Abstract
Sleep abnormalities have widespread and costly public health consequences, yet we have only a rudimentary understanding of the events occurring at the cellular level in the brain that regulate sleep. Several key signaling molecules that regulate sleep across taxa come from the family of neuropeptide transmitters. For example, in Drosophila melanogaster, the neuropeptide Y (NPY)-related transmitter short neuropeptide F (sNPF) appears to promote sleep. In this study, we utilized optogenetic activation of neuronal populations expressing sNPF to determine the causal effects of precisely timed activity in these cells on sleep behavior. Combining sNPF-GAL4 and UAS-Chrimson transgenes allowed us to activate sNPF neurons using red light. We found that activating sNPF neurons for as little as 3 seconds at a time of day when most flies were awake caused a rapid transition to sleep that persisted for another 2+ hours following the stimulation. Changing the timing of red light stimulation to times of day when flies were already asleep caused the control flies to wake up (due to the pulse of light), but the flies in which sNPF neurons were activated stayed asleep through the light pulse, and then showed further increases in sleep at later points when they would have normally been waking up. Video recording of individual fly responses to short-term (0.5–20 second) activation of sNPF neurons demonstrated a clear light duration-dependent decrease in movement during the subsequent 4-minute period. These results provide supportive evidence that sNPF-producing neurons promote long-lasting increases in sleep, and show for the first time that even brief periods of activation of these neurons can cause changes in behavior that persist after cessation of activation. We have also presented evidence that sNPF neuron activation produces a homeostatic sleep drive that can be dissipated at times long after the neurons were stimulated. Future studies will determine the specific roles of sub-populations of sNPF-producing neurons, and will also assess how sNPF neurons act in concert with other neuronal circuits to control sleep.
Keywords: Sleep, short neuropeptide F (sNPF), Drosophila melanogaster, optogenetic
1. Introduction
Sleep is a critical and nearly universal behavioral state in which humans spend approximately one third of their lives. Sleep disturbances due to insomnia, sleep apnea, psychiatric and neurological disorders, jetlag, and shift work, and daily pressures are also highly prevalent and disruptive in today’s society [1–4]. However, the cellular and molecular mechanisms that regulate sleep are still not well understood [5–7]. Due to the complexity of human sleep systems, as well as their inaccessibility for experimental manipulation, studying animals with simpler nervous systems can be a powerful approach [8]. The fruit fly Drosophila melanogaster in particular has emerged as a useful system in which to study sleep. Drosophila shares many behavioral and neurochemical characteristics of sleep with mammalian species [9–12], but has a much simpler nervous system and a vast capacity for genetic manipulations [13]. Behaviorally, sleep in Drosophila has circadian rhythmicity, is associated with a preferred posture and sleep location, and sleeping animals have an increased arousal threshold. Additionally, sleep deprivation in Drosophila impairs vigilance and produces a subsequent homeostatic sleep rebound [9, 11, 14]. In terms of neural circuits, we have learned that sleep in Drosophila is regulated by multiple brain regions, including circadian clock neurons, the mushroom bodies, the pars intercerebralis, the ellipsoid body, and the fan-shaped body [15–21]. At the molecular level, many genes and neurotransmitters have been linked to sleep control in the fly [reviewed in 10, 22, 23–25]. Among these are several neuropeptides, such as short neuropeptide F (sNPF), pigment-dispersing factor (PDF), neuropeptide F (NPF), SIFamide, myoinhibitory peptide (MIP), CCHa1, allatostatin-A, and ion transport peptide (ITP) [26–38].
Neuropeptides are protein-based neurotransmitters of particular interest as potential targets of drug treatments due to their diversity and selectivity [39]. For example, the multi-billion dollar pharmaceutical sleep aid market targets primarily classical neurotransmitters such as norepinephrine and, in particular, GABA. However, since GABA plays a role in virtually all behaviors, drugs targeting GABA signaling have pervasive issues with side effects [40, 41]. There exists a wide array of neuropeptides (>100 in humans, >80 identified in Drosophila), with each transmitter having a relatively specific role in regulating behavior, such that a given neuropeptide might only influence a few types of behavior (sleep and feeding, e.g.) [42]. Their diversity and their specificity of function make neuropeptides intriguing targets for pharmaceutical development of sleep drugs [40].
In this study, we focused on the neuropeptide short neuropeptide F (sNPF), which acts through a single G-protein-coupled receptor (GPCR) called NPFR76F, which is related to mammalian neuropeptide Y (NPY) Y2 receptors [43–45]. NPY in vertebrates appears to modulate a handful of behaviors, including feeding and sleep [reviewed in 46, 47]. Some studies in rodents have found that NPY or agonist administration promotes sleep [48, 49], while others have found the opposite [50]. This may be due to the location of NPY administration, dose, the brain state at the time of NPY administration, and competition between different behavioral drives (NPY signals that induce feeding may counteract sleep drive, e.g.). NPY in zebrafish was found to promote sleep [51], and NPY administration in human males reduced the latency to fall asleep and increased sleep time [52]. Thus, the overall balance of the literature seems to be in favor of NPF being a sleep-promoting signal. sNPF signaling in Drosophila is also thought to promote sleep. The first piece of evidence for this comes from a constitutive mutation of the sNPF gene, which was found to reduce daytime sleep and nighttime sleep, especially during the first half of the night [35]. However, another research group reported nearly opposite results, using the same mutant as well as other constitutive genetic alterations [27]. This highlights some of the caveats with relying on constitutive mutations to study the roles of molecules in adult behavior; namely, that the mutations may influence developmental processes, thus influencing adult behavior indirectly, and that the animal has its entire life to compensate for the lack of the mutated gene. Shang et al [35] also provided additional evidence that sNPF is sleep-promoting, by expressing the heat-activated cation channel dTRPA1 [53, 54] in sNPF neurons using the GAL4/UAS system [55]. Indeed, when the temperature was adjusted from 21°C to 27°C for a 24-hour period, experimental animals appeared to spend nearly the entire day asleep, and when the temperature was again lowered the next day, returned to a normal sleep pattern [35]. However, even with this conditional thermogenetic activation paradigm, stimulation of sNPF neurons still took place over a long time-frame, making it impossible to determine how behavior would be affected by more acute activation of sNPF neurons.
Therefore, in this study we used optogenetic activation of sNPF neurons, which allowed for briefer stimulation periods. In this approach, light-sensitive ion channels can be expressed in neurons of interest, allowing them to be activated by pulses of external light. Researchers have now designed several light sensors that respond to different spectral ranges [56–58]. Since we wanted to activate neurons in adult fruit flies, which have a protective cuticle that blocks much ultraviolet and blue light, we chose to use the red light-sensitive cation channel Chrimson [57], which has been used successfully in other recently published studies of neuronal activation [59–62]. Specifically, our goals were to determine the following: (1) Does optogenetic stimulation confirm that activity in sNPF neurons promotes sleep? (2) If so, for how long is sleep promoted following brief activation of sNPF neurons? (3) Does sNPF neuron activation have similar effects on sleep at different times of day?
2. Materials & Methods
2.1. Fly Lines and Maintenance
We used the following transgenic Drosophila melanogaster strains: w−;sNPF-GAL4/CyO (from Yuhua Shang, Michael Rosbash lab, Brandeis University), w−;;UAS-CsChrimson (Bloomington Drosophila Stock Center, # 55136). Both of these strains had been backcrossed with our control w1118 CS line (from the lab of Leslie Griffith, Brandeis University). Flies were maintained in plastic bottles containing fly food at 25° C, on a 12 hour light/12 hour dark (LD) cycle. Fly food batches consisted of 4 L distilled H2O, 400 g cornmeal (Quaker yellow), 24 g agar (Drosophila agar, 100 mesh), 72 g yeast (Red Star active dry yeast), 240 g dextrose, 120 g sucrose, 2 g methyl paraben (Josh’s Frogs), and 40 mLs of a mix of propionic acid (41.8%) and phosphoric acid (4.15%). Cornmeal, agar, and yeast were all from Genesee Scientific, and dextrose, sucrose, propionic acid, and phosphoric acid were all from Fisher Scientific. All incubators used for housing and experimentation were from Percival Scientific. All experimental crosses were set up between male and virgin female flies on food of the type described above, which had been melted and combined with 1 mM all-trans-retinal (ATR; Toronto Research Chemicals, prepared as a 100 mM stock in ethanol). Preliminary tests found that the presence of ATR in the food did not alter sleep patterns on its own, but it was required for successful activation of the optogenetic sensor (data not shown). Crosses were raised in dark:dark (DD) conditions in a separate 25° C incubator. DD conditions were used in order to prevent unwanted optogenetic activation during development, because even moderate ambient white light can be sufficient to activate the Chrimson sensor.
2.2. Optogenetic Activation
We employed 2 distinct approaches to activate neurons optogenetically. The first approach utilized a 225 LED, 12.5” × 12.5” red light grid (HQRP) placed directly below or above Drosophila activity monitors (DAM; Trikinetics, Inc.) in an incubator kept otherwise in DD conditions. The timing of lighting was controlled by free DAM software. In the second “acute” approach, a single red-orange Luxeon Rebel 1-Up LED (LEDSupply, Part # 07040-PH000-G) attached to a TO-220 heat sink (RadioShack – now not available, but we have used Part # 129242 from Jameco as a substitute) was used for optogenetic stimulation, with current supplied by a 1000mA Wired BuckPuck (LEDSupply, Part # 3023-D-E-1000). The timing and intensity of lighting was controlled in this case by an Arduino Uno using homemade circuits and scripts (circuit diagram and scripts freely available on request). Spectral analysis of LED emissions was carried out using a Red Tide USB650 spectrometer (Ocean Optics) and Logger Pro software (Vernier).
2.3. Behavioral Assays
Drosophila sleep behavior was measured as follows. Experimental flies (3–10 days post-eclosion) were anesthetized by CO2 and loaded into small polycarbonate tubes (5 mm diameter × 65 mm length) containing a small plug of 5% sucrose / 1% agarose / 1 mM ATR food at one end. Tubes were capped with parafilm with a hole for air supply. The tubes were subsequently loaded into DAM2 boards and placed into a 25 °C incubator in DD conditions. Through Trikinetics DAMsystem310X software, the DAM2 boards measured fly activity, tallying the number of times each individual fly crossed an infrared beam running down the center of each glass tube. Sleep was operationally defined as 5 minutes without a beam-break, a well-established threshold that is the standard in Drosophila sleep studies [9, 63].
