Abstract
Genetic aberrations among uterine epithelioid leiomyosarcomas are unknown. Following identification of an index case with NR4A3-PGR fusion demonstrating monomorphic morphologic features, we interrogated additional uterine tumors demonstrating similar histology and sought to describe the morphologic and immunohistochemical characteristics of PGR-rearranged sarcomas. Targeted next-generation RNA sequencing was performed on RNA extracted from formalin-fixed paraffin-embedded tissue of the index case. Fluorescence in situ hybridization (FISH) using custom probes flanking PGR and NR4A3 genes was applied to 17 epithelioid leiomyosarcomas, 6 endometrial stromal tumors, and 3 perivascular epithelioid cell tumors. NR4A3-PGR fusion (n=4) and PGR rearrangement (n=2) were detected in six (35%) epithelioid leiomyosarcomas. Median patient age was 45 years, and all presented with FIGO stage I or II tumors, two being alive with disease at 75 and 180 months. All tumors were centered in the cervical stroma or myometrium and consisted of cells with abundant eosinophilic cytoplasm (epithelioid) including many displaying dense intracytoplasmic inclusions (rhabdoid). Myxoid matrix and hydropic change imparted a microcystic growth pattern in four tumors. Five also showed a minor spindle cell component which was low-grade in three, consisting of bland spindle cells with low mitotic activity. High-grade spindle cell morphology was seen in two tumors, exhibiting a storiform pattern of atypical spindle cells associated with brisk mitotic activity. Desmin, ER and PR were positive in all six tumors, while CD10 and HMB45 were negative. PGR rearrangements define a genetic subset of epithelioid leiomyosarcomas with often biphasic morphology consisting of epithelioid and rhabdoid as well as spindle cell components.
Keywords: Epithelioid leiomyosarcoma, endometrial stromal sarcoma, PEComa, uterine sarcoma, rhabdoid, NR4A3, PGR
INTRODUCTION
Recurrent chromosomal translocations resulting in gene fusions have been well documented among uterine sarcomas, particularly those of endometrial stromal derivation. JAZF1-SUZ12 fusion represents the most common genetic aberration described among low-grade endometrial stromal sarcomas, followed by less frequent JAZF1-PHF1 (1), EPC1-PHF1 (1), MEAF6-PHF1 (2), MBTD1-CXorf67 (3), and EPC2-PHF1 fusions (4). YWHAE-NUTM2 (5, 6), ZC3H7B-BCOR (7),(8, 9), EPC1-SUZ12 (10), and EPC1-BCOR (10) gene fusions have also been identified among high-grade endometrial stromal sarcomas with distinctive morphologic and immunophenotypic features. Endometrial stromal sarcoma associated fusions have also been reported in undifferentiated uterine sarcomas, which in retrospect likely represent misdiagnosed or underrecognized high-grade endometrial stromal sarcomas or high-grade endometrial stromal sarcomas that arose from low-grade tumors (11).
Fusions have also been identified in less common uterine mesenchymal tumors. These include inflammatory myofibroblastic tumors which often harbor ALK1 rearrangements (12–15) and fibrosarcoma-like uterine sarcomas underpinned by NTRK fusions (16). Patients with ALK1-rearranged tumors who develop recurrent disease may benefit from treatment with ALK directed therapies such as crizotinib or alectinib (17) (18) and those with NTRK fusion may benefit from larotrectinib and repotrectinib (19, 20). Patients may not need access to these drugs by clinical trial after an anticipated FDA approval of larotrectinib in October 2018. TFE3 and RAD51B rearrangements have also been described in a subset of uterine perivascular epithelioid cell tumors (PEComas) (21–24).
Genetic aberrations among uterine epithelioid leiomyosarcomas are unknown likely due to the rarity of such tumors; whether gene fusions are involved in the pathogenesis of epithelioid smooth muscle tumors has yet to be elucidated. We recently encountered a group of uterine epithelioid leiomyosarcoma that harbors novel PGR gene rearrangements. In this study, we characterized the fusion partner gene by targeted next-generation RNA sequencing (RNAseq) and confirmed PGR rearrangement by fluorescence in situ hybridization (FISH). We also sought to describe the morphologic and immunohistochemical features of this distinctive clinicopathologic entity.
