Martin M, Richard AM, Knudsen TB, Shah I, Little S, Wambaugh J, Setzer RW, Kothiya P, et al. (2016). Analysis of the effects of cell stress and cytotoxicity on in vitro assay activity across a diverse chemical and assay space. Toxicol. Sci 152, 323–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan MM, and Utiger RD (1978). Iodothyronine metabolism in rat liver homogenates. J. Clin. Invest 61, 459–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koehrle J, Auf’mkolk M, Rokos H, Hesch R-D, and Cody V (1986). Rat liver iodothyronine monodeiodinase. J. Biol. Chem 261, 11613–11622. [PubMed] [Google Scholar]
- Köhrle J (1999). Local activation and inactivation of thyroid hormones: the deiodinase family. Mol. Cell. Endocrinol 151, 130–119. [DOI] [PubMed] [Google Scholar]
- Kuiper GGJM, Kester MHA, Peeters RP, and Visser TJ (2005). Biochemical mechanisms of thyroid hormone deiodination. Thyroid 15, 787–798. [DOI] [PubMed] [Google Scholar]
- Lecat-Guillet N, Merer G, Lopez R, Pourcher T, Rousseau B, and Ambroise Y (2007). A 96-well automated radioiodide uptake assay for sodium/iodide symporter inhibitors. Assay Drug Devel. Tech 5, 535–540. [DOI] [PubMed] [Google Scholar]
- Leghait J, Gayrard V, Picard-Hagen N, Camp M, Perdu E, Toutain PL, and Viguié C (2009). Fipronil-induced disruption of thyroid function in rates is mediated by increased total and free thyroxine clearances concomitantly to increased activity of hepatic enzymes. Toxicology 255, 38–44. [DOI] [PubMed] [Google Scholar]
- Maechler M, Rousseeuw P, Struyf A, Hubert M, and Hornik K (2016). cluster: Cluster Analysis Basics and Extensions. R package version 2.0.5 Available at: https://CRAN.R-project.org/package=cluster, last accessed November 28, 2018. [Google Scholar]
- Marsili A, Aguayo-Mazzucato C, Chen T, Kuman A, Chung M, Lunsford EP, Harney JW, Van-Tran T, Gianetti E, Ramadan W, et al. (2011). Mice with a targeted deletion of the type 2 deiodinase are insulin resistant and susceptible to diet induced obesity. PLoS ONE 6, e20832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin MT, Dix DJ, Judson RS, Kavlock RJ, Reif DM, Richard AM, Rotroff DM, Romanov S, Medvedev A, Poltoratskaya N, et al. (2010). Impact of environmental chemicals on key transcription regulators and correlation to toxicity end points within EPA’s ToxCast Program. Chem. Res. Toxicol 23, 578–590. [DOI] [PubMed] [Google Scholar]
- Murk AJ, Rijntjes E, Blaauboer BJ, Clewell R, Crofton KM, Dingemans MM, Furlow JD, Kavlock R, Köhrle J, Opitz R, et al. (2013). Mechanism-based testing strategy using in vitro approaches for identification of thyroid hormone disrupting chemicals. Toxicol. In Vitro 27, 1320–1346. [DOI] [PubMed] [Google Scholar]
- OECD (Organisation for Economic Co-operation and Development). (2014). New Scoping Document on In Vitro and Ex Vivo Assays for the Identification of Modulators of Thyroid Hormone Signalling. In OECD Environment, Health and Safety Publications, Series on Testing and Assessment, No. 207; Paris, France: ENV/JM/MONO(2014)23. Available at: 10.1787/9789264274716-en, last accessed November 28, 2018. [DOI] [Google Scholar]
- Orozco A, Valverde-R C, Olvera A, and García-G C (2012). Iodothyronine deiodinases: a functional and evolutionary perspective. J. Endocrinol 215, 207–219. [DOI] [PubMed] [Google Scholar]
- Paul KB, Hedge JM, Rotroff DM, Hornung MW, Crofton KM, and Simmons SO (2014). Development of a thyroperoxidase inhibition assay for high-throughput screening. Chem. Res. Toxicol 27, 387–399. [DOI] [PubMed] [Google Scholar]
