Abstract
The terminal cells of the larval Drosophila tracheal system extend dozens of branched cellular processes, most of which become hollow intracellular tubes that support gas exchange with internal tissues. Previously, we undertook a forward genetic mosaic screen to uncover the pathways regulating terminal cell size, morphogenesis, and the generation and maintenance of new intracellular tubes. Our initial work identified several mutations affecting terminal cell size and branch number, and suggested that branch complexity and cell size are typically coupled but could be genetically separated. To deepen our understanding of these processes, we have further characterized and determined the molecular identities of mutations in the genes sprout, denuded and asthmatic, that had been implicated in our initial screen. Here we reveal the molecular identity of these genes and describe their function in the context of the TOR and Hippo pathways, which are widely appreciated to be key regulators of cell and organ size.
INTRODUCTION:
Over the course of normal development in the Drosophila trachea, increases in organ size correlate with increases in branch number. In prior work, we determined that mutations that affect cell size tend to have a corresponding effect on branch number (Ghabrial et al., 2011). Nearly all larval tracheal cells are post-mitotic, hence, the dramatic increase in organ size and number of tracheal tubes from embryonic stages to the end of the third larval instar must be mediated almost exclusively through pathways that regulate cell size. Among these are the target of Rapamycin (TOR) and Hippo pathways (Tumaneng et al., 2012a). Increased signaling through the Hippo and TOR pathways, as seen when their respective inhibitors, Warts and TSC1, are removed, result in increased terminal cell size and ectopic branching (Ghabrial et al., 2011). The Hippo pathway acts principally through controlling the subcellular localization of transcriptional factor Yorkie/YAP (Johnson and Halder, 2014; Tumaneng et al., 2012b). The TOR pathway integrates Insulin signaling and other nutritional information, such as amino acid availability, to promote growth and proliferation through increased rates of protein synthesis (Edgar, 2006; Hietakangas and and Cohen, 2009). Both Hippo and TOR pathways also regulate DNA replication and ploidy (Jiang et al., 2014; Pierce et al., 2004; Zielke et al., 2011). For both pathways, downstream effectors remain to be fully elucidated
In post-mitotic tissues, endoreplication is a commonly used strategy to promote growth, with DNA replication controlled by genes downstream of TOR and Hippo pathways (Zhang et al., 2000). For example, both pathways have been shown to regulate the transcription factor, E2F1, which initiates S-phase by transcribing Cyclin E (Bayarmagnai et al., 2012; Duronio and O'Farrell, 1995; Duronio et al., 1995; Reddy et al., 2010; Zhang et al., 2017). Endoreplication occurs in most larval cells during the 5 days of growth leading up to pupariation. Many cells in the tracheal system have been shown to endoreplicate (Guha and Kornberg, 2005; Zhou et al., 2016), while a pool of undifferentiated tracheoblasts remain diploid and are activated to divide and populate much of the pupal and adult tracheal system during the third larval instar (Guha and Kornberg, 2005; Weaver and Krasnow, 2008). However, some differentiated tracheal cells that contribute to smaller tubes, such as the anterior dorsal branch stalk cells, maintain their mitotic potential, as do a small subset of cells (tr2) in the larger dorsal trunk tubes. During larval stages, these differentiated cells enter S-phase, label with phospho-histone H3 antibody, alter their morphology, and proliferate, ultimately contributing multiple cell types to the pupal tracheal system (Weaver and Krasnow, 2008). Tracheal terminal cells do not contribute to the pupal tracheal system; however, whether they endoreplicate and if that is important for cell size and branch complexity has not been explicitly addressed.
In addition to examining known regulators of cell size, we have continued to follow-up on our unbiased genetic approach towards identifying novel factors, some of which could be specific to the tracheal system. Previously, we found that mutations in essential house-keeping genes, which result in clone loss in mitotic tissues such as the eye imaginal disc, appear to be better tolerated in post-mitotic cells, perhaps due to perdurance of mRNA and/or protein present in the mother cell. For example, while eye imaginal disc clones mutant for glutamyl-prolyl tRNA synthetase are lost or restricted to a few cells in size, tracheal cells mutant for glutamyl-prolyl-tRNA synthetase were recovered at a relatively high frequency but found to decrease both cell size and branch number (Ghabrial et al., 2011). Mutations in other genes likewise had a strong effect on terminal cell size and branch number including mutations in the tracheal master transcription factor, trachealess (trh), and the terminal cell-specific transcription factor blistered/SRF (Ghabrial et al., 2011) (Guillemin et al., 1996).
On the other hand, mutations in tumor suppressor genes, which cause dramatic over-proliferation defects in eye clones, were recovered as mutant tracheal terminal cells with abnormally large cell bodies and higher numbers of branches (Ghabrial et al., 2011). Such mutants included the Hippo pathway member warts/lats1 (miracle-gro), and the TOR pathway inhibitor, Tsc1 (Jolly green giant). Consistent with this trend, activation of the FGF Receptor (Breathless) pathway in terminal cells, which results in an increase in cell size, also gives rise to ectopic branches (Jarecki et al., 1999; Lee et al., 1996). Expression of a forced FGFR dimer (λ-breathless) resulted in a warts-like phenotype, with increased tube diameter, tortuosity, and the generation of tubes coursing through the cell soma (Schottenfeld-Roames and Ghabrial, 2012). It is striking that ectopic branches arise around the terminal cell nucleus, perhaps reflecting that the elevated growth signal lacks a specific spatial cue such as might be provided by a hypoxic tissue secreting the FGFR ligand, Branchless.
