Abstract
Phenol-soluble modulin α3 (PSMα3) is a cytotoxic peptide secreted by virulent strains of Staphylococcus aureus. We used a stereochemical strategy to examine the mechanism of PSMα3-mediated toxicity. One hypothesis is that PSMα3 toxicity requires fibril formation; an alternative is that toxicity is caused by soluble forms of PSMα3, possibly oligomeric. We find that the unnatural enantiomer (D residues) displays cytotoxicity comparable to that of L-PSMα3. Racemic PSMα3 is similarly toxic to enantiopure PSMα3 (L or D) under some conditions, but the toxicity is lost under conditions that cause racemic PSMα3 to aggregate. A crystal structure of racemic PSMα3-NH2 displays α-helical secondary structure and a packing pattern that is reminiscent of the cross-α arrangement recently discovered in crystals of L-PSMα3. Our data suggest that the cytotoxicity of PSMα3 does not depend on stereospecific engagement of a target protein or other chiral macromolecule, an observation that supports a mechanism based on membrane disruption. In addition, our data support the hypothesis that toxicity is exerted by a soluble form rather than an insoluble fibrillar form.
Methicillin-resistant strains of the pathogen Staphylococcus aureus (MRSA) are a substantial health risk. Such strains were initially encountered in health care facilities (hospital-associated MRSA, or HA-MRSA), where they are particularly problematic for immunocompromised patients.1,2 Recently, however, highly virulent community-associated strains (CA-MRSA) have emerged outside hospitals.3,4 CA-MRSA can cause severe, tissue-destroying infections in otherwise healthy individuals.5 CA-MRSA strains produce high levels of a family of peptide toxins known as phenol-soluble modulins (PSMs), which seem to be important for virulence.6,7 PSMs induce inflammatory responses and, at low levels, recruit neutrophils via interaction with the formyl peptide receptor 2 (FPR2).8,9 After CA-MRSA cells are phagocytosed, PSM expression increases, and the neutrophil is killed.1,11
PSM-mediated toxicity toward eukaryotic cells has been proposed to arise from membrane disruption.12–14 This hypothesis is consistent with reports that lysis can be induced for many cell types, including S. aureus itself,15 and that synthetic lipid vesicles can be permeabilized by PSMs.16,17 PSMα3 (MEFVA KLFKF FKDLL GKFLG NN), the most toxic among the PSMs produced by S. aureus,6 is rich in hydrophobic residues and cationic residues. This composition is reminiscent of that of host-defense peptides (HDPs), which are produced by eukaryotes in response to microbial infection.18 Many HDPs access α-helical secondary structure that is globally amphiphilic (hydrophobic and cationic residues segregated on opposite sides of the helix). These HDPs can induce bacterial cell lysis,19–23 although they likely have other modes of action, too.24,25 The similarity between PSMα3 and helix-forming HDPs is consistent with the hypothesis that PSMα3 disrupts cell membranes.
The phenol-soluble modulins can self-assemble to form extracellular amyloid-like fibrils that are thought to stabilize S. aureus biofilms.26,27 Thus, the capacity for fibril formation by PSMs may support virulence in ways that are not directly related to toxicity manifested toward eukaryotic cells. Although PSM peptides are produced with N-terminal formyl groups, deformylated peptides are prevalent in the fibrils.27
Recently, an atomic-resolution structure was reported for PSMα3 (no formyl group; PDB 5I55).28 PSMα3 forms a globally amphiphilic α-helix, with the helix axis perpendicular to the long axis of the fibrillar crystals and side-by-side packing of helices along the long axis. This assembly pattern was designated "cross-α" by analogy to the term "cross-β" that is widely employed to describe other amyloids.29,30 Peptides that form cross-β assemblies display mostly extended conformations and form intermolecular β-sheets, with β-strands oriented perpendicular to the long fibril axis. The striking deviation of the PSMα3 packing pattern28 from all previously characterized peptide fibril structures represents a significant expansion in our understanding of amyloid assemblies. Related structures have subsequently been described.31
The discovery of cross-α structure in PSMα3 fibrillar crystals led to the suggestion that cross-α fibrillar assembly of PSMα3 is required for cytotoxicity.28,32 In contrast, extensive studies of neurotoxic peptides that form cross-β fibrils, such as the Aβ(1-42) peptide associated with Alzheimer's Disease, suggest that toxicity arises from a soluble form, perhaps oligomeric, rather than from fibrils.33–37 This view motivated the recent discovery that the enantiomer of Aβ(1-42) can suppress the toxicity of the natural form of the peptide.38 Racemic Aβ(1-42) was shown to form fibrils more rapidly than does pure L- Aβ(1-42) or pure D-Aβ(1-42), consistent with the general trend that racemates tend to be less soluble than the corresponding pure enantiomers.39 Suppression of L-Aβ(1-42) toxicity by mixture with D-Aβ(1-42) is consistent with earlier demonstrations that β-sheet assembly of racemic peptides is more favorable than β-sheet assembly of pure enantiomers.40 The hypothesis that cross-α fibril formation is required for PSMα3 toxicity28,32 differs from mechanisms proposed to explain the membrane-disrupting effects of HDPs,21–23 and the link between fibril formation and PSMα3 toxicity has been challenged.41 We have used a stereochemical strategy to evaluate the relationship between fibrillar aggregation and cytotoxicity of PSMα3.