MATERIALS AND METHODS
Case Selection
The index case was previously diagnosed as an epithelioid leiomyosarcoma with nuclear isomorphism and a distinctive rhabdoid component that was found to harbor a NR4A3-PGR gene fusion by RNASeq. Retrospective archival searches of the surgical pathology files at Memorial Sloan Kettering Cancer Center (New York, NY, USA), Massachusetts General Hospital (Boston, MA, USA), and King Edward Memorial Hospital (Perth, Australia), as well as the consult files of three of the authors (E.O., C.R.A., C.J.R.S.) were conducted for uterine epithelioid leiomyosarcomas, endometrial stromal sarcomas with epithelioid change, and uterine PEComas (Table 1). These entities were selected based on the distinctive monotonous morphology of the index case that may lead to classification as epithelioid leiomyosarcoma, PEComa, or endometrial stromal sarcoma with epithelioid features. All available hematoxylin-and-eosin (H&E) and immunohistochemical stained slides as well as pathology reports were reviewed by two pathologists (S.C., C.R.A.). Inclusion in the study required rhabdoid and epithelioid morphology with only mild to moderate nuclear pleomorphism; tumors with marked nuclear pleomorphism were excluded. Gross, morphologic, and immunophenotypic features were recorded. Clinical data, including age, stage, and outcome, was obtained through the electronic medical records and pathology reports at Memorial Sloan Kettering Cancer Center, Massachusetts General Hospital, and King Edward Memorial Hospital. This study received institutional research board approval.
Table 1.
Summary of Study Cohort
| Tumor type | n | PgR rearrangement (n) | NR4A3 rearrangement (n) |
|---|---|---|---|
| Epithelioid leiomyosarcoma | 17 | 6 | 4 |
| Endometrial stromal tumor | 6 | 0 | 0 |
| Low-grade endometrial stromal sarcoma | 1 | 0 | 0 |
| High-grade endometrial stromal sarcoma | 4 | 0 | 0 |
| Endometrial stromal nodule | 1 | 0 | 0 |
| Perivascular epithelioid cell tumor | 3 | 0 | 0 |
Immunohistochemistry
Staining was performed on four μm formalin-fixed paraffin-embedded tissue sections for desmin, CD10, estrogen receptor (ER), progesterone receptor (PR), and HMB45 (Supplementary Table 1). Appropriate on-slide positive controls were used. Immunohistochemical stains were evaluated using a semi-quantitative scoring method based on the percentage of positive tumor cells as follows: negative (0), 1–25% (1), 26–50% (2), 51–75% (3), and 76–100% (4).
Next-generation targeted RNA sequencing
RNA was extracted from formalin-fixed paraffin-embedded tissue cut onto positively charged glass slides (four μm section, 7–10 per case) using the ExpressArt FFPE Clear RNA Ready kit (Amsbio, Cambridge, MA). RNAseq libraries were prepared using 20–100 ng total RNA with the TruSight RNA Fusion Panel (Illumina, San Diego, CA), an enrichment-based assay that targets 507 known fusion-associated genes. The sample was sequenced with 76 base-pair paired-end reads on an Illumina MiSeq. Results were analyzed using the STAR aligner and Manta fusion caller as well as the JAFFA fusion caller utilizing BOWTIE2 aligner (25, 26). mRNA expression level of NR4A3 was evaluated in the index case and compared to >100 sarcoma types available on the same RNAseq platform. The expression of PGR could not be evaluated since this gene was not represented on the targeted RNAseq gene list.
Fluorescence in situ hybridization
As previously described, FISH was performed on interphase nuclei from formalin-fixed paraffin-embedded four μm tissue sections using custom bacterial artificial chromosomes (BAC) flanking NR4A3 and PGR (Supplementary Table 2). Briefly, BAC clones were chosen according to the UCSC genome browser (http://genome.ucsc.edu) and obtained from BACPAC sources of Children’s Hospital of Oakland Research Institute (Oakland, CA; http://bacpac.chori.org). BAC DNA was isolated, labeled with fluorochromes by nick translation, denatured, and hybridized to de-paraffinized pre-treated slides. Slides were incubated overnight, washed, and mounted with DAPI using a Zeiss fluorescence microscope (Zeiss Axioplan, Oberkochen, Germany) and Isis 5 software (Metasystems). Gene rearrangement was confirmed when at least 20% of 200 successive tumor nuclei showed a break apart signal.
RESULTS
Clinical features
The entire cohort consisted of 26 tumors, including 17 epithelioid leiomyosarcomas, 6 endometrial stromal tumors with epithelioid features (1 low-grade endometrial stromal sarcoma, 4 high-grade endometrial stromal sarcomas, 1 endometrial stromal nodule), and 3 PEComas identified from institutional pathology and consultation archives (Table 1). The six patients with PGR fusion-positive tumors classified as epithelioid leiomyosarcoma had a median age of 45 (range, 41 to 56) years (Table 2). Four patients presented with a uterine or pelvic mass associated with painful menses or abdominal pain in two of them. Two other patients presented with symptomatic fibroids. All patients were treated with total hysterectomy and bilateral salpingo-oophorectomy and found to have either FIGO stage I or II disease. Only one patient received post-operative chemoradiation. Two patients with FIGO stage II tumors were alive with disease 75 and 180 months after initial presentation. Three patients with FIGO stage I or II tumors had no evidence of disease at last follow-up, 96, 9, and 1 month after initial presentation, respectively. One patient with FIGO stage I disease was lost to follow up.
Table 2.