Sleep and circadian parameters were extracted from the aggregate behavioral data using the Sleep and Circadian Analysis MATLAB Program (SCAMP), which was published originally in Donelson et al. [64] and is now available in an updated version for free download at www.trikinetics.com. Sleep studies were performed for a minimum of 1 baseline day, an experimental day during which red light activation was performed, and 1 recovery day. We ran multiple assays in which red light was provided at different circadian times (CT). To help visualize the effect of optogenetic stimulation within a given strain of flies, subtraction plots were created by subtracting sleep amounts for each animal during half hour bins during the baseline day from the same half hour bins during the experimental day.
Acute assessment of the effects of optogenetic neuronal activation was carried out during subjective daytime (CT0–8) as follows. Wells in a plastic 24-well plate were filled with agarose gel until only a millimeter of space was left at the top. A 48-LED, 1000mA infrared light source (Olymstore) was used to illuminate the preparation. The light sensor built into the infrared light source was covered with clay to prevent the illuminator from turning off due to ambient lighting. Flies were aspirated into the chamber individually and the chamber was covered with a glass cover slip. Fly behavior was assessed by video recordings over a 30-second baseline, the red light stimulation period, and for 4 minutes following the stimulation, using a Sony CCD/R XC-E150 camera equipped with a M6Z1212–3S Manual Zoom lens (Computar) in tandem with a VM-100 extension tube (Computar) and equipped with an 850 nm Near-IR Longpass M55 filter (MidOpt). The camera was suspended directly above the preparation on a tripod, and was attached to a camera adaptor and recorded to a MacBook Pro using EasyCap Viewer software. The LED described in the Optogenetic Activation section was held in a clamp attached to a ring stand and oriented so that its beam of light just covered the entire arena. The intensity and timing of red LED pulses were controlled by an Arduino Uno board. Output pulses also turned on an infrared LED that was placed in the frame of the camera so that the exact timing of light could be visualized in the video. The intensity of light used in these experiments was tested using a light meter (Thor Labs). Light intensity at 650 nm due to red LED stimulation was measured at 12.5 mW, whereas ambient light was measured at 0.1 mW. Videos were hand-scored by a experimenter, blind to the condition of the animal, for the number of seconds that each fly was walking, grooming, or not moving during the 4 minutes following red light stimulation.
2.4. Arousal Testing
To test the ability of mechanical stimulation to arouse animals that were asleep naturally or asleep due to optogenetic activation of sNPF neurons, we used a VWR vortexer modified to hold 4 DAM monitors. The timing of shaking was controlled using the DAM software, and the intensity was controlled on the shaker itself. A short (0.5 second) burst of shaking was delivered during 6 consecutive hours, with the first burst being delivered 5 minutes into the red light period. Arousal tests were performed using 1 hour of optogenetic red light stimulation, which was timed to occur at either CT 7/8 or CT 19. Out of the total number of live flies, we marked flies as being awake or asleep at the time of the shaking burst based on the usual criterion of having 0 beam crosses during the previous 5 minutes. Then we calculated a percentage of sleeping flies that were aroused by the stimulus based on whether they had at least 1 beam cross during the minute following shaking.
2.5. Statistical Analysis
“Stimulation day – Baseline day” subtractions from sleep studies were analyzed separately for male and female flies using mixed-model ANOVAs, with 30-minute bin as a within-subject factor and genotype as a between-subject factor, followed by Tukey post-hoc tests. Results from acute optogenetic stimulation were analyzed using an ANOVA, with genotype, sex, and stimulation duration as factors, followed by Tukey post-hoc tests. Differences were only deemed significant in the experiments above if the experimental group was significantly different from both of the control groups. In arousal experiments, activity data averaged across all 6 arousal events were analyzed using a mixed-model ANOVA, with time point (pre- vs. post-arousal minute) as the within-subject factor and genotype as a between-subject factor, followed by Tukey post-hoc tests. A p < 0.05 was the cutoff for significance for all post-hoc tests and ANOVAs. Statistical analyses were conducted using JMP13.
3. Results
3.1. Optogenetic activation of sNPF neurons during wakefulness promotes long-term increases in sleep
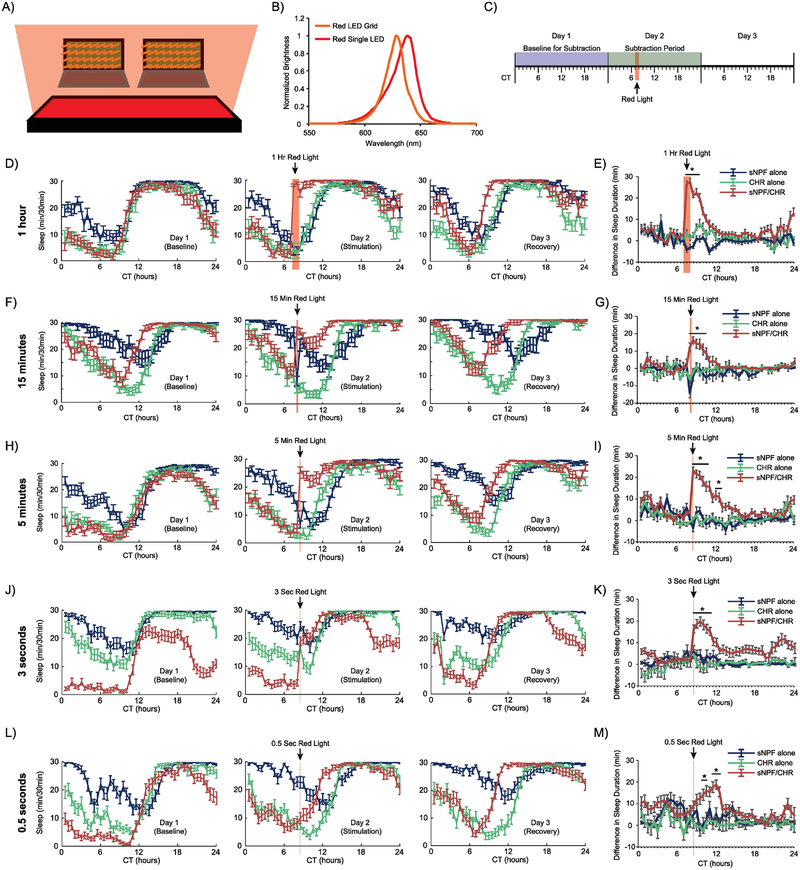
Experimental flies containing both sNPF-GAL4 and UAS-CsChrimson transgenes (sNPF/CHR) and control flies containing one transgene or the other were stimulated optogenetically with red light (Fig. 1A–B) during the second day of sleep monitoring. In separate experiments, the duration of this stimulation was 1 hour, 15 minutes, 5 minutes, 3 seconds, and 0.5 seconds. The longer durations were chosen so that they mimicked more chronic activation paradigms, but were short enough that they could be applied at different times of day. The shorter durations were chosen to mimic the durations of activation used in our acute, single-fly stimulation studies (see Results section 3.5). We hoped that this range of stimulation duration would allow us to determine a dose-response curve for the effects of sNPF neuron activation on sleep. Although there was no ambient light, red light stimuli in these experiments were timed to occur during the subjective daytime (circadian time (CT) 7 or 8). Both male and female flies were tested, and sections 3.1 and 3.2 present data from females, and sections 3.3 and 3.4 present data from males. In female flies, the red light stimulus caused a sharp increase in sleep during the stimulus, and this effect persisted decrementally for hours afterward, as can be seen in plots of minutes of sleep/30-minute bins (Fig. 1D, F, H, J, and L), or in plots subtracting the amount of sleep in each 30-minute bin during the stimulation day – the baseline day (Fig. 1E, G, I, K, and M). In each experiment, experimental flies eventually woke up and returned to a similar sleep pattern to the baseline day, with the exception that their sleep onset was typically shifted earlier in the day by an hour or two. This effect appears to reflect a slightly shorter circadian period in experimental flies, which can still be observed in experiments in which no red light pulse is provided (data not shown).
Figure 1. Optogenetic activation of sNPF neurons when female flies are active promotes long-lasting increases in sleep.
(A) Schematic representing the experimental setup. Female flies were placed in DAM2 monitors just above a red LED illuminator grid. (B) Plots of the spectral ranges of the red LED grid used for all sleep experiments and the single red LED used in the experiments whose results are shown in Figure 5. (C) A general timeline of the experiments whose data are shown in this figure. Red light stimulation for 1 hour, 15 minutes, 5 minutes, 3 seconds, or 0.5 seconds was provided at CT7–8 in conditions of otherwise total darkness. Minutes of sleep per 30-min bin were recorded. To determine how much sleep was altered by red light stimulation, subtraction plots were created by subtracting the amount of sleep during each 30-minute bin during the Baseline for Subtraction from the amount of sleep during the 30-minute bin from the same circadian time of day during the Subtraction Period. (D, F, H, J, L) Sleep patterns across the 3 experimental days for the 1-hour, 15-minute, 5-minute, 3-second, and 0.5-second stimulation experiments, respectively. (E, G, I, K, M) Subtraction plots for the 1-hour, 15-minute, 5-minute, 3-second, and 0.5-second stimulation experiments, respectively. 1 hour of stimulation caused the greatest magnitude initial increase in sleep, but all stimulation paradigms resulted in significantly increased sleep that took hours to decay back to baseline. Error bars represent the standard error of the mean, and * represents a post-hoc comparison with p < 0.05. The numbers of experimental animals were as follows, for sNPF alone, CHR alone, and sNPF/CHR groups, respectively: 1-hour experiment – 28, 29, 27; 15-minute experiment – 20, 30, 32; 5-minute experiment – 32, 31, 30; 3-second experiment – 31, 32, 32; and 0.5-second experiment – 31, 31, 31.
For the 1-hour stimulation (Fig. 1D–E, there was a significant overall effect of genotype (F(2,81)=51.6, p<.0001), bin, (47,3807)=5.9, p <.0001), and interaction effect between genotype and bin (94,3807)=5.3, p<.0001). Post-hoc analysis found that the sNPF/CHR flies were significantly different than both control groups at CT7.5-CT9.5, which corresponded to the two 30-minute periods containing the stimulation and the three 30-minute periods following (Fig. 1E).