Here we examine endoreplication specifically in tracheal terminal cells, determining that they do endoreplicate, primarily during the second larval instar, and that this correlates with increases in cell size and branch complexity. We test a requirement for E2f1, which is essential for endoreplication, in terminal cell growth and branching, and then go on to determine the molecular identities of sprout, denuded and asthmatic, and to characterize the cell size and branch complexity defects of null mutations in those genes. Our data appear to converge on the central role of the Hippo and TOR pathways in the regulation of cell size and branch complexity in tracheal terminal cells.
MATERIALS AND METHODS
Fly strains and mosaic analysis.
Mutations in sprout, denuded and asthmatic were previously described in (Ghabrial et al., 2011). FRT82B E2FrM729 was from the Bloomington stock center. For mosaic analysis, males carrying the mutation of interest were crossed to virgins of the following genotype: y, w, hsFLP122; btl-GAL4, UAS-GFP, DsRED2nls; FRT82B cu, GFP RNAi. For Grim and Hid mis-expression experiments, a classic MARCM approach was used; briefly, males carrying UAS-Grim or UAS-Hid and FRT82B Tub-Gal80 were crossed to virgins of the genotype y, w, hsFLP122; btl-GAL4, UAS-GFP; FRT2A,FRT82B. For both mosaic strategies, eggs were collected for 4 hours and then embryos were subjected to a 1 hour heat shock at 38.5 C. Crosses were maintained at 25 C. On day 5, wandering third instar larvae were collected for study. For visualization of gross terminal cell morphology and gas filling, larvae were heat killed (5 – 10 seconds at 70 C), mounted in glycerol and imaged. For antibody staining, larvae were filleted in ice-cold PBS supplemented with Tween20 and TritonX100 (PBSTw,Tr) and fixed for 15 – 20 minutes in 4% formaldehyde.
Immunofluorescence.
Fixed fillets were rinsed 3x in PBSTw,Tr (0.3%, 0.3%) and then incubated with primary antibody in PBSTw,Tr supplemented with 4% horse serum overnight at 4 C. Fillets were then rinsed 3x in PBSTw,Tr (0.3%, 0.3%) and washed 3x 30 minutes. Fillets were incubated with secondary antibody in PBSTw,Tr for 2 hours at room temperature, and then were rinsed 3x in PBSTw,Tr (0.3%, 0.3%) and washed 3x 30 minutes. Larvae were mounted in aquapolymount and imaged on Leica compound fluorescent microscopes. Images were deconvolved using the Leica 3D deconvolution software.
Antibodies
Mouse α-Armadillo (DSHB, AB_528089); chicken α-GFP (Invitrogen, AB_2534023); Rabbit α-PKC-zeta (Santa Cruz, sc-216); Rabbit α-Wkdpep (Ghabrial lab, AB_2737402). Secondary antibodies (Invitrogen) were conjugated to Alexa dyes (A488, A594, or A647) and used at 1:1000 dilutions.
Edu labeling
0.1mM Edu was mixed with standard fly food, accompanied by a few pellets of bromophenol blue. Larvae sorted under the dissecting microscope based on size and spiracles and transferred to Edu food. Late wandering 3rd instar larvae that ingested bromophenol blue were collected 24-48 hours later. Larvae were filleted and Edu was developed according to the Click-it Edu labeling kit followed by standard immunostaining (Invitrogen, C-10419).
Sholl analysis
2nd , early 3rd and late 3rd instar btl-GAL4, UAS-GFP; FRT 2A FRT82B larvae were identified based on size and spiracle morphology. Larvae were heat killed and z-stacks of intact terminal cells from the 5th metamere were imaged on the Leica DMR microscope. Terminal cells were processed using the Simple Neurite tracer and the Sholl analysis plug-in using Fiji/imageJ software. Statistical significance was determined was by t-Test.
RESULTS AND DISCUSSION
Tracheal terminal cells endoreplicate.
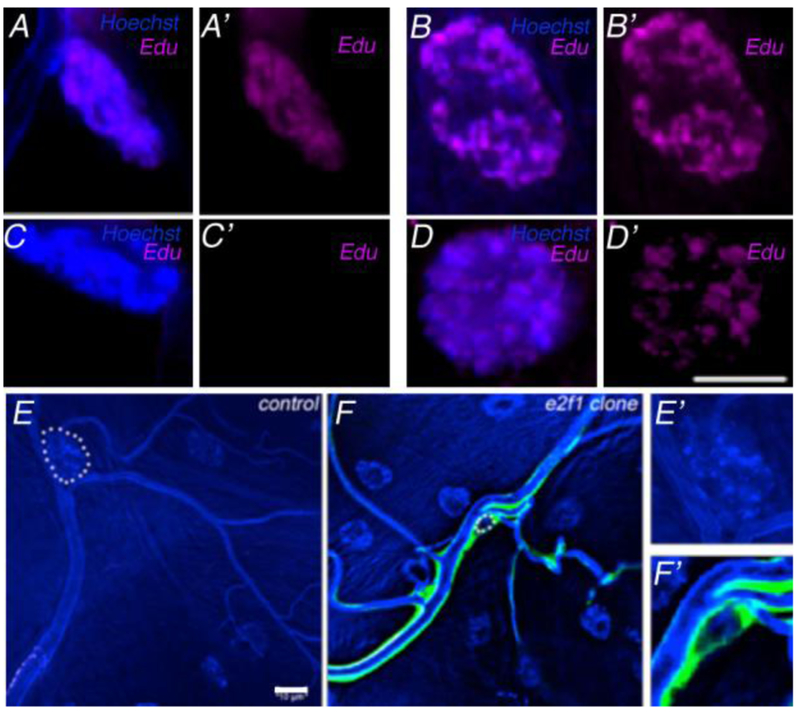
To test whether terminal cells undergo endoreplication, we fed 2nd and early 3rd instar larvae food laced with Edu, (5-ethynyl-2’-deoxyuridine) a nucleoside analog to thymidine, and then examined Edu staining at late third larval instar. While larvae fed at 2nd instar incorporated Edu throughout the trachea including terminal cells (Figure 1 A-B’, n=7 larvae), and also other tissues, larvae fed at early 3rd instar lacked terminal cell labeling (Fig. 1 C-C’, n= 3 larvae). Indeed, with the exception of occasional fusion cells and of cycling tracheoblasts (data not shown), no tracheal Edu incorporation occurred during the third larval instar. As an internal control, we note that Edu labeling was observed in muscle (Figure 1 D-D’), as well as all other larval tissue examined in larval fillets (data not shown). These data indicate that tracheal DNA replication in larvae mostly occurs prior to 3rd instar. Mutations in genes essential for endoreplication, such as e2f1, considered in more detail below, affect nuclear size of third instar terminal cells (Figure 1E-F’), as expected.