Clinical features of PGR fusion-positive uterine epithelioid leiomyosarcomas
| Case | Age, y | Signs and symptoms | Treatment | FIGO stage | Follow-up, mo |
|---|---|---|---|---|---|
| 1 | 43 | Painful menses, pelvic mass | Surgery, chemoradiation | II | AWD, 180 |
| 2 | 46 | Abdominal pain, pelvic mass | Surgery | II | NED, 9 |
| 3 | 56 | Symptomatic fibroids | Surgery | I | LTF |
| 4 | 42 | Uterine mass | Surgery | I | NED, 96 |
| 5 | 41 | Uterine mass | Surgery | II | AWD, 75 |
| 6 | 55 | Symptomatic fibroids | Surgery | II | NED, 1 |
AWD indicates alive with disease; LTF, lost to follow up; NED, no evidence of disease.
Limited clinical data was also available in 14 of the remaining 20 patients with PGR fusion-negative uterine tumors (Supplementary Table 3). Eleven patients presented with a uterine or pelvic mass, two of whom also had abdominal pain. Three patients had abnormal vaginal bleeding. The median age was 56 (range, 27 to 71) years. All were treated surgically, with three patients also receiving chemotherapy and one receiving chemoradiation. Among the 12 patients with available staging and follow-up data, 4, 2, 3, and 3 patients presented with stage I, II, III, and IV disease, respectively. One died of disease 4 months after initial presentation, and three recurred 3 to 29 months later. Two patients had no evidence of disease 5 and 60 months from presentation. The remaining six patients were lost to follow-up.
Pathologic features
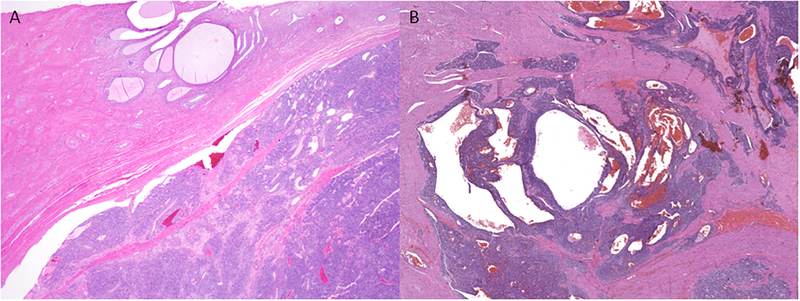
PGR fusion-positive sarcomas ranged from 3 to 27 (median, 9) cm in size (Table 3). Three were centered in the cervix, one of which extended into the corpus and another into the ovary. Two tumors were limited to the corpus. One tumor arose in the corpus and extended into the vaginal wall. Tumors displayed a variably brown, red, yellow-tan to white, friable, hemorrhagic cut surface. One was multi-cystic, while another was predominately necrotic with an adjacent firm, tan whorled rim. Myometrial invasion was pushing in four tumors (Figure 1A) and irregular and tongue-like in two (Figure 1B). No involvement of the endometrium was seen.
Table 3.
Pathologic features of PGR fusion-positive uterine sarcomas
| Morphology | Immunohistochemical profile | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Case | Size, cm | Spindled | Rhabdoid | Myxoid | MI | LVI | Necrosis | CD10 | Desmin | ER | PR | HMB45 | Fusion status |
| 1* | 5 | + | + | + | 19 | − | − | 0 | 4 | 4 | 4 | 0 | NR4A3-PGR |
| 2 | 12 | + | + | + | 9 | − | − | 0 | 2 | 4 | 4 | NP | NR4A3-PGR |
| 3 | 3 | + | + | + | 4 | − | − | 0 | 4 | 4 | 4 | 0 | NR4A3-PGR |
| 4 | 15 | − | + | − | 12 | + | − | 0 | 3 | 3 | 4 | 0 | PGR rearrangement only |
| 5 | 4 | + | + | − | 10 | + | − | 0 | 3 | 4 | 4 | NP | NR4A3-PGR |
| 6 | 27 | + | + | − | 10 | − | + | NP | 3 | 4 | 4 | 0 | PGR rearrangement only |
F indicates focal; HPF, high power field; LVI, lymphovascular invasion; MI, mitotic index (# per 10 high power fields); NP, not performed.
Index case subjected to RNA sequencing.
Figure 1. Tumor interface of PGR-rearranged sarcomas.
Tumors may exhibit a (A) circumscribed, pushing border or (B) infiltration into adjacent tissues.
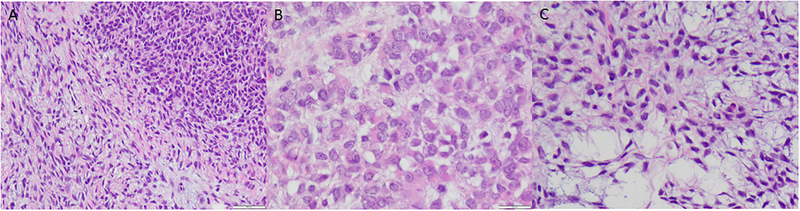
All six tumors were composed of an epithelioid and rhabdoid component consisting of uniformly cellular sheets of cells with abundant eosinophilic granular cytoplasm and centrally or eccentrically located nuclei with irregular membranes, occasional grooves, vesicular chromatin, and mostly one to two prominent nucleoli (Figures 2A and 2B, Table 3). Median mitotic index was 10 (range, 4 to 19) per 10 high power fields (HPF). Myxoid matrix and hydropic change were seen in three and one tumors, respectively, imparting a microcystic growth pattern.