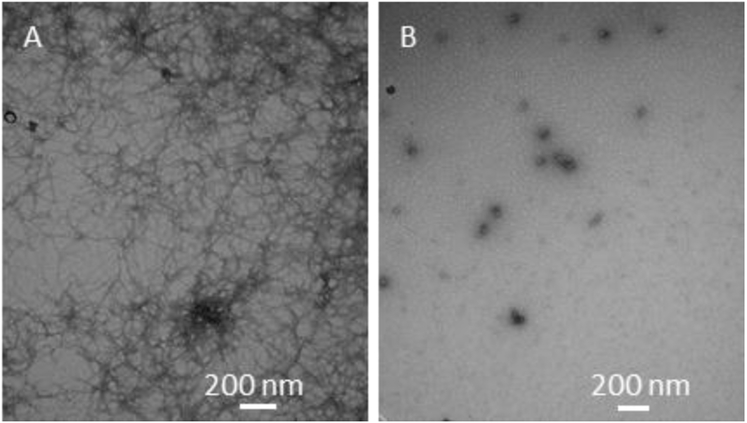
When a solution of 800 μM racemic PSMα3 (no formyl group) in pure water was allowed to stand for 24 hr at room temperature, extensive fibril formation was observed by transmission electron microscopy (TEM) (Figure 1A). However, no fibrils could be detected by TEM when a solution of 800 μM L- or D-PSMα3 in pure water was treated in the same way (Figure 1B). In addition, no fibrils were detected after 24 hr incubation for 25 μM racemic PSMα3, L-PSMα3 or D-PSMα3 in pure water (Figure S1). These comparisons suggest that racemic PSMα3 is more prone to fibril formation relative to a pure enantiomer, as expected.39 For L- or D-PSMα3, high concentrations are required for fibril formation in pure water; the fibrils previously described for L-PSMα3 were grown from a 10,000 μM solution over 1-2 weeks,28 and we observed fibrils after incubation of 6,000 μM L- or D-PSMα3 in pure water for 20 days (Figure S2).
Figure 1.
Representative negative-stain TEM images of PSMα3 after incubation at 800 μM for 24 h. A: Racemic PSMα3. Extensive fibril formation is evident. B: L- PSMα3; no fibrils could be observed in multiple TEM images.