Figure 1: Endoreplication of Tracheal Terminal Cells.
Terminal cells (A, C) and muscle cells (B, D) whose nuclei were visualized by Hoechst staining (blue), were examined for incorporation of Edu (magenta) subsequent to feeding during 2nd (A,B) or early 3rd (C,D) larval instars. All larvae were scored at third wandering instar regardless of the time of feeding. Merged images are shown in A-D, Edu staining only is displayed in A’-D’). Nuclear size is dependent on endoreplication, as cells mutant for e2f1 show dramatically smaller nuclei (compare E, F). In mosaic larvae stained with Hoechst, control terminal cell nuclei (E) were much larger than e2f1 clones marked by GFP expression (F). Terminal cell nuclei are outlined in white dashed circles. Autofluorescence in the UV channel also mark the tubes of the tracheal system. (E’) and (F’) show enlarged nuclei from (E) and (F), respectively. Scale bars =10 μm.
Terminal cell branch complexity increases dramatically between 2nd and 3rd instar.
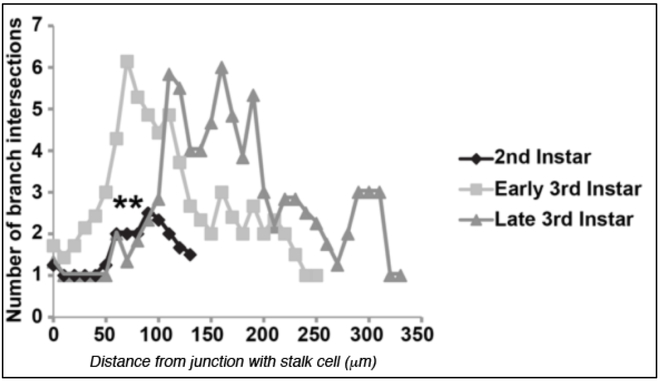
We performed a careful analysis of terminal cell growth during second and third larval instars. Significant increases in branch complexity, quantified as the number of branch intersections at any given distance from the center of the cell (using Sholl analysis as described previously (Schottenfeld-Roames et al., 2014)), was seen between 2nd and early 3rd instar (Figure 2). Cells continued to extend new branches, and to cover larger areas, during the third instar, but without a similar jump in complexity as seen between 2nd and early 3rd instars. Larvae scored during the second instar had between 3 and 8 branch tips (mean of 4.9), which by early third instar had increased to between 10 - 17 branch tips (mean of 13.3), and by the end of third instar to 18 - 31 branch tips (mean of 25.1), numbers largely in agreement with those previously reported by JayaNandanan and colleagues (JayaNandanan et al., 2014).
Figure 2: Branch complexity increases dramatically between 2nd and early 3rd instar.
A Sholl analysis of branching complexity was performed at 2nd, early 3rd, and late 3rd instar. In all cases, terminal cells from the 5th tracheal metamere were examined (tr5). The distance from the stalk cell junction at which the circles intersect terminal cell branches are indicated on the X-axis while the number of branches crossed by the circles are indicated on the Y-axis. ** indicates a T Test value of p<0.01 for the comparison of 2nd instar data with that of early and of late 3rd instar, with 7, 7 and 6 terminal cells scored for each stage, respectively.
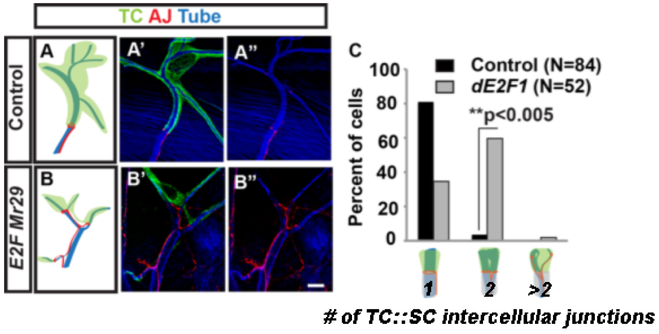
To test whether endoreplication is required for normal terminal cell growth and branching, we examined cells mutant for the transcription factor E2f1, which is known to be required for S phase during endoreplication in post-mitotic tissues (Orr-Weaver, 2015). Terminal cells mutant for E2f1 had fewer branches and also showed gas-filling defects (Figure 3A-B, 100% penetrant phenotype, n = 52, and data not shown). The number of branches per terminal cell ranged from 8 to 18 with a mean of 12 +/− 1 SEM (n =9). Moreover, we found that the smaller E2f1 mutant terminal cells induced neighboring wild type stalk cells to increase in size and to invade the small mutant terminal cells (Figure 3C), much as we had previously observed for mutations in the TOR pathway (Francis and Ghabrial, 2015). Compensatory stalk cell branching, as described in Francis and Ghabrial, results in an increase in stalk cell size and branch complexity, as the stalk cell extends into the terminal cell, much like fingers pushing into a balloon. When this occurs, an increase in the number of intercellular connections between the stalk cell and the terminal cell are observed: in wild type, there is almost always a single intercellular junction connecting the terminal and stalk cell; in the case of compensatory branching, there may be two intercellular junctions (a “mild” phenotype) or more (a “severe” phenotype). Taken together, these data suggest that endoreplication is an important driver of terminal cell growth and branching.
