Abstract
The major genetic cause of frontotemporal dementia (FTD) and amyotrophic lateral sclerosis (ALS) is a C9orf72 G4C2 repeat expansion1,2. Proposed mechanisms by which the expansion causes c9FTD/ALS include toxicity from repeat-containing RNA and from dipeptide repeat proteins translated from these transcripts. To investigate the contribution of poly(GR) dipeptide repeat proteins to c9FTD/ALS pathogenesis in a mammalian in vivo model, we generated mice that expressed GFP-(GR)100 in the brain. GFP-(GR)100 mice developed age-dependent neurodegeneration, brain atrophy, and motor and memory deficits through the accumulation of diffuse, cytoplasmic poly(GR). Poly(GR) co-localized with ribosomal subunits and the translation initiation factor eIF3η in GFP-(GR)100 mice and, of importance, in c9FTD/ALS patients. Combined with the differential expression of ribosome-associated genes in GFP-(GR)100 mice, these findings demonstrate poly(GR)-mediated ribosomal distress. Indeed, poly(GR) inhibited canonical and non-canonical protein translation in HEK293T cells, and also induced the formation of stress granules and delayed their disassembly. These data suggest that poly(GR) contributes to c9FTD/ALS by impairing protein translation and stress granule dynamics, consequently causing chronic cellular stress and preventing cells from mounting an effective stress response. Decreasing poly(GR) and/or interrupting interactions between poly(GR) and ribosomal and stress granule-associated proteins may thus represent potential therapeutic strategies to restore homeostasis.
G4C2 repeat expansions in chromosome 9 open reading frame 72 (C9orf72) are the most common genetic cause of amyotrophic lateral sclerosis (ALS), a motor neuron disease, and frontotemporal dementia (FTD), characterized by personality, behavior and/or language changes1,2. This genetic overlap, along with shared clinical and neuropathological features, suggests that common pathological mechanisms drive both diseases. Among putative pathomechanisms of ‘c9FTD/ALS’ are loss of C9ORF72 function and toxicity caused by repeat-containing transcripts3. Repeat RNAs disrupt nucleocytoplasmic transport4 and impair RNA metabolism by sequestering RNA-binding proteins5–10. Furthermore, these transcripts undergo repeat-associated non-ATG (RAN) translation, producing dipeptide repeat (DPR) proteins (GP, GA, GR, PA or PR)11–15. Poly(GR) inclusions correlate with neurodegeneration in c9ALS disease-affected regions16, and arginine-rich DPRs are especially toxic to cultured cells and neurons, as well as Drosophila17–26. While these investigations often used short poly(GR) peptides17–21,24, which may influence its intracellular localization and toxicity, studies suggest that poly(GR) causes nucleolar stress19–21, impairs mitochondrial function23, nucleocytoplasmic transport17,24 and protein translation18,20, and induces stress granule formation20–22. Nevertheless, the consequences of poly(GR) expression have not been investigated in a mammalian in vivo model. Here, we generated mice that express poly(GR) in the brain and found that poly(GR), which caused neurodegeneration and behavioral deficits, co-localized with ribosomal proteins and altered expression of ribosome-associated genes, thus implicating ribosomal dysfunction in c9FTD/ALS.
GFP–(GR)100 or GFP (control) expression in the mouse brain was achieved by neonatal intracerebroventricular injections of adeno-associated virus serotype 1 (AAV1) vectors. To study poly(GR) toxicity in the absence of repeat RNA, a codon-optimized vector encoding GFP–(GR)100 was used. Poly(GR) exhibited a predominantly cytoplasmic, diffuse distribution in the cortex of GFP–(GR)100 mice (Fig. 1a) and throughout their brain, but was rarely detected in the spinal cord (Supplementary Fig. 1). In c9 FTD/ALS patient brains, diffuse poly(GR) was observed but poly(GR) predominantly formed cytoplasmic inclusions (Fig. 1b), suggestive of a temporal transition from a diffuse to an aggregated state. Such a transition was observed in HEK293T cells expressing high levels of GFP–(GR)100 (Supplementary Fig. 2a–d). Cytoplasmic poly(GR) was largely diffuse 24 h post-transfection but formed aggregates by 48 h, likely due to the higher poly(GR) levels at this time point. Notably, compared to GFP–(GR)100-expressing HEK293T cells, poly(GR) in six-month-old GFP–(GR)100 mice was markedly lower, as assessed by immunoassay (Supplementary Fig. 2e,f). These data are consistent with poly(GR) needing to meet a threshold level to aggregate. We thus compared poly(GR) in the brain of GFP–(GR)100 mice to mice expressing (G4C2)66 or (G4C2)149 and c9FTD/ALS patients (Fig. 1c). Poly(GR) in our mouse models was statistically lower than in c9FTD/ALS patients but nonetheless within the lower range of levels among patients. Of interest, poly(GR) levels were comparable between GFP–(GR)100 mice, in which poly(GR) remained diffuse, and (G4C2)66 mice, in which poly(GR) forms aggregates27 (Fig. 1c and Supplementary Fig. 3a), indicating that, in addition to poly(GR) concentration, other factors influence its aggregation. Poly(GR) is reportedly recruited to poly(GA) inclusions28, and we observed this phenomenon in (G4C2)66 and (G4C2)149 mouse brains; poly(GR) co-aggregated with poly(GA) in cells containing poly(GA), but remained diffuse in cells devoid of poly(GA) (Supplementary Fig. 3a).
Fig. 1 |. GFP-(GR)100 mice exhibited neurodegeneration and behavioral deficits.
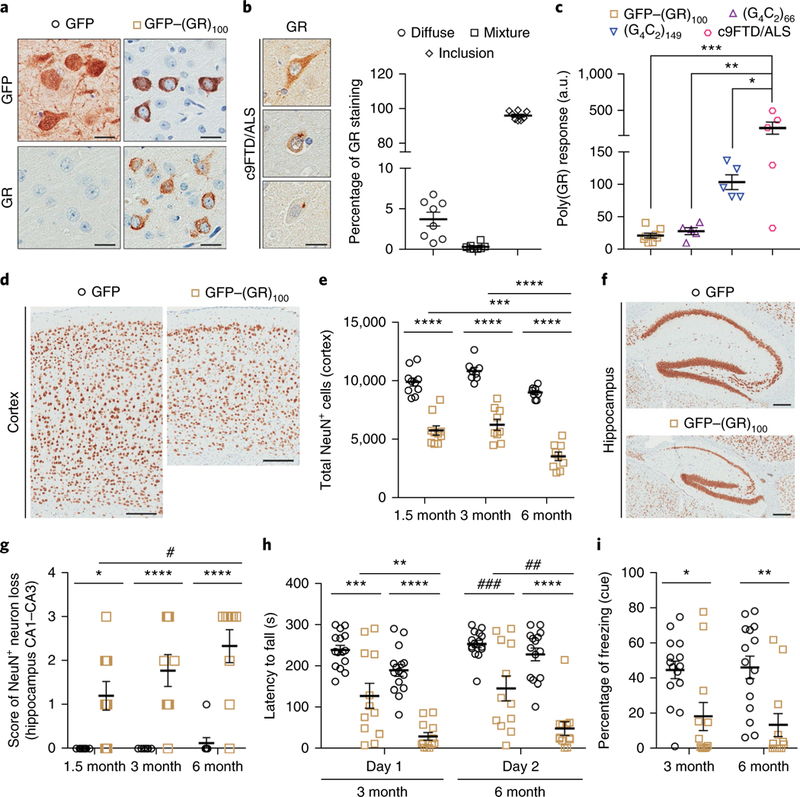
a, Immunohistochemical analysis with anti-GFP or anti-GR antibodies in the cortex of 6-month-old mice expressing GFP (n = 12) or GFP−(GR)100 (n = 9). Scale bars, 20 μm. b, Immunohistochemical (left) and quantitative (right) analyses of poly(GR) in c9FTD/ALS patient frontal cortex (n = 8). Scale bar, 20 μm. c, An immunoassay was used to compare poly(GR) levels in GFP−(GR)100 (n = 8), (G4C2)66 (n = 5) and (G4C2)149 (n = 5) mice, and c9FTD/ALS patients (n = 5). d-g, Representative images of NeuN-labeled cells in the cortex (d) and hippocampus (f) of 6-month-old GFP (n = 8) or GFP-(GR)100 (n = 9) mice (scale bars, 200 μm), and quantification of NeuN-labeled cells in the cortex (e) and hippocampus (g) of GFP mice at 1.5 (n = 10), 3 (n = 8) and 6 (n = 8) months of age, or GFP−(GR)100 mice at 1.5 (n = 10), 3 (n = 9) or 6 (n = 9) months of age. h, Day 1 and day 2 results from a 4-day rotarod test used to determine motor deficits of 3- and 6-month-old mice expressing GFP (n = 15 per group) or GFP−(GR)100 (n = 12 per group) by evaluating latency to fall from a rotating rod (see also Supplementary Fig. 6d for day 3 and day 4 results). i, Results from the fear-conditioning test used to determine associative learning and memory of 3- and 6-month-old mice expressing GFP (n = 15 per group) or GFP-(GR)100 (n = 12 per group) by evaluating the percentage of time frozen in response to a conditioned (cued) stimulus. Data are presented as mean±s.e.m. In c, ***P= 0.0006, **P = 0.0023 and *P = 0.0462, one-way analysis of variance (ANOVA), Tukey’s post hoc analysis. In e, ****P< 0.0001 and ***P = 0.0008, two-way ANOVA, Tukey’s post hoc analysis. In g, ****P<0.0001, ***P= 0.0004, *P = 0.0165 and #P = 0.0343, two-way ANOVA, Tukey’s post hoc analysis. In h, ****P<0.0001, ***P= 0.0001, **P= 0.0020, ###P = 0.0003 and ##P= 0.0021, two-way ANOVA, Tukey’s post hoc analysis. In i, **P = 0.0043 and *P = 0.0286, two-way ANOVA, Tukey’s post hoc analysis.