For the 15-minute stimulation (Fig. 1F–G), there were also significant overall effects of genotype (F(2,79)=15.1, p<.0001) and bin (47,3713)=2.1, p <.0001), and a significant interaction effect between genotype and bin (94,3713)=4.0, p<.0001) (Fig. 1E). Post-hoc tests found significant differences between the sNPF/CHR flies and both control groups at CT8.5–10.5, which again contained the illumination period and the 3 subsequent 30-minute periods (Fig. 1G).
In the 5-minute stimulation experiment, results were similar, although as expected, the maximum sleep induction was slightly lower than with longer durations of optogenetic activation of sNPF neurons (Fig. 1H–I). In females there was a significant overall effect of genotype (F(2,91)=55.4, p<.0001), bin, (47,4277)=6.9, p <.0001), and interaction effect between genotype and bin (94,4277)=6.0, p<.0001). Post-hoc tests found significant differences between the sNPF/CHR flies and both control groups during CT8.5-CT11 and CT12–12.5 (Fig. 1I), where the light stimulation occurred during the first 5 minutes after CT8.
Interestingly, stimulation with red light for just 3 seconds also induced a robust sleep induction in female sNPF/CHR flies (Fig. 1J–K). There were significant overall effects of genotype (F(2,92)=80.6, p<.0001) and bin (47,4324)=4.9, p <.0001), and a significant interaction effect between genotype and bin (94,4324)=3.4, p<.0001). Post-hoc tests found significant differences between the sNPF/CHR flies and both control groups during CT8.5-CT10.5 (Fig. 1K), where the light stimulation occurred during the first 3 seconds after CT8.
Stimulation for 0.5 seconds resulted in a somewhat different behavioral effect. Sleep was still significantly induced, but without the initial sharp increase (Fig. 1L–M). There were still significant overall effects of genotype (F(2,90)=61.7, p <0.0001) and bin (F(47,4230)=3.7, p <0.0001), and a significant interaction between genotype and bin (F(94,4230)=2.6, p <0.0001). Post-hoc tests found significant differences between the sNPF/CHR flies and both control groups during CT10–10.5 and CT 11.5–12.5 (Fig. 1M). Interestingly, these significant points were all delayed relative to the light stimulation, which occurred during the first 0.5 seconds after CT8.
3.2. Optogenetic activation of sNPF neurons at times when flies are already sleeping produces delayed sleep increases.
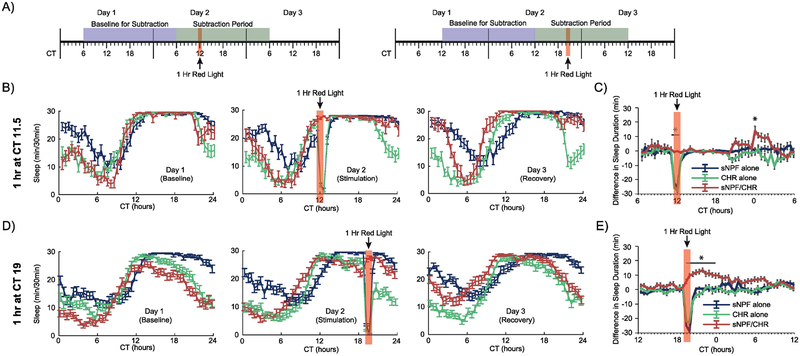
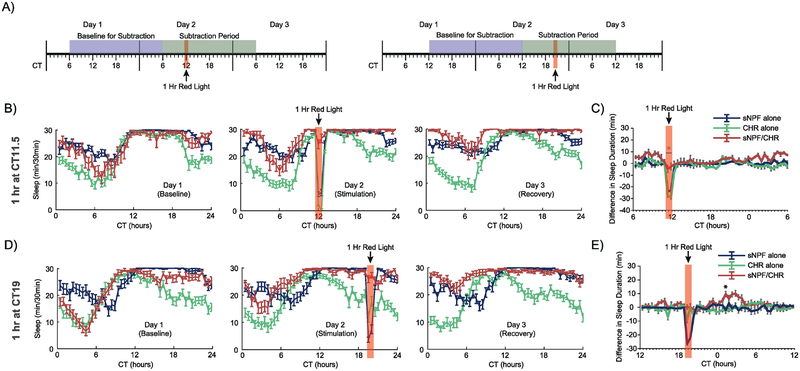
We next set out to test whether sNPF neurons had similar roles in regulating sleep at different times of day. Thus, we altered the timing of the red light stimulation to fall at a point when flies had just fallen asleep (CT 11.5), and at a later point during the subjective night when flies were still mostly asleep (CT 19) (Fig. 2A). In the previous daytime activation studies, most flies were awake at the time of stimulation, but in these nighttime stimulation experiments, female flies in all 3 groups were mostly asleep at the time when red light was applied. This changed the overall pattern of response to the red light – both groups of control flies were awakened briefly by the red light, but went back to sleep immediately after the light was turned out (Fig. 2B and 2D). The experimental sNPF/CHR flies, on the other hand, were kept asleep by the red light stimulation – they did not show the same transient arousal. They also did not show the dramatic, sudden increase in sleep seen in the daytime stimulation studies in Figure 1. This appears to be because most flies were asleep already, and thus were hitting a ceiling in terms of how much they could possibly sleep in each 30-minute bin. What was evident in these experiments is that the drive to sleep induced by activating sNPF neurons was maintained, and became evident at later points when the flies would have been waking up during the following morning. This delayed sleep augmentation pattern can be seen best in the subtraction plots in Fig. 2C and 2E.
Figure 2. Optogenetic activation of sNPF neurons at times when female flies are already sleeping.
(A) Timelines of the experiments whose data are shown in this figure. Red light stimulation for 1 hour was provided at either CT11.5 or CT19 in conditions of otherwise total darkness. All experimental animals in these experiments were females. Data were analyzed as in Figure 1, with the exception that the 24-hour Subtraction Period and Baseline for Subtraction were shifted based on the timing of the red light stimulation. (B, D) Sleep patterns across the 3 experimental days for the CT11.5 and CT19 experiments, respectively. (C, E) Subtraction plots for the CT11.5 and CT19 experiments, respectively. Red light stimulation in these experiments woke up control flies, but experimental flies were kept asleep. Further, they showed delayed increases in sleep at points when they would normally have started waking up. Error bars represent the standard error of the mean, and * represents a post-hoc comparison with p < 0.05. The numbers of experimental animals were as follows, for sNPF alone, CHR alone, and sNPF/CHR groups, respectively: CT11.5 experiment – 32, 32, 19; and CT19 experiment – 61, 58, 65.
In the CT 11.5 stimulation study, subtraction analysis (Fig. 2C) was carried out over a 24-hour period starting at CT6, in order to capture the delayed sleep enhancement evident in this experiment. There were significant overall effects of genotype (F(2,79)=56.8, p<.0001) and bin (F(47,3713)=12.9, p<.0001), and a significant interaction between genotype and bin (F(94,3713)=4.0, p<.0001). Post-hoc analysis found that the experimental animals differed from both control groups during the two 30-minute bins of the red light stimulation and also during a single bin at CT 24.
In the CT 19 stimulation experiment, subtraction analysis (Fig. 2E) was carried out over a 24-hour period starting at CT12, again to capture the full period during which sleep induction occurred. There were significant overall effects of genotype (F(2,181)=132.3, p<.0001) and bin (F(47,8507)=21.9, p<.0001), and a significant interaction between genotype and bin (F(94,8507)=10.5, p<.0001). Post-hoc analysis found that the experimental animals differed from both control groups at each of the two bins during the red light stimulation as well as the following 8 bins.
3.3. Daytime optogenetic activation of sNPF neurons in male flies
We also tested the effects of optogenetic activation of sNPF neurons on sleep in male flies. However, there were some technical issues that made it difficult to determine how males were influenced by this stimulation. Primarily, we found that experimental males in many experiments had such high baseline levels of sleep that further sleep induction was not possible to observe consistently. Please see the Discussion for more commentary on this issue. However, in sections 3.3 and 3.4 we have reported below the statistical results for male flies in all of the experiments reported above for females in sections 3.1 and 3.2. In this section, we report the results of daytime optogenetic activation experiments using light stimuli durations ranging from 1 hour down to 0.5 seconds.
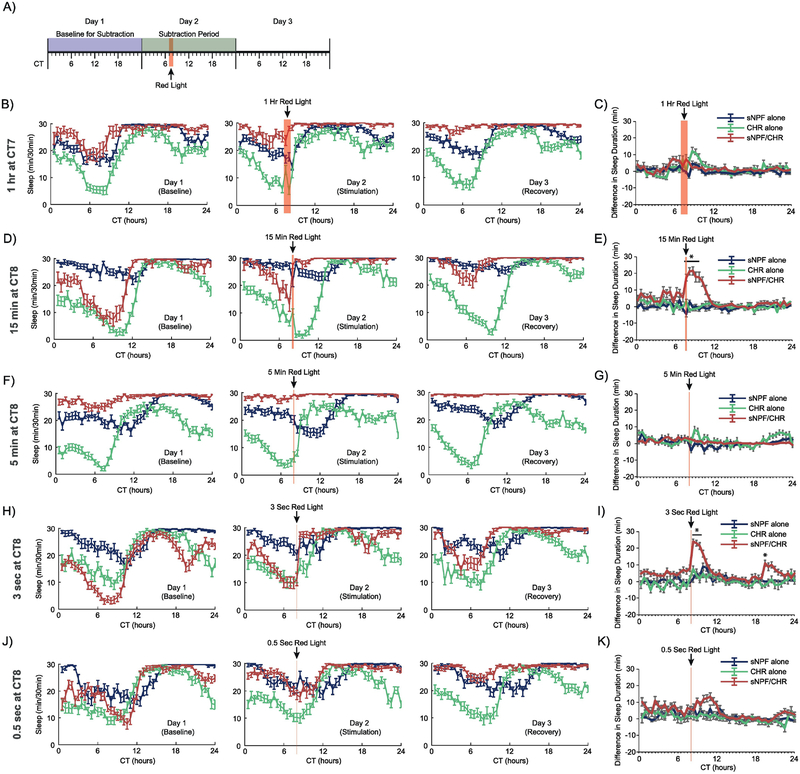
For the 1-hour stimulation at CT7 in males (Fig. 3B–C), there were significant overall effects of genotype (F(2,69)=5.2, p<.008) and bin (47,3243)=3.2, p <.0001), and there was a significant interaction effect between genotype and bin (94,3243)=1.9, p<.0001). Post-hoc analysis found that male sNPF/CHR flies were not different from both controls during any single bin.