- Paul Friedman K, Watt ED, Hornung MW, Hedge JM, Judson RS, Crofton KM, Houck KA, and Simmons SO (2016). Tiered high-throughput screening approach to identify thyroperoxidase inhibitors within the ToxCast phase I and II chemical libraries. Toxicol. Sci 151, 160–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team (2016). R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria: Available at: https://www.R-project.org/, last accessed November 28, 2018. [Google Scholar]
- Renko K, Hoefig CS, Hiller F, Schomburg L, and Köhrle J (2012). Identification of iopanoic acid as substrate of type 1 deiodinase by a novel nonradioactive iodide-release assay. Endocrinology 153, 2506–2513. [DOI] [PubMed] [Google Scholar]
- Renko K, Schäche S, Hoefig CS, Welsink T, Schwiebert C, Braun D, Becker NP, Köhrle J, and Schomburg L (2015). An improved nonradioactive screening method identifies genistein and xanthohumol as potent inhibitors of iodothyronine deiodinases. Thyroid 25, 962–968. [DOI] [PubMed] [Google Scholar]
- Richard AM, Judson RS, Houck KA, Grulke CM, Volarath P, Thillainadarajah I, Yang C, Rathman J, Martin M, Wambaugh JF, et al. (2016). ToxCast chemical landscape: Paving the road to 21st century toxicology. Chem. Res. Toxicol 29, 1225–1251. [DOI] [PubMed] [Google Scholar]
- Roberts SC, Bianco AC, and Stapleton HM (2015). Disruption of type 2 iodothyronine deiodinase activity in cultured human glial cells by polybrominated diphenyl ethers. Chem. Res. Toxicol 28, 1265–1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosene ML, Wittmann G, Arrojo e Drigo R, Singru PS, Lechan RM, Bianco AC (2010). Inhibition of the type 2 iodothyronine deiodinase underlies the elevated plasma TSH associated with amiodarone treatment. Endocriology 151, 5961–5970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvatore D, Low SC, Berry M, Maia AL, Harney JW, Croteau W, St Germain DL, Larsen PR (1995). Type 3 lodothyronine deiodinase: cloning, in vitro expression, and functional analysis of the placental selenoenzyme. J Clin Invest. 96, 2421–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandell EB, and Kolthoff IM (1937). Micro determination of iodine by a catalytic method. Michrochim. Acta, 1, 9–25. [Google Scholar]
- Schweizer U, and Steegborn C (2015). New insights into the structure and mechanism of iodothyronine deiodinases. J. Mol. Endocrin 55, R37–R52. [DOI] [PubMed] [Google Scholar]
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, and Knudsen TB (2013). Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem. Res. Toxicol 26, 878–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spanka M, Hesch R-D, Irmscher K, and Köhrle J (1990). 5’-Deiodination in rat hepatocytes: Effects of specific flavonoid inhibitors. Endocrinology 126, 1660–1667. [DOI] [PubMed] [Google Scholar]
- Thorne N, Auld DS, and Inglese J (2010). Apparent activity in high-throughput screening: origins of compound-dependent assay interference. Curr. Opin. Chem. Biol 14, 315–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuarog A, Dorris ML, Guziec LJ, and Guziec FS Jr. (1994). The selenium analog of methimazole. Measurement of its inhibitory effect on type 1 5’-deiodinase and of its antithyroid activity. Biochem. Pharmacol 48, 1447–1453. [DOI] [PubMed] [Google Scholar]
- U.S. Environmental Protection Agency (U.S. EPA). (2012). U.S. Environmental Protection Agency Endocrine Disruptor Screening Program Universe of Chemicals and General Validation Principles. U.S. Environmental Protection Agency; Available at: https://www.epa.gov/sites/production/files/2015-08/documents/edsp_chemical_universe_and_general_validations_white_paper_11_12.pdf, last accessed November 28, 2018. [Google Scholar]