Figure 3: E2f1 is required for normal terminal cell growth and branching.
Terminal cell clones were positively labeled by GFP expression (green) while tracheal tubes were visualized by autofluorescence in the UV channel (blue), and cell junctions were marked by staining against adherens junction (AJ) components (Arm, red). In control terminal cells (A-A”), a ring of AJ staining marks a single intercellular junction with a neighboring (unlabeled) autocellular stalk cell. In E2f1 mutant cells (B-B”), smaller and less branched terminal cells were connected to single stalk cells by two or more cell junctions 61% (n=28) of the time (C). Data are representative of marked homozygous mutant tracheal terminal cells examined in multiple animals from three replicate crosses to generate genetic mosaic animals. Scale bar = 10 μm.
sprout encodes the Drosophila InR.
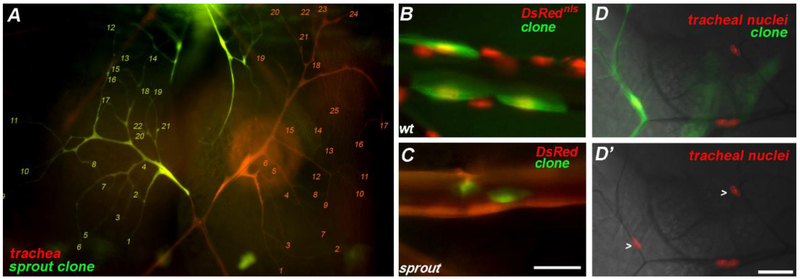
We next turned to mutations in sprout, which caused a similar decrease in cell size, but did not appear to strongly affect branch number. For instance, the number of branch tips for a sprout mutant dorsal branch terminal cell and the control dorsal branch terminal cell on the contralateral side were roughly the same (Figure 4; 22 +/− 2.3 SEM and 25 +/− 1.9 SEM, respectively; n = 7 sprout terminal cells and 7 control terminal cells, which were representative of dozens of terminal cell clones examined in larvae from more than 4 replicate crosses) despite the smaller size of the sprout cells. Terminal cells mutant for sprout were estimated to have a soma of about 2/3 the size of neighboring terminal cells and to likewise ramify on a target area of about 2/3 the size (n = 5 sprout terminal cells, 5 control terminal cells which were representative of the dozens of terminal cell clones examined from more than 3 replicate crosses). In the same mosaic animals, dorsal trunk cells, which were easier to measure due to their simpler hexagonal shapes, sprout cells covered areas of ~ 1/4 the size of control dorsal trunk cells (n=3 sprout, 5 control dorsal trunk cells). We also note that sprout mutant dorsal trunk cells tended to be oval in shape rather than hexagonal.
Figure 4: Terminal cells mutant for sprout are reduced in size but not branch complexity.
A) A mosaic larva with a sprout terminal cell clone (green) is shown; the branch tips for the sprout mutant and contralateral control terminal cell are enumerated (21 and 25, respectively). In B-C), control and sprout clones, respectively, in the dorsal trunk are shown. Note that in A and C, DsRed counterstains all tracheal cells, while in B and D DsRednls is employed in the same capacity. In D and D’ compare the similar size of sprout and heterozygous control terminal cell nuclei (> points to terminal cell nuclei); this is in contrast to staining in terminal cells where endoreplication is blocked. Scale bars = 50 microns. Bar for A, B and C, shown in C, for D and D; shown in D’.