Figure 3. Optogenetic activation of sNPF neurons in male flies during subjective day.
This figure depicts the results from male animals from four separate optogenetic activation experiments. (A) A general timeline of the experiments whose data are shown in this figure. (B, D, F, H, J) Sleep patterns across the 3 experimental days for the 1 hr at CT7, 15 min at CT8, 5 min at CT8, 3 sec at CT8, and 0.5 sec at CT8 experiments, respectively. (C, E, G, I, K) Subtraction plots for the same four experiments, respectively. In some of these experiments, the high levels of sleep in male animals made it difficult to observe sleep induction during the red light stimulation (but see the 15 min and 3 sec experiments, in which males were awake enough to show a significant increase in sleep). Error bars represent the standard error of the mean, and * represents a post-hoc comparison with p < 0.05. The numbers of experimental animals were as follows, for sNPF alone, CHR alone, and sNPF/CHR groups, respectively: 1-hour experiment – 29, 29, 14; 15-minute experiment –32, 30, 23; 5-minute experiment – 32, 32, 30; 3-second experiment – 31, 30, 31; and 0.5-second experiment – 31, 28, 32.
For the 15-minute stimulation at CT8 in males (Fig. 3D–E), there were significant overall effects of genotype (F(2,82)=70.5, p<.0001) and bin (47,3854)=6.9, p <.0001), and there was a significant interaction between genotype and bin (94,3854)=6.3, p<.0001). In this experiment, males had low enough baseline sleep that a significant sleep induction was observed in sNPF/CHR flies relative to both control groups, from CT8.5–10.5.
For the 5-minute stimulation at CT8 in males (Fig. 3F–G), there were significant overall effects of genotype (F(2,91)=10.2, p<.0001) and bin (F(47,4277)=4.5, p<.0001), and there was a significant interaction effect between genotype and bin (F(94,4277)=2.9, p<.0001). Post-hoc tests found no bins where sNPF/CHR male flies were significantly different from controls.
For the 3-second stimulation at CT8 in males (Fig. 3H–I), there were significant overall effects of genotype (F(2,89)=71.6, p<.0001) and bin (F(47, 4183)=9.3, p< .0001), and there was a significant interaction effect between genotype and bin (F(94, 4183)=3.4, p<.0001). Post-hoc tests found significantly higher sleep in sNPF/CHR flies compared with both controls over three 30-minute bins from CT8.5–9.5, and during a single bin at CT19 (Fig. 3I).
For the 0.5-second stimulation at CT8 in males (Fig. 3J–K), there were significant overall effects of genotype (F(2,88)=46.3, p<0.0001) and bin (F(47,4136)=5.9, p<0.0001), and a significant interaction between genotype and bin (F(94,4136)=2.0, p<0.0001). Post-hoc tests found no single bin where experimental sNPF/CHR flies differed from both control groups (Fig.3K).
3.4. Nighttime optogenetic activation of sNPF neurons in male flies.
In this section, we report the results of nighttime optogenetic activation experiments in male flies. As in female flies, 1 hour of red light was delivered either at CT11.5 or at CT19.
For the experiment using 1 hour of red light stimulation at CT11.5 (Fig. 3G–H), there were significant overall effects of genotype (F(2,76)=51.5, p<.0001) and bin (47,3572)=23.0, p <.0001), and there was a significant interaction effect between genotype and bin (94,3572)=3.6, p<.0001). Post-hoc tests found that male experimental flies differed from controls only during the two bins of the red light stimulus.
For the experiment using 1 hour of red light stimulation at CT19 (Fig. 3I–J), there were significant overall effects of genotype (F(2,89)=27.4, p<.0001) and bin (47,4183)=7.0, p <.0001), and there was a significant interaction effect between genotype and bin (94,4183)=4.3, p<.0001). Post-hoc tests found that male experimental flies differed from both controls only during a single bin at CT1.5.
3.5. Brief optogenetic activation of sNPF neurons decreases movement
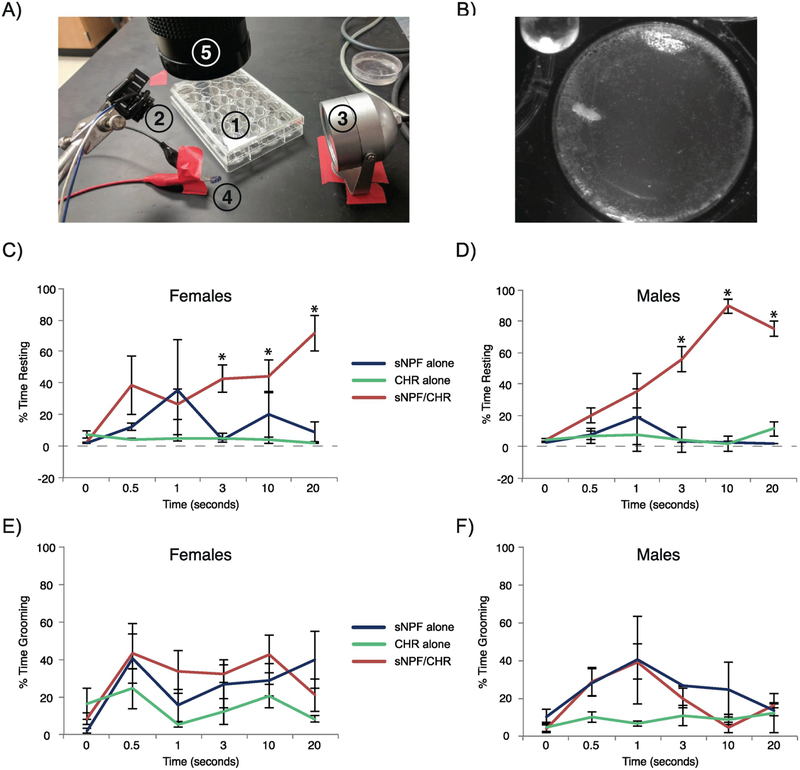
Previous studies had used manipulations of sNPF neurons that ranged from 12 hours in duration to the entire lifespan of the animal [27, 35], so it remained unclear whether brief activation of these neurons would also have an effect on behavior. Therefore, we took advantage of the rapid onset and offset of optogenetic activation and, following a 30-second baseline, used red light pulses ranging from 0.5–20 seconds in duration to activate sNPF neurons in individual flies, and videotaped their behavior over the next 4 minutes (Fig. 5A–B). Videos were hand-scored for time that the fly was moving (including grooming, walking, etc.) by experimenters who were blind to the genotype of the fly, and the percentage of time that each fly spent moving during the period following stimulation was calculated (Fig. 5C–F). An ANOVA examining the fraction of time spent resting following optogenetic stimulation found no significant effect of sex (F(1,88)=0.11, p=0.7), or any significant interaction between sex and genotype (F(2,88)=2.6, p=0.08) or sex and stimulation duration (F(5,88)=0.5, p=0.8) or sex x genotype x stimulation duration (F(10,88)=1.4, p=0.2). Therefore, the sex of the animal did not seem to be a factor in how sNPF neuron activation altered behavior. However, there were significant overall effects of genotype (F(2,88)=61.7, p<0.0001) and stimulation duration (F(5,88)=6.8, p<0.0001), and there was a significant interaction between genotype and stimulation duration (F(10,88)=6.8, p<0.0001). Post-hoc testing found that, for optogenetic stimulation durations of 3, 10, or 20 seconds, the sNPF/CHR experimental flies had significantly greater amounts of inactive resting time across the 4 minutes following red light stimulation when compared with both control groups (Fig. 5C–D). No significant effect was observed for stimulation durations of 0, 0.5, or 1 second, showing that the magnitude of this reduction in movement increased progressively with longer stimulation periods. In the absence of optogenetic stimulation, movement patterns in male and female experimental flies were not different than the patterns in control flies, showing that expression of Chrimson in sNPF neurons did not alter baseline behavior and that ambient light was not sufficient to activate Chrimson.
Figure 5. Brief optogenetic activation of flies causes similar reductions in movement in female and male flies.
Individual flies were placed in a small chamber and videotaped under infrared light. After a 30-second baseline, a red light pulse was delivered for 0, 0.5, 1, 3, 10, or 20 seconds. Videos were subsequently scored for movement across the subsequent four 1-minute bins. (A) Image showing the elements of the experimental setup. #1 is a 24-well plate containing recording chambers partially filled with agarose. #2 is the red LED for optogenetic stimulation, #3 is an infrared illuminator for background light, #4 is an infrared-emitting indicator light, and #5 is a camera for video recording, fitted with an long-pass filter. (B) A sample image of a recording, showing the size of the chamber relative to that of the fly. The infrared indicator light can be seen in the top left corner. (C-D) Plots showing the percentage of time spent resting during the 4 minutes following optogenetic stimulation in female and male flies, respectively. (E-F) Plots showing the percentage of time spent grooming during the 4 minutes following optogenetic stimulation in female and male flies, respectively. Error bars represent the standard error of the mean, * represents post-hoc comparisons between genotypes at a given stimulation duration for which male and female sNPF/CHR flies combined were significantly different than both control groups at p < 0.05. All experimental groups had 3–5 animals, except the male, sNPF alone, 1-second stimulation group, in which only 2 videos were usable.
We also assessed the fraction of time following optogenetic stimulation that flies spent grooming (Fig. 5E–F). An ANOVA found significant overall effects of genotype (F(2,88)=8.0, p=0.0006), stimulation duration (F(5,88)=4.2, p=0.002), and a marginal effect of sex (F(1,88)=4.1, p=0.05). There were no significant interaction effects, including for genotype x stimulation duration (F(10,88)=1.1, p=0.4) and for sex x genotype x stimulation duration (F(10,88)=0.9, p=0.6). Thus, unlike for time spent resting, optogenetic stimulation did not significantly affect time spent grooming.
During the period of red light stimulation, both male and female sNPF/CHR flies showed uncoordinated movement responses that ceased as soon as the red light turned off, whereas controls showed very little reaction to the red light (Supplementary Videos 1 and 2). See the Discussion for commentary on the possible cause and significance of this acute response to sNPF neuron activation. Videos from 3–4 minutes after optogenetic stimulation show the striking reduction in activity in sNPF/CHR flies compared with control flies (Supplementary Videos 3 and 4).