- U.S. Environmental Protection Agency (U.S. EPA). (2014). U.S. Environmental Protection Agency Endocrine Disruptor Screening Program Comprehensive Management Plan. U.S. Environmental Protection Agency; Available at: https://www.epa.gov/sites/production/files/2015-08/documents/edsp_comprehesive_management_plan_021414_f.pdf, last accessed November 28, 2018. [Google Scholar]
- U.S. Environmental Protection Agency (U.S. EPA). (2015). ToxCast Assay Summary Information. Data released October 2015. U.S. Environmental Protection Agency; Available at: https:/s/www.epa.gov/chemical-research/toxicitv-forecaster-toxcasttm-data. last accessed November 28, 2018. [Google Scholar]
- U.S. Environmental Protection Agency (U.S. EPA). (2017a). Continuing Development of Alternative High-Throughput Screens to Determine Endocrine Disruption, Focusing on Androgen Receptor, Steroidogenesis, and Thyroid Pathways. White paper from Federal Insecticide, Fungicide, and Rodenticide Act Scientific Advisory Panel, November 28-30,2017. U.S. Environmental Protection Agency; Available at: https://cfpub.epa.gov/si/si_public_record_report.cfm?dirEntryId=338571, last accessed November 28, 2018. [Google Scholar]
- U.S. Environmental Protection Agency (U.S. EPA). (2017b). ECOlogical Structure-Activity Relationship Model (ECOSAR) Class Program. Version 2.0 U.S. Environmental Protection Agency; Available at: https://www.epa.gov/tsca-screening-tools/ecological-structure-activity-relationships-ecosar-predictive-model. See also https://www.epa.gov/tsca-screening-tools/ecological-structure-activity-relationships-program-ecosar-methodology-document, last accessed November 28, 2018. [Google Scholar]
- Visser TJ, Kaptein E, and Aboul-Enein Y (1992). Selenouracil derivatives are potent inhibitors of the selenoenzyme type I iodothyronine deiodinase. Biochem. Biophys. Res. Comm 189, 1362–1367. [DOI] [PubMed] [Google Scholar]
- Wang J, Hallinger DR, Murr AS, Buckalew AR, Simmons SO, Laws SC, and Stoker TE (2018). High-throughput screening and quantitative chemical ranking for sodium-iodide symporter inhibitors in ToxCast phase I chemical library. Environ. Sci. Technol 52, 5417–5426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warnes GR, Bolker B, Bonebakker L, Gentleman R, Liaw WHA, Lumley T, Maechler M, Magnusson A, Moeller S, Schwartz M, and Venables B (2016). gplots: Various R Programming Tools for Plotting Data. R package version 3.0.1 Available at: https://CRAN.R-proiect.org/package=gplots, last accessed November 28,2018. [Google Scholar]
- Wassen FWJS, Klootwijk W, Kaptein E, Duncker DJ, Visser TJ, and Kuiper GGJM (2004). Characteristics and thyroid state-dependent regulation of iodothyronine deiodinases in pigs. Endocrinology 145, 4251–4263. [DOI] [PubMed] [Google Scholar]
- Zhang J-H, Chung TDY, and Oldenburg KR (1999). A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J. Biomolec. Screening 4, 67–73. [DOI] [PubMed] [Google Scholar]
- Zoeller RT (2005). Environmental chemicals as thyroid hormone analogues: New studies indicate that thyroid hormone receptors are targets of industrial chemicals? Mol. Cell. Endocrinol 242, 10–15. [DOI] [PubMed] [Google Scholar]
- Zoeller RT, Tan SW, and Tyl RW (2007). General background on the hypothalamic-pituitary-thyroid (HPT) axis. Crit. Rev. Toxicol 37:11–53. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.