Figure 2. Histology of PGR-rearranged sarcomas.
(A) Most tumors have a biphasic appearance with a (B) high-grade rhabdoid and epithelioid component that may be associated with myxoid matrix. (C) The spindle cell component when present demonstrates bland fusiform cells that may impart a microcystic pattern.
Five tumors also exhibited a spindle cell component (Table 3). Three consisted of densely cellular foci of small cells with fusiform nuclei, inconspicuous nucleoli, and scant wavy eosinophilic cytoplasm (Figure 2C) with a mitotic index of <1 per 10 HPF. This component was predominately distinct from foci of epithelioid cells (Figure 3A and 3B). The spindle cell component in the other two tumors consisted of cells with intermediate size ovoid to elongated nuclei with one or two prominent nucleoli, irregular nuclear membranes, and abundant eosinophilic cytoplasm arranged in a storiform pattern (Figure 3C). The mitotic index was 8 per 10 HPF in one of the two tumors (Figure 3D).
Figure 3. Distinctive patterns of spindle cell components of PGR-rearranged sarcomas.
Spindle cells may be (A) admixed with (left and bottom) or (B) form a distinct nodule (right) separate from the high-grade rhabdoid and epithelioid component. (C) The spindle cell component may show increased nuclear pleomorphism.
Numerous small arterioles without perivascular tumor cell whorling were seen throughout the tumors. Scattered large thick-walled blood vessels were also present. Tumor and infarct necrosis was present in only one tumor, while lymphovascular invasion was seen in two (Table 3).
Among the PGR fusion-negative tumors, the median tumor size was 12 (range, 3 to 28) cm (Supplementary Table 4). Rhabdoid morphology was seen in all 20 tumors, 15 of which also showed a spindle cell component. Myxoid stroma was seen in only four tumors, three of which showed both rhabdoid and spindle cell features. Median mitotic index was 9 (range, 1 to 55) per 10 HPF. Lymphovascular invasion and tumor cell necrosis was seen in nine and seven tumors, respectively.
Immunohistochemical features
Desmin was positive in all six PGR fusion-positive tumors (Table 3, Figure 4A). Expression was present in both epithelioid and rhabdoid and spindle cell components. In three tumors, immunoreactivity was seen throughout the spindle cell component and only scattered cells in the epithelioid and rhabdoid component (Figure 4B). All six tumors expressed PR and ER (Figures 4C and 4D). HMB45 was performed on four tumors and was negative. Five tumors tested for CD10 were also negative. H-caldesmon was positive in one tumor, but negative in four others; myogenin was negative in three tumors (data not shown).
Figure 4. Immunoprofile of PGR-rearranged sarcomas.
Desmin may be (A) diffuse or (B) focal). All tumors are ER-positive and may demonstrate (C) diffuse ER immunoreactivity. (D) All tumors are diffusely and strongly positive for PR.
Among the PGR fusion-negative tumors, desmin expression was seen in 11 epithelioid leiomyosarcomas, 2 endometrial stromal tumors, and 3 PEComas (Supplementary Table 4). CD10 was positive in three epithelioid leiomyosarcomas, two endometrial stromal tumors, and two PEComas. ER and PR expression were seen in nine epithelioid leiomyosarcomas, one endometrial stromal tumor, and three PEComas. HMB45 staining was present in ≤25% of cells in three epithelioid leiomyosarcomas and in >25% of cells in all three PEComas.
Molecular features
Targeted RNA sequencing of the index case (case 1) demonstrated a PGR-NR4A3 fusion, resulting from a t(9;11)(q22.3;q22.1) translocation (Fig. 5A). The fusion junction reads showed that PGR exon 2 was fused in frame to the entire coding sequence of NR4A3, starting with exon 2 (Fig. 5B). Thus, the projected fusion protein would include the progesterone receptor domain of PGR and the entire NR4A3 protein. A marked mRNA upregulation of NR4A3 was observed compared to other sarcoma types (Fig. 5C). NR4A3 and PGR rearrangements were also confirmed subsequently by FISH. The remaining study group was screened by FISH for abnormalities in both NR4A3 and PGR genes. A NR4A3-PGR fusion was detected in three additional tumors previously diagnosed as epithelioid leiomyosarcoma (Figure 6). PGR rearrangement with no identifiable fusion partner was observed in two tumors also previously diagnosed as epithelioid leiomyosarcoma. PGR and NR4A3 rearrangements were not detected by FISH in the remaining 11 epithelioid leiomyosarcomas, 5 endometrial stromal tumors, or 3 PEComas.