Brain weight, but not body weight, of GFP–(GR)100 mice decreased from 1.5 to 6 months of age (Supplementary Fig. 4a,b). NeuN staining revealed cortical thinning and CA1–CA3 hippocampal cell loss in GFP–(GR)100 mice (Fig. 1d–g). Cerebellar Purkinje cell loss was also observed (Supplementary Fig. 4c). Transgene RNA and protein levels were significantly higher in GFP mice than GFP–(GR)100 mice, arguing against neuronal loss being caused by high transgene expression (Supplementary Figs. 1a and 4d). Consistent with minimal poly(GR) expression in GFP–(GR)100 mouse spinal cord, no spinal cord neuron loss was seen (Supplementary Fig. 4e,f).
On testing mouse brains for reactive astrocytes and microglia, transcript levels of Gfap, an astrogliosis marker, and Iba1, a microgliosis marker, were found to be significantly increased in 1.5-month-old GFP–(GR)100 mice (Supplementary Fig. 5a). Gfap and Iba1 RNA gradually decreased thereafter, but remained significantly higher in GFP–(GR)100 mice than control mice at 3 and 6 months. A similar profile was observed for Gfap and Iba1 proteins (Supplementary Fig. 5b,c).
Examination of c9FTD/ALS behavioral features in three- and six-month-old GFP–(GR)100 mice revealed locomotive impairment, demonstrated by reduced distance travelled in the open-field test at six months (Supplementary Fig. 6a), and a progressive worsening of motor skills, including increased falls during the rod walk and wire hang tests (Supplementary Fig. 6b,c), and a decreased latency to fall in the rotarod test (Fig. 1h and Supplementary Fig. 6d). GFP–(GR)100 mice also displayed decreased associative memory, evidenced by a decrease in cued (Fig. 1i) but not contextual (Supplementary Fig. 6e) freezing in the fear-conditioning task. The progressive behavioral deterioration of GFP–(GR)100 mice mirrored their age-dependent neurodegeneration.
We next investigated the pathological mechanisms that underlie the neurotoxic consequences of poly(GR) expression in vivo. Poly(GR) binds ribosomal proteins18,20,23, and we found that diffuse poly(GR) co-localized with S6, L21 and S25 ribosomal proteins and the translation initiation factor eIF3n in GFP–(GR)100 mouse brains (Fig. 2a). Diffuse and aggregated RAN-translated poly(GR) in (G4C2)66 or (G4C2)149 mice also co-localized with S6 (Fig. 2b and Supplementary Fig. 3b). Of particular importance, both diffuse and aggregated cytoplasmic poly(GR) co-localized with S6, L21 and eIF3n in the cortex of c9FTD/ALS patients (Fig. 2c). Poly(GA) failed to co-localize with S6 in brains of c9FTD/ALS patients or GFP–(GA)50-expressing mice (Fig. 2d), demonstrating specificity of poly(GR) co-localization with ribosomes. Although poly(GR) reportedly interacts with mitochondrial ribosomal proteins23, it rarely co-localized with the mitochondrial marker, Tom20, in GFP–(GR)100 mice but did partially co-localize with the endoplasmic reticulum (ER) marker, KDEL, likely through its binding to ribosomes on the rough ER (Fig. 2e).
Fig. 2 |. Poly(GR) proteins co-localized with ribosomal proteins in GFP–(GR)100 mice and c9FTD/ALS patients.
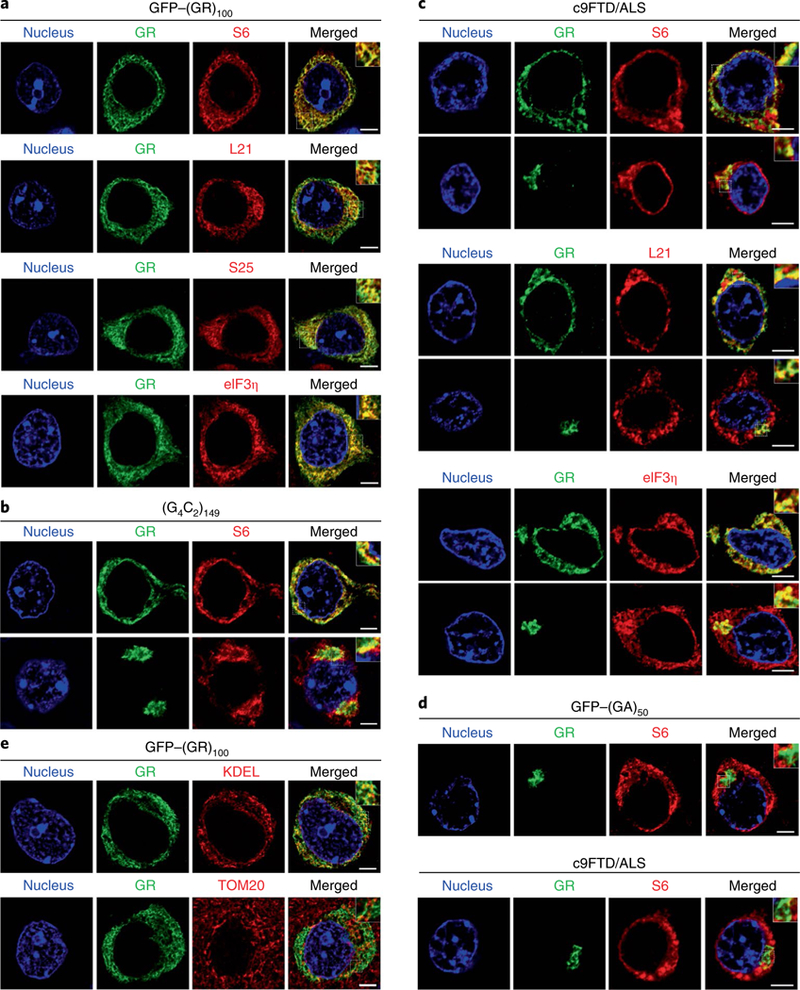
a, Double-immunofluorescence staining for GFP–(GR)100 and ribosomal proteins (S6, L21 or S25) or the translation initiation factor eIF3η in the cortex of 6-month-old GFP–(GR)100 mice (n = 5). b, Double-immunofluorescence staining for poly(GR) and S6 in the cortex of 6-month-old (G4C2)149-expressing mice (n = 3). c, Double-immunofluorescence staining for poly(GR) and S6, L21 or eIF3η in c9FTD/ALS patient frontal cortex (n = 3). d, Double-immunofluorescence staining for poly(GA) and S6 in the cortex of 6-month-old GFP–(GA)50-expressing mice (top) (n = 3) or c9FTD/ALS patients (bottom) (n = 3). e, Double-immunofluorescence staining for GFP–(GR)100 and the endoplasmic reticulum marker KDEL (top) or the mitochondrial marker Tom20 (bottom) in the cortex of 6-month-old GFP–(GR)100 mice (n = 5). All scale bars, 5 μ m.