3.6. Flies can be aroused from optogenetically induced sleep
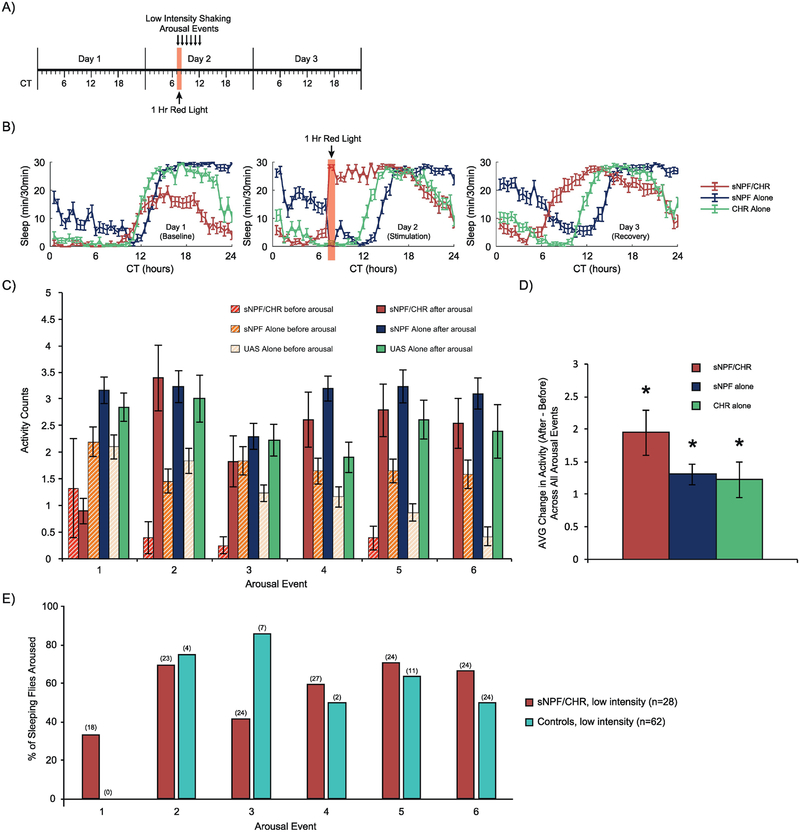
We next sought out to determine if sleep induced through the optogenetic activation of sNPF neurons was readily reversible, a defining feature of natural sleep [8, 9, 11]. To accomplish this, we stimulated female flies with red light for an hour and determined what percentage of flies were aroused by brief pulses of mechanical stimulation, the first given 5 minutes into the red light stimulus and then each hour during the 5 subsequent hours. In the first experimental protocol (Fig. 6A–B), the red light and subsequent arousal stimuli were presented during the subjective day (starting at CT 7). Activity (# of beam-crosses) during the pre-arousal minute was compared with activity during the post-arousal minute, across all 6 arousals, for all animals. During the first arousal, sNPF/CHR flies were not capable of being aroused, presumably due to ongoing activation of sNPF neurons, but in all subsequent arousals, sNPF/CHR flies were readily aroused (Fig. 6C). Averaged across all 6 arousal events (Fig. 6D), there was a significant overall induction of activity in the 3 groups (F(1,87)=93.3, p<0.0001), and there were no significant differences between groups in the magnitude of the change in activity induced by mechanical stimulation (F(2,87)=2.1, p=0.13). We also focused specifically on sleeping flies, and categorized them based on whether they had at least one beam cross during the first post-arousal minute (aroused) or if they did not (still asleep). For graphical simplicity, and because there was no significant difference in activity induction between the two control groups, we combined the data for the sNPF alone and CHR alone controls. We found that the arousal rates in the sNPF/CHR population were comparable to the rates in control animals, although for several of the arousal events at this time of day, very few control flies were sleeping at the time of stimulation (Fig. 6E).
Figure 6. Mechanical arousal of female flies following optogenetic activation during subjective daytime.
(A) Timeline for the experiment. Red light stimulation for 1 hour was provided at CT7 in conditions of otherwise total darkness. Mechanical arousal stimuli at Low Intensity were provided for 0.5 seconds every hour for 6 hours, with the first stimulation taking place 5 minutes into the red light stimulation. All experimental animals in this experiment were females. (B) Sleep patterns across the 3 experimental days. (C) Quantification of activity counts (beam breaks) during the minute pre-arousal compared with the first minute post-arousal, separately for each arousal event. (D) Comparison of the change in activity induced by arousal, averaged across all 6 arousal events. All groups significantly increased their activity due to arousal. (E) Visualization of the % of sleeping flies that were awoken by the mechanical stimulation. For clarity, the sNPF alone and CHR alone control groups were combined into a single Controls group. sNPF/CHR flies appeared to be aroused from sleep at similar rates to Controls, but there were very few sleeping Control animals when arousal was tested at this time of day. Error bars in panel B-D represent the standard error of the mean, and the numbers in parentheses in panel E represent the number of sleeping flies in each group. The total numbers of experimental animals were as follows, for sNPF alone, CHR alone, and sNPF/CHR groups, respectively: 31, 31, 28.
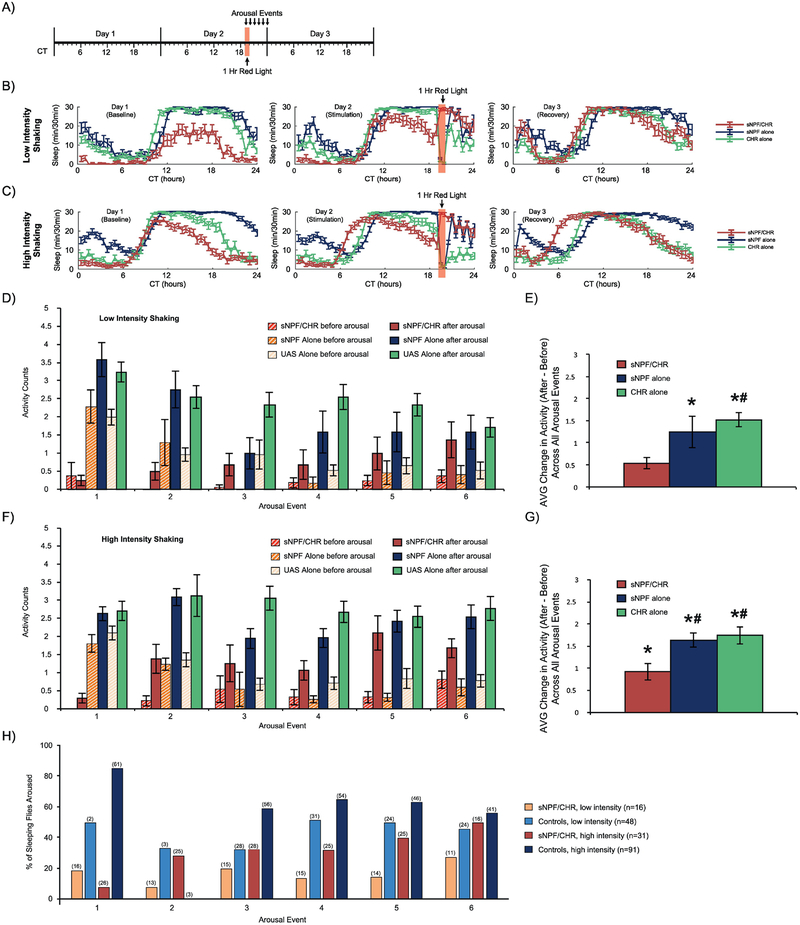
We therefore provided red light illumination for 1 hour at CT19, when more of the control flies were naturally sleeping (Fig. 7A). We ran two separate experiments, in which the intensity of shaking was set for either “3” or “5” on the shaker. In both studies, it was clear that the experimental group was put to sleep by the red light stimulus, whereas the controls were actually awakened (Fig. 7B–C). Activity in the post-arousal minute was compared with activity in the pre-arousal minute for each of the 6 arousal events (Fig. 7D, 7F). As in the CT7 arousal experiment, activity in the sNPF/CHR group was not increased during the first arousal occurring during the red light stimulation, but was augmented during each of the next 5 arousal events after the cessation of the red light. Averaging across all 6 arousal events, in the “Low Intensity” shaking experiment there was significantly more activity post-arousal than pre-arousal (F(1,62)=69.4, p<0.0001), and there was a significant relationship between group and how much activity was increased (F(2,62)=5.0, p=0.01). Post-hoc testing found that there was a significant overall post-arousal increase in activity in both controls, but not in the sNPF/CHR group. The CHR Alone group also had a significantly greater increase in activity than did the sNPF/CHR group (Fig. 7E). In the “High Intensity” shaking experiment, there was again an overall increase in activity across all 6 arousal events (F(1,116)=173.8, p<0.0001) and a significant relationship between group and the increase in activity post-arousal (F(2,116)=5.1, p=0.008). Post-hoc testing found that, with higher stimulation, sNPF/CHR flies now also showed a significant increase in activity, although both control groups still had significantly greater induction of activity than did the sNPF/CHR group (Fig. 7G).
Figure 7. Flies in optogenetically induced sleep can be aroused by mechanical stimulation.
(A) Timeline for both of the experiments whose data are shown in this figure. Red light stimulation for 1 hour was provided at CT19 in conditions of otherwise total darkness. Mechanical arousal stimuli at either Low Intensity or High Intensity were provided for 0.5 seconds every hour for 6 hours, with the first stimulation taking place 5 minutes into the red light stimulation. All experimental animals in these experiments were females. (B-C) Sleep patterns across the 3 experimental days for the Low Intensity and High Intensity experiments, respectively. (D-E) Results from the Low Intensity shaking experiment. (F-G) Results from the High Intensity shaking experiment. (D, F) Quantification of activity counts (beam breaks) during the minute pre-arousal compared with the first minute post-arousal, separately for each arousal event. (E, G) Comparison of the change in activity induced by arousal, averaged across all 6 arousal events. (H) Visualization of the % of sleeping flies that were awoken by the mechanical stimulation. For clarity, the sNPF alone and CHR alone control groups were combined into a single Controls group for each stimulation intensity. Although the % of sleeping flies that were aroused was typically smaller for the sNPF/CHR flies than for the Controls, sNPF/CHR flies could be aroused from sleep, and were aroused more frequently with greater shaking intensity. Error bars in panels B-C represent the standard error of the mean, and the numbers in parentheses in panel D represent the number of sleeping flies in each group. The total numbers of experimental animals were as follows, for sNPF alone, CHR alone, and sNPF/CHR groups, respectively: Low Intensity shaking experiment – 31, 17, 16; High Intensity shaking experiment – 25, 31, 31.