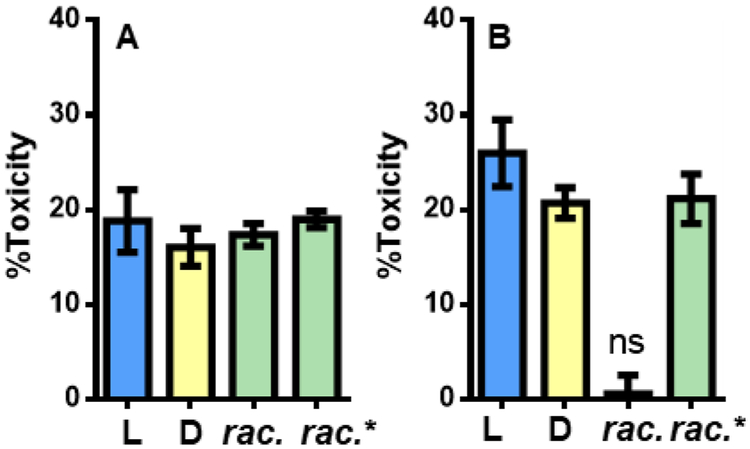
We compared toxicities of different stereochemical forms of PSMα3 toward HEK293FT cells. Solutions containing 25 μM L-PSMα3, D-PSMα3 or racemic PSMα3 in pure water were incubated at room temperature for 24 hr and then diluted with pure water to 5 μM and applied to HEK293FT cells. In each case, toxicity was observed (Figure 2A), and the extents of toxicity were similar. In a second experiment (Figure 2B), the initial solutions contained 800 μM L-, D- or racemic PSMα3 in pure water. After 24 hr, each solution was diluted to 5 μM for the toxicity test. Under these conditions, both L- and D-PSMα3 displayed toxicity comparable to that observed in the first experiment (incubation at 25 μM). However, racemic PSMα3 was not toxic after incubation at 800 μM; this was the only sample in pure water that displayed extensive fibril formation. As part of this experiment, aliquots of L- and D-PSMα3 solutions incubated (separately) at 800 μM for 24 hours were applied in a 1:1 ratio to cells to generate 5 μM racemic PSMα3. In this case, the toxicity matched that observed for L- or D-PSMα3 alone.
Figure 2.
Cytotoxicity of PSMα3 (5 μM total peptide) against HEK293FT cells as measured by an ATP-based luminescent viability assay. Rac. indicates preincubated in racemic form; rac* indicates L- and D- PSMα3 pre-incubated separately and mixed when added to cells. A: Peptides pre-incubated at 25 μM for 24 h before addition to cells. B: Peptides pre-incubated at 800 μM for 24 h before addition to cells. 0% and 100% toxicity represent averaged luminescence values of cells treated with vehicle or 400 μM Triton X-100, respectively, n = 3 independent experiments with at least three technical replicates per condition. Error bars represent standard error. For all conditions, p < 0.0001 compared to vehicle by One-way ANOVA with Bonferroni’s test unless denoted with not-significant (ns) in which case p > 0.05.
Changes in solution conditions can affect the propensity of a peptide to self-assemble. To probe this possibility with PSMα3, we conducted a series of toxicity studies analogous to those described above but employing buffer (10 mM sodium phosphate, pH 8,150 mM NaCl) rather than pure water for incubation of various forms of PSMα3 at 800 μM before exposure to cells (at 5 μM; Figure S3). The toxicity pattern among different stereochemical forms in the buffer experiments mirrored the pattern observed after peptide incubation in pure water: L- and D-PSMα3 were toxic to similar extents after incubation in buffer, but racemic PSMα3 was much less toxic. However, the physical behavior observed in the buffer experiments differed from that observed after incubation in pure water. Extensive fibril formation was detected via TEM for L- or D-PSMα3 after incubation at 800 μM in buffer (Figure S4), while no fibrils had been detected after incubation of L- or D-PSMα3 at 800 μM in pure water (Figures 1B and S5). The similar levels of toxicity exerted after incubation in either buffer or water (Figure S3), despite the difference in fibril formation (detected in buffer only), is consistent with the conclusion that fibril formation is not required for PSMα3 toxicity toward HEK293FT cells.
We interpret our results to indicate that toxicity toward HEK293FT cells arises from a soluble form of PSMα3 (monomeric and/or oligomeric). This conclusion differs from a previous proposal regarding the role of fibrils in cell killing.28 The observation that L and D forms of PSMα3 are equally toxic is consistent with trends reported for several HDPs,42–45 which have been interpreted to support a membrane disruption mechanism. In contrast, if an HDP mechanism required binding to a specific protein, one would expect substantial activity differences between L and D forms of the peptide. Indeed, we observed that in transfected HEK293FT cells, FPR2 activation was significantly reduced for N-formyl D-PSMα3 relative to the L enantiomer (Figure S6). This result is consistent with the hypothesis that the biological activities of PSMα3 arise from at least two modes of action: (1) PSMα3 activates the FPR2 receptor at nanomolar concentrations via stereospecific interactions;8 (2) PSMα3 lyses cell membranes at micromolar concentrations via non-stereospecific interactions.