The above data implicate ribosomal dysfunction in c9FTD/ALS, and RNA sequencing studies support this. We compared transcriptome profiles between 1.5-month-old GFP and GFP–(GR)100 mice. Clustering the 1,000 most variable genes showed distinct expression profiles for GFP–(GR)100 and GFP mice, as did principal component analysis (Supplementary Fig. 7a,b). We identified 3,287 up- and 3,402 downregulated transcripts using DESeq2 (Supplementary Data Set 1a,b); samples clustered by group, showing that the poly(GR) effect on differentially expressed genes was consistent (Fig. 3a). The MA plot highlights genes that were up- or downregulated with a false-discovery rate (FDR) < 0.01 when comparing GFP–(GR)100 to GFP mice (Fig. 3b). Weighted gene co-expression network analysis (WGCNA) using a merging distance of 0.01 identified 60 modules, 56 of which were significantly associated with genotype after adjusting for multiple tests (Supplementary Fig. 7c and Supplementary Data Set 1c). Gene Ontology enrichment analysis across the 60 modules identified the skyblue3 and green modules as the most significantly enriched. Biological process analysis of the skyblue3 module identified immune response as the top term (P = 5.85 × 10–13, Bonferroni = 6.79 × 10–9; Supplementary Data Set 1d), consistent with the inflammation observed in GFP–(GR)100 mice (Supplementary Fig. 5). Within the green module, molecular function, cellular component and biological process analyses respectively identified top terms specific to ribosomal constituents (P = 3.12 × 10−13, Bonferroni= 1.26X 10−9; Supplementary Data Set 1e), cytosolic ribosomal activity (P = 1.32 X10−11, Bonferroni = 2.09 × 10–8; Supplementary Data Set 1f) and protein translation (P = 2.22 × 10–5, Bonferroni = 0.26; Supplementary Data Set 1d). A GeNets InWeb3 network for the green module showed that the primary cluster of protein-protein interactions consisted exclusively of ribosomal proteins, the gene expression of which was elevated in GFP–(GR)100 mice (Fig. 3c). Interestingly, while poly(GR) did not localize to mitochondria, several genes encoding mitochondrial ribosomal proteins were differentially expressed in GFP–(GR)100 mice (Supplementary Data Set 1a,b).
Fig. 3 |. Transcriptome analyses revealed altered ribosome pathways in GFP–(GR)100 mice.
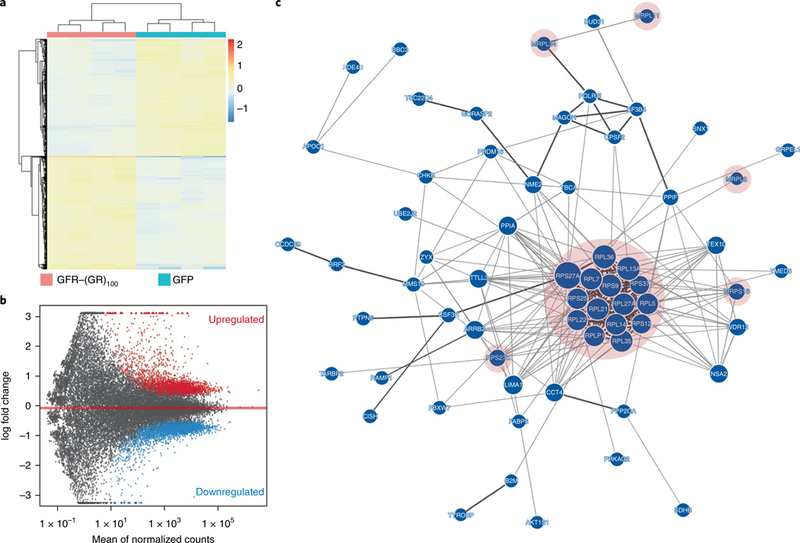
a, Hierarchical clustering of differentially expressed genes (FDR < 0.01) in brains of 1.5-month-old mice expressing GFP or GFP–(GR)100 (n = 4 per group). b, MA plots of up- and downregulated genes (FDR < 0.01) in brains of 1.5-month-old mice expressing GFP–(GR)100 compared to GFP controls (n = 4 per group). c, Network diagram of the green module identified as the top module by WGCNA. The primary cluster of protein–protein interactions, highlighted in pink, consisted entirely of ribosomal proteins (n = 4 per group).
The altered expression of genes encoding ribosomal proteins could reflect a compensatory response caused by poly(GR)-induced ribosomal inhibition. We thus sought to examine protein translation in HEK293T cells expressing GFP–(GR)100 but first confirmed that diffuse and aggregated cytoplasmic poly(GR) co-localized with S6 and eIF3n in these cells (Fig. 4a), as observed for diffuse poly(GR) in GFP–(GR)100 mice (Fig. 2a) and both diffuse and aggregated poly(GR) in c9fTd/ALS patients (Fig. 2c). We also found that aggregated but not diffuse poly(GR) co-localized with the stress granule marker TIA-1 (Fig. 4b–e). Significantly more GFP–(GR)100-expressing HEK293T cells harbored TIA-1 aggregates than GFP-expressing cells (Fig. 4b,c), suggesting that poly(GR) inclusions induce stress granule formation. In support of this, we observed cytoplasmic TIA-1 aggregates that co-localized with poly(GR) inclusions but not diffuse poly(GR) in the cortex and hippocampus of (G4C2)66 and (G4C2)149 mice, in contrast to its diffuse, predominantly nuclear distribution in (G4C2)2 and GFP–(GR)100 mice (Fig. 4d,e and Supplementary Fig. 8a,b). These data show that only aggregated poly(GR) induces spontaneous stress granule formation. To examine whether diffuse poly(GR) is nonetheless recruited to TIA-1-positive granules in response to stress, GFP- and GFP–(GR)100-expressing HEK293T cells were heat shocked. Cells expressing diffuse GFP- (GR)100 did not form more stress granules than GFP-expressing cells, but they did maintain a greater number of stress granules after the heat shock was terminated, indicating that poly(GR) impaired stress granule disassembly (Supplementary Fig. 8c,d).
Fig. 4 |. Expression of GFP–(GR)100 impaired canonical and non-canonical translation.