We then determined the percentage of sleeping flies that were awoken by mechanical stimulation (Fig. 7H). The overall patterns in the data were the following: (1) a portion of experimental animals could be awoken, even during the red light stimulation itself, (2) experimental flies tended to be more difficult to awake than control animals, and (3) all groups were awoken to a greater extent by the high intensity stimulation than the low intensity stimulation.
4. Discussion
We present evidence using optogenetic stimulation that suggests that activity in sNPF neurons promotes sleep. These results support the conclusions from previous findings [35] using a hypomorphic sNPF mutant and thermogenetic sNPF neuron activation, and contradict the previous findings using loss-of-function alleles for sNPF and its receptor that suggested that sNPF signaling promotes wake [27]. Our system of neuronal activation also allowed us to activate sNPF neurons for shorter periods of times than in previous studies.
Using optogenetic stimuli ranging from 0.5 seconds to 1 hour in duration, we found that even brief periods of sNPF neuron activation are capable of inducing sleep, and that the sleep induction could persist for hours following stimulation. We did find that 1 hour of stimulation caused the greatest immediate increase in sleep, but even 0.5 seconds of stimulation led to a long-lasting sleep increase. This is an intriguing result, because it suggests that a mild treatment that enhances signaling in sNPF neurons even briefly could cause significant and enduring sleep promotion. It will be interesting to determine if many neuropeptidergic neurons involved in sleep regulation have this capacity, or if this is a unique characteristic of sNPF neurons.
The longevity of neuropeptide action is one of the major draws of exploring neuropeptide signaling pathways as targets for pharmaceutical sleep treatments – namely, that long-lasting sleep changes can be induced by a single, mild dose of drug. Another potential benefit of neuropeptides as pharmaceutical targets is that their specificity in regulating just a few behaviors potentially allows neuropeptide-regulating drugs to influence the desired behavior with minimal side effects. Of note, the first sleep drug (Suvorexant, marketed by Merck as Belsomra) targeting a neuropeptide system (orexin/hypocretin) came on the market in 2014 [65, 66], and appears to be moderately effective at improving sleep in people with insomnia [67, 68]. This highlights the need for more in-depth understanding of how neuropeptide systems regulate sleep so that we can design new classes of effective and targeted drugs to help the vast number of people with sleep difficulties.
Using brief periods of activation also allowed us to stimulate sNPF neurons at different times of day. We were interested in assessing this because of findings using the hypomorphic sNPF mutant, in which sleep was reduced mostly during the daytime and early nighttime, with less of an effect later in the night [35]. Thus, we had hypothesized that sNPF signaling might have more relevance for sleep induction at certain times of day. However, we found that sNPF neuron activation was capable of promoting sleep at all times of day that we tested (CT7/8, CT11.5, and CT19). However, the patterns of responses to activation at these times differed considerably, and uncovered interesting features of sleep promotion by sNPF neurons.
When activation was carried out at CT7/8, a point when animals were predominantly awake, sleep was induced immediately in sNPF/CHR flies but not in controls, and this increase in sleep decayed over the next approximately 2 hours. In contrast, when activation took place at CT11.5, a point when animals had just reached a high level of sleep, the pattern was quite different. Control animals were awoken by the red light stimulus at this time of day, whereas animals expressing Chrimson in sNPF neurons were kept asleep, although they did not show an increase in sleep relative to baseline because their sleep was already at a ceiling at this time of day. The arousal in control animals shows that flies are naturally capable of seeing bright red light, and that it can awaken them at times when they are sleeping. However, we consistently observed that control flies immediately went back to sleep and resumed normal sleep/wake patterns after the red light stimulus ceased. Importantly, we saw that sNPF/CHR flies showed a further increase in sleep (relative to the baseline day) the following morning, when they should have been waking up. This meant that there was a delay of more than 6 hours between the optogenetic activation of sNPF neurons and the expression of sleep induction.
A similar effect was observed following activation of sNPF neurons at CT19, a time point when flies were mostly asleep but were beginning to wake up. In this situation, control flies were again awoken by the red light and returned to normal sleep patterns afterward. sNPF/CHR flies were again kept asleep during the red light stimulus, although in this case, because sleep was not at a ceiling at this time, their sleep were able to increase a bit above baseline levels. Most likely because only a mild increase in sleep could be expressed above baseline levels, the sleep increase took longer to dissipate than in daytime stimulation experiments. Sleep was significantly increased above baseline levels for 5 consecutive hours, whereas the same duration of red light stimulation during the day only increased sleep for 2.5 hours. Together, these findings suggests that sNPF neuron signaling does not simply shift flies into a sleep state for a set amount of time, but instead may create a stored homeostatic sleep drive, which can be expressed immediately or with a delay, depending on when sNPF neurons are stimulated relative to when flies are awake.
Using optogenetic stimulation also allowed us to test the behavioral effects of even briefer periods of sNPF neuron activation. By videotaping individual flies, we could observe more details of the nature of their responses to red light stimulation. We found that stimulation periods of 3 to 20 seconds were all capable of causing a significant reduction in movement during the subsequent 4 minutes. There was a dose-response relationship between the duration of stimulation and the amount of reduction in movement. Time spent grooming was unchanged, suggesting that sNPF neuron activation does not reduce locomotion (beam crossing due to walking is the metric used to tabulate sleep in DAM monitors used in our population studies) by increasing time spent grooming. These findings demonstrate that even quite brief periods of activity in sNPF neurons can produce an induction in rest-like behavior that lasts minutes beyond the period of stimulation. We did see that, during the red light illumination, all experimental flies showed uncoordinated movement, which in some cases resulted in the fly falling onto its back. In case s like these, flies sometimes became quiescent in this position once the red light illumination ceased, before ultimately (the delay here was variable, from almost instantaneous to several seconds) flipping themselves back over and resuming movement. However, even once flies began moving again, they would soon rest again, resulting in consistent reductions in time spent walking in sNPF/CHR flies compared with controls. The rapidity of the movement response when the red light was turned on suggests a fast neurotransmitter action, rather than the typically slower responses due to neuropeptides like sNPF [35, 45]. Indeed, many sNPF-positive cells are thought to co-express other transmitters, including fast transmitters such as acetylcholine, glutamate, GABA, and glycine ([69–72]. Thus, it is possible that the fast-onset, fast-offset effects of sNPF-GAL4 x UAS-Chrimson activation are mediated by other neurotransmitters, whereas the more delayed and long-lasting reduction in movement and increase in sleep is due to neuromodulatory effects.
We found that flies could be aroused by mechanical stimulation from sleep induced by sNPF neuron activation. This was important because it showed that the increase in rest that we observed was not due to an inability to move. Instead, we found that flies could be rapidly awoken from the induced rest, which is a critical feature of natural sleep. Overall, sNPF/CHR flies had lower levels of arousal than control flies. This may be because, as we have seen, sNPF neuron activation augments sleep drive. Thus, flies that are asleep following sNPF neuron activation may be somewhat more difficult to awake than controls. sNPF/CHR flies were able to be awoken more readily when the intensity of mechanical stimulation was increased, showing that these flies were not incapable of being aroused, but instead that a greater arousal stimulus was required to do so.
We have focused most of our analysis on female flies. This is because in many experiments, male sNPF/CHR flies showed a progressive increase in baseline levels of sleep from day to day, such that their levels of sleep were so high at the time of optogenetic stimulation that no further increase in sleep could be observed. We do not believe that this reduced effect in male flies indicates that sNPF has no role in promoting sleep in male flies, for the following reasons. First, in our acute analysis shown in Fig. 3, male and female sNPF/CHR flies showed a very similar decrease in movement following optogenetic stimulation. Second, in the few experiments when males showed relatively low baseline levels of sleep, they showed similar patterns of sleep induction as females (see for example Fig. 1E–F, 15-minute at CT8 experiment). Third, in experiments where red light was applied at night, control males were awoken but sNPF/CHR flies were kept asleep, just as was the case for female flies. Because levels of sleep were clearly occurring even before red light was administered, this effect is likely a result of having high levels of the Cs-Chrimson ion channel expressed in sNPF neurons. It is possible that these channels are “leaky”, and when highly expressed allow small depolarizing currents to occur. Alternatively, high expression levels of Chrimson may simply disrupt other membrane mechanisms, causing gradual changes in the activity of the neurons in which it is expressed. Why this effect would be stronger in males than in females is unclear. Based on studies targeting Chrimson to other neural populations, the specific nature of the non-specific effect observed seems to depend on the neurons being targeted (data not shown). That is to say, Chrimson does not inherently seem to promote sleepiness in male animals – it only does so when it is expressed in sleep-promoting neurons.
Many of our experiments found that the sNPF-GAL4 control line had high levels of baseline sleep, especially in female flies (Figures 1, 2, 6, and 7). Our fly lines were back-crossed to a common background strain when we first obtained them, and in our preliminary experiments no major differences in baseline sleep were observed between lines. However, it is possible that the sNPFGAL4 line acquired some random sleep-modifying polymorphisms during the last few years, which drove its sleep higher. The fact that there were baseline differences in sleep between strains is one reason why we chose to focus our statistical analysis on the difference between the period after optogenetic stimulation as compared with baseline, rather than directly comparing raw numbers of minutes of sleep. We also required that there be significant differences between the sNPFGAL4/UAS-Chrimson experimental group and both control lines in order for the differences to be deemed significant. And ultimately, despite higher baseline sleep in sNPF-GAL4 controls, it was clear that neither the sNPF-GAL4 nor UAS-Chrimson control lines had any sleep induction response to red light stimulation during periods of wake, and both were awoken in very similar ways by red light stimulation during periods of sleep.
We focused primarily on the effects of sNPF neuron activation on overall sleep levels, but it is worth pointing out that red light stimulation did not substantially alter circadian rhythms. A recent study found that sNPF-positive small ventrolateral neurons (sLNvs) within the central clock produce glycine, and that manipulating glycinergic signaling in those neurons and their targets could disrupt both circadian period length and rhythmicity [72]. In our study, glycine is presumably being released from those sNPF-positive neurons during optogenetic stimulation, but we did not observe clear circadian disturbances. However, the manipulations in the previous study were more long-term than ours, which may account for the minimal circadian effects observed here.
