Abstract
Dynein adaptor proteins such as Bicaudal D2 (BicD2) are integral components of the dynein transport machinery, as they recognize cargoes for cell cycle-specific transport and link them to the motor complex. Human BicD2 switches from selecting secretory and Golgi-derived vesicles for transport in G1 and S phase (by recognizing Rab6GTP), to selecting the nucleus for transport in G2 phase (by recognizing nuclear pore protein Nup358), but the molecular mechanisms governing this switch are elusive. Here, we have developed a quantitative model for BicD2/cargo interactions that integrates affinities, oligomeric states, and cellular concentrations of the reactants. BicD2 and cargo form predominantly 2:2 complexes. Furthermore, the affinity of BicD2 toward its cargo Nup358 is higher than that toward Rab6GTP. Based on our calculations, an estimated 1000 BicD2 molecules per cell would be recruited to the nucleus through Nup358 in the absence of regulation. Notably, RanGTP is a negative regulator of the Nup358/BicD2 interaction that weakens the affinity by a factor of 10 and may play a role in averting dynein recruitment to the nucleus outside of the G2 phase. However, our quantitative model predicts that an additional negative regulator remains to be identified. In the absence of negative regulation, the affinity of Nup358 would likely be sufficient to recruit BicD2 to the nucleus in G2 phase. Our quantitative model makes testable predictions of how cellular transport events are orchestrated. These transport processes are important for brain development, cell cycle control, signaling, and neurotransmission at synapses.
Graphical Abstract
Cytoplasmic dynein is the predominant motor complex that mediates the transport of almost all cargo that is transported toward the minus end of microtubules.1 It orchestrates a vast number of cellular transport events, including RNA/protein complexes, chromosomes, vesicles, mitochondria, proteins, and organelles, but general principles of how the correct cargo is selected at the correct time have not been established. Understanding how cargo selection for dyneindependent transport is regulated is important as it facilitates transport events that are essential for faithful chromosome segregation, signaling, signal transmission at synapses in the brain, cell migration, phagocytosis, muscle development, and brain development.1
Adaptor proteins such as Bicaudal D2 (BicD2) are integral parts of the dynein transport machinery, as they recognize cargo and link it to the motor complex.2−7 In metazoans, dynein adaptors like BicD2 are required to activate dynein for processive transport once cargo is bound.2−7 BicD2/cargo complexes directly link dynein with its activator dynactin.2−8 This mechanism couples cargo loading to activation of processive transport and prevents unproductive transport events that would cause futile hydrolysis of ATP.
In the absence of cargo, full-length BicD2 exists in an autoinhibited state, in which the N-terminal dynein/dynactin recruitment site (NTD) is inaccessible and likely masked by the C-terminal cargo binding domain (CTD) (Figure 1A–C).2,5,9,10 Binding of cargo to the CTD releases autoinhibition. The CTD of BicD2 is essential for autoinhibition, since a transport competent ternary complex can be assembled from dynein, dynactin, and the BicD2-NTD (i.e., without the CTD).2−6
Figure 1.
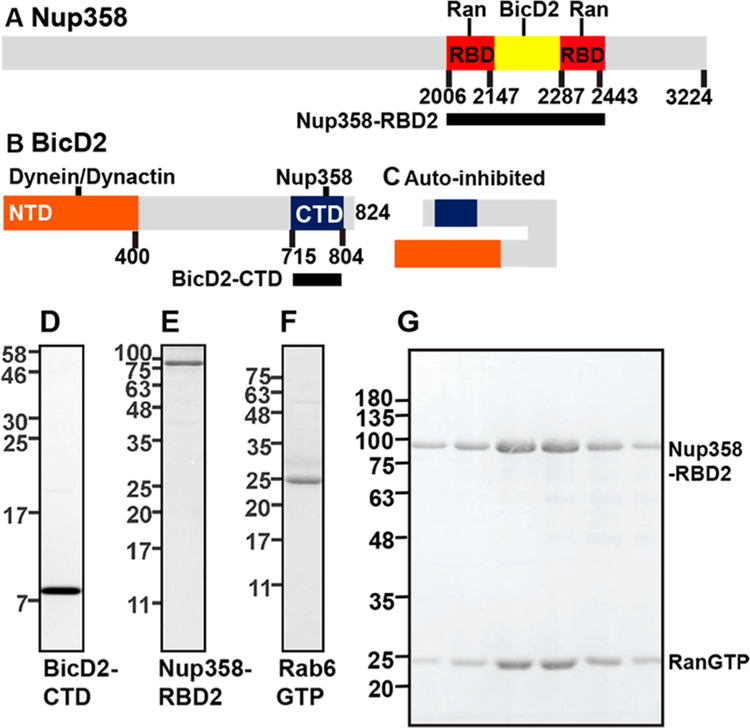
Purification of the cargo binding domain of BicD2 and its cargoes. (A) For biophysical studies, Nup358-RBD2 (black bar), a fragment of Nup358 that includes the BicD2 binding site and the two adjacent Ran binding domains (RBD red), was purified. (B) The N-terminal domain (NTD, orange) of BicD2 interacts with dynein/dynactin, while the CTD (blue) recruits cargo.2,11 For biophysical studies, the BicD2-CTD was purified (black bar). (C) In absence of cargo, the NTD and CTD of BicD2 form an autoinhibited state that cannot recruit dynein/dynactin.2,5,9,10 (D−F) SDS-PAGE analysis of purified proteins is shown. Masses of molecular weight standards in kDa are indicated on the left. (D) BicD2-CTD. (E) Nup358-RBD2. (F) Rab6GTP. (G) Purification of a RanGTP/Nup358 complex. Purified Nup358-RBD2 and RanGTP were mixed. Nup358-RBD2 with RanGTP bound was separated by gel filtration. SDS-PAGE analysis of the elution fractions is shown.
BicD2 recognizes multiple cargoes, which bind to its CTD. The structures of two homologues of the human BicD2-CTD were recently determined.9,10 Binding assays confirmed that several cargoes bind to essentially the same binding site on the BicD2-CTD and therefore likely compete.9,10 In humans, the two predominant known BicD2 cargoes are Rab6GTP, which recruits BicD2 to Golgi-derived and secretory vesicles, and the nuclear pore complex protein Nup358, which recruits BicD2 to the nucleus.11,12 Additional cargoes were identified for BicD2 homologues in flies and worms, including Egalitarian,13−15 Fragile X mental retardation protein,16 the clathrin heavy chain,17 the lamin Dm0,18 and Unc-83.19 Whether such interactions are also formed by human homologues remains to be investigated.
It is unknown how the cargo selectivity of BicD2 is regulated to switch between the transport of Rab6-positive vesicles in G1 and S phase12,20−23 and transport of the nucleus in G2 phase (by recognizing Nup358).11 Throughout the cell cycle, Nup358, Rab6GTP, and BicD2 are expressed and accessible from the cytosol.11,24 Several phosphorylation sites specific for the G2 phase-specific kinase cyclin-dependent kinase 1 (Cdk1) were mapped in the BicD2 binding domain of Nup358, which strengthen the interaction in a pull-down assay.25 However, additional regulatory mechanisms remain to be identified.
BicD2 recruits dynein/dynactin to Rab6-positive secretory and Golgi-derived vesicles and promotes their minus-end directed transport along microtubules.12 These interactions are important for promoting the dynein-dependent sorting of these vesicles. Rab6 is a GTPase,26 which cycles between GTP- and GDP-bound states, but only the GTP-bound state binds BicD2 with high affinity.12,27 The transport of secretory and Golgi-derived vesicles is important for signaling pathways and for neurotransmission at synapses.
Notably, BicD2 also recruits dynein/dynactin to the nucleus. The cell nucleus is transported and positioned in a cell cyclespecific manner, a process that is important for cell cycle control, as well as brain and muscle development.11,28−30 However, it remains largely elusive how the timely transport of the nucleus is initiated and orchestrated. Dynein recruitment sites at the nuclear envelope are provided by two proteins, Nup133 and Nup358, that are part of the nuclear pore complex (NPC).11,25,28−30
Nup358 (also known as Ran binding protein 2, RanBP2) is a component of the cytoplasmic filaments of the NPC.31,32 In the G2 phase, an interaction is formed between Nup358 and BicD2, which recruits dynein and dynactin to the nucleus (Nup358/BicD2 pathway).11 Cdk1 promotes activation of this pathway,25,33,34 which is essential for apical nuclear migration in brain progenitor cells, a fundamental process in brain development that is required for the differentiation of these cells.30 In the G2 phase, nuclei of brain progenitor cells migrate in a dynein-powered movement along microtubule tracks. Once the nuclei reach their destination at the apical surface of the brain, the brain progenitor cells divide, which ultimately leads to their differentiation. In all cells, dynein recruitment is required for positioning the nucleus respective to the centrosome in initial stages of mitosis and thus is needed for proper spindle assembly and faithful chromosome segregation.11,28 Dynein is also important for muscle development, as it positions nuclei in postmitotic, multinucleated myotubes.35 The importance of BicD2-dependent transport events in brain and muscle development is reflected in the fact that mutations in BicD2 cause a subset of spinal muscular atrophy (SMA) cases, a neuromuscular disease which is the most common genetic cause of death in infants.36−39
Here, we present new insights on how BicD2 switches between its predominant cargoes Rab6GTP and Nup358 in a cell cycle-dependent manner. We have determined the affinities of two BicD2/cargo complexes. Unexpectedly, the affinity of BicD2/Nup358 is higher than that of BicD2/Rab6GTP. Based on these data, we developed a quantitative model for BicD2/cargo interactions that allows identifying testable regulatory mechanisms for effective switching between distinct cargoes. Our model predicts that a negative regulator remains to be identified, which prevents untimely dynein recruitment to the nucleus outside of the G2 phase. Of note, we identified RanGTP as a negativeregulator of the Nup358/BicD2 interaction.
MATERIALS AND METHODS
Protein Expression and Purification.