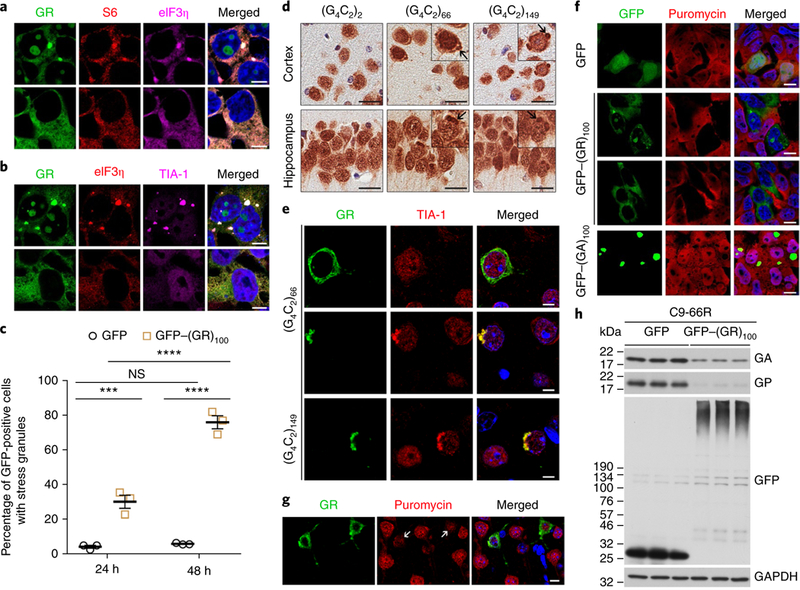
a,b, Triple-immunofluorescence staining for GFP–(GR)100, eIF3η and either S6 or TIA-1 in HEK293T cells expressing cytoplasmically aggregated or diffuse GFP–(GR)100 under basal conditions (n = 3 independent experiments). Scale bars, 5 μ m. c, Quantification of the percentage of GFP- or GFP–(GR)100-expressing HEK293T cells containing TIA-1-positive stress granules 24 h and 48 h post-transfection (n = 3 independent experiments). d, Immunohistochemical analysis of TIA-1 in the cortex and hippocampus of six-month-old (G4C2)2, (G4C2)66 and (G4C2)149 mice (n = 3 per group). Insets, higher magnification showing examples of cytoplasmic TIA-1 inclusions, which are indicated by the arrows. Scale bars, 20 μ m. e, Double-immunofluorescence staining for poly(GR) and TIA-1 in the cortex of 6-month-old (G4C2)66 and (G4C2)149 mice (n = 3 per group). Scale bars, 10 μ m. f, Double-immunofluorescence staining for GFP and puromycin in HEK293T cells expressing GFP, GFP–(GR)100 or GFP–(GA)100 to examine the production of newly translated proteins (n = 3 independent experiments). Scale bars, 5 μ m. g, Double-immunofluorescence staining for poly(GR) and puromycin in the cortex of 1.5-month-old mice expressing GFP–(GR)100 (n = 3). Scale bars, 20 μ m. The arrows indicate poly(GR)-expressing cells. h, Immunoblot of the indicated proteins to examine the effect of GFP or GFP–(GR)100 on (G4C2)66 RAN translation of poly(GA) and poly(GP) in HEK293T cells co-expressing (G4C2)66 and either GFP or GFP–(GR)100 (n = 3 independent experiments). Cropped blots are shown in full in Supplementary Fig. 10a. Data are presented as mean ± s.e.m. In c, ****P < 0.0001; ***P = 0.0007; NS, P = 0.9710; two-way ANOVA; Tukey’s post hoc analysis.
Since stress granule formation is associated with translation inhibition, these findings provided yet another reason to examine the effect of poly(GR) on protein translation. Thus, GFP- and GFP–(GR)100-expressing HEK293T cells were treated with puro-mycin, which is incorporated into newly translated proteins29. Immunofluorescence studies revealed a striking decrease of puromycin in cells expressing diffuse or aggregated cytoplasmic GFP–(GR)100 (Fig. 4f), which was confirmed by western blot (Supplementary Fig. 9). GFP–(GA)100, in contrast, did not inhibit protein translation (Fig. 4f and Supplementary Fig. 9). Notably, in the cortex of GFP–(GR)100 mice, a reduction in puromycin-labeling was observed in poly(GR)-expressing cells compared to poly(GR)–negative cells (Fig. 4g).
Given that poly(GR) co-localized with S25, a regulator of noncanonical translation30,31, we examined whether GFP–(GR)100 influences (G4C2)66 RAN translation in HEK293T cells. RAN translation of (G4C2)66 in cells co-expressing GFP produced poly(GA) and poly(GP). However, in the presence of GFP–(GR)100, levels of (G4C2)66 RAN-translated poly(GA) and poly(GP) were markedly lower despite (G4C2)66 RNA and the percentage of RNA foci-bearing cells being comparable between GFP–(GR)100- and GFP-expressing cells (Fig. 4h and Supplementary Fig. 10).
Here, we show that GFP-(GR)100 mice developed age-dependent neuron loss and impaired motor and memory skills. Compared to AAV1-GFP–(GA)50 mice32, GFP–(GR)100 mice developed an earlier disease onset and more severe neurodegeneration, consistent with the notion that poly(GR) is more toxic than poly(GA)33 (Supplementary Discussion). The neurodegeneration and behavioral deficits in GFP–(GR)100 mice were accompanied by poly(GR) co-localization with ribosomal subunits and the translation initiation factor eIF3n, which was also observed in c9FTD/ALS patients. These data, combined with our RNA-Seq analysis demonstrating differentially expressed ribosome-associated genes in GFP–(GR)100 mice, our studies in HEK293T cells showing that diffuse and aggregated cytoplasmic GFP–(GR)100 inhibit protein translation, and proteomic studies identifying ribosomal proteins as top poly(GR) interactors18,20,23, implicate impaired ribosomal activity in poly(GR)- mediated neurotoxicity. We also show that aggregated cytoplasmic poly(GR) induced the formation of stable stress granules, and diffuse poly(GR) was recruited to stress-induced granules and impaired their disassembly, consistent with reports that poly(GR) alters stress granule dynamics by disturbing the liquid–liquid phase transition of stress granule proteins with low-complexity sequence domains20–22,34. These findings and reports that cellular stress enhances G4C2 repeat RAN translation35,36 raise the following possible scenario: during the early stage of cellular stress, RAN translation of expanded G4C2 repeats is enhanced and promotes the production of DPRs. As poly(GR) accumulates and aggregates, it interacts with ribosomal subunits and impairs protein translation. It also promotes stress granule assembly and impairs their disassembly, ultimately creating a state of chronic cellular stress. Nevertheless, through a negativefeedback loop in which poly(GR) inhibits RAN translation, it may abate the harmful events induced by its expression.
TDP-43 pathology is a hallmark feature of c9FTD/ALS, but inclusions of phosphorylated TDP-43 were rarely observed in GFP–(GR)100 mice (Supplementary Figs. 11a and 12d and Supplementary Discussion). Furthermore, no evidence of disrupted nucleocytoplas-mic trafficking, impaired nuclear membrane integrity or nucleolar stress was observed in six-month-old GFP–(GR)100 mice nor in one-week-old mice, an age that preceded neuronal loss (Supplementary Figs. 11b–f and 12 and Supplementary Discussion), demonstrating that poly(GR) caused neurodegeneration in vivo independently of TDP-43 pathology, nucleocytoplasmic transport defects and nucleolar stress.
Overall, our studies provide compelling evidence that, through its interaction with ribosomal subunits, translation initiation factors and stress granule-associated proteins, poly(GR) impairs ribosomal activity, canonical and non-canonical protein translation, and stress granule dynamics, thus contributing to the neurodegeneration seen in c9FTD/ALS patients. These data are consistent with a previous report showing that arginine-rich DPRs interact with RNA-binding proteins and proteins with low-complexity sequence domains, which promote assembly of membrane-less organelles20,34. Reducing poly(GR) levels, and blocking interactions between poly(GR) and ribosomal and stress granule-associated proteins, thus represent therapeutic strategies worthy of investigation. By providing a model that mimics features of c9FTD/ALS, GFP–(GR)100 mice would facilitate such studies.