This study leads to several areas of future research. First, because sNPF is expressed in many neuronal populations, it will be important to determine which neural clusters are driving the increases in sleep observed following sNPF neuron activation. sNPF is found within two different groups of central clock neurons (small ventrolateral neurons (sLNvs) and a subset of dorsolateral neurons (LNds)), as well as in mushroom body Kenyon cells [69–71], and the central clock and mushroom bodies play important roles in regulating sleep [15, 18, 20]. Therefore, these are particular neural clusters of interest to examine. Second, many peptidergic neurons also release other transmitters, and sNPF-expressing neurons are no exception. sNPF-expressing sLNvs co-express the neuropeptide PDF and the small-molecule transmitters glycine, the sNPF-expressing LNds co-express acetylcholine, and mushroom body sNPF-expressing neurons likely express other neurotransmitters as well, although their identities have not been established [69–72]. Thus, it is possible that the effects of optogenetic (or thermogenetic, as in Shang et al [35]) activation of sNPF neurons induces sleep not solely through the release of sNPF but via other transmitter signals as well. Although the focus of this work was the neurons themselves, and how activity in those neurons affected sleep, future studies could use RNAi or other genetic strategies to knock down levels of sNPF or other known co-transmitters to determine the role of specific signals in mediating sleep promotion. Third, because our data suggest that sNPF neuron activation creates stored sleep drive, a question is therefore: where is this sleep drive stored? Recently, certain circuits within the Drosophila central complex have been shown to be a locus of homeostatic sleep drive [17, 19, 34, 73]. It would be interesting if sNPF neurons are upstream of these circuits, such that high levels of sNPF neuron activity might cause a build-up of sleep drive in central complex circuits that can then be discharged gradually as the animal sleeps.
Overall, we have confirmed that sNPF neurons are sleep-promoting, and have shown for the first time that even brief stimulation of these neurons can cause long-lasting increases in sleep. This is particularly interesting in terms of the utility of targeting neuropeptide circuits with sleep-promoting drugs. It would be ideal for a single dose of such a compound to promote sleep over an hours-long period, with sleep behavior returning to normal the following day, and this is exactly what we observed following brief optogenetic activation of sNPF-expressing neurons. Because there is homology between the Drosophila sNPF and mammalian NPY systems in terms of the DNA sequences coding for their receptors [44], these transmitters may be positioned similarly across species within sleep-regulating neural circuits. NPY may therefore represent a worthwhile target to explore for sleep-promoting treatments in humans.
Supplementary Material
Supplementary Video 1. Lack of immediate reaction to red light stimulation in a UAS alone control female fly. The infrared light in the corner of the video indicates when the red light was on (10 seconds in this case). This control fly should not have expressed the Chrimson sensor in sNPF-producing neurons, and showed very little obvious response to the red light being turned on or off.
Supplementary Video 2. Immediate reaction to red light stimulation in an sNPF/CHR female fly. This fly expressing Chrimson in its sNPF-producing neurons showed an immediate uncoordinated movement reaction when the red light was turned on. This reaction also turnsed off rapidly as soon as the red light was turned off. In this case, the fly took several seconds before it returned to its feet.
Supplementary Video 3. Continued movement in a UAS alone control female fly later in recording. The same fly as in Video 1 shows almost constant movement or grooming. This video begins approximately 2 minutes following red light stimulation.
Supplementary Video 4. Minimal movement in an sNPF/CHR female fly later in recording. The same experimental fly as in Video 2 spends almost no time walking, but does show grooming behavior. This video begins approximately 2 minutes following red light stimulation.
Figure 4. Optogenetic activation of sNPF neurons in male flies during subjective night.
(A) Timelines of the experiments whose data are shown in this figure. Red light stimulation for 1 hour was provided at either CT11.5 or CT19 in conditions of otherwise total darkness. All experimental animals in these experiments were males. (B, D) Sleep patterns across the 3 experimental days for the 1 hr at CT11.5 and 1 hr at CT19 experiments, respectively. (C, E) Subtraction plots for the same two experiments. As in the previous figure, high levels of sleep in male animals made it difficult to assess sleep induction due to the red light stimulation. Error bars represent the standard error of the mean, and * represents a post-hoc comparison with p < 0.05. The numbers of experimental animals were as follows, for sNPF alone, CHR alone, and sNPF/CHR groups, respectively: CT11.5 experiment – 32, 32 15; and for CT19 experiment – 32, 29, 31.
Highlights for Juneau et al.
Optogenetic activation of short neuropeptide F (sNPF) neurons induces sleep.
Even brief sNPF neuron activation induces long-lasting increases in sleep.
sNPF neuron activity appears to cause an increase in homeostatic sleep drive.
Flies can be aroused from sleep induced by sNPF neuron activation.
Acknowledgments
We would like to express thanks to the Bloomington Stock Center for maintaining and providing fly stocks used in this study, to Tami Gura and Zoë Brasher for their help making fly food and flipping fly stocks, to Dilhan Sirtalan and Emily Perkins for help scoring video data, and to Jill Linz for help analyzing LED spectral properties. Funding to support this research was provided by NIH AREA Grant R15NS101692 (to C.G.V.), and by Swarthmore College, in the form of a Faculty Development Grant (to C.G.V.), and by Skidmore College, in the form of a Faculty Development Grant (to C.G.V.) and Faculty-Student Summer Collaborative Research Fellowships (to Z.C.J., J.M.S., and R.F.T.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of Interest
The authors declare no conflict of interest.
References Cited
- 1.Hublin C, et al. , Insufficient sleep--a population-based study in adults. Sleep, 2001. 24(4): p. 392–400. [DOI] [PubMed] [Google Scholar]
- 2.Guilleminault C and Daniel D, Major sleep disorders, in Sleep deprivation: clinical issues, phramacology, and sleep loss effects, Kushida C, Editor. 2005, Marcel Dekker: New York. p. 71–80. [Google Scholar]
- 3.Wulff K, et al. , Sleep and circadian rhythm disruption in psychiatric and neurodegenerative disease. Nat Rev Neurosci, 2010. 11(8): p. 589–99. [DOI] [PubMed] [Google Scholar]
- 4.Ford ES, Cunningham TJ, and Croft JB, Trends in Self-Reported Sleep Duration among US Adults from 1985 to 2012. Sleep, 2015. 38(5): p. 829–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Joiner WJ, Unraveling the Evolutionary Determinants of Sleep. Curr Biol, 2016. 26(20): p. R1073–R1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Krueger JM, et al. , Sleep function: Toward elucidating an enigma. Sleep Med Rev, 2016. 28: p. 46–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sehgal A and Mignot E, Genetics of sleep and sleep disorders. Cell, 2011. 146(2): p. 194–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hendricks JC, Sehgal A, and Pack AI, The need for a simple animal model to understand sleep. Prog Neurobiol, 2000. 61(4): p. 339–51. [DOI] [PubMed] [Google Scholar]
- 9.Hendricks JC, et al. , Rest in Drosophila is a sleep-like state. Neuron, 2000. 25(1): p. 129–38. [DOI] [PubMed] [Google Scholar]
- 10.Potdar S and Sheeba V, Lessons from sleeping flies: insights from Drosophila melanogaster on the neuronal circuitry and importance of sleep. J Neurogenet, 2013. 27(1–2): p. 23–42. [DOI] [PubMed] [Google Scholar]
- 11.Shaw PJ, et al. , Correlates of sleep and waking in Drosophila melanogaster. Science, 2000. 287(5459): p. 1834–7. [DOI] [PubMed] [Google Scholar]
- 12.Zimmerman JE, et al. , Conservation of sleep: insights from non-mammalian model systems. Trends Neurosci, 2008. 31(7): p. 371–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Venken KJ, Simpson JH, and Bellen HJ, Genetic manipulation of genes and cells in the nervous system of the fruit fly. Neuron, 2011. 72(2): p. 202–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huber R, et al. , Sleep homeostasis in Drosophila melanogaster. Sleep, 2004. 27(4): p. 628–39. [DOI] [PubMed] [Google Scholar]
- 15.Cavanaugh DJ, et al. , The Drosophila Circadian Clock Gates Sleep through Time-of-Day Dependent Modulation of Sleep-Promoting Neurons. Sleep, 2016. 39(2): p. 345–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Crocker A, et al. , Identification of a neural circuit that underlies the effects of octopamine on sleep:wake behavior. Neuron, 2010. 65(5): p. 670–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Donlea JM, Pimentel D, and Miesenbock G, Neuronal machinery of sleep homeostasis in Drosophila. Neuron, 2014. 81(4): p. 860–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Joiner WJ, et al. , Sleep in Drosophila is regulated by adult mushroom bodies. Nature, 2006. 441(7094): p. 757–60. [DOI] [PubMed] [Google Scholar]
- 19.Liu S, et al. , Sleep Drive Is Encoded by Neural Plastic Changes in a Dedicated Circuit. Cell, 2016. 165(6): p. 1347–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pitman JL, et al. , A dynamic role for the mushroom bodies in promoting sleep in Drosophila. Nature, 2006. 441(7094): p. 753–6. [DOI] [PubMed] [Google Scholar]
- 21.Artiushin G and Sehgal A, The Drosophila circuitry of sleep-wake regulation. Curr Opin Neurobiol, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Crocker A and Sehgal A, Genetic analysis of sleep. Genes Dev, 2010. 24(12): p. 1220–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nall A and Sehgal A, Monoamines and sleep in Drosophila. Behav Neurosci, 2014. 128(3): p. 264–72. [DOI] [PubMed] [Google Scholar]
- 24.Sehgal A, et al. , Molecular analysis of sleep: wake cycles in Drosophila. Cold Spring Harb Symp Quant Biol, 2007. 72: p. 557–64. [DOI] [PubMed] [Google Scholar]
- 25.Tomita J, Ban G, and Kume K, Genes and neural circuits for sleep of the fruit fly. Neurosci Res, 2017. [DOI] [PubMed] [Google Scholar]