We turned to crystallography to assess the assembly of PSMα3 at atomic resolution. We could not grow diffraction-quality crystals of racemic PSMα3, which bears a C-terminal acid group; however, racemic PSMα3 with a C-terminal primary amide provided needle-shaped crystals that were suitable for x-ray diffraction (Figure S7). Studies of fibril formation and HEK293FT cell toxicity described above for different stereochemical forms of PSMα3 were conducted with peptides bearing a C-terminal acid group; these studies were repeated with L-, D- and racemic PSMα3-NH2; C-terminal amide and acid analogues behaved similarly (Figures S8-S10).
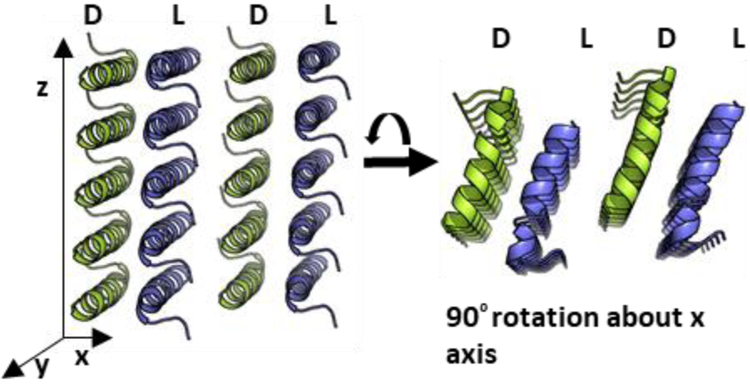
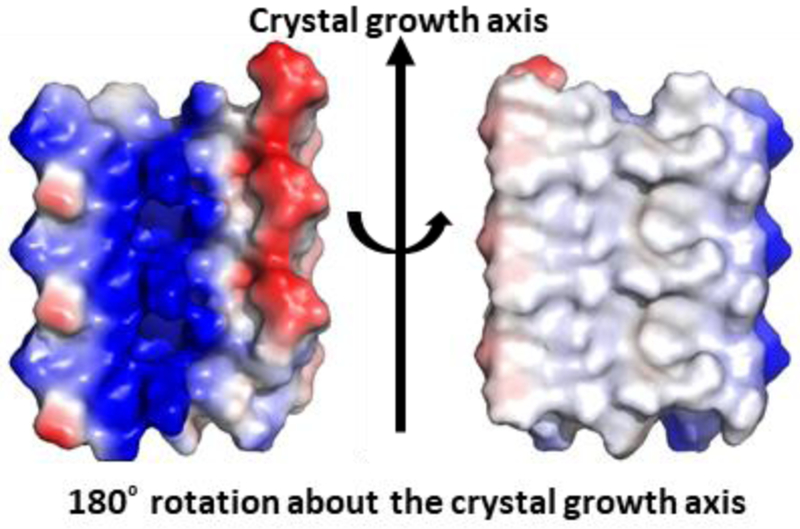
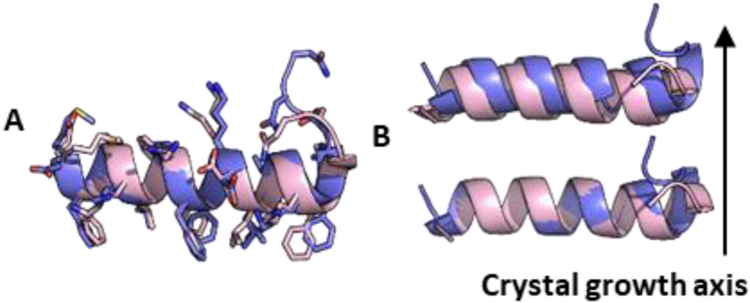
The structure of racemic PSMα3-NH2 was solved at 1.45 Å and revealed a cross-α packing pattern (Figure 3). Each molecule adopts an amphiphilic α-helical conformation very similar to that found in the crystal of L-PSMα3.28 Helices of the same chirality stack alongthe long axis of the crystals; stacks of L-PSMα3-NH2 helices alternate with stacks of D-PSMα3-NH2 helices. The homochiral helix-helix interface involves a parallel orientation and features both non-polar and polar side chain contacts (Figure 4). The peptide conformations are very similar in the two structures (Figure 5A), with small differences at the termini. Overlay of a homochiral nearest-neighbor pair from the racemic PSMα3-NH2 structure with a nearest-neighbor pair from the L-PSMα3 structure reveals the same orientations of helix axes and very similar spacing between the helices in the two structures (Figures 5B and S11). In each crystal, the helices stack along the crystal growth axis. However, there is a 1.9 Å shift of the helices relative to one another in the pair from the racemic PSMα3-NH2 structure relative to the pair from the L-PSMα3 structure. The heterochiral interface involves antiparallel helix orientation. Fiber diffraction data for racemic PSMα3-NH2 (Figure S12) are consistent with the packing observed in the crystalline form.