Methods
Methods, including statements of data availability and any associated accession codes and references, are available at https://doi.org/10.1038/s41591–018-0071–1.
Generation of GFP–(GR)100 plasmids.
The (GR)100 plasmid was generated as previously described37. A DNA fragment coding for 50 GR dipeptides was synthesized by GeneArt and used as a template in PCR reactions to generate two fragments of 50 GR dipeptides. These contained either a HindIII site at the 5′ end and a BsmBI site at the 3′ end, or a BsmBI site at the 5′ end and a BamHI site at the 3′ end. BsmBI is a Type IIS restriction enzyme that recognizes an asymmetric DNA sequence and cleaves outside of its recognition sequence, allowing for seamless sub-cloning of DNA coding sequences. These fragments were cloned into HindIII and BamHI restriction sites of the mammalian expression vector pEGFP–C1 (Clontech Laboratories). The pEGFP–C1–(GR)100 plasmid was digested using HindIII and BamHI, and then the HindIII–GR100–BamHI DNA fragment was sub-cloned into a modified AAV packaging vector (pAM/CBA-pl-WPRE-BGH (‘pAAV’)) containing the CMV-enhanced chicken β-actin promoter and the EGFP coding sequence with a multiple cloning site that allows for cloning the (GR)100 fragment in frame with the EGFP coding sequence. The sequence of all plasmids was verified by sequence analysis. The sequence of GFP–(GR)100 is shown in Supplementary Table 1.
Mouse studies.
All procedures using mice were performed in accordance with the National Institutes of Health Guide for Care and Use of Experimental Animals and approved by the Mayo Clinic Institutional Animal Care and Use Committee (Protocol numbers A42014 and A47214).
Intracerebroventricular injections of virus in neonatal mouse brain.
Intracerebroventricular injections of AAV1–GFP–(GR)100 virus in post-natal day 0 (P0) C57BL/6J pups were performed as previously described using 2 μl (1 × 1010 genomes μl–1) of AAV1–GFP, AAV1–GFP–(GR)100, AAV9–(G4C2)2, AAV9–(G4C2)66 or AAV9–(G4C2)149 solution per cerebral ventricle27,32.
Behavioral tests.
Three- and six-month-old mice expressing GFP (n = 15) or GFP–(GR)100 (n = 12) were subjected to behavioral analysis on two consecutive weeks by the Mayo Mouse Behavior Core. On five consecutive days of week 1, the mice were subjected to the open-field assay on day 1, contextual and cued fear-conditioning tests on days 2 and 3, and the rod walk and wire hang tests on day 4 and 5, respectively. On four consecutive days of week 2, the mice underwent a rotarod test. All mice were acclimated to the room 1 h before testing, and then returned to their home cage and home room after each test.
Open-field, context and cue fear-conditioning, and rotarod tests.
The open-field test, context and cue fear-conditioning tests, and rotarod test were performed as described previously27,32. For the open-field test, each mouse was placed in a box and behavioral activity was recorded and tracked for 10 min using an automated camera tracking system, and then analyzed using Anymaze software. For context and cue fear-conditioning, on day 1, each mouse was placed into the chamber and baseline freezing behavior was recorded during the first 2 min. Then, mice were exposed to white noise as the conditioned stimulus, and subsequently received a mild foot shock as the unconditioned stimulus. On day 2, each mouse was returned to the test chamber and freezing behavior was recorded for 5 min (context test). Afterwards, each mouse was placed in a modified chamber, and then exposed to the auditory conditioned stimulus and freezing behavior was recorded for another 3 min (cue test). Baseline freezing behavior obtained during training was subtracted from the context or cue tests to control for variability in each animal. For the rotarod test, each mouse was habituated to stay on the spindle with slow constant speed for 1 min, and then the spindle was accelerated from 4 r.p.m. to 40 r.p.m. The latency to fall time was recorded when the mouse fell off the spindle.
Rod walking test.
The apparatus consists of a narrow rod (60 cm long, 0.75 cm wide) that was suspended between 2 poles 40 cm above a layer of bedding material to prevent injury to the animal when it falls. Each mouse was gently lowered onto the rod and allowed to stabilize footing before being released. The ability of the mouse to navigate the rod within 5 min was measured. During the test, if a mouse fell down it was returned to the rod, and the total number of falls was counted.
Hanging wire test.
The apparatus consists of a wire (55 cm wide, 2 mm thick) secured tightly to two vertical stands. The wire is maintained 35 cm above a layer of bedding material to prevent injury to the mouse when it falls. Each mouse was picked up by the tail and brought close to the wire so they could grip the wire with their forelimbs. The ability of the animal to suspend itself on the rod was measured for 2 min. If a mouse fell during these 2 min, it was returned to the wire, and the total number of falls was counted.
Tissue processing.
Sagittal half-brains were immersion-fixed in 10% formalin or 4% paraformaldehyde, embedded in paraffin, sectioned (5 μm thick) and then mounted on glass slides for immunofluorescence or immunohistochemical staining. The cortex and hippocampus of the other half-brain were dissected and frozen on dry ice. Frozen mouse cortex and hippocampus, or frozen human post-mortem cerebellum, were homogenized in ice-cold buffer (50 mM Tris pH 7.4, 50 mM NaCl, 1 mM EDTA) with 2 × protease and phosphatase inhibitors. Homogenates were used for RNA or protein extractions.
Preparation of brain protein lysates.
To prepare protein lysates, Triton X-100 and 10% sodium dodecyl sulfate (SDS) were added to the homogenates at a final concentration of 1% and 2%, respectively. Homogenates were sonicated on ice, and then centrifuged at 16,000g for 20 min. Supernatants were saved and protein concentrations were determined by BCA assay (Thermo Fisher Scientific).
Human tissues.
Post-mortem cerebellar and frontal cortical tissues from FTD and ALS patients with or without C9orf72 repeat expansion were obtained from the Mayo Clinic Florida Brain Bank. Information on human patients is provided in Supplementary Table 2. Written informed consent was obtained before study entry from all subjects or their legal next of kin if they were unable to give written consent, and biological samples were obtained with Mayo Clinic Institutional Review Board approval.
Immunohistochemistry staining.
Paraffin sections of mouse and human brain were subjected to immunohistochemical staining as previously described27,32. Sections were deparaffinized, rehydrated, steamed for 30 min in distilled H2O, incubated with Dako Peroxidase Block for 5 min, blocked with Dako All Purpose Blocker for 1 h and then incubated with primary antibody for 45 min (Supplementary Table 1). After washing, sections stained with anti-mouse or anti-rabbit primary antibodies were incubated for 30 min in Dako Envision-Plus anti-mouse or anti-rabbit labeled HRP polymer, respectively. For anti-goat TIA-1 staining, sections were incubated for 15 min in anti-goat probe, and then for 15 min in anti-goat labeled HRP polymer. Peroxidase labeling was visualized with the chromogen solution 3,3′-diaminobenzidine (DAB-Plus). Sections were counterstained with Lerner 1 hematoxylin (Thermo Fisher Scientific) and coverslipped with Cytoseal mounting medium (Thermo Fisher Scientific).
Quantification of neuropathology.
Aperio ePathology technology (Leica Biosystems) was used to quantify neuropathology as previously described27,32. High-resolution digitized images of immunostained slides were obtained using a ScanScope AT2 (Leica Biosystems). The distribution patterns of poly(GR) staining in c9FTD/ALS cases were quantified manually, and calculated as the percentage of poly(GR)-positive cells with diffuse poly(GR), poly(GR) inclusions or a combination of both staining types.