DNA sequences encoding human Rab6a (which is referred to as Rab6 in the remaining text), BicD2, Nup358, and Ran were cloned into expression vectors as described.33 The GTPases Rab6a and Ran were expressed with point mutations that lock them in the GTP-bound state (i.e., GTPase deficient). Rab6aGTP Q72L and RanGTP Q69L are stabilized in the GTP-bound state and are referred to as Rab6GTP and RanGTP in the remaining text.12,40 Full-length cDNAs were either purchased from OpenBiosystems or obtained from researchers as described under Acknowledgments. Expression constructs were generated by PCR from full-length DNA and cloned into an expression vector by two restriction enzymes. To create the expression constructs for Nup358-RBD2 (Figure 1A) and Rab6GTP, the sequences encoding for residues 2006−2443 of human Nup358 (NCBI database entry XM_005264002.2) and full-length human Rab6a Q72L (NCBI BC096818.1) were each cloned into the pGEX-6P1 vector (GE Healthcare) with the BamHI and XhoI restriction sites. This plasmid was used to express N-terminal glutathione S-transferase (GST) fusion proteins. Note that the N-terminal GST tag can be cleaved off by PreScission protease (GE Healthcare).
To create expression constructs of the human BicD2-CTD (Figure 1B, NCBI NM_029791.4) and RanGTP Q69L (NCBI NM_006325.4), sequences corresponding to residues 715−804 of human BicD2 and human Ran Q69L were each cloned into the pET28a vector with the NdeI and XhoI restriction sites. To create an expression construct for Rab6minGTP, the sequence corresponding to residues 13−174 of human Rab6a Q72L was cloned into a pET28a-pres vector with NdeI and XhoI restriction sites. This is a modified ET28a vector, where the thrombin cleavage site was replaced by a PreScission protease cleavage site (same site as in the pGEX-6P1 vector). These constructs were used to express fusion proteins with an N-terminal His6-tag, which can be cleaved off by either thrombin (pET28a) or PreScission protease (pET28a-pres).
All proteins were expressed in the E. coli Rosetta 2(DE3)-pLysS strain at 37 °C as described,33 with the following modifications: RanGTP was expressed in the E. coli RIL (DE3)pLysS strain. For expression of Nup358-RBD2, after the cells were induced with IPTG, the temperature was changed to 25 °C and cells were harvested after 14−18 h.
Protein purification was performed using protocols as described,33,41,42 using the following strategies: His6-tagged BicD2-CTD, His6-tagged Rab6GTP (residues 13−174), and His6-tagged RanGTP were purified by Ni-NTA affinity chromatography, followed by protease cleavage of the His6-tag (using thrombin or PreScission protease depending on the vector), and a second affinity chromatography step postcleavage. GST-tagged Nup358-RBD2 was purified by glutathione affinity chromatography and eluted by glutathione. GST-tagged Rab6GTP was purified by glutathione affinity chromatography and eluted by PreScission protease cleavage of the tag. Then, 1 mM GTP and 2 mM MgCl2 were added to purified Rab6GTP and RanGTP. All proteins were further purified by gel filtration chromatography as described33 using a HiLoad 16/600 Superdex 200 pg column (GE Healthcare) and the following gel filtration buffer: 20 mM HEPES pH 7.5, 150 mM NaCl, 0.5 mM TCEP. For purification of Rab6GTP and RanGTP, the buffer was supplemented with 2 mM MgCl2. In addition, 1 mM GTP was added to these proteins after purification. Protein concentrations were determined by spectrophotometry with the peptide method and concentrated proteins were flash-frozen in liquid nitrogen as described.33 Purified proteins were analyzed by SDS-PAGE, using 10% and 16% acrylamide gels, and stained by Coomassie Blue.
A Nup358-RBD2/RanGTP complex was assembled from purified Nup358-RBD2 and RanGTP at a 1:4 molar ratio, 0.5 mM GTP was added, and the mixture was incubated for 30 min on ice. The complex was purified by size exclusion chromatography.
For analytical size exclusion chromatography, 2.5 mM GTP and 7.5 mM MgCl2 were added to all samples. Proteins were mixed in an equimolar ratio (0.2 mg of BicD2-CTD, 0.5 mg of Rab6GTP and 1.6 mg of Nup358-RBD2). Samples (400 μL) were injected onto a Superdex 200 Increase 10/300 GL column (GE Healthcare) that was equilibrated with the following buffer: 20 mM HEPES pH 7.5, 150 mM NaCl, 0.5 mM TCEP, and 0.5 mM MgCl2.
Size Exclusion Chromatography Coupled to Multiangle Light Scattering (SEC-MALS).
Purified proteins were subjected to analytical size exclusion chromatography as described41−43 on a Superdex 200 10/300 GL column (GE Healthcare), using 20 mM HEPES pH 7.5, 150 mM NaCl, 0.5 mM TCEP as a gel filtration buffer, at ambient room temperature. The size exclusion chromatography column was connected to multi-angle light scattering and refractive index detectors (DAWN 8+ and Optilab TrEX; Wyatt Technology). Weight-averaged molar masses were determined by multi-angle light scattering using the ASTRA 6 software (Wyatt Technology) as described42,43 and averaged from three experiments. Representative experiments are shown. The error of the mass determination was calculated at 5%, which was in all cases greater than the standard deviation of these experiments.
Isothermal Calorimetry Titration (ITC) Experiments.
Purified proteins were dialyzed in the following buffer: 150 mM NaCl, 30 mM HEPES pH 7.5, 1 mM TCEP, and 1 mM MgCl2. For Rab6GTP constructs, the MgCl2 concentration was increased to 2 mM and 0.7 mM GTP was added. In all ITC experiments, the BicD2-CTD was placed in the cell of a MicroCal auto-ITC200 calorimeter (GE Healthcare) and titrated with a binding partner in the syringe at a 10-fold higher concentration. Protein concentrations used are listed in Table S1. Experiments were performed at 25 °C. As interacting partners, Nup358-RBD2, Nup358-RBD2/RanGTP complex, Rab6GTP, andRab6GTPmin were used.
Each BicD2 titration was corrected by subtracting the curves of the respective interacting partners into buffer. The titration curve was fitted with the one-site model to determine the equilibrium binding constant KA, the number of sites N, and the change in enthalpy ΔH. The equilibrium dissociation constant KD (affinity) is the inverse of the equilibrium binding constant KA. The Origin software (OriginLab) was used for data analysis.
Calculation of Ratios of Complexed and Unbound Reactants.
In order to calculate ratios of complexed and unbound reactants, Mathematica (Wolfram) was used to numerically solve a system of five eqs (eqs 1–5, see Results) for five variables ([BicD2], [Nup358], [Rab6GTP], [BicD2/Nup358], and [BicD2/Rab6GTP], which describe the thermodynamic equilibria for interactions of BicD2 with Nup358 and Rab6GTP. While analytic solutions to this system of equations were found, the equations were too complex for rational analysis.
To calculate total cellular concentrations of reactants, the number of molecules per cell was calculated by multiplying the abundance ratios from Table 2A with the number of molecules of Nup358 in HeLa cells in G1 phase (32000). This number was obtained from the number of NPCs per cell44 and a copy number of 16 Nup358 molecules per NPC.45 Cytosolic concentrations of reactants were calculated from the number of molecules per cell, Avogadro’s constant, and the average volume of a HeLa cell cytosol (1310 μm3 or 1.3 × 10−12 L). The average volume of a HeLa cell cytosol was determined by subtracting the average volume of a HeLa cell nucleus (690 μm3)46 from the average volume of a HeLa cell (2000 μm3).47 A similar volume was obtained in studies that determined cytosolic volumes of HeLa cells, which also excluded volumes of smaller organelles (mitochondria, endoplasmic reticulum, vesicles).48
Table 2a.
| Table 2A Abundance of Nup358, Bicd2 and Rab6 in human organisms, normalized to Nup358 | |||||
|---|---|---|---|---|---|
| Human dataset (quality score) | Nup358 | Rab6 | BicD2 | ||
| Human peptide atlas 2014 (26.0) | 1 | 2.5 | 0.4 | ||
| Global proteome machine 2014 (23.9) | 1 | 11.0 | 0.4 | ||
| Integrated Human Proteome (15.5) | 1 | 6.4 | 0.4 | ||
| Average ratio | 1 | 6.6 ± 4.3 | 0.4 ± 0 | ||
| BicD1 | BicDRI | BicDR2 | α&β Tubulin | ||
| Human peptide atlas 2014 (26.0) | 0.05 | 0 | 0.08 | 307 | |
| Global proteome machine 2014 (26.0) | 0.11 | 0 | 0.05 | ||
| Integrated human proteome (26.0) | 0.03 | 0 | 0.06 | 698 | |
| Average ratio | 0.06 ± 0.04 | 0 | 0.06 ± 0.02 | 503 ± 200 | |
| Table 2B Cytosolic concentration of human Nup358, BicD2 and Rab6 | |||||
| Nup358 G1 phase | Rab6 | BicD2 | BicD1 & BicDR2 | α&β Tubulin (Control) | |
| Molecules per cell | 32000 | 211000 | 13000 | 4000 | 1.5*107 |
| Cytosolic conc. (nM) | 40 | 268 | 16 | 5 | 20000 |
Data was obtained from the protein abundance database.55 The protein abundance data quality score is based on the assumption that interacting proteins from a complex have a tendency to have similar abundance, and a large score indicates high data quality. All available data sets from whole human organisms were used, which reported abundance for all three proteins.55,67,68 As a control, the cellular concentration of αβ-tubulin combined was calculated using the same method, which fits well with the cellular concentration of tubulin reported in the literature (e.g., 25 μM and 12−18 μM58–60).
RESULTS
Reconstitution of Nup358/BicD2 Cargo Complexes for Biophysical Characterization.
In order to characterize BicD2/cargo interactions using biophysical methods, we established protein purification protocols that yielded highly pure BicD2 cargoes (Figure 1). To this end, we purified the C-terminal cargo binding domain of human BicD2 (BicD2-CTD, residues 715−804) (Figure 1B,D), based on previous studies of a homologue.9−11
In addition, we purified two BicD2 cargoes: Rab6GTP and a Nup358 fragment. To this end, human full-length Rab6aGTP Q72L was purified, which is stabilized in the GTP-bound state by the Q72L mutation (Figure 1F),12 and is referred to as Rab6GTP in the remaining text. The stabilization is necessary because GTP-bound Rab6 has a much higher affinity to BicD2 than the GDP-bound form.27 In addition, we purified a truncated version (Rab6GTP Q72L, residues 13−174), which is referred to as Rab6GTPmin.23
The second BicD2 cargo was a human Nup358 fragment that included the previously mapped BicD2 binding site (residues 2147−2287, Figure 1A, yellow)11 and in addition the two Ran binding domains (RBDs) framing the binding site (Figure 1A, red), which bind RanGTP with high affinity.31,49 The purified fragment is referred to as Nup358-RBD2 and contains residues 2006−2443 of human Nup358 (Figure 1E).