Figure 5. Fusion structure and molecular correlates.
(A) Diagrammatic representation of the NR4A3-PGR gene fusion, resulting from a t(9;11)(q22.3;q22.1) translocation. The gene loci are indicated with vertical red lines on the horizontal chromosomes, while the exonic breakpoints are indicated by a red arrow. Green and orange arrows indicate the direction of transcription of the individual genes. (B) Detailed RNA sequencing fusion junction reads, exon composition of the fusion transcripts and protein domain structures of each protein. (C) Box plot showing high levels of NR4A3 mRNA expression for the index case compared to other sarcoma types.
Figure 6. NR4A3-PGR fusion by FISH fusion assay.
NR4A3 signal (green, telomeric) comes together with PGR signal (yellow, telomeric), in keeping with an unbalanced NR4A3-PGR fusion; the red centromeric signal of PGR being lost (left). In comparison, two normal cells showing red-yellow signals of PGR, separate from the green NR4A3 signal (right).
DISCUSSION
In this study, we identified a molecular subset of uterine epithelioid leiomyosarcoma exhibiting distinctive isomorphic epithelioid and rhabdoid features and harboring PGR rearrangements, often in association with an NR4A3 fusion partner. All six tumors affected pre- or perimenopausal women and arose from either the wall of the uterine corpus or cervix. Most tumors exhibited a biphasic appearance with a high-grade epithelioid and rhabdoid component alongside or intermixed with a low- or high-grade spindle cell component. Myxoid matrix and hydropic change often imparted a microcystic and reticular pattern. Diffuse PR, diffuse or focal ER, and at least focal desmin expression was seen in all tumors, while CD10 and HMB45 were negative in all tumors tested. Two of the six patients developed recurrent disease, confirming the malignant behavior of these tumors.
The progesterone receptor gene (PGR) is located on 11q22.1 and encodes a member of the steroid receptor superfamily. Expressed ubiquitously in human tissues, the encoded protein mediates the physiological effects of progesterone involved in the establishment and maintenance of pregnancy. Differential expression of PGR may occur in tumorigenesis; however, PGR mutations and polymorphisms are infrequent in cancer. Somatic PGR mutations are usually missense and occur in approximately 1% of all cancer types. According to The Cancer Genome Atlas, PGR mutations were found in only 2.4% of endometrial carcinomas and were enriched among endometrioid tumors (27); PGR mutations comprise only 0.2% of ovarian high-grade serous carcinomas (28). No PGR gene fusions have been identified by TCGA studies in various common cancer types, including breast and ovarian malignancies (28, 29). Thus, our findings suggest that this small subset of leiomyosarcomas with distinctive rhabdoid and epithelioid morphology are characterized by novel PGR rearrangements, not previously identified in other tumors types.
Fusion of PGR with the nuclear receptor subfamily 4, group A, member 3 (NR4A3) was seen in two-thirds of our uterine leiomyosarcomas with epithelioid and rhabdoid morphology. NR4A3 gene located at 9q22 encodes a member of the steroid-thyroid hormone-retinoid receptor superfamily that activates peroxisome proliferator-activated receptor-gamma (PPAR-gamma) (30). NR4A3 gene fusions, mostly commonly with EWSR1, have been detected in extraskeletal myxoid chondrosarcomas (30–32). Other gene fusion partners such as TAF15, TCF12, TFG, FUS, and HSPA8 have also been reported with less frequency among these rare soft tissue neoplasms (31, 33, 34). While our uterine leiomyosarcomas harboring NR4A3-PGR fusions are morphologically distinct from extraskeletal myxoid chondrosarcomas, it is of interest to note that rhabdoid features are characteristic of extraskeletal myxoid chondrosarcomas with non-EWSR1 NR4A3 fusions (31). Furthermore, myxoid stroma was seen in three of four NR4A3-PGR fusion-positive uterine sarcomas, and not in the remaining two tumors with PGR rearrangement only.
These tumors are morphologically distinct from typical uterine epithelioid leiomyosarcomas. In general, epithelioid leiomyosarcomas are defined by round and polygonal cells comprising more than 50% of the overall tumor (35–37). Since epithelioid smooth muscle tumors are rare, criteria predictive of malignant behavior are currently not well established. Based on the largest reported studies, malignancy may be assigned in the setting of any cytologic atypia and five or more mitotic figures per 10 HPF in the absence of tumor necrosis; or five or more mitotic figures per 10 HPF and tumor cell necrosis with any degree of cytologic atypia (35–37). In practice, most epithelioid smooth muscle tumors fulfilling these criteria usually demonstrate marked nuclear pleomorphism, similar to the vast majority of conventional spindle cell leiomyosarcomas. While mild to moderate cytologic atypia was present among our tumors harboring PGR rearrangement, overt nuclear pleomorphism was lacking.