Purkinje cells in the cerebellum of mice were quantified in a blinded fashion. Using the ruler function of ImageScope software (v12.1; Leica Biosystems), a line totaling 10 mm along the Purkinje cell layer was delineated for each mouse. The number of Purkinje cells within this distance was counted. Data were presented as the average number of Purkinje cells per 1 mm within each cohort. To quantify the NeuN-positive neuronal density, and percentage of Gfap and Iba1 burden in a blinded fashion, the whole cortex was annotated on mid-sagittal serial sections stained for NeuN, Gfap or Iba1 from each mouse using ImageScope software. The rounded appearance and staining intensity of NeuN-positive neurons was used to design a custom-designed nuclear algorithm. A NeuN-positive neuronal density measure was calculated as the total number of neurons in the annotated area. Gfap-positive astrocytes and Iba1-positive microglia were quantified using custom-designed positive pixel count algorithms. The number of positively stained pixels as a proportion of all pixels was used as the output parameter for both Gfap and Iba1 in the annotated area. The numbers of NeuN-positive cells in the anterior horn of the spinal cord were counted manually. The NeuN staining was also used to rate the severity of hippocampal CA1–CA3 neuronal loss using a four-point scale: none (0: no neuronal loss), mild (1: slightly disrupted neuronal layers of CA1–CA3 region), moderate (2: medium loss of neuronal layers), severe (3: extensive loss of neurons). To quantify GFP-positive cells in various neuroanatomical regions of mid-sagittal sections stained for GFP, the motor cortex, hippocampal CA1–CA3 regions and cerebellar Purkinje cell layer for each mouse were delineated using ImageScope software. The number of GFP-positive cells and hematoxylin-stained nuclei (for a count of total cells) were manually counted, and the percentage of GFP-positive was calculated for each region.
Immunofluorescence staining in mouse and human brains.
Paraffin sections (5 μm) of mouse and human brains were subjected to immunofluorescence staining, as described previously27,32. Sections were deparaffinized, rehydrated, steamed for 30 min in Dako antigen retrieval solution, blocked with Dako All Purpose Blocker for 1 h and incubated with primary antibody (Supplementary Table 1). After washing, sections were incubated with corresponding Alexa Fluor 488- or 568-conjugated donkey anti-species (1:500, Molecular Probes) for 2 h. Hoechst 33258 (1 |agml−1, Thermo Fisher Scientific) was used to stain cellular nuclei. Images were obtained on a Zeiss LSM 700 laser scanning confocal microscope.
RNA extraction, PCR with reverse transcription (RT-PCR) and quantitative PCR (qPCR).
To extract total RNA in mouse brain, one volume of brain homogenate was mixed with three volumes of Trizol LS Reagent (Thermo Fisher Scientific), and frozen on dry ice. One day later, total RNA was extracted using the Direct-zol RNA MiniPrep kit (Zymo Research) according to the manufacturer’s instructions, combined with an in-column DNase I digestion step. To extract total RNA from cultured cells, cell pellets were lysed in the TRI Reagent and then total RNA was extracted using Direct-zol RNA MiniPrep kit (Zymo Research). cDNA was obtained after RT-PCR using 1,000 ng of RNA with random primers and the High Capacity cDNA Transcription Kit (Applied Biosystems) according to the manufacturer’s instructions.
To quantify RNA levels of the indicated transcripts in mouse brain or cultured cells, qPCR was conducted in triplicate for all samples using SYBR green assay (Thermo Fisher Scientific) on an ABI Prism 7900HT Fast Real-Time PCR System (Applied Biosystems). Primer sequences are listed in Supplementary Table 1. Relative RNA expression of Gfap and Iba1, as well as C9–66R, was normalized to Gapdh or GAPDH values, an endogenous transcript control.
RNA-Seq and Gene Ontology analysis.
The library preparation and RNA-Seq was performed by the Mayo Clinic Sequencing Core Facility (Rochester, MN) as previously described38. In brief, a total of 200 ng of RNA in a 50 μl volume was used for preparation of paired-end sequencing libraries using the TruSeq RNA Library Prep Kit v2 (Illumina). Afterward, samples were subjected to quality control, cluster generation and sequencing on the Illumina HiSeq 2000 platform. The reads were de-multiplexed and converted to FASTQ format using CASAVA software from Illumina by the Mayo Clinic Bioinformatics Core (Rochester, MN). Alignment and QC were performed using the latest version of Mayo Clinic’s Map-RSeq (v. 1.2.1.5) RNA-Seq pipeline39.
Transcript counts were normalized using conditional quantile normalization for transcript length and GC content40. We then normalized for hippocampal and cortical brain weight to account for cell death between GFP and GFP–(GR)100 mice. We then tested for differential expression between the GFP and GFP–(GR)100 mice using a two-sided Wald test in DESeq241, after performing the regularized logarithm (rlog) transformation. We used a strict FDR < 0.01. We followed up with a WGCNA42 and Gene Ontology over-representation test to provide biologically meaningful context for gene sets using only the significantly differentially expressed genes. To make WGCNA modules as specific as possible, we set WGCNA ‘mergeCutHeight’ to 0.01. Clusters were made using ‘pheatmap’ in R (v. 3.4.1) with Euclidean clustering distance. Complete results for differentially expressed genes and WGCNA module associations are available in Supplementary Data Set 1. Differentially expressed gene P values were adjusted using the Benjamini-Hochberg method (also known as the FDR). WGCNA module associations to genotype were tested using Pearson correlation, and P values were adjusted using the Holm procedure. All data with P < 0.05 were considered statistically significant.
Cell culture and treatments.
HEK293T cells were grown in Opti-Mem plus 10% FBS and 1% penicillin-streptomycin. To examine nascent translated proteins, SUnSET assays were performed as previously described29. In brief, cells grown in 6-well plates or on glass coverslips in 24-well plates were transfected with 1 μg (6-well plates) or 0.3 μg (24-well plates) of an expression vector (GFP, GFP–(GR)100 or GFP–(GA)100) using Lipofectamine 2000 (Thermo Fisher Scientific). After 24 h, the medium was replaced with fresh medium containing 1 μg ml–1 puromycin. After 1 h, cells were collected for western blot analysis and fixed for immunofluorescence staining. To determine the effect of poly(GR) on RAN translation, cells grown in 6-well plates were transfected with 2 μg C9–66R and 1 μg of either GFP or GFP–(GR)100 using Lipofectamine 2000 (Thermo Fisher Scientific). After 48 h, cells were collected for western blot analysis. To determine the effect of poly(GR) on RNA foci formation, cells grown on glass coverslips in 24-well plates were transfected with 0.5 μg C9–66R and 0.25 μg GFP or GFP–(GR)100 using Lipofectamine 2000 (Thermo Fisher Scientific). After 48 h, cells were fixed for RNA fluorescence in situ hybridization (FISH).
In vivo SUnSET assay.
The in vivo SUnSET assay was performed by stereotaxic intracerebroventricular administration of puromycin into the brain of mice. In brief, 1.5-month-old GFP–(GR)100 mice were anesthetized with isoflurane (3% for induction, 1–2% for maintenance). Using sterile instruments and gloves, a midsagittal longitudinal incision was made in the scalp to expose the skull. Then, 10 μl of puromycin at a concentration of 1 μg μl–1 was administrated per hemibrain using the following coordinates: 0 mm anterior/posterior (anterior to bregma), 1.0 mm to the right or left medial/lateral and 1.9 mm dorsal/ventral. One hour post-injection, the mice were collected and brains were immersion-fixed in 10% formalin for further immunofluorescence staining.
Preparation of cell lysates.
To prepare cell lysates for SUnSET studies, cell pellets were lysed in radio-immunoprecipitation assay buffer (25 mM Tris-HCl pH 7.6, 150 mM NaCl, 1% sodium deoxycholate, 1% NP-40, 0.1% sodium dodecyl sulfate, protease and phosphatase inhibitors), sonicated on ice and then centrifuged at 16,000g for 20 min. Supernatants were saved as cell lysates. To prepare cell lysates for poly(GP) immunoassays, cell pellets were lysed in co-immunoprecipitation buffer (50 mM Tris-HCl, pH 7.4, 300 mM Nad, 1% Triton X-100, 5 mM EDTA) plus 2% SDS, and both protease and phosphatase inhibitors, sonicated on ice and then centrifuged at 16,000g for 20 min. Supernatants were saved as cell lysates. The protein concentration of lysates was determined by BCA assay (Thermo Fisher Scientific), and samples were then subjected to western blot or immunoassay analyses.