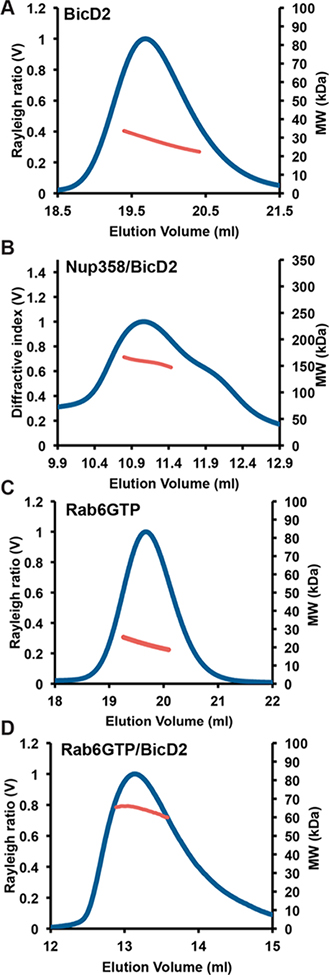
To determine the molar mass of the purified proteins, we used size exclusion chromatography coupled to multi-angle light scattering (SEC-MALS). This method allows for determination of the molar mass across a size exclusion chromatography elution peak with high accuracy (5% error). The molar mass of the purified BicD2-CTD was determined to be MW = 23.3 ± 1.3 kDa, which closely matches the mass of a BicD2 dimer (20.8 kDa) (Figure 2A). Of note, two BicD2 homologues exists as dimers in crystal structures.9,10
Figure 2.
BicD2/cargo complexes predominantly form 2:2 oligomers.(A) The purified BicD2-CTD was analyzed by SEC-MALS (at 2 mg/mL). Rayleigh ratio (blue) and molar mass MW (red) versus the elution volume are shown. The determined molar mass is MW = 23.3 ± 1.3 kDa, which closely matches the mass of a BicD2 dimer (20.8 kDa). (B) Purified Nup358-RBD2/BicD2-CTD complex was analyzed by SEC-MALS (at 14 mg/mL). Refractive index (blue) and molar mass MW (red) versus the elution volume are shown. The determined molar mass is MW = 155.4 ± 7.8 kDa, which is slightly below the molar mass of a 2:2 complex of Nup358-RBD2/BicD2-CTD (174.6 kDa). (C) The purified Rab6GTPmin was analyzed by SEC-MALS (at a concentration of 6 mg/mL). Rayleigh ratio (blue) and molar mass MW (red) versus the elution volume are shown. The determined molar mass is MW = 21.5 ±1.0 kDa, closely matching the molar mass of a Rab6GTPmin monomer (19.0 kDa). (D) The Rab6GTP/BicD2-CTD complex was analyzed at a protein concentration of 7 mg/mL by SEC-MALS. The molar mass of the major peak is MW= 61.0 ± 3 kDa, which is slightly less than the molar mass of a 2:2 complex (69.2 kDa).
To analyze the oligomeric state, we assembled a Nup358/BicD2 complex by mixing Nup358-RBD2 and BicD2-CTD. Subsequently the complex was purified by gel filtration. The molar mass of the major peak of the complex was determined to be 155.4 ± 7.8 kDa, which is lower than the molar mass of a 2:2 complex of Nup358-RBD2/BicD2-CTD (174.6 kDa) (Figure 2B). For comparison, the calculated molar mass of one Nup358-RBD2 protomer is 76.9 kDa, and that of one BicD2-CTD protomer is 10.4 kDa. A shoulder at a lower molecular weight indicates the presence of other oligomeric states (such as 1:1 and/or 1:2 complexes). We conclude that the Nup358-RBD2/BicD2-CTD complex forms various oligomeric states in a concentration-dependent manner. However, the predominant state is a 2:2 complex.
Next, the molar mass of Rab6GTPmin was determined to be MW = 21.5 ± 1.0 kDa, closely matching the molar mass of a Rab6GTPmin monomer (19.0 kDa) (Figure 2C). Note that in the X-ray structure, Rab6GTP forms a dimer.23,22min These data suggest that Rab6GTP predominantly forms a monomer in solution, but is able to dimerize at higher protein concentrations required for crystallization.
Finally, to analyze the oligomeric state of a Rab6GTP/BicD2 complex, we assembled a complex by mixing full-length Rab6GTP and BicD2-CTD, which was subsequently purified by gel filtration. The molar mass of the major peak of the complex was determined to be MW = 61.0 ± 3 kDa (Figure 2D), which is slightly less than the theoretical molar mass of a 2:2 complex of a Rab6GTP/BicD2-CTD complex (69.2 kDa; the calculated molar masses of the Rab6GTP and BicD2-CTD protomers are 23.6 kDa and 10.4 kDa, respectively). Of note, the observed molar mass of the complex at a lower protein concentration (2 mg/mL) is lower (43.1 ± 2.1 kDa; data not shown). We conclude that the Rab6GTP/BicD2-CTD complex predominantly forms a 2:2 complex, and other oligomeric states to a lesser degree.
In conclusion, BicD2-CTD forms a dimer in solution. The molar masses of the assembled BicD2/cargo complexes (Nup358-RBD2/BicD2-CTD and Rab6GTP/BicD2-CTD) indicate that they form multiple oligomeric states. Based on the molar masses, the predominant species are consistent with 2:2 complexes of BicD2 and cargo.
Nup358 Binds to BicD2 with High Affinity.
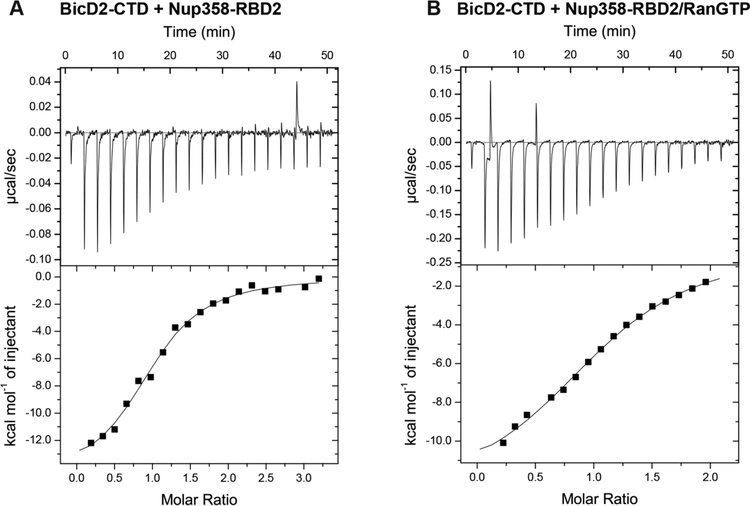
Throughout the cell cycle, Nup358 and Rab6GTP are accessible from the cytosol where BicD2 resides.11 Therefore, we designed experiments in order to investigate whether the affinity of BicD2 toward its cargo is a major determinant for cargo selection. Thus, we determined the affinities of BicD2 toward two cargoes by isothermal titration calorimetry (ITC): Rab6GTP and Nup358. For all ITC experiments, BicD2-CTD was placed in the cell of a calorimeter and titrated with the binding partner in the syringe at a 10-fold higher concentration. The affinity of Nup358-RDB2 toward the BicD2-CTD was determined to be 0.5 ± 0.07 μM (Figure 3A, Table 1A).
Figure 3.
Nup358 binds to BicD2 with high affinity. (A) The ITC titration curve of BicD2-CTD with Nup358-RBD2 is shown, from which the affinity was determined to 0.5 ± 0.07 μM. (B) ITC titration curve of the BicD2-CTD with the Nup358-RBD2/RanGTP complex. The affinity was determined to be 3.8 ± 0.4 μM. Note that RanGTP is a negative regulator of the interaction that lowers the affinity of Nup358 toward BicD2 by a factor of 10.
Table 1.
| Table 1A Affinity (KD) measurements by ITC establish RanGTP as a regulator for the Nup358/BicD2 interaction | |||
|---|---|---|---|
| Cargo | BicD2 | Affinity (μM) | # Sites |
| Nup358-RBD2 | BicD2-CTD | 0.5 ± 0.07 | 1.0 |
| RanGTP/Nup358-RBD2 | BicD2-CTD | 3.8 ± 0.4 | 1.1 |
| Table 1B Affinity of Rab6GTP to BicD2 | |||
| Cargo | BicD2 | Affinity (μM) | # Sites |
| Rab6GTP | BicD2-CTD | 12.6 ± 1.2 | 1.1 |
| Rab6GTPmin | BicD2-CTD | 12 ± 1.2 | 1.5 |
| Table 1C Determination of the enthalpy change ΔH and the entropy change ΔS by ITC | |||
| Cargo | BicD2 | ΔH (kcal/mol) | ΔS (cal/mol/deg) |
| Nup358-RBD2 | BicD2-CTD | −14.9 | −21 |
| RanGTP/Nup358-RBD2 | BicD2-CTD | −12.8 | −18 |
| Rab6GTP | BicD2-CTD | −11.0 | −13 |
The titration curve is consistent with a 1:1 molar ratio of Nup358 to BicD2 and the titration curve fits well to a one-site model. This molar ratio is consistent with a 2:2 complex based on the molar masses of BicD2-CTD and the BicD2/cargo complex obtained in our SEC-MALS data. However, we cannot distinguish between the following two possibilities of complex formation: (1) two individual cargo molecules could bind to two binding sites on the BicD2 dimer, assuming that the two sites have the same affinity; (2) a dimeric cargo could bind to a single site on the BicD2 dimer.