The nuclear monomorphism seen in PGR fusion-positive leiomyosarcomas resembles that seen in low-grade endometrial stromal sarcomas with epithelioid change (38), high-grade endometrial stromal sarcomas of various genotypes (5, 6, 8, 9, 39, 40), and undifferentiated uterine sarcomas of uniform type (41). The biphasic appearance of PGR fusion-positive uterine leiomyosarcomas may be confused with low-grade endometrial stromal sarcomas with epithelioid change (38) and high-grade endometrial stromal sarcomas harboring YWHAE-NUTM2 fusion (5, 6) or BCOR internal tandem duplication (39, 40). However, the complete lack of CD10 expression, typical low-grade endometrial stromal sarcoma morphology, and endometrial involvement in PGR fusion-positive sarcomas all argue against endometrial stromal derivation and are helpful features in their distinction from low-grade endometrial stromal sarcomas with epithelioid change and high-grade endometrial stromal sarcomas with YWHAE or BCOR genetic abnormalities. Undifferentiated uterine sarcoma of either uniform or pleomorphic type remains a diagnosis of exclusion. The presence of desmin expression and PGR rearrangement allow distinction of PGR fusion-positive sarcomas from undifferentiated uterine sarcoma.
Uterine epithelioid leiomyosarcomas, including PGR fusion-positive tumors, share morphologic overlap with lesions classified as uterine PEComas. Both smooth muscle tumors and PEComas may exhibit epithelioid and spindle cell features as well as melanocytic differentiation by immunohistochemistry. Some studies report that the presence of delicate capillaries surrounding tumor cells and nests favors a diagnosis of PEComa over smooth muscle neoplasia (21); in addition, thick-walled blood vessels, perinuclear vacuoles, and diffuse eosinophilic cytoplasm without granularity are more common among smooth muscle tumors rather than PEComas (21, 42). Expression of two or more melanocytic markers, such as HMB45 and Melan A, even if present in ≤5% of tumor cells, is considered diagnostic of PEComa in the appropriate morphologic context according to another study (22). However, melanocytic marker expression has also been reported in a subset of uterine smooth muscle tumors (43–45). With the exception of identifying TSC1/TSC2 mutations (24) and TFE3 or RAD51B rearrangements (21, 23, 24, 46), there is no gold standard in the diagnosis of uterine PEComas at this time. No PGR rearrangements were detected by FISH in the three PEComas tested in our cohort, nor in the cases tested by whole transcriptome RNAseq in our prior study (24). In addition, none of our PGR fusion-positive sarcomas showed any HMB45 staining, suggesting that these tumors are in fact distinct from tumors classified as uterine PEComas.
While clinical outcome data is limited, the observed survival among these six patients suggests that PGR fusion-positive sarcomas share malignant behavior. Multi-institutional studies with larger number of patients and longer follow-up are required to more accurately describe the clinical behavior of these rare lesions. While none of the patients in our cohort received hormonal therapy, PR expression as a result of PGR rearrangement suggests hormonal therapy may be a reasonable approach for treatment of advanced/recurrent disease..
In summary, PGR gene rearrangements define a novel molecular subset of epithelioid leiomyosarcomas. These tumors exhibit a high-grade but isomorphic rhabdoid and epithelioid component that may be associated with a low- or high-grade spindle cell component. Myogenic differentiation and lack of CD10 and HMB45 expression distinguish these tumors from mimickers such as endometrial stromal sarcoma and PEComa. Further studies are needed to evaluate the relationship between this genetic subset and the remaining group of epithelioid leiomyosarcoma with similar morphology and clinical features.