Immunofluorescence staining, quantification of poly(GR) aggregates and TIA-1-positive stress granules in GFP- or GFP–(GR)100-expressing HEK293T cells.
Cells were fixed with 4% paraformaldehyde for 15 min, permeabilized with 0.5% Triton X-100 for 10 min, blocked with 10% normal horse serum or 5% non-fat dry milk in PBS for 1 h, and then incubated with primary antibody overnight at 4 °C (Supplementary Table 1). After washing, the cells were incubated with the corresponding Alexa Fluor 568- or 633-conjugated donkey anti-species secondary antibodies (1:500, Molecular Probes) for 2 h. Hoechst 33258 (1 μg ml–1, Life Technologies) was used to stain cellular nuclei. Images were obtained on a Zeiss LSM 700 laser scanning confocal microscope. To quantify the percentage of cells with cytoplasmic diffuse or aggregated GFP–(GR)100, the number of GFP-positive cells containing diffuse or aggregated GFP was counted in a blinded fashion from three independent experiments (~180–350 cells were counted per experiment for the 24 h time point, and ~170–500 cells per experiment were counted for the 48 h time point). To quantify the percentage of GFP- or GFP–(GR)100-expressing cells containing TIA-1-positive stress granules under basal conditions, the number of GFP-positive cells with or without TIA-I aggregates was counted in a blinded fashion from three independent experiments (~210–340 cells were counted per experiment for the 24 h time point, and ~250–460 cells per experiment were counted for the 48 h time point). To quantify the percentage of cells with TIA-1-positive stress granules under heat stress and recovery conditions, the number of GFP-positive cells with or without TIA-I aggregates was counted in a blinded fashion from three independent experiments (~200–300 cells per experiment were counted per treatment).
Western blot analysis.
Western blot analysis was performed as previously described32,37. In brief, lysates were diluted with 2 × SDS-loading buffer at a 1:1 ratio (v/v), and then heated at 95 °C for 5 min. Afterward, equal amounts of protein were loaded into 12-well or 20-well 4–20% Tris-glycine gels (Novex). After transferring proteins to membranes, membranes were blocked with 5% non-fat dry milk in Tris-buffered saline (TBS) plus 0.1% Tween 20 (TBST) for 1 h, and then incubated with a mouse monoclonal anti-puromycin antibody (MABE342, 1:2,000, EMD Millipore), rabbit polyclonal anti-GFP antibody (A-6455, 1:4,000, Life Technologies), rabbit polyclonal anti-GA antibody37, rabbit polyclonal anti-GP antibody11 or mouse monoclonal GAPDH antibody (H86504M, 1:10,000, Meridian Life Science) overnight at 4 °C. Membranes were washed in TBST and incubated with donkey anti-rabbit or anti-mouse IgG antibodies conjugated to horseradish peroxidase (1:5,000; Jackson ImmunoResearch) for 1 h. Protein expression was visualized by enhanced chemiluminescence treatment and exposure to film. The intensity of bands was quantified by FUJI FILM MultiGauge Software, and then normalized to the corresponding controls.
Immunoassay analysis of poly(GP) and poly(GR) proteins.
Levels of poly(GP) proteins in HEK293T cell lysates were measured using a previously described MSD-based sandwich immunoassay43. In brief, HEK293T cell lysates were diluted in TBS and an equal amount of protein for all samples was tested in duplicate wells. Serial dilutions of recombinant (GP)8 peptide in TBS were used to prepare the standard curve. Response values corresponding to the intensity of emitted light on electrochemical stimulation of the assay plate using the MSD QUICKPLEX SQ120 were acquired. Responses were background-corrected using the average response from lysates obtained from non-transfected cells.
Levels of poly(GR) proteins in HEK293T cell lysates, as well as in mouse and human brain lysates, were similarly measured using a newly developed MSD-based sandwich immunoassay that utilizes affinity-purified rabbit polyclonal antibodies that detect poly(GR) as capture and detection antibodies. Lysates were diluted in TBS and added to wells (50 μl per well) at a final concentration of 65 μg per well. In addition, to confirm specificity of the assay for poly(GR), peptides representing each of the 5 dipeptide repeat proteins translated from C9orf72 repeat expansions ((GA)8, (GP)8, (GR)8, (PA)8 and (PR)8) were diluted in TBS and added to wells (50 μl per well) at final concentrations of 4, 20 and 100 ng per well. Response values corresponding to the intensity of emitted light on electrochemical stimulation of the assay plate using the MSD QUICKPLEX SQ120 were acquired and background-corrected using responses from lysates of respective controls.
RNA FISH and quantification of RNA foci.
RNA FISH was performed as previously described44 with slight modifications. In brief, cells grown on coverslips in 24-well plates were fixed with 4% paraformaldehyde in diethyl pyrocarbonate (DEPC)-PBS. Afterward, cells were permeabilized with 0.2% Triton X-100/DEPC–PBS. Cells were then incubated with hybridization buffer (50% formamide, 10% dextran sulfate, 2 × saline-sodium citrate buffer (SSC), 50 mM sodium phosphate buffer pH 7.0) for 30 min at 66 °C. In the meantime, LNA probes to detect sense G4C2 repeats (/5TYE563/CCCCGGCCCCGGCCCC) were heated at 85°C for 75 s, and then diluted to 40 nM with hybridization buffer. After hybridization, cells were incubated with the diluted LNA probes at 66 °C for 24 h in a dark, humidified chamber. Cells were then washed once with 2 × SSC/0.1% Tween-20 for 5 min at room temperature, and twice with 0.2 × SSC for 10 min at 65 °C in the dark. The cells were counterstained with Hoechst 33258 (1 μg ml–1, Thermo Fisher Scientific), and the coverslips were mounted. Images were obtained on a Zeiss LSM700 laser scanning confocal microscope. To quantify foci-bearing cells, ~1,200–1,800 cells per coverslip and 3 coverslips per treatment were used for counting the number of foci-positive cells. These counts were used to determine the average percentage of foci-positive cells.
Statistics.
Data are presented as mean ± standard error of mean (s.e.m.), and analyzed with two-tailed unpaired t-test, one-way or two-way ANOVA followed by Tukey’s post hoc analysis (Prism statistical software). Statistical analysis of RNA-Seq data is described in RNA-Seq and Gene Ontology analysis above.
Reporting Summary.
Further information on experimental design is available in the Nature Research Reporting Summary linked to this article.
Data availability.
The data that support the findings of this study are available from the corresponding author upon reasonable request. The GEO accession number of the RNA-Seq data is GSE112931.
Supplementary Material
Acknowledgements
We are grateful to all patients who agreed to donate post-mortem tissue. This work was supported by the National Institutes of Health/National Institute of Neurological Disorders and Stroke (R35NS097273 (L.P.); P01NS084974 (L.P., D.W.D., R.R. and B.O.); P01NS099114 (T.F.G. and L.P.); R01NS088689 (L.P.); R35NS097263(10) (A.D.G.)); the Mayo Clinic Foundation (L.P.); the Amyotrophic Lateral Sclerosis Association (T.F.G., L.P., Y.-J.Z. and M.P.), the Robert Packard Center for ALS Research at Johns Hopkins (A.D.G. and L.P.) and the Target ALS Foundation (T.F.G., A.D.G., L.P. and Y.-J.Z.). We would like to thank J. N. Stankowski, E. A. Perkerson, L. Rousseau and V. Phillips for technical support. This manuscript is dedicated to Dr Antimo D’Aniello.
Footnotes
Competing interests
The authors declare no competing interests.