The BicD2 binding site on Nup358 has been previously mapped to residues 2147−2287 (Figure 1A yellow), and a recombinant minimal Nup358/BicD2 complex was previously reconstituted.11 The binding site is flanked by two Ran binding domains (RBDs, residues 2013−2142 and residues 2310−2339, Figure 1A, red). Ran is a GTPase that cycles between GTP- and GDP-bound states. RBDs have highly conserved sequence motifs and can be recombinantly purified. They bind RanGTP with high affinity (4.3 nM) and RanGDP with a 10-fold lower affinity.49 It has not been investigated if RanGTP regulates dynein recruitment to the nucleus, even though this idea is intriguing due to its role in regulation of cell cycle events.50,51 Due to the spatial proximity of the RanGTP binding sites to the BicD2 binding site,11 we hypothesized that RanGTP modulates the affinity of Nup358 toward BicD2. To address this question, we purified a complex of RanGTP Q69L and Nup358-RBD2 (Figure 1G). RanGTP Q69L is referred to as RanGTP in the remaining text, as the mutation stabilizes Ran in the GTP-bound form.40 Stabilization was necessary since RanGTP binds more tightly to the RBDs of Nup358. Due to the high affinity, the complex remains stable during gel filtration (Figure 1G). Next, the affinity of the RanGTP/Nup358-RBD2 complex toward the BicD2-CTD was determined by ITC (Table 1A, Figure 3B). Notably, the affinity of this RanGTP/Nup358 complex for BicD2 is weaker by almost 1 order of magnitude (3.8 ± 0.4 μM), compared to Nup358 alone (0.5 ± 0.07 μM) (Table 1A, Figure 3). Our results suggest that RanGTP may be a novel negative regulator of the Nup358/BicD2 interaction and decreases the affinity of Nup358 toward BicD2 by a factor of 10.
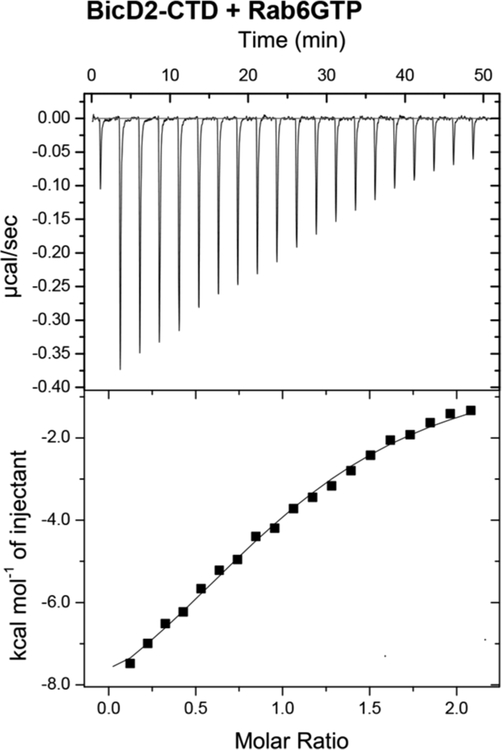
BicD2 recognizes various cargoes sharing the same binding site.9,10 In order to establish whether these cargoes compete for binding, we have determined the affinity of a second BicD2 cargo, Rab6GTP, by ITC. In previous studies, an affinity of 1 μM was reported for Rab6GTP toward BicD2,52 similar to what we found for the Nup358-RBD2 interaction with BicD2 (0.5 ± 0.07 μM, Table 1A). However, we consistently obtained a lower affinity of Rab6GTP to BicD2 of 12.6 ± 1.2 μM (Figure 4, Table 1B). Entropy changes upon binding also vary for both cargoes (Table 1C). Notably, an N- and C-terminally truncated version of Rab6GTP Q72L (residues 13−174), which was used in these previous studies,52 essentially yielded the same affinity as full-length Rab6GTP (12.6 ± 1.2 vs 12 ± 1.2 μM, respectively; Figure S1, Table 1B). In addition, we purified and characterized a slightly longer fragment of BicD2 that was used in these previous studies (residues 706−824), and we removed the affinity tag. Again, the affinity of the longer BicD2 fragment to Rab6GTP was omparable to the shorter BicD2-CTD (data not shown). It should be noted that the longer BicD2 fragment was more prone to aggregation as judged by size exclusion chromatography, compared to our BicD2-CTD.
Figure 4.
Affinity between Rab6GTP and BicD2-CTD was determined by ITC as 12.6 ± 1.2 μM. The ITC titration curve of BicD2-CTD with Rab6GTP is shown.
Therefore, the discrepancy between these previously obtained affinities and our results is likely due to subtle differences in the experimental procedure, such as higher ionic strength of the reaction buffer (150 mM NaCl compared to 80 mM LiCl used to the previous study) or the fact that the BicD2 fragment in the previous study was GST-tagged, which may alter the affinity.52 It should be emphasized that all of our ITC experiments were performed under identical conditions, which allow for direct comparison of the affinities of BicD2 toward distinct cargoes. Thus, our results showed that Rab6GTP has a lower affinitytoward BicD2-CTD compared to Nup358.
To conclude, in the absence of regulators, BicD2 binds Nup358 with a higher affinity than Rab6GTP. Of note, we did identify a negative regulator, RanGTP, which lowers the affinity of Nup358 to BicD2 by a factor of 10.
Nup358 Competes Efficiently with Rab6GTP for Binding of BicD2-CTD.
Based on the affinities from the ITC experiments, one would expect that in the presence of all three proteins, Nup358 would compete efficiently with Rab6GTP for binding of BicD2-CTD. In order to test this, we designed a competition assay with all three proteins (Figure 5).
Figure 5.
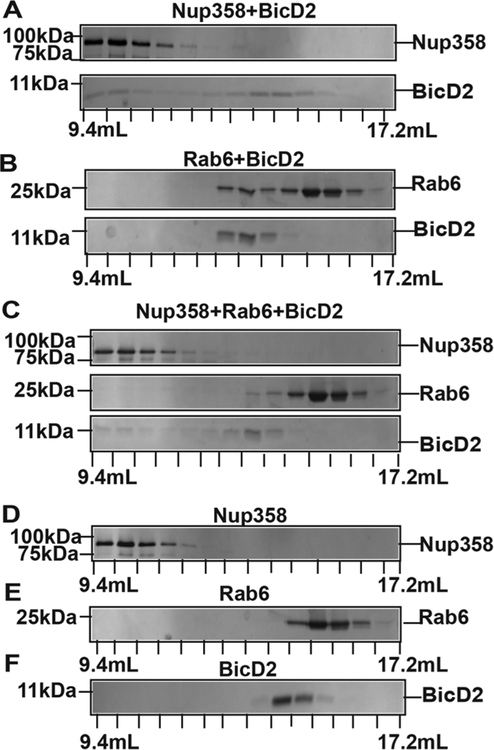
Nup358 competes efficiently with Rab6GTP for binding of BicD2-CTD. (A) Purified Nup358-RBD2 and BicD2-CTD were mixed in a 1:1 molar ratio and analyzed by size exclusion chromatography. An SDS-PAGE of the elution fractions is shown. Masses of molecular weight standards are shown on the left and elution volumes on the bottom. (B) The same analysis was performed for Rab6GTP and BicD2-CTD. (C) In a competition assay, Nup358-RBD2, Rab6GTP, and BicD2-CTD were mixed in a 1:1:1 molar ratio and the mixture was analyzed by size exclusion chromatography. (D−F) As controls, the individual proteins were analyzed: (D) Nup358-RBD2, (E) Rab6GTP, and (F) BicD2-CTD.
First, we assessed binding of Nup358-RBD2 and Rab6GTP to BicD2-CTD individually. To this end, Nup358-RBD2 was mixed with BicD2-CTD and analyzed by size exclusion chromatography (Figure 5A). In this experiment, the majority of the BicD2-CTD peak is shifted toward a higher mass compared to BicD2-CTD alone (Figure 5A,F), and a large fraction of BicD2-CTD coelutes with Nup358-RBD2. During gel filtration, complexes dynamically associate and dissociate. Since free BicD2-CTD has a much lower mass compared to Nup358/BicD2, it migrates more slowly, which leads to a gradual separation and loss of BicD2 from Nup358/BicD2 complexes. This likely results in the observed large fraction of BicD2 that elutes at a volume corresponding to higher mass compared to BicD2 alone, but yet does not coelute in the fractions with Nup358-RBD2. These results suggest that Nup358-RBD2 interacts strongly with the BicD2-CTD.
Next, we mixed Rab6GTP with BicD2-CTD and analyzed the mixture by size exclusion chromatography (Figure 5B). In this experiment, the majority of the BicD2-CTD peak is shifted toward higher mass compared to BicD2-CTD alone (Figure 5B,F). Notably, a large fraction of the Rab6GTP elution peak is also shifted toward a higher mass (Figure 5B,E). These results indicate a strong interaction of Rab6GTP with BicD2-CTD.
Next, in a competition assay, Nup358-RBD2, Rab6GTP, and BicD2-CTD were mixed in a 1:1:1 molar ratio and analyzed by size exclusion chromatography (Figure 5C). In this experiment, a large pool of BicD2-CTD coelutes with Nup358-RBD2, as expected. A very small pool of BicD2-CTD coelutes with Rab6GTP, and a sizable pool of BicD2-CTD elutes between both peaks. Notably, in the competition experiment, the majority of Rab6GTP elutes at the same volume as Rab6GTP alone (Figure 5C,E), and only a very small fraction of the elution peak is shifted toward a higher mass, which suggests that only a small fraction of Rab6GTP is bound to BicD2. To compare, when Rab6GTP is mixed with BicD2-CTD in the absence of Nup358, a large fraction of the Rab6GTP elution peak is shifted toward a higher mass (Figure 5B,F). It should also be noted that during gel filtration, complexes dynamically associate and dissociate. In the case of Nup358-RBD2 and BicD2-CTD, the molar masses of the interaction partners are very different, leading to a larger degree of separation and loss of BicD2 from the complex, compared to Rab6GTP/ BicD2 complexes, which are subjected to a lesser degree of separation. This explains the observed elution profile, where a small population of BicD2 coelutes with Nup358, but a large pool of BicD2 elutes at a higher mass compared to BicD2 alone or compared to Rab6GTP.
These data suggest that in the presence of all three components, a larger fraction of BicD2-CTD is recruited to Nup358-RBD2 compared to Rab6GTP.
To conclude, these data suggest that Rab6GTP and Nup358-RBD2 interact with the BicD2-CTD individually. Notably, in the presence of all three proteins, Nup358-RBD recruits a larger fraction of BicD2-CTD compared to Rab6GTP. These results areconsistent with the binding affinities obtained by ITC experiments and confirm that Nup358-RBD2 efficiently competes with Rab6GTP for binding of BicD2-CTD.