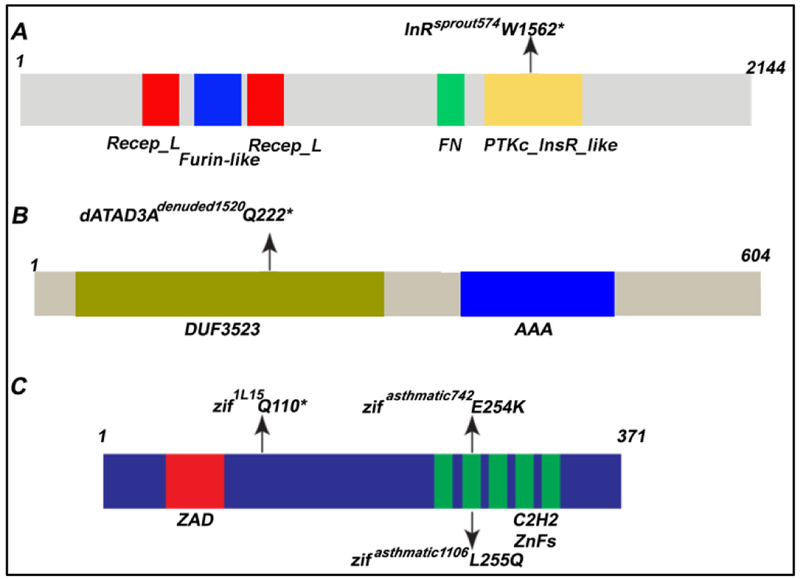
To gain insight into how sprout regulates terminal cell size and branching, we decided to determine the molecular nature of the gene. The mutation in sprout mapped distal to FRT82B, and whole genome sequencing identified a nonsense mutation in the Drosophila Insulin-like Receptor (Fernandez-Almonacid and Rosen, 1987) (InR; Figure 5A). The sprout allele of InR is likely a null, as the mutation is predicted to truncate the receptor within the active site of the kinase domain (W1562Stop). Because InR activation antagonizes PTEN (Oldham et al., 2002), which in turn acts as an upstream inhibitor in the TOR pathway we speculate that the decrease in sprout cell size is due to decreased TOR signaling. Interestingly, prior work (Dekanty et al., 2005) identified a role of InR, mediated by signaling through the TOR pathway, in modulating Sima protein levels. Sima is the Drosophila Hifα ortholog, which regulates terminal cell branching in response to oxygen (Bacon et al., 1998; Centanin et al., 2008). Loss of InR is thus predicted to result in decreased Hifα in the nucleus, and might have been expected to lead to even greater reductions in terminal cell branching given that Sima positively regulates the Breathless-FGFR. Why mutations in InR do not appear to strongly affect branch complexity remains a question for future study, but may reflect that Sima/Hifα exerts a negative feedback on TOR activity – Hifα-target gene, Scylla, activates TSC1-TSC2 (Dekanty et al., 2005) – and additionally, that loss of InR does not impact endoreplication. Indeed, we find no detectable effect on nuclear size or DNA content in sprout terminal cell clones as compared to neighboring heterozygous terminal cells in the same animal (Figure 4D). The lack of effect on endoreplication may also account for the limited ability of mutant InR terminal cells to induce compensatory branching of neighboring stalk cells (73% of sprout/InR terminal cells shared a single intercellular junction with the neighboring stalk cell, while 27% had shared two intercellular junctions, which was elevated from the 4% occurrence observed in wild type).
Figure 5: The molecular identities of sprout, denuded and asthmatic.
A) Mapping and whole genome sequencing revealed a nonsense mutation within the coding sequence of InR in sprout mutant animals. The mutation is predicted to cause a truncation of the protein within the kinase domain and is therefore likely to be an amorphic allele. B) Mapping and whole genome sequencing of mutations in denuded revealed a nonsense mutation within the coding sequence of dATAD3A. The mutation is predicted to truncate the protein after 222 amino acids, completely deleting the AAA domain. C) Mutations previously assigned to the asthmatic complementation group were mapped by meiotic recombination and failed to complement mutations in zif. Sequencing of zif revealed missense mutations in the second zinc finger domain of the two of the mutations in the asthmatic complementation group – see details below. For further analysis of zif/asthmatic, we used the putative null allele, 1L15, that was previously shown to carry a non-sense mutation predicted to truncate the protein such that all zinc finger domains would be lacking.
denuded encodes the Drosophila ATAD3.
Tracheal terminal cells mutant for denuded frequently formed primary terminal cell branches with seamless tubes of normal morphology, but subsequent generations of branches were pruned or altogether absent (Figure 6). Through a combination of meiotic recombination mapping and whole genome sequencing, we determined that the denuded mutations (Figure 5B) were in the previously identified gene belphegor (bor)/dATAD3A (Harel et al., 2016). ATAD3A is a nuclear-encoded mitochondrial membrane protein that is a downstream effector of TOR – indeed, bor was identified as one of the genes most sensitive to rapamycin treatment. ATAD3A and is required for mitochondrial biogenesis and for ER-Mitochondrial interaction (Baudier, 2018; Guertin et al., 2006). Consistent with a role downstream of Tor, we find that terminal cells mutant for denuded induced compensatory stalk cell branching (57% of terminal cells shared a single intercellular junction with the adjacent stalk cell, while 41% displayed a mild phenotype with two intercellular junctions and 2% displayed severe defects, with more than 2 intercellular junctions; n = 51). Loss of function in the human ATAD3A is implicated in developmental defects and neurological syndromes, such as hereditary spastic paraplegia.
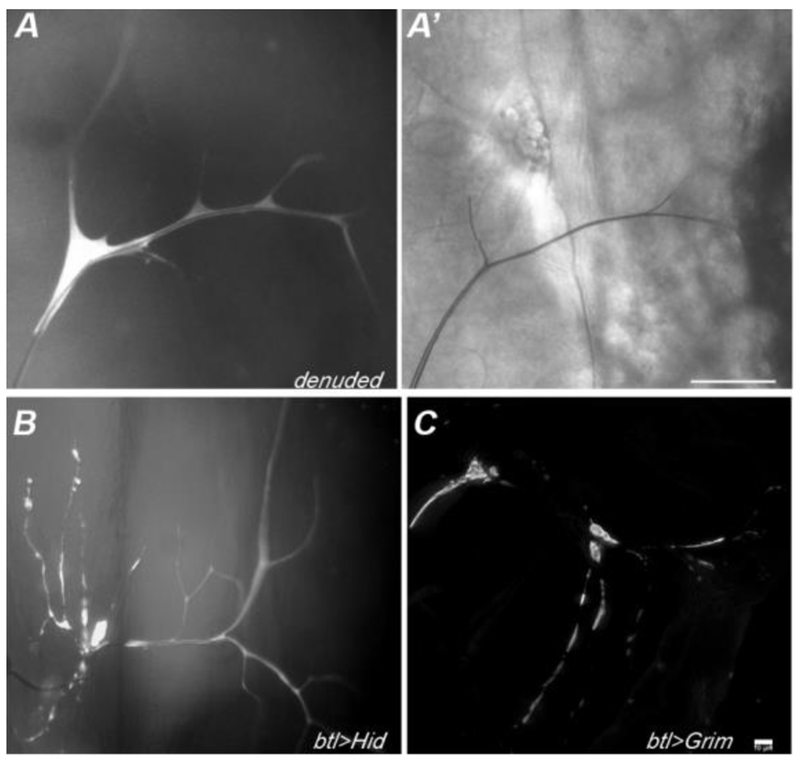
Figure 6: Terminal cells mutant for denuded show reduced size and branch complexity.