Additional information
Supplementary information is available for this paper at https://doi.org/10.1038/s41591–018-0071–1.
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.DeJesus-Hernandez M et al. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron 72, 245–256 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Renton AE et al. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron 72, 257–268 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gendron TF, Belzil VV, Zhang YJ & Petrucelli L Mechanisms of toxicity in C9FTLD/ALS. Acta Neuropathol. 127, 359–376 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang K et al. The C9orf72 repeat expansion disrupts nucleocytoplasmic transport. Nature 525, 56–61 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Donnelly CJ et al. RNA toxicity from the ALS/FTD C9ORF72 expansion is mitigated by antisense intervention. Neuron 80, 415–428 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cooper-Knock J et al. Sequestration of multiple RNA recognition motif-containing proteins by C9orf72 repeat expansions. Brain 137, 2040–2051 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sareen D et al. Targeting RNA foci in iPSC-derived motor neurons from ALS patients with a C9ORF72 repeat expansion. Sci. Transl. Med. 5, 208ra149 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Conlon EG et al. The C9ORF72 GGGGCC expansion forms RNA G-quadruplex inclusions and sequesters hnRNP H to disrupt splicing in ALS brains. Elife 5(2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee YB et al. Hexanucleotide repeats in ALS/FTD form length-dependent RNA foci, sequester RNA binding proteins, and are neurotoxic. Cell Rep. 5, 1178–1186 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mori K et al. hnRNP A3 binds to GGGGCC repeats and is a constituent of p62-positive/TDP43-negative inclusions in the hippocampus of patients with C9orf72 mutations. Acta Neuropathol. 125, 413–423 (2013). [DOI] [PubMed] [Google Scholar]
- 11.Ash PE et al. Unconventional translation of C9ORF72 GGGGCC expansion generates insoluble polypeptides specific to c9FTD/ALS. Neuron 77, 639–646 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gendron TF et al. Antisense transcripts of the expanded C9ORF72 hexanucleotide repeat form nuclear RNA foci and undergo repeat-associated non-ATG translation in c9FTD/ALS. Acta Neuropathol. 126, 829–844 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mori K et al. Bidirectional transcripts of the expanded C9orf72hexanucleotide repeat are translated into aggregating dipeptide repeat proteins. Acta Neuropathol. 126, 881–893 (2013). [DOI] [PubMed] [Google Scholar]
- 14.Mori K et al. The C9orf72GGGGCC repeat is translated into aggregating dipeptide-repeat proteins in FTLD/ALS. Science 339, 1335–1338 (2013). [DOI] [PubMed] [Google Scholar]
- 15.Zu T et al. RAN proteins and RNA foci from antisense transcripts in C9ORF72 ALS and frontotemporal dementia. Proc. Natl Acad. Sci. USA 110, E4968–E4977 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saberi S et al. Sense-encoded poly-GR dipeptide repeat proteins correlate to neurodegeneration and uniquely co-localize with TDP-43 in dendrites of repeat-expanded C9orf72 amyotrophic lateral sclerosis. Acta Neuropathol. 135, 459–474 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jovicic A et al. Modifiers of C9orf72 dipeptide repeat toxicity connect nucleocytoplasmic transport defects to FTD/ALS. Nat. Neurosci. 18, 1226–1229 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kanekura K et al. Poly-dipeptides encoded by the C9ORF72 repeats block global protein translation. Hum. Mol. Genet. 25, 1803–1813 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kwon I et al. Poly-dipeptides encoded by the C9orf72 repeats bind nucleoli,impede RNA biogenesis, and kill cells. Science 345, 1139–1145 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee KH et al. C9orf72 dipeptide repeats impair the assembly, dynamics, and function of membrane-less organelles. Cell 167, 774–788.e17 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tao Z et al. Nucleolar stress and impaired stress granule formation contribute to C9orf72 RAN translation-induced cytotoxicity. Hum. Mol. Genet. 24, 2426–2441 (2015). [DOI] [PubMed] [Google Scholar]
- 22.Yamakawa M et al. Characterization of the dipeptide repeat protein in the molecular pathogenesis of c9FTD/ALS. Hum. Mol. Genet. 15, 1630–1645 (2014). [DOI] [PubMed] [Google Scholar]
- 23.Lopez-Gonzalez R et al. Poly(GR) in C9ORF72-related ALS/FTD compromises mitochondrial function and increases oxidative stress and DNA damage in iPSC-derived motor neurons. Neuron 92, 383–391 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Freibaum BD et al. GGGGCC repeat expansion in C9orf72 compromises nucleocytoplasmic transport. Nature 525, 129–133 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boeynaems S et al. Drosophila screen connects nuclear transport genes to DPR pathology in c9ALS/FTD. Sci. Rep. 6, 20877 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shi KY et al. Toxic PRn poly-dipeptides encoded by the C9orf72 repeat expansion block nuclear import and export. Proc. Natl Acad. Sci. USA 114, E1111–E1117 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chew J et al. Neurodegeneration. C9ORF72 repeat expansions in mice cause TDP-43 pathology, neuronal loss, and behavioral deficits. Science 348, 1151–1154 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang D et al. FTD/ALS-associated poly(GR) protein impairs the Notch pathway and is recruited by poly(GA) into cytoplasmic inclusions. Acta Neuropathol. 130, 525–535 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schmidt EK, Clavarino G, Ceppi M & Pierre P SUnSET, a nonradioactive method to monitor protein synthesis. Nat. Methods 6, 275–277 (2009). [DOI] [PubMed] [Google Scholar]
- 30.Landry DM, Hertz MI & Thompson SR RPS25 is essential for translation initiation by the Dicistroviridae and hepatitis C viral IRESs. Genes Dev. 23, 2753–2764 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hertz MI, Landry DM, Willis AE, Luo G & Thompson SR Ribosomal protein S25 dependency reveals a common mechanism for diverse internal ribosome entry sites and ribosome shunting. Mol. Cell. Biol. 33, 1016–1026 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang YJ et al. C9ORF72 poly(GA) aggregates sequester and impair HR23 and nucleocytoplasmic transport proteins. Nat. Neurosci. 19, 668–677 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mizielinska S et al. C9orf72 repeat expansions cause neurodegeneration in Drosophila through arginine-rich proteins. Science 345, 1192–1194 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lin Y et al. Toxic PR poly-dipeptides encoded by the C9orf72 repeat expansion target LC domain polymers. Cell 167, 789–802.e12 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cheng W et al. C9ORF72 GGGGCC repeat-associated non-AUG translation is upregulated by stress through eIF2alpha phosphorylation. Nat. Commun. 9, 51 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Green KM et al. RAN translation at C9orf72-associated repeat expansions is selectively enhanced by the integrated stress response. Nat. Commun. 8, 2005 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang YJ et al. Aggregation-prone c9FTD/ALS poly(GA) RAN-translated proteins cause neurotoxicity by inducing ER stress. Acta Neuropathol. 128, 505–524 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Prudencio M et al. Distinct brain transcriptome profiles in C9orf72- associated and sporadic ALS. Nat. Neurosci. 18, 1175–1182 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kalari KR et al. MAP-RSeq: Mayo Analysis Pipeline for RNA sequencing. BMC Bioinformatics 15, 224 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hansen KD, Irizarry RA & Wu Z Removing technical variability in RNA-seq data using conditional quantile normalization. Biostatistics 13, 204–216 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Love MI, Huber W & Anders S Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Langfelder P & Horvath S WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9, 559 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gendron TF et al. Cerebellar c9RAN proteins associate with clinical and neuropathological characteristics of C9ORF72 repeat expansion carriers. Acta Neuropathol. 130, 559–573 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kramer NJ et al. Spt4 selectively regulates the expression of C9orf72 sense and antisense mutant transcripts. Science 353, 708–712 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request. The GEO accession number of the RNA-Seq data is GSE112931.