Figure 3.
Ciystal packing of racemic PSMα3-NH2. L-peptides blue; D-peptides yellow. The long axis of the fibril-like crystals corresponds to the z axis.
Figure 4.
Sheets formed by homochiral peptides in the racemic PSMα3-NH2 crystal display functionally different surfaces. In these electrostatic potential maps, regions of positive potential are blue, regions of negative potential are red, and neutral regions are gray. Three L-peptide molecules are shown.
Figure 5.
Comparison of racemic PSMα3-NH2 (blue) and L-PSMα3 (pink, PDB ID 5155). A: Overlay of L-peptides from the racemic and chiral structures. For Cα atoms of residues 1-22, RMSD = 1.93 Å, and for Cα atoms of residues 2-19, RMSD = 0.51 Å. B: Comparison ofthe homochiral nearest-neighbor dimer for racemic PSMα3-NH2 (blue) with the analogous nearest-neighbor dimer for L-PSMα3 (pink; PDB 5155), backbone only (see Fig. S11 for more detail).
The similarity of distances and orientations between neighboring homochiral helices in the racemic PSMα3-NH2 structure and the L-PSMα3 structure (PDB 5155) suggests a structural similarity between the amyloid fibrils formed by the racemic and L peptides. Therefore, our observation that fibril formation by racemic PSMα3 causes a loss of toxicity is consistent with our conclusion that the fibrillar form of L-PSMα3 is not toxic. However, we note the caveat that fibrils formed from racemic PSMα3 might differ in internal structure and/or surface properties, relative to fibrils formed from L-PSMα3, and such differences could lead to variations in toxic effects exerted by these fibrils. Variations in ThT assays results (Figure S13) and TEM images (Figures S4 and S14) between L-PSMα3 and racemic PSMα3 suggest physical differences between homochiral and heterochiral aggregates, although it is not clear whether these physical differences affect biological activity.
We have used a stereochemistry-based strategy to probe the mechanism of toxicity exerted by the CA-MRSA toxin PSMα3 toward HEK293FT cells. The two enantiomeric forms of this peptide are equally toxic, which is consistent with the hypothesis that PSMα3 can kill cells without engaging a specific protein receptor, via membrane disruption. Comparisons of toxic effects exerted by L-, D- or racemic PSMα3 under different conditions support the hypothesis that toxicity requires a soluble form of the peptide. Our conclusions are consistent with those of Zheng et al.41 The atomic-resolution structure of racemic PSMα3-NH2 disclosed here provides a new example of cross-α packing.28
Supplementary Material
ACKNOWLEDGMENT
This research was supported by the NIH (R01 GM061238, to SHG) and the NSF (IOS 1353674, to KTF), B.P.C. was supported in part by an NSF graduate research fellowship (DGE-1747503) and by NIH biotechnology training grant T32GM008349. Storage and distribution provided by the PlasmID Repository at Harvard Medical School is funded in part by NCI Cancer Center Support Grant (NIH 5 P30 CA06516). Crystallization and structure solution were supported by the Collaborative Crystallography Core in the Department of Biochemistry, UW-Madison. GM/CA@APS is supported by funds from the National Cancer Institute (ACB-12002) and the National Institute of General Medical Sciences (AGM-12006). This research used resources of the Advanced Photon Source, a U.S. Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE-AC02-06CH11357. The Eiger 16M detector was funded by an NIH-Office of Research Infrastructure Programs, High-End Instrumentation Grant (1S10OD012289-01A1). We thank Nagarajan Venugopalan and Craig Ogata for beamline support. Fibril diffraction data was collected at the Life Sciences Collaborative Access Team (LS-CAT) beamline 21-ID-D at APS. Use of the LS-CAT Sector 21 was supported by the Michigan Economic Development Corporation and the Michigan Technology Tri-Corridor (Grant 085P1000817).The data analysis of fibril diffraction is facilitated by Dr. Weikang Ma and supported by grant 9 P41 GM103622 from the National Institute of General Medical Sciences of the National Institutes of Health.
Footnotes
Supporting Information
Model coordinates and structure factors have been deposited in the Protein Data Bank as entry 6NIV.