The Affinity of BicD2 toward Nup358 Is Strong Enough to Recruit BicD2 to the Nucleus in the Absence of Regulation.
In order to predict the outcome of cellular transport pathways, we developed a quantitative model for BicD2/cargo interactions that integrates the affinities obtained by ITC. This mechanism will allow us to assess how many of the binding sites for BicD2 in the cell are likely to be saturated at distinct time points and to determine whether regulatory mechanisms are effective to switch between different types of cargo. The quantitative model includes the predominant BicD2 cargoes Nup358 and Rab6GTP, but additional minor cargoes can be added in the future. In human tissue culture cells, endogenous BicD2 predominantly colocalizes with Rab6-positive vesicles in G1 and S phase. In the G2 phase, BicD2 mainly colocalizes with Nup358 at the nuclear envelope.11 In addition, cytoplasmic staining of endogenous BicD2 suggests a cytoplasmic pool at both cell cycle stages.11 Nup358 and Rab6GTP are also the two predominant cargoes in humans based on pull-down assays.11 These BicD2/cargo interactions can be described with the following five equations:
| (1) |
| (2) |
| (3) |
| (4) |
| (5) |
These equations are based on the assumption that Nup358 and Rab6GTP bind to BicD2 in a 1:1 ratio, which was directly obtained from our ITC experiments (Figures 3 and 4) and which was also confirmed by the molar masses of the BicD2/cargo complexes (Figure 2). The initial model has the following parameters: concentrations of the reactants [BicD2], [Nup358], [Rab6GTP], [BicD2/Nup358], and [BicD2/Rab6GTP] (i.e., the unbound and complexed fractions of these proteins), the total concentration of these proteins in the cell ([BicD2total], [Nup358total], and [Rab6GTPtotal]), and equilibrium dissociation constants KD1 (for binding of Nup358) and KD2 (for binding of Rab6GTP) for the reactions.
KD1 and KD2 were obtained from ITC experiments (Tables 1A, B). To calculate total cellular concentrations of the reactants, the number of molecules of Nup358 in the cell was determined from the published number of NPCs in HeLa cells44 and the copy number of Nup358 per NPC, which is 16 according to recent structural models.45,53 The number of NPCs per cell is well established in the field;44 however, it should be noted that a minor fraction of additional Nup358 molecules may be localized in cytoplasmic pools.54 Based on these data, there are 32000 molecules of Nup358 in NPCs in G1 phase and 64000 molecules at the onset of mitosis per cell.44 The average volume of the cytoplasm of a HeLa cell is 1310 μm3 (1.3 × 10−12 L) (see Materials and Methods). With this volume and the Avogadro’s constant, the molar concentration of Nup358 in G1 phase is 40 nM (Table 2B). It should be noted that, at the onset of mitosis, the number of NPCs in a cell doubles;44 however, the cytosolic volume sees a roughly two-fold increase, as well,48 and therefore the cellular concentration of Nup358 is expected to be similar in the interphase and G2 phase.
Cellular concentrations of BicD2 and Rab6GTP were calculated from abundances reported in the protein abundance database.55 These abundances come from proteomic mass spectrometry data sets and are reported in parts per million, which is the number of molecules of the target protein in respect to the entire proteome. To scale these data, we calculated the relative ratios of the abundances of the target proteins respective to Nup358 for each data set and converted the abundance ratios to cellular concentrations based on the concentration of Nup358. Three human data sets were selected and averaged (Table 2A). Based on these data, the cellular concentration of Rab6 is 268 nM (Table 2B). It should be noted that the cellular pool of Rab6 consists of GTP- and GDP-bound states. Since only the GTP-bound state has a strong affinity toward BicD2,52 the concentration of Rab6 that is available for binding to BicD2 will be lower than 268 nM. While the ratio of cellular Rab6GTP/Rab6GDP is unknown, the pool of Rab6 that is sequestered to membranes is expected to be in the GTP-bound state. It has been established that, in cells, a larger fraction of Rab6 is sequestered to membranes than that to the cytosol; therefore, we conclude that our concentration estimate is reasonable.12,56
Based on our calculation, the cellular concentration of BicD2 is 16 nM, and the abundance is similar in all data sets. Furthermore, Western blots of human tissue culture cell extracts at distinct cell cycle stages have shown that the cellular concentrations of BicD224 and Rab6aGTP57 do not change throughout the cell cycle. As discussed above, the concentration of Nup358 is also kept at the same level throughout the cell cycle.44 The number of all three proteins, however, is expected to roughly double, as the cellular volume roughly doubles in G2 phase.48
In addition to BicD2, human cells have three paralogues that likely bind the same cargoes: BicD1, BicDR1, and BicDR2. The cellular concentration of these paralogues combined is 5 nM. As this is lower than the concentration of BicD2 (16 nM), these paralogues are currently not considered in subsequent calculations.
As a control, we used the same approach to calculate the cellular concentration of tubulin, which is very well established in the literature. A concentration of 20 μM was obtained for tubulin (including all isoforms of human α- and β-tubulin), which fits well with the cellular concentration reported in the literature (25 and 12−18 μM58−60).
Notably, the cellular concentration of BicD2 is more than an order of magnitude lower than the concentration of the cargoes Nup358 and Rab6GTP combined, which therefore represents a limiting factor for cargo transport and selection. A limiting amount of BicD2 is also in line with the published observation that overexpression of BicD1 leads to increased recruitment of dynein to Rab6-positive vesicles.12
Eqs 1–5 can be applied to simulate several scenarios, which are characterized by distinct input values. Notably, the number of BicD2 molecules that are recruited to Rab6-positive vesicles and to the nucleus through Nup358 can be calculated. The values for KD1 and KD2 were obtained from ITC experiments (Table 1A) and inserted into these equations together with the cellular concentrations of the reactants (Table 2B). As a result, only five variables remained, which allowed us to numerically solve eqs 1–5. Table 3A summarizes the resulting concentrations of [BicD2/Nup358], [BicD2/Rab6GTP], and the unbound reactants for various input values.
Table 3A.
Calculation of concentrations of Nup358/BicD2 and Rab6/BicD2 complexes formed, as well as of the free reactants (BicD2free, Nup358free, Rab6free).
| Condition | Input parameters (nM) | BÌCD2free (nM) | Nup358/BicD2 (nM) | Rab6/BicD2 (nM) | Nup358free (nM) | Rab6free (nM) |
|---|---|---|---|---|---|---|
| 1. Standard Condition (SC)] | KD1a=500 | 15 | 1.2 | 0.3 | 39 | 267 |
| KD2b=12,600 | ||||||
| Nup358tot=40 | ||||||
| Rab6tot=268 | ||||||
| BicD2totc=16 | ||||||
| 2. RanGTP present | SC KD1=3,800 | 16 | 0.2 | 0.3 | 40 | 267 |
| 3. Nup358 absent | SC Nup358tot=0 | 16 | 0 | 0.3 | 0 | 267 |
| 4. Rab6 absent | SC Rab6tot=0 | 15 | 1.2 | 0 | 39 | 0 |
| 5. Rab6 absent | SC Rab6tot=0 KD1=3,800 | 16 | 0.2 | 0 | .40 | 0 |
KD1 is the dissociation equilibrium constant for binding of Nup358 to BicD2.
KD2 is the dissociation equilibrium constant for binding of Rab6GTP to BicD2.
the subscripttot denotes total cellular concentration.
The first scenario that was tested assumed that RanGTP is not bound to Nup358 (i.e., KD1 = 0.5 μM) and that the cellular concentration of Nup358total is 40 nM. In addition, it was assumed that KD2 = 12.6 μM, [Rab6GTPtotal] = 268 nM, and [BicD2total] = 16 nM. With these values, 1.2 nM BicD2 is recruited to Nup358, 0.3 nM BicD2 is recruited to Rab6GTP, and 15 nM BicD2 is not complexed (Table 3A). This corresponds to ~1000 molecules of BicD2 bound to Nup358 and ~250 molecules of BicD2 bound to Rab6GTP(Table 3B). These results suggest that in the absence of regulation, Nup358 strongly binds to BicD2 and can efficiently compete with Rab6GTP, although the BicD2 binding sites are far from being saturated.
Table 3B.
Number of BicD2 molecules recruited to cargo
| Condition (Table 3A) | # BicD2 bound to Nup358 | # Rab6GTP bound to BicD2 |
|---|---|---|
| 1 | 1000 | 250 |
| 2 | 150 | 250 |
| 3 | 0 | 250 |
| 4 | 1000 | 0 |
| 5 | 150 | 0 |
The total number of BicD2 molecules per cell was calculated to be 13000 (16nM) (Table 2).
Because the affinity of Nup358 toward BicD2 is comparatively high, we sought to identify negative regulators for the interaction that could prevent BicD2 recruitment outside of the G2 phase. One such negative regulator is RanGTP, which lowers the affinity by a factor of 10. If the same calculation is performed in this second scenario with a KD1 of 3.8 μM, 0.2 nM BicD2 is recruited to Nup358 and 0.3 nM BicD2 is recruited to Rab6GTP (Table 3A). In this scenario, much less BicD2 is recruited to the nucleus (~150) (Table 3B). This suggests that RanGTP could potentially be an important negative regulator for the Nup358/BicD2 interaction.
A third alternative scenario for cargo switching would be that the accessibility of these BicD2 binding sites is regulated in a cell cycle-specific manner. This mechanism can be simulated by assuming that one of the BicD2/cargo complexes is absent. If Nup358 was absent, 0.3 nM BicD2 would be recruited to Rab6GTP (roughly the same amount as with Nup358 present). In the absence of Rab6GTP, 0.2−1.2 nM BicD2 would be recruited to the nucleus through Nup358 (Table 3A).
To conclude, we have developed a quantitative model for BicD2/cargo interactions. Our results suggest that the number of BicD2 molecules is lower than the number of cargo molecules, making it a limiting factor under certain conditions. Due to the high affinity of Nup358 to BicD2, the concentration of Nup358/BicD2 complexes formed in various scenarios is ~0.2−1.2 nM, which corresponds to ~50−1000 molecules of BicD2 that are recruited to the nucleus.