- 26.Agosto J, et al. , Modulation of GABAA receptor desensitization uncouples sleep onset and maintenance in Drosophila. Nat Neurosci, 2008. 11(3): p. 354–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen W, et al. , Regulation of sleep by the short neuropeptide F (sNPF) in Drosophila melanogaster. Insect Biochem Mol Biol, 2013. 43(9): p. 809–19. [DOI] [PubMed] [Google Scholar]
- 28.Crocker A and Sehgal A, Octopamine regulates sleep in drosophila through protein kinase A-dependent mechanisms. J Neurosci, 2008. 28(38): p. 9377–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.He C, et al. , Regulation of sleep by neuropeptide Y-like system in Drosophila melanogaster. PLoS One, 2013. 8(9): p. e74237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hermann-Luibl C, et al. , The ion transport peptide is a new functional clock neuropeptide in the fruit fly Drosophila melanogaster. J Neurosci, 2014. 34(29): p. 9522–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oh Y, et al. , A homeostatic sleep-stabilizing pathway in Drosophila composed of the sex Peptide receptor and its ligand, the myoinhibitory Peptide. PLoS Biol, 2014. 12(10): p. e1001974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Parisky KM, et al. , PDF cells are a GABA-responsive wake-promoting component of the Drosophila sleep circuit. Neuron, 2008. 60(4): p. 672–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Park S, et al. , SIFamide and SIFamide receptor defines a novel neuropeptide signaling to promote sleep in Drosophila. Mol Cells, 2014. 37(4): p. 295–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pimentel D, et al. , Operation of a homeostatic sleep switch. Nature, 2016. 536(7616): p. 333–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shang Y, et al. , Short Neuropeptide F is a Sleep-Promoting Inhibitory Modulator. Neuron, 2013. 80(1): p. 171–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yuan Q, Joiner WJ, and Sehgal A, A sleep-promoting role for the Drosophila serotonin receptor 1A. Curr Biol, 2006. 16(11): p. 1051–62. [DOI] [PubMed] [Google Scholar]
- 37.Fujiwara Y, et al. , The CCHamide1 Neuropeptide Expressed in the Anterior Dorsal Neuron 1 Conveys a Circadian Signal to the Ventral Lateral Neurons in. Front Physiol, 2018. 9: p. 1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Donlea JM, et al. , Recurrent Circuitry for Balancing Sleep Need and Sleep. Neuron, 2018. 97(2): p. 378–389.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Burbach JP, What are neuropeptides? Methods Mol Biol, 2011. 789: p. 1–36. [DOI] [PubMed] [Google Scholar]
- 40.Nishino S and Fujiki N, Neuropeptides as possible targets in sleep disorders. Expert Opin Ther Targets, 2007. 11(1): p. 37–59. [DOI] [PubMed] [Google Scholar]
- 41.Zisapel N, Drugs for insomnia. Expert Opin Emerg Drugs, 2012. 17(3): p. 299–317. [DOI] [PubMed] [Google Scholar]
- 42.Nassel DR and Winther AM, Drosophila neuropeptides in regulation of physiology and behavior. Prog Neurobiol, 2010. 92(1): p. 42–104. [DOI] [PubMed] [Google Scholar]
- 43.Garczynski SF, Brown MR, and Crim JW, Structural studies of Drosophila short neuropeptide F: Occurrence and receptor binding activity. Peptides, 2006. 27(3): p. 575–82. [DOI] [PubMed] [Google Scholar]
- 44.Mertens I, et al. , Characterization of the short neuropeptide F receptor from Drosophila melanogaster. Biochem Biophys Res Commun, 2002. 297(5): p. 1140–8. [DOI] [PubMed] [Google Scholar]
- 45.Vecsey CG, Pirez N, and Griffith LC, The Drosophila neuropeptides PDF and sNPF have opposing electrophysiological and molecular effects on central neurons. Journal of Neurophysiology, 2014. 111(5): p. 1033–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dyzma M, et al. , Neuropeptide Y and sleep. Sleep Med Rev, 2010. 14(3): p. 161–5. [DOI] [PubMed] [Google Scholar]
- 47.Yulyaningsih E, et al. , NPY receptors as potential targets for anti-obesity drug development. Br J Pharmacol, 2011. 163(6): p. 1170–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Akanmu MA, et al. , Neuropeptide-Y Y2-receptor agonist, PYY3–36 promotes non-rapid eye movement sleep in rat. Neurosci Res, 2006. 54(3): p. 165–70. [DOI] [PubMed] [Google Scholar]
- 49.Tóth A, et al. , Effect of basal forebrain neuropeptide Y administration on sleep and spontaneous behavior in freely moving rats. Brain Res Bull, 2007. 72(4–6): p. 293–301. [DOI] [PubMed] [Google Scholar]
- 50.Szentirmai E and Krueger JM, Central administration of neuropeptide Y induces wakefulness in rats. Am J Physiol Regul Integr Comp Physiol, 2006. 291(2): p. R473–80. [DOI] [PubMed] [Google Scholar]
- 51.Singh C, Rihel J, and Prober DA, Neuropeptide Y Regulates Sleep by Modulating Noradrenergic Signaling. Curr Biol, 2017. 27(24): p. 3796–3811.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Antonijevic IA, et al. , Neuropeptide Y promotes sleep and inhibits ACTH and cortisol release in young men. Neuropharmacology, 2000. 39(8): p. 1474–81. [DOI] [PubMed] [Google Scholar]
- 53.Hamada FN, et al. , An internal thermal sensor controlling temperature preference in Drosophila. Nature, 2008. 454(7201): p. 217–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bernstein JG, Garrity PA, and Boyden ES, Optogenetics and thermogenetics: technologies for controlling the activity of targeted cells within intact neural circuits. Curr Opin Neurobiol, 2012. 22(1): p. 61–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jones WD, The expanding reach of the GAL4/UAS system into the behavioral neurobiology of Drosophila. BMB Rep, 2009. 42(11): p. 705–12. [DOI] [PubMed] [Google Scholar]
- 56.Deisseroth K, Optogenetics: 10 years of microbial opsins in neuroscience. Nat Neurosci, 2015. 18(9): p. 1213–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Klapoetke NC, et al. , Independent optical excitation of distinct neural populations. Nat Methods, 2014. 11(3): p. 338–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Inagaki HK, et al. , Optogenetic control of Drosophila using a red-shifted channelrhodopsin reveals experience-dependent influences on courtship. Nat Methods, 2014. 11(3): p. 325–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tabuchi M, et al. , Clock-Generated Temporal Codes Determine Synaptic Plasticity to Control Sleep. Cell, 2018. 175(5): p. 1213–1227.e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Guo F, et al. , A Circadian Output Circuit Controls Sleep-Wake Arousal in Drosophila. Neuron, 2018. 100(3): p. 624–635.e4. [DOI] [PubMed] [Google Scholar]
- 61.Yap MHW, et al. , Oscillatory brain activity in spontaneous and induced sleep stages in flies. Nat Commun, 2017. 8(1): p. 1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Troup M, et al. , Acute control of the sleep switch in. Elife, 2018. 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Andretic R and Shaw PJ, Essentials of sleep recordings in Drosophila: moving beyond sleep time. Methods Enzymol, 2005. 393: p. 759–72. [DOI] [PubMed] [Google Scholar]
- 64.Donelson N, et al. , High-resolution positional tracking for long-term analysis of Drosophila sleep and locomotion using the “tracker” program. PLoS One, 2012. 7(5): p. e37250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lee-Iannotti JK and Parish JM, Suvorexant: a promising, novel treatment for insomnia. Neuropsychiatr Dis Treat, 2016. 12: p. 491–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Herring WJ, et al. , Orexin receptor antagonism for treatment of insomnia: a randomized clinical trial of suvorexant. Neurology, 2012. 79(23): p. 2265–74. [DOI] [PubMed] [Google Scholar]
- 67.Citrome L, Suvorexant for insomnia: a systematic review of the efficacy and safety profile for this newly approved hypnotic - what is the number needed to treat, number needed to harm and likelihood to be helped or harmed? Int J Clin Pract, 2014. 68(12): p. 1429–41. [DOI] [PubMed] [Google Scholar]
- 68.Herring WJ, et al. , Effects of suvorexant on the Insomnia Severity Index in patients with insomnia: analysis of pooled phase 3 data. Sleep Med, 2018. [DOI] [PubMed] [Google Scholar]
- 69.Johard HA, et al. , Intrinsic neurons of Drosophila mushroom bodies express short neuropeptide F: relations to extrinsic neurons expressing different neurotransmitters. J Comp Neurol, 2008. 507(4): p. 1479–96. [DOI] [PubMed] [Google Scholar]
- 70.Johard HA, et al. , Peptidergic clock neurons in Drosophila: ion transport peptide and short neuropeptide F in subsets of dorsal and ventral lateral neurons. J Comp Neurol, 2009. 516(1): p. 59–73. [DOI] [PubMed] [Google Scholar]
- 71.Nassel DR, et al. , A large population of diverse neurons in the Drosophila central nervous system expresses short neuropeptide F, suggesting multiple distributed peptide functions. BMC Neurosci, 2008. 9: p. 90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Frenkel L, et al. , Organization of Circadian Behavior Relies on Glycinergic Transmission. Cell Rep, 2017. 19(1): p. 72–85. [DOI] [PubMed] [Google Scholar]
- 73.Donlea JM, Neuronal and molecular mechanisms of sleep homeostasis. Curr Opin Insect Sci, 2017. 24: p. 51–57. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Video 1. Lack of immediate reaction to red light stimulation in a UAS alone control female fly. The infrared light in the corner of the video indicates when the red light was on (10 seconds in this case). This control fly should not have expressed the Chrimson sensor in sNPF-producing neurons, and showed very little obvious response to the red light being turned on or off.
Supplementary Video 2. Immediate reaction to red light stimulation in an sNPF/CHR female fly. This fly expressing Chrimson in its sNPF-producing neurons showed an immediate uncoordinated movement reaction when the red light was turned on. This reaction also turnsed off rapidly as soon as the red light was turned off. In this case, the fly took several seconds before it returned to its feet.
Supplementary Video 3. Continued movement in a UAS alone control female fly later in recording. The same fly as in Video 1 shows almost constant movement or grooming. This video begins approximately 2 minutes following red light stimulation.
Supplementary Video 4. Minimal movement in an sNPF/CHR female fly later in recording. The same experimental fly as in Video 2 spends almost no time walking, but does show grooming behavior. This video begins approximately 2 minutes following red light stimulation.