Experimental details including peptide synthesis and characterization, X-ray crystallography, cell toxicity assay, FPR2 activation assay, Thioflavin T assay, transmission electron microscopy, atomic force microscopy; X-ray diffraction data collection, fibril diffraction data and analysis, structure solution, and refinement; additional comparison of homochiral packing along crystal growth axis; additional cell assay result; additional transition electron microscope result.
The authors declare no competing financial interests.
REFERENCES
- (1).Chambers HF; DeLeo FR Waves of Resistance: Staphylococcus Aureus in the Antibiotic Era. Nat. Rev. Microbiol. 2009, 7 (9), 629–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Klevens RM; Morrison MMAM; Nadle J; Petit S; Gershman K; Ray S; Harrison LH; Lynfield R; Craig AS; Zell ER; Fosheim GE; Mcdougal LK; Carey RB; Fridkin SK Invasive Methicillin-Re-sistant Staphylococcus Aureus Infections in the United States. J. Am. Med. Assoc. 2007, 298 (15), 1763–1771. [DOI] [PubMed] [Google Scholar]
- (3).DeLeo FR; Otto M; Kreiswirth BN; Chambers HF Community-Associated Meticillin-Resistant Staphylococcus Aureus. Lancet 2010, 375 (9725), 1557–1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Klein EY; Sun L; Smith DL; Laxminarayan R The Changing Epidemiology of Methicillin-Resistant Staphylococcus Aureus in the United States: A National Observational Study. Am. J. Epidemiol. 2013, 177 (7), 666–674. [DOI] [PubMed] [Google Scholar]
- (5).Miller LG; Perdreau-Remington Francoise Rieg G; Mehdi S; Perlroth J; Bayer AS; Tang AW; Phung TO; Spellberg B Necrotizing Fasciitis Caused by Community-Associated Methicillin-Resistant. N Engl J Med 2005, 352 (14), 1445–1453. [DOI] [PubMed] [Google Scholar]
- (6).Wang R; Braughton KR; Kretschmer D; Bach T-HL; Queck SY; Li M; Kennedy AD; Dorward DW; Klebanoff SJ; Peschel A; DeLeo FR; Otto M Identification of Novel Cytolytic Peptides as Key Virulence Determinants for Community-Associated MRSA. Nat. Med. 2007,13 (12), 1510–1514. [DOI] [PubMed] [Google Scholar]
- (7).Otto M Basis of Virulence in Community-Associated Methicillin-Resistant Staphylococcus Aureus. Annu. Rev. Microbiol. 2010, 64 (1), 143–162. [DOI] [PubMed] [Google Scholar]
- (8).Kretschmer D; Gleske A-K; Rautenberg M; Wang R; Köberle M; Bohn E; Schöneberg T; Rabiet M-J; Boulay F; Klebanoff SJ; van Kessel KA; van Strijp JA;Otto M;Peschel A Human Formyl Peptide Receptor 2 Senses Highly Pathogenic Staphylococcus Aureus. Cell Host Microbe 2010, 7 (6), 463–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Rautenberg M; Joo H-S; Otto M; Peschel A Neutrophil Responses to Staphylococcal Pathogens and Commensals via the Formyl Peptide Receptor 2 Relates to Phenol-Soluble Modulin Release and Virulence. FASEB J. 2011, 25 (4), 1254–1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Surewaard BGJ; De Haas CJC; Vervoort F; Rigby KM; Deleo FR; Otto M; Van Strijp JAG; Nijland R Staphylococcal Alpha-Phenol Soluble Modulins Contribute to Neutrophil Lysis after Phagocytosis. Cell. Microbiol. 2013,15 (8), 1427–1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Otto M Staphylococcus Aureus Toxins. Curr. Opin. Microbiol. 