In mosaic larvae, terminal cells homozygous for denuded (15-20 allele shown) have reduced cell size and branch number. Terminal cell clones were identified as GFP (white) positive cells. In particular, while one or two main branches appear of normal size and gas-filled (A’), side branches are reduced in number and appear thin and whispy. The defects observed in denuded clones are not due to apoptosis, as terminal cells induced to apoptose by expression of Hid (B) or Grim (C), using the MARCM system, show blebbing of the cytoplasm and branch loss. Note that extensive branching occurs in these terminal cells prior to the onset of branch loss, this may reflect the delay in GAL4-induced expression associated with turn-over of GAL80 subsequent to clone induction. Scale bar in A’ = 50 microns, Scale bar for B,C (in C) = 10 microns.
One of our denuded alleles (denuded1520) is likely to be a molecular null as we determined that a nonsense mutation is present, converting Q222 into a stop codon. The pruning defect is likely to be one associated with reduced growth rather than cell death, since forced expression of pro-apoptic genes in tracheal terminal cells results in dramatic blebbing of the cell, loss of gas-filling, and degeneration of existing branches (Figure 6 B,C, and data not shown).
The transcription factor Zif is required for maintenance of apical membrane polarity.
Mutations in asthmatic were initially identified based on their gas-filling defects – terminal cells and autocellular tubes were affected (Figure 7). Further analysis showed that the tubes formed by asthmatic mutant terminal cells were frequently discontinuous and that the number of side branches were reduced as compared to internal controls. Mapping, complementation tests, and sequence analysis determined that two of the three asthmatic mutations are allelic to zinc finger protein (zif) (Figure 5C). The third allele of asthmatic, 1530, complemented zif and was subsequently found to complement 742 (but not 1106). Accordingly, we rename the 1106 and 742 mutations as zif alleles, and retain the name asthmatic for 1530; whether 1106 also carries a mutation in asthmatic, or if 1106 and 1530 carry mutations in a third essential gene has not yet been determined.
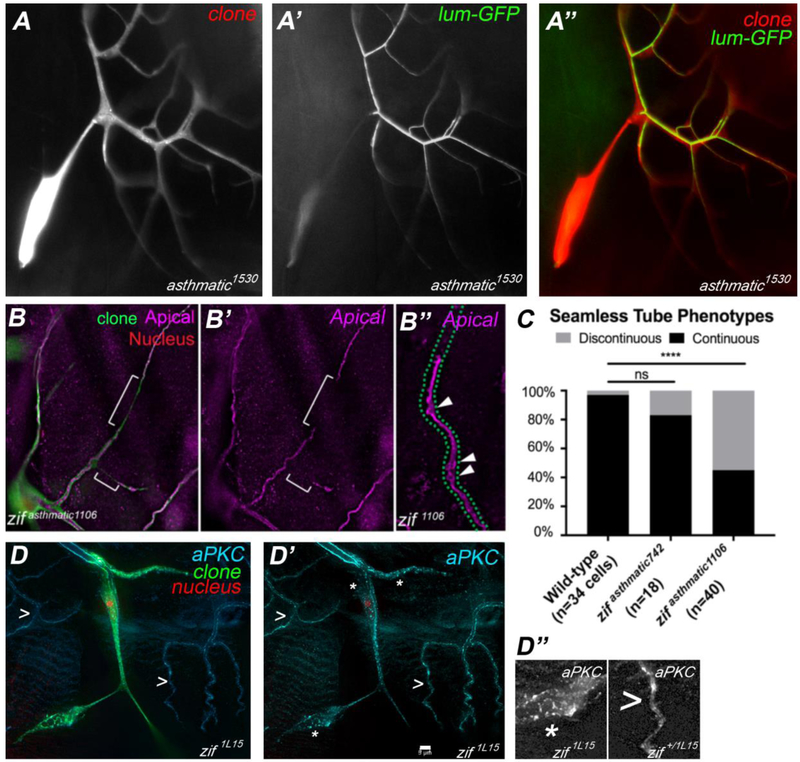
Figure 7: Mutations in zif disrupt maintenance of apical polarity in seamless tubes.
Terminal cells mutant for asthmatic (A, red in A”) lack gas-filled tubes; however, they do make tubes that lack gas-filling, as revealed by the localization of a secreted GFP – lum-GFP – in the lumenal space (A’, green in A”). Mapping studies revealed that asthmatic1530 (shown in A-A”) is not part of the same complementation group as asthmatic742 and 1106, which we now identify as new alleles of zif. We will henceforth refer to only 1530 as asthmatic and rename the 742 and 1106 as alleles of zif. Staining of apical membranes in zif1106 terminal cells (B-B”), reveals the presence of tube shape irregularities (arrowheads in B”) and discontinuities (indicated by [ in B and B’). Discontinuity defects were quantified in (C). To determine the null phenotype, we examined zif1L15 mutant terminal cells (green, D). We found that seamless tubes were largely absent or were severely truncated and that apical membrane antigens such as aPKC (cyan) and Wkdpep (not shown) were no longer enriched on the lumenal membrane, but were dispersed throughout the cell (D and D’). The location of the terminal cell nucleus is revealed by expression of DsRED2nls (red, D). Note that apical membrane localization of aPKC in neighboring heterozygous terminal cells is intact (> in D, D’). In D” adjacent sections of the panel in D’ are shown enlarged (*, portion of zif clone, > portion of zif+/− control cell). Scale Bar (D’) = 5 microns, applies to A-D’.