Therefore, based on our calculation, the affinity of Nup358/BicD2 is likely high enough to recruit BicD2 to the nucleus at any point of the cell cycle. Notably, currently available data do not explain how dynein recruitment to the nucleus is averted outside of G2 phase. Of note, we have established RanGTP as a negative regulator of the Nup358/BicD2 interaction.
DISCUSSION
BicD2 recognizes cargo for cell cycle-specific, dynein-dependent transport events that are important for brain development, cell cycle control, signaling, and neurotransmission at synapses. In cell biology studies, it has been observed that BicD2 switches from the transport of Rab6-positive vesicles in G1 and S phase12,20−23 to the transport of the nucleus in G2 phase (by recognizing Nup358).11 Our results provide new mechanistic insights into how the dynein adaptor human BicD2 selects between its predominant cargoes Rab6GTP and Nup358 in a cell cycle-dependent manner. Here, we have determined the affinities of two BicD2/cargo complexes, and we have developed a quantitative model for BicD2/cargo interactions. BicD2/Nup358 has a higher affinity compared to the BicD2/Rab6GTP complex, which would be sufficient to recruit a significant number of BicD2 molecules to the nucleus at any cell cycle stage. Notably, RanGTP is a negative regulator of the BicD2/Nup358 interaction, which may potentially play a role in preventing dynein recruitment to the nucleus outside of G2 phase.
Based on our data, we propose the following hypothesis for how BicD2 selects between its predominant cargoes Rab6GTP and Nup358 (Figure 6): The Nup358 domain binds BicD2 with high affinity, which in the absence of regulation would result in an untimely recruitment of dynein to the nucleus. Of note, a single dynein motor can produce a force of 4.3 pN,61 which is likely more than sufficient to create the force required to transport the nucleus at the average velocity, which is observed during apical nuclear migration of brain progenitor cells.30 We calculated the force required for transport of the nucleus (F = 2.6 × 10−13 N) by applying Stokes law (F = 6πrηv), with 1.4 nm/s as average velocity v,30 5.5 μm as radius r,46 and 1.8 Pa s as cellular viscosity η.62
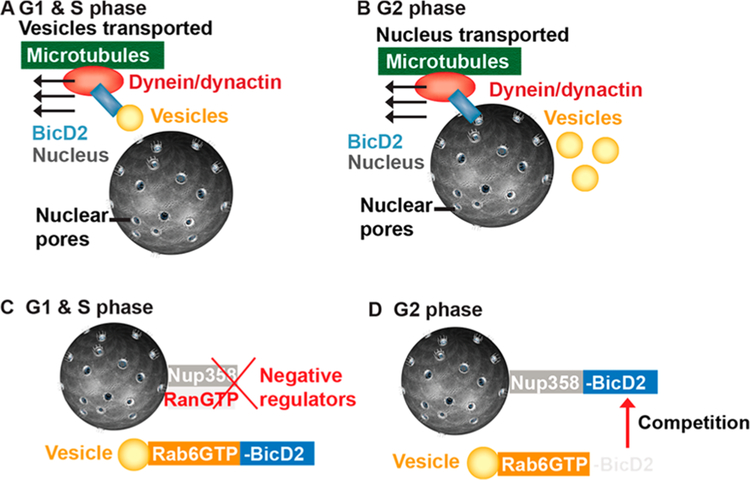
Figure 6.
Proposed regulatory mechanisms for cargo selection for BicD2/dynein-dependent transport events. (A, B) BicD2 switches between recognizing vesicles and the nucleus for transport in a cell cycle-specific manner. (A) During the G1 and S phases, BicD2 mainly recognizes Rab6-positive secretory and Golgi-derived vesicles as cargo for transport.11 (B) The cell nucleus is recognized as cargo for dynein by BicD2 specifically in G2 phase.11 (C) In the G1 and S phases, BicD2 predominantly recruits dynein to Rab6-positive vesicles. Since the affinity of Nup358 toward BicD2 is high, a negative regulatory mechanism is likely required to avert BicD2 recruitment to the nucleus outside of G2 phase. A candidate is RanGTP; however, an additional negative regulator likely remains to be identified. (D) Our results suggest that the affinity of Nup358 is strong enough to efficiently compete with Rab6aGTP for BicD2. Therefore, in the absence of negative regulation, Nup358 is expected to recruit ~1000 BicD2 molecules to the nucleus.
Since a single dynein motor is likely sufficient to create the force needed for the transport of the nucleus, we propose that this transport pathway is turned off during the G1 and S phases by a yet-to-be-identified negative regulatory mechanism. In the absence of negative regulation, the affinity of Nup358 would likely be sufficient to recruit BicD2 to the nucleus in G2 phase. Additional G2 phase-specific positive regulatory mechanisms could shift the equilibrium even further toward recruitment of BicD2 to Nup358, such as phosphorylation of Nup358 by Cdk125 and potentially dissociation of RanGTP from the RBDs. Our quantitative model makes testable predictions on how cellular transport events are orchestrated, which are important for brain development, cell cycle control, signaling, and neurotransmission.
It remains to be established whether autoinhibited full-length BicD2 has an altered affinity toward cargoes compared to the BicD2-CTD used in our study. In cells, overexpressed BicD2-CTD can compete with endogenous BicD2 for binding of Rab6.63 It should be emphasized that the main point of our study is to compare affinities of BicD2 toward Rab6GTP and Nup358, and since the autoinhibition mechanism is likely the same for both cargoes, it is not expected that the ratio of the affinities or the cargo selection would be different for full-length BicD2 compared to BicD2-CTD.
Interestingly, we identified a negative regulator of the Nup358/BicD2 interaction, which may potentially play a regulatory role: RanGTP, which lowers the affinity by a factor of 10. Since Nup358/BicD2 and Nup358/RanGTP complexes can be reconstituted from nonoverlapping Nup358 fragments,11,31,49 it is unlikely that BicD2 and RanGTP compete for binding of Nup358, although that possibility cannot fully be excluded. A more likely mode of regulation is that binding of RanGTP leads to structural changes in Nup358, which result in a reduced affinity of BicD2 to Nup358. RanGTP and RanGTP/transport factor complexes bind to the RBDs of Nup358 with high affinity and RanGDP with a 10-fold lower affinity.49 In an invivo context, RanGTP is likely bound transiently to the RBDs of Nup358 and not always associated. As GTP-bound nuclear transport receptors arrive from the nucleus at the RBDs of Nup358, Ran GTPase-activating protein 1 (RanGAP1) triggers RanGTP hydrolysis, which triggers the release of RanGDP and transport factors to the cytoplasm. While RanGTP may be an important regulator of the Nup358/BicD2 interaction, RanGTP binding alone is not sufficient to prevent untimely dynein recruitment (Table 3B), and therefore, additional negative regulators likely remain to be identified.
Candidates for negative regulators include post-translational modifications, such as phosphorylation (Table S2). Alternatively, a putative negative regulator may be a yet-to-be-identified interaction partner that conceals the BicD2 binding site of Nup358 outside of the G2 phase. Another compelling hypothesis is that the BicD2 binding sites on Nup358 are buried in the scaffold of the NPC during most of the interphase and only become accessible for BicD2 in G2 phase through phosphorylation-induced structural changes of the NPC. This hypothesis is in line with experiments, in which a Nup358 domain that contained the BicD2 binding site was ectopically anchored at the plasma membrane in HeLa cells.11 Putative regulators such as kinases and interacting partners were still present, but the binding site was accessible and not integrated in the NPC scaffold.11 Notably, in this experiment, the BicD2-CTD was recruited to the plasma membrane, and cell cyclespecific regulation was lost,11 which is in line with the idea of cell cycle-specific regulation of the accessibility of the BicD2 binding sites.
Importantly, our quantitative model allowed us to calculate how many BicD2 molecules are recruited to the nucleus via Nup358. It is important to establish how many motors are recruited in order to gain a mechanistic understanding of the transport of the nucleus and to quantify the generated forces. Based on our quantitative model, ~1000 BicD2 molecules are recruited to the nucleus via Nup358 in the absence of regulation, which would in turn recruit up to one dynein motor per BicD2 dimer.6,7 Additional dynein complexes are recruited though the Nup133/CENP-F pathway.28−30 Our model also predicts a large pool of free BicD2, which would likely be autoinhibited and unable to recruit dynein/dynactin in the absence of cargo. A pool of free BicD2 is in line with results from immunostaining of endogenous BicD2 in G2 phase, where cytoplasmic staining indicates a cytoplasmic pool in addition to BicD2 that localizes to the nuclear envelope.11
Our quantitative model is based on rigorous biophysical experiments, using purified, recombinant protein domains. Such a reconstituted system is advantageous because it allows us to dissect functions of individual components of a more complex system. Future experiments will assess whether our minimal system can successfully predict in vivo functions. In order to improve the model further, affinities of less prominent BicD2 cargoes could be incorporated, although none are currently identified for human BicD2. It should be noted that our model assumes a homogeneous distribution of BicD2, Nup358, and Rab6GTP in the cytosol, which is a simplification, given that Nup358 and Rab6GTP are enriched on membrane surfaces. In the future, the model can be improved by modeling local concentrations of these proteins in distinct cellular compartments as well as diffusion processes between them.64,65 While our quantitative model could be improved by using a more accurate method to determine cellular concentrations of the reactants in the future, it should be emphasized that only large deviations in these concentrations (~two orders of a magnitude) would result in different transport outcomes.
The importance of BicD2-dependent transport events in brain and muscle development is reflected in the fact that mutations in BicD2 and a subunit of dynein cause a subset of spinal muscular atrophy (SMA) cases, a devastating neuromuscular disease36−39 that is the most common genetic cause of death in infants. While the pathogenesis is unknown, several of the disease mutations are located either in the BicD2/cargo interface or in the BicD2/dynein/dynactin interface, suggesting that transport defects cause these diseases.27 Therefore, regulatory mechanisms established through our work can be targeted to help devise therapies for SMA.