2014,17(1), 32–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Cheung GYC; Kretschmer D; Queck SY; Joo HS; Wang R; Duong AC; Nguyen TH; Bach THL; Porter AR; DeLeo FR; Peschel A; Otto M Insight into Structure-Function Relationship in Phenol-Soluble Modulins Using an Alanine Screen of the Phenol-Soluble Modulin (PSM) A3 Peptide. FASEB J. 2014, 28 (1), 153–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Duong AC; Cheung GYC; Otto M Interaction of Phenol-Soluble Modulins with Phosphatidylcholine Vesicles. Pathogens 2012,1 (1), 3–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Chatterjee SS; Joo H-S; Duong AC; Dieringer TD; Tan VY; Song Y; Fischer ER; Cheung GYC; Li M; Otto M Essential Staphylococcus Aureus Toxin Export System. Nat. Med. 2013, 19 (3), 364–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Ebner P; Luqman A; Reichert S; Hauf K; Popella P; Forchham-mer K; Otto M; Götz F Non-Classical Protein Excretion Is Boosted by PSMα-Induced Cell Leakage. Cell Rep. 2017, 20 (6), 1278–1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Otto M Phenol-Soluble Modulins. Int. J. Med. Microbiol. 2014, 304 (2), 164–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Laabei M; Jamieson WD; Yang Y; Van Den Elsen ); Jenkins ATA Investigating the Lytic Activity and Structural Properties of Staphylococcus Aureus Phenol Soluble Modulin (PSM) Peptide Toxins. Bio-chim. Biophys. Acta - Biomembr. 2014,1838 (12), 3153–3161. [DOI] [PubMed] [Google Scholar]
- (18).Yeaman M; Yount N Mechanisms of Antimicrobial Peptide Action and Resistance. 2003, 55 (1), 27–55. [DOI] [PubMed] [Google Scholar]
- (19).Powers JPS; Hancock REW The Relationship between Peptide Structure and Antibacterial Activity. Peptides 2003, 24 (11), 1681–1691. [DOI] [PubMed] [Google Scholar]
- (20).Zasloff M Antimicrobial Peptides of Multicellular Organisms. Nature 2002, 415 (January), 389–395. [DOI] [PubMed] [Google Scholar]
- (21).Yang L; Weiss TM; Lehrer RI; Huang HW Crystallization of Antimicrobial Pores in Membranes: Magainin and Protegrin. Biophys. J. 2000, 79 (4), 2002–2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Matsuzaki K Why and How Are Peptide-Lipid Interactions Utilized for Self-Defense Magainins and Tachyplesins as Archetypes. Bio-chim. Biophys. Acta 1999,1462,1–10. [DOI] [PubMed] [Google Scholar]
- (23).Shai Y Mechanism of the Binding, Insertion and Destabilization of Phospholipid Bilayer Membranes by α-Helical Antimicrobial and Cell Non-Selective Membrane-Lytic Peptides. Biochim. Biophys. Acta - Biomembr. 1999,1462 (1–2), 55–70. [DOI] [PubMed] [Google Scholar]
- (24).Brown KL; Hancock REW Cationic Host Defense (Antimicrobial) Peptides. Curr. Opin. Immunol. 2006,18 (1), 24–30. [DOI] [PubMed] [Google Scholar]
- (25).Brogden KA Antimicrobial Peptides: Pore Formers or Metabolic Inhibitors in Bacteria? Nat. Rev. Microbiol. 2005, 3 (3), 238–250. [DOI] [PubMed] [Google Scholar]
- (26).Marinelli P; Pallares I; Navarro S; Ventura S Dissecting the Contribution of Staphylococcus Aureus α-Phenol-Soluble Modulins to Biofilm Amyloid Structure. Sci. Rep. 2016, 6 (September), 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Schwartz K; Syed AK; Stephenson RE; Rickard AH; Boles BR Functional Amyloids Composed of Phenol Soluble Modulins Stabilize Staphylococcus Aureus Biofilms. PLoS Pathog. 2012, 8 (6), el002744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Tayeb-Fligelman E; Tabachnikov O; Moshe A; Goldshmidt-Tran O; Sawaya MR; Coquelle N; Colletier J-P; Landau M The Cytotoxic Staphylococcus Aureus PSMα3 Reveals a Cross-α Amyloid-like Fibril. Science. 2017, 355 (6327), 831–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Pauling L; Corey RB Configurations of Polypeptide Chains With Favored Orientations Around Single Bonds: Two New Pleated Sheets. Proc. Natl. Acad. Sci. 1951, 37 (11), 729–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Geddes AJ; Parker KD; Atkins EDT; Beighton E Cross β Conformation in Proteins. J. Mol. Biol. 1968, 32, 343–358. [DOI] [PubMed] [Google Scholar]