Intriguingly, zif was previously characterized based on its role in the regulation of the apical domain of neurons, with Zif shown to repress expression of aPKC (Chang et al., 2010). We examined the putative null allele of zif (1L15) and found that it showed a more extreme terminal cell phenotype than 742 or 1106, which carry point mutations in the second zinc finger. Terminal cells null for zif were reduced in size, showed little or no branching, and formed discontinuous tubes. Strikingly, the zif null terminal cells also showed apical protein localization defects, with aPKC, as well as another apical marker (anti-Wkdpep, (Schottenfeld-Roames and Ghabrial, 2012)), distributed throughout the cell and no longer enriched on the lumenal membrane. The mislocalization of apical proteins was reminiscent of the defects we reported for TOR pathway and vATPase mutants (Francis and Ghabrial, 2015); consistent with that, we observed induction of stalk cell compensatory branching associated with zif mutant terminal cells (data not shown). Pan-tracheal RNAi of zif resulted in developmental arrest, with only a few unusually small larvae progressing to third instar, and none to pupal stages. Terminal cells were not gas-filled, and other branches were subject to liquid filling under stress. Terminal cells appeared to undergo apoptosis as judged by blebbing of their cytoplasm (similar to the cells shown in Figure 6B,C, data not shown). Taken together, these data suggest that Zif is required for terminal cells to maintain apical polarity. Interestingly, this requirement in the tracheal system was specific for terminal cells as zif clones in other positions in the tracheal system did not affect localization of aPKC (data not shown). In the absence of a proper apical domain, terminal cell growth and branching appear compromised with subsequent induction of compensatory branching in the adjacent stalk cell.
Interestingly, the three complementation groups we chose to further characterize all impacted related growth pathways, with sprout and denuded acting upstream and downstream, respectively, of the TOR pathway, and zif showing apical polarity maintenance defects similar to those we reported with loss of TOR pathway signaling, raising the possibility that Zif may also factor into Tor signaling. Future studies examining the epistatic relationship between Zif and Tor (or Hippo) will establish if these genes are acting with a single pathway or are acting in parallel but converging on an overlapping set of downstream targets. Going forward, it will be important to understand how the FGF pathway, which drives all tracheal branching events, intersects with the Hippo and Tor cell growth pathways. In particular, it will be very interesting to see how, or if, local activation of the FGFR pathway in tracheal terminal cells provokes a growth and branching response that differs from the global activation of the FGFR pathway, or the global de-repression of Tor or Hippo pathways. All of the latter result in the induction of ectopic seamless tubes around the terminal cell nucleus, whereas a response to FGF point sources (hypoxia-induced branchless expression) is thought to sculpt the unique shapes (branching pattern) of each terminal cell without eliciting ectopic seamless tube growth in the soma.
Highlights.
Terminal cell size and branching require E2F1-dependent endoreplication
The terminal cell size mutant sprout is an allele if InR
The terminal cell size and branch number mutant denuded is an allele of ATAD3A
Mutations in zif also affect terminal cell size and branch number
ACKNOWLEDGEMENTS.
SB would like to acknowledge funding from the Taub Institute. JR would like to acknowledge funding from NIH pre-doctoral fellowship (F31 HL 132501). ASG would like to acknowledge funding from NIH 2R01GM089782 and the Dean’s office at Columbia University Medical Center.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Bacon NC, Wappner P, O'Rourke JF, Bartlett SM, Shilo B, Pugh CW, Ratcliffe PJ, 1998. Regulation of the Drosophila bHLH-PAS protein Sima by hypoxia: functional evidence for homology with mammalian HIF-1 alpha. Biochem Biophys Res Commun 249, 811–816. [DOI] [PubMed] [Google Scholar]
- Baudier J, 2018. ATAD3 proteins: brokers of a mitochondria-endoplasmic reticulum connection in mammalian cells. Biol Rev Camb Philos Soc 93, 827–844. [DOI] [PubMed] [Google Scholar]
- Bayarmagnai B, Nicolay BN, Islam AB, Lopez-Bigas N, Frolov MV, 2012. Drosophila GAGA factor is required for full activation of the dE2f1-Yki/Sd transcriptional program. Cell Cycle 11, 4191–4202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centanin L, Dekanty A, Romero N, Irisarri M, Gorr TA, Wappner P, 2008. Cell autonomy of HIF effects in Drosophila: tracheal cells sense hypoxia and induce terminal branch sprouting. Dev Cell 14, 547–558. [DOI] [PubMed] [Google Scholar]
- Chang KC, Garcia-Alvarez G, Somers G, Sousa-Nunes R, Rossi F, Lee YY, Soon SB, Gonzalez C, Chia W, Wang H, 2010. Interplay between the transcription factor Zif and aPKC regulates neuroblast polarity and self-renewal. Dev Cell 19, 778–785. [DOI] [PubMed] [Google Scholar]