Supplementary Material
REFERENCES
- 1.Micci F, Panagopoulos I, Bjerkehagen B, et al. Consistent rearrangement of chromosomal band 6p21 with generation of fusion genes JAZF1/PHF1 and EPC1/PHF1 in endometrial stromal sarcoma. Cancer Res. 2006;66:107–112. [DOI] [PubMed] [Google Scholar]
- 2.Panagopoulos I, Micci F, Thorsen J, et al. Novel fusion of MYST/Esa1-associated factor 6 and PHF1 in endometrial stromal sarcoma. PLoS One. 2012;7:e39354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dewaele B, Przybyl J, Quattrone A, et al. Identification of a novel, recurrent MBTD1-CXorf67 fusion in low-grade endometrial stromal sarcoma. Int J Cancer. 2014;134:1112–1122. [DOI] [PubMed] [Google Scholar]
- 4.Brunetti M, Gorunova L, Davidson B, et al. Identification of an EPC2-PHF1 fusion transcript in low-grade endometrial stromal sarcoma. Oncotarget. 2018;9:19203–19208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee CH, Ou WB, Marino-Enriquez A, et al. 14-3-3 fusion oncogenes in high-grade endometrial stromal sarcoma. Proc Natl Acad Sci U S A. 2012;109:929–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee CH, Marino-Enriquez A, Ou W, et al. The clinicopathologic features of YWHAE-FAM22 endometrial stromal sarcomas: a histologically high-grade and clinically aggressive tumor. Am J Surg Pathol. 2012;36:641–653. [DOI] [PubMed] [Google Scholar]
- 7.Panagopoulos I, Thorsen J, Gorunova L, et al. Fusion of the ZC3H7B and BCOR genes in endometrial stromal sarcomas carrying an X;22-translocation. Genes Chromosomes Cancer. 2013;52:610–618. [DOI] [PubMed] [Google Scholar]
- 8.Hoang LN, Aneja A, Conlon N, et al. Novel high-grade endometrial stromal sarcoma: a morphologic mimicker of myxoid leiomyosarcoma. Am J Surg Pathol. 2017;41:12–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lewis N, Soslow RA, Delair DF, et al. ZC3H7B-BCOR high-grade endometrial stromal sarcomas: a report of 17 cases of a newly defined entity. Mod Pathol. 2018;31:674–684. [DOI] [PubMed] [Google Scholar]
- 10.Dickson BC, Lum A, Swanson D, et al. Novel EPC1 gene fusions in endometrial stromal sarcoma. Genes Chromosomes Cancer. 2018. [DOI] [PubMed] [Google Scholar]
- 11.Sciallis AP, Bedroske PP, Schoolmeester JK, et al. High-grade endometrial stromal sarcomas: a clinicopathologic study of a group of tumors with heterogenous morphologic and genetic features. Am J Surg Pathol. 2014;38:1161–1172. [DOI] [PubMed] [Google Scholar]
- 12.Mohammad N, Haimes JD, Mishkin S, et al. ALK is a specific diagnostic marker for inflammatory myofibroblastic tumor of the uterus. Am J Surg Pathol. 2018;42:1353–1359. [DOI] [PubMed] [Google Scholar]
- 13.Devereaux KA, Kunder CA, Longacre TA. ALK-rearranged tumors are highly enriched in the STUMP subcategory of uterine tumors. Am J Surg Pathol. 2018. [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- 14.Bennett JA, Nardi V, Rouzbahman M, et al. Inflammatory myofibroblastic tumor of the uterus: a clinicopathological, immunohistochemical, and molecular analysis of 13 cases highlighting their broad morphologic spectrum. Mod Pathol. 2017;30:1489–1503. [DOI] [PubMed] [Google Scholar]
- 15.Haimes JD, Stewart CJR, Kudlow BA, et al. Uterine inflammatory myofibroblastic tumors frequently harbor ALK fusions with IGFBP5 and THBS1. Am J Surg Pathol. 2017;41:773–780. [DOI] [PubMed] [Google Scholar]
- 16.Chiang S, Cotzia P, Hyman DM, et al. NTRK fusions define a novel uterine sarcoma subtype with features of fibrosarcoma. Am J Surg Pathol. 2018;42:791–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saiki M, Ohyanagi F, Ariyasu R, et al. Dramatic response to alectinib in inflammatory myofibroblastic tumor with anaplastic lymphoma kinase fusion gene. Jpn J Clin Oncol. 2017;47:1189–1192. [DOI] [PubMed] [Google Scholar]
- 18.Pickett JL, Chou A, Andrici JA, et al. Inflammatory myofibroblastic tumors of the female genital tract are under-recognized: a low threshold for ALK immunohistochemistry is required. Am J Surg Pathol. 2017;41:1433–1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Drilon A, Laetsch TW, Kummar S, et al. Efficacy of larotrectinib in TRK fusion-positive cancers in adults and children. N Engl J Med. 2018;378:731–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Drilon A, Ou SI, Cho BC, et al. Repotrectinib (TPX-0005) is a next-generation ROS1/TRK/ALK inhibitor that potently inhibits ROS1/TRK/ALK solvent-front mutations. Cancer Discov. 2018. [DOI] [PubMed] [Google Scholar]
- 21.Bennett JA, Braga AC, Pinto A, et al. Uterine PEComas: a morphologic, immunohistochemical, and molecular analysis of 32 tumors. Am J Surg Pathol. 2018;42:1370–1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schoolmeester JK, Howitt BE, Hirsch MS, et al. Perivascular epithelioid cell neoplasm (PEComa) of the gynecologic tract: clinicopathologic and immunohistochemical characterization of 16 cases. Am J Surg Pathol. 2014;38:176–188. [DOI] [PubMed] [Google Scholar]