- (31).Zhang S; Huang H; Yang J; Kratochvil HT; Lolicato M; Liu Y; Shu X; Liu L; Degrado WF Designed Peptides That Seemble into Cross-α Amyloid-like Structures. Nat. Chem. Biol. 2018, 14 (September), 870–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Malishev R; Tayeb-fligelman E; David S; Meijler MM; Landau M ; Jelinek R Reciprocal Interactions between Membrane Bilayers and S . Aureus PSM α 3 Cross- α Amyloid Fibrils Account for Species-Specific Cytotoxicity. J. Mol. Biol. 2018, 430 (10), 1431–1441. [DOI] [PubMed] [Google Scholar]
- (33).Shankar GM; Li S; Mehta TH; Garcia-Munoz A; Shepardson NE; Smith I; Brett FM; Farrell MA; Rowan MJ; Lemere CA; Regan CM;Walsh DM; Sabatini BL Selkoe DJ Amyloid-β Protein Dimers Isolated Directly from Alzheimer’s Brains Impair Synaptic Plasticity and Memory. Nat. Med. 2008,14 (8), 837–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Dahlgren KN; Manelli AM; Stine WB; Baker LK; Krafft GA; LaDu MJ Oligomeric and Fibrillar Species of Amyloid-β Peptides Differentially Affect Neuronal Viability. J. Biol. Chem. 2002, 277 (35), 32046–32053. [DOI] [PubMed] [Google Scholar]
- (35).Walsh DM; Klyubin I; Fadeeva JV; Cullen WK; Anwyl R; Wolfe MS; Rowan MJ; Selkoe DJ Naturally Secreted Oligomers of Amyloid β Protein Potently Inhibit Hippocampal Long-Term Potentiation in Vivo. Nature 2002, 416 (6880), 535–539. [DOI] [PubMed] [Google Scholar]
- (36).Ono K; Condron MM; Teplow DB Structure-Neurtoxicity Relationships of Amyloid β-Protein Oligomers. Proc. Natl. Acad. Sci. 2009, 106 (35), 14745–14750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Haass C; Selkoe DJ Soluble Protein Oligomers in Neurodegeneration: Lessons from the Alzheimer’s Amyloid β-Peptide. Nat. Rev. Mol. Cell Biol. 2007, 8 (2), 101–112. [DOI] [PubMed] [Google Scholar]
- (38).Dutta S; Foley AR; Warner CJA; Zhang X; Rolandi M; Abrams B; Raskatov JA Suppression of Oligomer Formation and Formation of Non-Toxic Fibrils upon Addition of Mirror-Image ΑM42 to the Natural 1-Enantiomer. Angew. Chemie - Int. Ed. 2017, 56 (38), 11506–11510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Brock CP; Schweizer WB; Dunitz JD On the Validity of Wallaces Rule: On the Density and Stability of Racemic Crystals Compared with Their Chiral Counterparts. J. Am. Chem. Soc. 1991,113 (26), 9811–9820. [Google Scholar]
- (40).Swanekamp RJ; DiMaio JTM; Bowerman CJ; Nilsson BL Coassembly of Enantiomeric Amphipathic Peptides into Amyloid-Inspired Rippled β-Sheet Fibrils. J. Am. Chem. Soc. 2012,134 (12), 5556–5559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Zheng Y; Joo HS; Nair V; Le KY; Otto M Do Amyloid Structures Formed by Staphylococcus Aureus Phenol-Soluble Modulins Have a Biological Function? Int. J. Med. Microbiol. 2017, 308 (6), 675–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Bessalle R; Kapitkovsky A; Gorea A; Shalit I; Fridkin M All-D-Magainin: Chirality, Antimicrobial Activity and Proteolytic Resistance. FEBS Lett. 1990, 274 (1–2), 151–155. [DOI] [PubMed] [Google Scholar]
- (43).Wade D; Bomant A; Wxhlint B; Drain CM; Andreut D; Bomant HG; Merrifield RB All-D Amino Acid-Containing Channel-Forming Antibiotic Peptides. 1990, 87 (June), 4761–4765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Hayouka Z; Mortenson DE; Kreitler DF; Weisblum B; Forest KT; Gellman SH Evidence for Phenylalanine Zipper-Mediated Dimerization in the X-Ray Crystal Structure of a Magainin 2 Analogue. J. Am. Chem. Soc. 2013,135 (42), 15738–15741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Yang H; Chen KH; Nowick JS Elucidation of the Teixobactin Pharmacophore. ACS Chem. Biol. 2016,11 (7), 1823–1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.