CONCLUSIONS
To conclude, BicD2/dynein-dependent transport events are crucial for normal brain and muscle development, cell cycle control, signaling pathways, and neurotransmission at synapses, and therefore, it is important to establish how these cellular transport events are orchestrated. The affinities of both a purified Nup358 domain and of Rab6GTP toward BicD2 are high. In the absence of regulation, a significant number of BicD2 molecules (~1000) would be recruited to the nucleus, which would provide binding sites for up to half as many dynein motors. In addition, phosphorylation by Cdk1 in G2 phase may further increase BicD2 recruitment to the nucleus.25 Of note, we have identified a negative regulator of the Nup358/BicD2 interaction, which may play a role in averting an untimely dynein recruitment; yet, an additional negative regulator is likely required to fully prevent dynein recruitment to the nucleus outside of the G2 phase. However, how exactly dynein and opposing motors are coordinated to facilitate correctly timed bidirectional transport of the nucleus at the appropriate velocity is an important biological problem that is currently not understood. It is also unknown how transport of the nucleus is differentially regulated in brain progenitor cells, muscle cells, and regular cells. Identifying regulatory mechanisms for the transport of the nucleus is a first step toward answering these important biological questions.
Supplementary Material
ACKNOWLEDGMENTS
Three plasmids for the expression of BicD2 and Nup358 fragments were provided by Anna Akhmanova (Utrecht University, The Netherlands).11 We also thank Anna Akhmanova for helpful comments regarding the manuscript. We thank Alexander Palazzo (University of Toronto, Canada) for a full-length Nup358 construct66 and Elias Coutavas from the Rockefeller University for a RanGTP expression construct. We thank David King, Howard Hughes Medical Institute, University of California at Berkeley, for mass spectrometry analysis. We thank Susan Bane and Brian Callahan (both from SUNY Binghamton) and the Chemistry Department for access to equipment. We also thank the Rockefeller University High-Throughput and Spectroscopy Resource Center for access to equipment.
Funding
This research was funded by NIH NIGMS (1R15GM128119-01) awarded to S.R.S. and by National Science Foundation Grant 1515028 awarded to C.G. Additional funding was provided by the Research Foundation of the State University of New York and the Department of Chemistry in SUNY Binghamton. K.M.L. was supported by summer fellowships from Harpur College and the Starzak fellowship from the Chemistry Department.
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.biochem.8b00987.
ITC titration curves, protein concentrations used for ITC experiments, and published post-translational modifications (PDF)
The authors declare no competing financial interest.
REFERENCES
- (1).Cianfrocco MA, DeSantis ME, Leschziner AE, and ReckPeterson SL (2015) Mechanism and regulation of cytoplasmic dynein. Annu. Rev. Cell Dev. Biol 31, 83–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Splinter D, Razafsky DS, Schlager MA, Serra-Marques A, Grigoriev I, Demmers J, Keijzer N, Jiang K, Poser I, Hyman AA, Hoogenraad CC, King SJ, and Akhmanova A (2012) BICD2, dynactin, and LIS1 cooperate in regulating dynein recruitment to cellular structures. Mol. Biol. Cell 23, 4226–4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Schlager MA, Hoang HT, Urnavicius L, Bullock SL, and Carter AP (2014) In vitro reconstitution of a highly processive recombinant human dynein complex. EMBO J 33, 1855–1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Schlager MA, Serra-Marques A, Grigoriev I, Gumy LF, Esteves da Silva M, Wulf PS, Akhmanova A, and Hoogenraad CC (2014) Bicaudal D family adaptor proteins control the velocity of dynein-based movements. Cell Rep 8, 1248–1256. [DOI] [PubMed] [Google Scholar]
- (5).McKenney RJ, Huynh W, Tanenbaum ME, Bhabha G, and Vale RD (2014) Activation of cytoplasmic dynein motility by dynactin-cargo adapter complexes. Science 345, 337–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Urnavicius L, Zhang K, Diamant AG, Motz C, Schlager MA, Yu M, Patel NA, Robinson CV, and Carter AP (2015) The structure of the dynactin complex and its interaction with dynein. Science 347, 1441–1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Urnavicius L, Lau CK, Elshenawy MM, Morales-Rios E, Motz C, Yildiz A, and Carter AP (2018) Cryo-EM shows how dynactin recruits two dyneins for faster movement. Nature 554, 202–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Grotjahn DA, Chowdhury S, Xu Y, McKenney RJ, Schroer TA, and Lander GC (2018) Cryo-electron tomography reveals that dynactin recruits a team of dyneins for processive motility. Nat. Struct. Mol. Biol 25, 203–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Liu Y, Salter HK, Holding AN, Johnson CM, Stephens E, Lukavsky PJ, Walshaw J, and Bullock SL (2013) Bicaudal-D uses a parallel, homodimeric coiled coil with heterotypic registry to coordinate recruitment of cargos to dynein. Genes Dev 27, 1233–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Terawaki S, Yoshikane A, Higuchi Y, and Wakamatsu K (2015) Structural basis for cargo binding and autoinhibition of Bicaudal-D1 by a parallel coiled-coil with homotypic registry. Biochem. Biophys. Res. Commun 460, 451–456. [DOI] [PubMed] [Google Scholar]
- (11).Splinter D, Tanenbaum ME, Lindqvist A, Jaarsma D, Flotho A, Yu KL, Grigoriev I, Engelsma D, Haasdijk ED, Keijzer N, Demmers J, Fornerod M, Melchior F, Hoogenraad CC, Medema RH, and Akhmanova A (2010) Bicaudal D2, dynein, and kinesin-1 associate with nuclear pore complexes and regulate centrosome and nuclear positioning during mitotic entry. PLoS Biol 8, e1000350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Matanis T, Akhmanova A, Wulf P, Del Nery E, Weide T, Stepanova T, Galjart N, Grosveld F, Goud B, De Zeeuw CI, Barnekow A, and Hoogenraad CC (2002) Bicaudal-D regulates COPI-independent Golgi-ER transport by recruiting the dyneindynactin motor complex. Nat. Cell Biol 4, 986–992. [DOI] [PubMed] [Google Scholar]
- (13).McClintock MA, Dix CI, Johnson CM, McLaughlin SH, Maizels RJ, Hoang HT, and Bullock SL (2018) RNA-directed activation of cytoplasmic dynein-1 in reconstituted transport RNPs. eLife 7, e36312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Dienstbier M, Boehl F, Li X, and Bullock SL (2009) Egalitarian is a selective RNA-binding protein linking mRNA localization signals to the dynein motor. Genes Dev 23, 1546–1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Sladewski TE, Billington N, Ali MY, Bookwalter CS, Lu H, Krementsova EB, Schroer TA, and Trybus KM (2018) Recruitment of two dyneins to an mRNA-dependent Bicaudal D transport complex. eLife 7, e36306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Bianco A, Dienstbier M, Salter HK, Gatto G, and Bullock SL (2010) Bicaudal-D Regulates Fragile X Mental Retardation Protein Levels, Motility, and Function during Neuronal Morphogenesis. Curr. Biol 20, 1487–1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Li X, Kuromi H, Briggs L, Green DB, Rocha JJ, Sweeney ST, and Bullock SL (2010) Bicaudal-D binds clathrin heavy chain to promote its transport and augments synaptic vesicle recycling. EMBO J 29, 992–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Stuurman N, Häner M, Sassea B, Hübner W, Suterd B, and Aebi U (1999) Interactions between coiled-coil proteins: Drosophila lamin Dm0 binds to the Bicaudal-D protein. Eur. J. Cell Biol 78, 278–287. [DOI] [PubMed] [Google Scholar]
- (19).Fridolfsson HN, Ly N, Meyerzon M, and Starr DA (2010) UNC-83 coordinates kinesin-1 and dynein activities at the nuclear envelope during nuclear migration. Dev. Biol 338, 237–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Martinez O, Schmidt A, Salamero J, Hoflack B, Roa M, and Goud B (1994) The small GTP-binding protein Rab6 functions in intra-Golgi transport. J. Cell Biol 127, 1575–1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Grigoriev I, Splinter D, Keijzer N, Wulf PS, Demmers J, Ohtsuka T, Modesti M, Maly IV, Grosveld F, Hoogenraad CC, and Akhmanova A (2007) Rab6 regulates transport and targeting of exocytotic carriers. Dev. Cell 13, 305–314. [DOI] [PubMed] [Google Scholar]
- (22).Eathiraj S, Pan X, Ritacco CJ, and Lambright DG (2005) Structural basis of family-wide Rab GTPase recognition by Rabenosyn-5. Nature 436, 415–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Bergbrede T, Pylypenko O, Rak A, and Alexandrov K (2005) Structure of the extremely slow GTPase Rab6A in the GTP bound form at 1.8 A resolution. J. Struct. Biol 152, 235–238. [DOI] [PubMed] [Google Scholar]
- (24).Holland PM, Milne A, Garka K, Johnson RS, Willis C, Sims JE, Rauch CT, Bird TA, and Virca GD (2002) Purification, cloning, and characterization of Nek8, a novel NIMA-related kinase, and its candidate substrate BicD2. J. Biol. Chem 277, 16229–16240. [DOI] [PubMed] [Google Scholar]
- (25).Baffet AD, Hu DJ, and Vallee RB (2015) Cdk1 activates pre-mitotic nuclear envelope dynein recruitment and apical nuclear migration in neural stem cells. Dev. Cell 33, 703–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Goud B, Zahraoui A, Tavitian A, and Saraste J (1990) Small GTP-binding protein associated with Golgi cisternae. Nature 345, 553–556. [DOI] [PubMed] [Google Scholar]