- 23.Schoolmeester JK, Dao LN, Sukov WR, et al. TFE3 translocation-associated perivascular epithelioid cell neoplasm (PEComa) of the gynecologic tract: morphology, immunophenotype, differential diagnosis. Am J Surg Pathol. 2015;39:394–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Agaram NP, Sung YS, Zhang L, et al. Dichotomy of genetic abnormalities in PEComas with therapeutic implications. Am J Surg Pathol. 2015;39:813–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu S, Tsai WH, Ding Y, et al. Comprehensive evaluation of fusion transcript detection algorithms and a meta-caller to combine top performing methods in paired-end RNA-seq data. Nucleic Acids Res. 2016;44:e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen X, Schulz-Trieglaff O, Shaw R, et al. Manta: rapid detection of structural variants and indels for germline and cancer sequencing applications. Bioinformatics. 2016;32:1220–1222. [DOI] [PubMed] [Google Scholar]
- 27.Cancer Genome Atlas Research N, Kandoth C, Schultz N, et al. Integrated genomic characterization of endometrial carcinoma. Nature. 2013;497:67–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cancer Genome Atlas Research N. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cancer Genome Atlas N. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Davis EJ, Wu YM, Robinson D, et al. Next generation sequencing of extraskeletal myxoid chondrosarcoma. Oncotarget. 2017;8:21770–21777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Agaram NP, Zhang L, Sung YS, et al. Extraskeletal myxoid chondrosarcoma with non-EWSR1-NR4A3 variant fusions correlate with rhabdoid phenotype and high-grade morphology. Hum Pathol. 2014;45:1084–1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Benini S, Cocchi S, Gamberi G, et al. Diagnostic utility of molecular investigation in extraskeletal myxoid chondrosarcoma. J Mol Diagn. 2014;16:314–323. [DOI] [PubMed] [Google Scholar]
- 33.Broehm CJ, Wu J, Gullapalli RR, et al. Extraskeletal myxoid chondrosarcoma with a t(9;16)(q22;p11.2) resulting in a NR4A3-FUS fusion. Cancer Genet. 2014;207:276–280. [DOI] [PubMed] [Google Scholar]
- 34.Urbini M, Astolfi A, Pantaleo MA, et al. HSPA8 as a novel fusion partner of NR4A3 in extraskeletal myxoid chondrosarcoma. Genes Chromosomes Cancer. 2017;56:582–586. [DOI] [PubMed] [Google Scholar]
- 35.Kurman RJ, Norris HJ. Mesenchymal tumors of the uterus. VI. Epithelioid smooth muscle tumors including leiomyoblastoma and clear-cell leiomyoma: a clinical and pathologic analysis of 26 cases. Cancer. 1976;37:1853–1865. [DOI] [PubMed] [Google Scholar]
- 36.Prayson RA, Goldblum JR, Hart WR. Epithelioid smooth-muscle tumors of the uterus: a clinicopathologic study of 18 patients. Am J Surg Pathol. 1997;21:383–391. [DOI] [PubMed] [Google Scholar]
- 37.Toledo G, Oliva E. Smooth muscle tumors of the uterus: a practical approach. Arch Pathol Lab Med. 2008;132:595–605. [DOI] [PubMed] [Google Scholar]
- 38.Oliva E, Clement PB, Young RH. Epithelioid endometrial and endometrioid stromal tumors: a report of four cases emphasizing their distinction from epithelioid smooth muscle tumors and other oxyphilic uterine and extrauterine tumors. Int J Gynecol Pathol. 2002;21:48–55. [DOI] [PubMed] [Google Scholar]
- 39.Chiang S, Lee CH, Stewart CJR, et al. BCOR is a robust diagnostic immunohistochemical marker of genetically diverse high-grade endometrial stromal sarcoma, including tumors exhibiting variant morphology. Mod Pathol. 2017;30:1251–1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marino-Enriquez A, Lauria A, Przybyl J, et al. BCOR internal tandem duplication in high-grade uterine sarcomas. Am J Surg Pathol. 2018;42:335–341. [DOI] [PubMed] [Google Scholar]
- 41.Kurihara S, Oda Y, Ohishi Y, et al. Endometrial stromal sarcomas and related high-grade sarcomas: immunohistochemical and molecular genetic study of 31 cases. Am J Surg Pathol. 2008;32:1228–1238. [DOI] [PubMed] [Google Scholar]
- 42.Folpe AL, Kwiatkowski DJ. Perivascular epithelioid cell neoplasms: pathology and pathogenesis. Hum Pathol. 2010;41:1–15. [DOI] [PubMed] [Google Scholar]
- 43.Zamecnik M, Michal M. HMB45+ hyalinized epithelioid tumor of the uterus is linked to epithelioid leiomyoma rather than to PEC-omas. Int J Surg Pathol. 2001;9:341–343. [DOI] [PubMed] [Google Scholar]
- 44.Zamecnik M, Voltr L, Chlumska A. HMB45+ cells in mixed stromal-smooth muscle tumour of the uterus. Histopathology. 2006;48:463–464. [DOI] [PubMed] [Google Scholar]
- 45.Simpson KW, Albores-Saavedra J. HMB-45 reactivity in conventional uterine leiomyosarcomas. Am J Surg Pathol. 2007;31:95–98. [DOI] [PubMed] [Google Scholar]
- 46.Maloney N, Giannikou K, Lefferts J, et al. Expanding the histomorphologic spectrum of TFE3 rearranged PEComas. Hum Pathol. 2018. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.