- (27).Hoang HT, Schlager MA, Carter AP, and Bullock SL (2017) DYNC1H1 mutations associated with neurological diseases compromise processivity of dynein−dynactin−cargo adaptor complexes. Proc. Natl. Acad. Sci. U. S. A 114, E1597–E1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Bolhy S, Bouhlel I, Dultz E, Nayak T, Zuccolo M, Gatti X, Vallee R, Ellenberg J, and Doye V (2011) A Nup133-dependent NPC-anchored network tethers centrosomes to the nuclear envelope in prophase. J. Cell Biol 192, 855–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Zuccolo M, Alves A, Galy V, Bolhy S, Formstecher E, Racine V, Sibarita JB, Fukagawa T, Shiekhattar R, Yen T, and Doye V (2007) The human Nup107/160 nuclear pore subcomplex contributes to proper kinetochore functions. EMBO J 26, 1853–1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Hu DJ, Baffet AD, Nayak T, Akhmanova A, Doye V, and Vallee RB (2013) Dynein recruitment to nuclear pores activates apical nuclear migration and mitotic entry in brain progenitor cells. Cell 154, 1300–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Wu J, Matunis MJ, Kraemer D, Blobel G, and Coutavas E (1995) Nup358, a cytoplasmically exposed nucleoporin with peptide repeats, RanGTP binding sites, zinc fingers, a cyclophilin A homologous domain, and a leucine-rich region. J. Biol. Chem 270, 14209–14213. [DOI] [PubMed] [Google Scholar]
- (32).Yokoyama N, Hayashi N, Seki T, Panté N, Ohba T, Nishii K, Kuma K, Hayashida T, Miyata T, Aebi U, Fukui M, and Nishimoto T (1995) A giant nucleopore protein that binds Ran/TC4. Nature 376, 184–188. [DOI] [PubMed] [Google Scholar]
- (33).Cui H, Loftus K, Noell C, and Solmaz S (2018) Identification of cyclin-dependent kinase 1 specific phosphorylation sites by an in vitro kinase assay. J. Visualized Exp 135, e57674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Loftus KM, Coutavas E, Cui H, King D, Ceravolo A, Pereiras D, and Solmaz S (2017) Mechanism for G2 phase-specific nuclear export of the kinetochore protein CENP-F. Cell Cycle 16, 1414–1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Wilson MH, and Holzbaur ELF (2012) Opposing microtubule motors drive robust nuclear dynamics in developing muscle cells. J. Cell Sci 125, 4158–4169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Neveling K, Martinez-Carrera LA, Holker I, Heister A, Verrips A, Hosseini-Barkooie SM, Gilissen C, Vermeer S, Pennings M, Meijer R, te Riele M, Frijns CJM, Suchowersky O, MacLaren L, Rudnik-Schoneborn S, Sinke RJ, Zerres K, Lowry RB, Lemmink HH, Garbes L, Veltman JA, Schelhaas HJ, Scheffer H, and Wirth B (2013) Mutations in BICD2, which encodes a golgin and important motor adaptor, cause congenital autosomal-dominant spinal muscular atrophy. Am. J. Hum. Genet 92, 946–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Oates EC, Rossor AM, Hafezparast M, Gonzalez M, Speziani F, MacArthur DG, Lek M, Cottenie E, Scoto M, Foley AR, Hurles M, Houlden H, Greensmith L, Auer-Grumbach M, Pieber TR, Strom TM, Schule R, Herrmann DN, Sowden JE, Acsadi G, Menezes MP, Clarke NF, Zuchner S, Muntoni F, North KN, and Reilly MM (2013) Mutations in BICD2 cause dominant congenital spinal muscular atrophy and hereditary spastic paraplegia. Am. J. Hum. Genet 92, 965–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Peeters K, Litvinenko I, Asselbergh B, Almeida-Souza L, Chamova T, Geuens T, Ydens E, Zimon M, Irobi J, De Vriendt E, De Winter V, Ooms T, Timmerman V, Tournev I, and Jordanova A (2013) Molecular defects in the motor adaptor BICD2 cause proximal spinal muscular atrophy with autosomal-dominant inheritance. Am. J. Hum. Genet 92, 955–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Synofzik M, Martinez-Carrera LA, Lindig T, Schöls L, and Wirth B (2014) Dominant spinal muscular atrophy due to BICD2: a novel mutation refines the phenotype. J. Neurol., Neurosurg. Psychiatry 85, 590–592. [DOI] [PubMed] [Google Scholar]
- (40).Klebe C, Bischoff FR, Ponstingl H, and Wittinghofer A (1995) Interaction of the Nuclear GTP-Binding Protein Ran with Its Regulatory Proteins RCC1 and RanGAP1. Biochemistry 34, 639–647. [DOI] [PubMed] [Google Scholar]
- (41).Solmaz SR, Chauhan R, Blobel G, and Melcak I (2011) Molecular architecture of the transport channel of the nuclear pore complex. Cell 147, 590–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Solmaz SR, Blobel G, and Melcak I (2013) Ring cycle for dilating and constricting the nuclear pore. Proc. Natl. Acad. Sci. U. S. A 110, 5858–5863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Sharma A, Solmaz SR, Blobel G, and Melcak I (2015) Ordered regions of channel nucleoporins Nup62, Nup54, and Nup58 form dynamic complexes in solution. J. Biol. Chem 290, 18370–18378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Maul GG, Maul HM, Scogna JE, Lieberman MW, Stein GS, Hsu BY-L, and Borun TW (1972) Time sequence of nuclear pore formation in phytohemagglutinin-stimulated lymphocytes and in HeLa cells during the cell cycle. J. Cell Biol 55, 433–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).von Appen A, Kosinski J, Sparks L, Ori A, DiGuilio AL, Vollmer B, Mackmull M-T, Banterle N, Parca L, Kastritis P, Buczak K, Mosalaganti S, Hagen W, Andres-Pons A, Lemke EA, Bork P, Antonin W, Glavy JS, Bui KH, and Beck M (2015) In situ structural analysis of the human nuclear pore complex. Nature 526, 140–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Monier K, Armas JCG, Etteldorf S, Ghazal P, and Sullivan KF (2000) Annexation of the interchromosomal space during viral infection. Nat. Cell Biol 2, 661–665. [DOI] [PubMed] [Google Scholar]
- (47).Zhao L, Kroenke CD, Song J, Piwnica-Worms D, Ackerman JJH, and Neil JJ (2008) Intracellular Water Specific MR of Microbead-adherent Cells: The HeLa Cell Intracellular Water Exchange Lifetime. NMR Biomed 21, 159–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Posakony J, England J, and Attardi G (1977) Mitochondrial growth and division during the cell cycle in HeLa cells. J. Cell Biol 74, 468–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Vetter IR, Nowak C, Nishimoto T, Kuhlmann J, and Wittinghofer A (1999) Structure of a Ran-binding domain complexed with Ran bound to a GTP analogue: implications for nuclear transport. Nature 398, 39–46. [DOI] [PubMed] [Google Scholar]
- (50).Kalab P, and Heald R (2008) The RanGTP gradient - a GPS for the mitotic spindle. J. Cell Sci 121, 1577–1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Kalab P, Weis K, and Heald R (2002) Visualization of a RanGTP gradient in interphase and mitotic Xenopus egg extracts. Science 295, 2452–2456. [DOI] [PubMed] [Google Scholar]
- (52).Bergbrede T, Chuky N, Schoebel S, Blankenfeldt W, Geyer M, Fuchs E, Goody RS, Barr F, and Alexandrov K (2009) Biophysical analysis of the interaction of Rab6a GTPase with its effector domains. J. Biol. Chem 284, 2628–2635. [DOI] [PubMed] [Google Scholar]
- (53).Kosinski J, Mosalaganti S, von Appen A, Teimer R, DiGuilio AL, Wan W, Bui KH, Hagen WJH, Briggs JAG, Glavy JS, Hurt E, and Beck M (2016) Molecular architecture of the inner ring scaffold of the human nuclear pore complex. Science 352, 363–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Joseph J, and Dasso M (2008) The nucleoporin Nup358 associates with and regulates interphase microtubules. FEBS Lett 582, 190–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Wang M, Herrmann CJ, Simonovic M, Szklarczyk D, and von Mering C (2015) Version 4.0 of PaxDb: Protein abundance data, integrated across model organisms, tissues, and cell-lines. Proteomics 15, 3163–3168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Matsuto M, Kano F, and Murata M (2015) Reconstitution of the targeting of Rab6A to the Golgi apparatus in semi-intact HeLa cells: A role of BICD2 in stabilizing Rab6A on Golgi membranes and a concerted role of Rab6A/BICD2 interactions in Golgi-to-ER retrograde transport. Biochim. Biophys. Acta, Mol. Cell Res 1853, 2592–2609. [DOI] [PubMed] [Google Scholar]
- (57).Young J, Ménétrey J, and Goud B (2010) RAB6C Is a Retrogene that Encodes a Centrosomal Protein Involved in Cell Cycle Progression. J. Mol. Biol 397, 69–88. [DOI] [PubMed] [Google Scholar]
- (58).Gard DL, and Kirschner MW (1987) A microtubuleassociated protein from Xenopus eggs that specifically promotes assembly at the plus-end. J. Cell Biol 105, 2203–2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Goldmacher VS, Audette CA, Guan Y, Sidhom E-H, Shah JV, Whiteman KR, and Kovtun YV (2015) High-Affinity Accumulation of a Maytansinoid in Cells via Weak Tubulin Interaction. PLoS One 10, e0117523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (60).Hiller G, and Weber K (1978) Radioimmunoassay for tubulin: a quantitative comparison of the tubulin content of different established tissue culture cells and tissues. Cell 14, 795–804. [DOI] [PubMed] [Google Scholar]
- (61).Belyy V, Schlager MA, Foster H, Reimer AE, Carter AP, and Yildiz A (2016) The mammalian dynein−dynactin complex is a strong opponent to kinesin in a tug-of-war competition. Nat. Cell Biol 18, 1018–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (62).Shtridelman Y, Cahyuti T, Townsend B, DeWitt D, and Macosko JC (2008) Force-Velocity Curves of Motor Proteins Cooperating In Vivo. Cell Biochem. Biophys 52, 19–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (63).Hoogenraad CC, Akhmanova A, Howell SA, Dortland BR, De Zeeuw CI, Willemsen R, Visser P, Grosveld F, and Galjart N (2001) Mammalian Golgi associated Bicaudal D2 functions in the dynein dynactin pathway by interacting with these complexes. EMBO J 20, 4041–4054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (64).Soula H, Caré B, Beslon G, and Berry H (2013) Anomalous versus Slowed-Down Brownian Diffusion in the Ligand-Binding Equilibrium. Biophys. J 105, 2064–2073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (65).Bers DM, and Peskoff A (1991) Diffusion around a cardiac calcium channel and the role of surface bound calcium. Biophys. J 59, 703–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (66).Mahadevan K, Zhang H, Akef A, Cui XA, Gueroussov S, Cenik C, Roth FP, and Palazzo AF (2013) RanBP2/Nup358 potentiates the translation of a subset of mRNAs encoding secretory proteins. PLoS Biol 11, e1001545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (67).Global Proteome Machine Organization (2013) The GPM (Global Proteome Machine) Database
- (68).Farrah T, Deutsch EW, Hoopmann MR, Hallows JL, Sun Z, Huang C-Y, and Moritz RL (2013) The State of the Human Proteome in 2012 as Viewed through PeptideAtlas. J. Proteome Res 12, 162–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.