- Dekanty A, Lavista-Llanos S, Irisarri M, Oldham S, Wappner P, 2005. The insulin-PI3K/TOR pathway induces a HIF-dependent transcriptional response in Drosophila by promoting nuclear localization of HIF-alpha/Sima. J Cell Sci 118, 5431–5441. [DOI] [PubMed] [Google Scholar]
- Duronio RJ, O'Farrell PH, 1995. Developmental control of the G1 to S transition in Drosophila: cyclin Eis a limiting downstream target of E2F. Genes Dev 9, 1456–1468. [DOI] [PubMed] [Google Scholar]
- Duronio RJ, O'Farrell PH, Xie JE, Brook A, Dyson N, 1995. The transcription factor E2F is required for S phase during Drosophila embryogenesis. Genes Dev 9, 1445–1455. [DOI] [PubMed] [Google Scholar]
- Edgar BA, 2006. How flies get their size: genetics meets physiology. Nat Rev Genet 7, 907–916. [DOI] [PubMed] [Google Scholar]
- Fernandez-Almonacid R, Rosen OM, 1987. Structure and ligand specificity of the Drosophila melanogaster insulin receptor. Mol Cell Biol 7, 2718–2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis D, Ghabrial AS, 2015. Compensatory branching morphogenesis of stalk cells in the Drosophila trachea. Development 142, 2048–2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghabrial AS, Levi BP, Krasnow MA, 2011. A systematic screen for tube morphogenesis and branching genes in the Drosophila tracheal system. PLoS Genet 7, e1002087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guertin DA, Guntur KV, Bell GW, Thoreen CC, Sabatini DM, 2006. Functional genomics identifies TOR-regulated genes that control growth and division. Curr Biol 16, 958–970. [DOI] [PubMed] [Google Scholar]
- Guha A, Kornberg TB, 2005. Tracheal branch repopulation precedes induction of the Drosophila dorsal air sac primordium. Dev Biol 287, 192–200. [DOI] [PubMed] [Google Scholar]
- Guillemin K, Groppe J, Ducker K, Treisman R, Hafen E, Affolter M, Krasnow MA, 1996. The pruned gene encodes the Drosophila serum response factor and regulates cytoplasmic outgrowth during terminal branching of the tracheal system. Development 122, 1353–1362. [DOI] [PubMed] [Google Scholar]
- Harel T, Yoon WH, Garone C, Gu S, Coban-Akdemir Z, Eldomery MK, Posey JE, Jhangiani SN, Rosenfeld JA, Cho MT, Fox S, Withers M, Brooks SM, Chiang T, Duraine L, Erdin S, Yuan B, Shao Y, Moussallem E, Lamperti C, Donati MA, Smith JD, McLaughlin HM, Eng CM, Walkiewicz M, Xia F, Pippucci T, Magini P, Seri M, Zeviani M, Hirano M, Hunter JV, Srour M, Zanigni S, Lewis RA, Muzny DM, Lotze TE, Boerwinkle E, Baylor-Hopkins Center for Mendelian, G., University of Washington Center for Mendelian, G., Gibbs RA, Hickey SE, Graham BH, Yang Y, Buhas D, Martin DM, Potocki L, Graziano C, Bellen HJ, Lupski JR, 2016. Recurrent De Novo and Biallelic Variation of ATAD3A, Encoding a Mitochondrial Membrane Protein, Results in Distinct Neurological Syndromes. Am J Hum Genet 99, 831–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hietakangas V, Cohen SM, 2009. Regulation of tissue growth through nutrient sensing. Annu Rev Genet 43, 389–410. [DOI] [PubMed] [Google Scholar]
- Jarecki J, Johnson E, Krasnow MA, 1999. Oxygen regulation of airway branching in Drosophila is mediated by branchless FGF. Cell 99, 211–220. [DOI] [PubMed] [Google Scholar]
- JayaNandanan N, Mathew R, Leptin M, 2014. Guidance of subcellular tubulogenesis by actin under the control of a synaptotagmin-like protein and Moesin. Nat Commun 5, 3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang N, Soba P, Parker E, Kim CC, Parrish JZ, 2014. The microRNA bantam regulates a developmental transition in epithelial cells that restricts sensory dendrite growth. Development 141, 2657–2668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson R, Halder G, 2014. The two faces of Hippo: targeting the Hippo pathway for regenerative medicine and cancer treatment. Nat Rev Drug Discov 13, 63–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T, Hacohen N, Krasnow M, Montell DJ, 1996. Regulated Breathless receptor tyrosine kinase activity required to pattern cell migration and branching in the Drosophila tracheal system. Genes Dev 10, 2912–2921. [DOI] [PubMed] [Google Scholar]
- Orr-Weaver TL, 2015. When bigger is better: the role of polyploidy in organogenesis. Trends Genet 31, 307–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce SB, Yost C, Britton JS, Loo LW, Flynn EM, Edgar BA, Eisenman RN, 2004. dMyc is required for larval growth and endoreplication in Drosophila. Development 131, 2317–2327. [DOI] [PubMed] [Google Scholar]
- Reddy BV, Rauskolb C, Irvine KD, 2010. Influence of fat-hippo and notch signaling on the proliferation and differentiation of Drosophila optic neuroepithelia. Development 137, 2397–2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schottenfeld-Roames J, Ghabrial AS, 2012. Whacked and Rab35 polarize dynein-motor-complex-dependent seamless tube growth. Nat Cell Biol 14, 386–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schottenfeld-Roames J, Rosa JB, Ghabrial AS, 2014. Seamless tube shape is constrained by endocytosis-dependent regulation of active Moesin. Curr Biol 24, 1756–1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tumaneng K, Russell RC, Guan KL, 2012a. Organ size control by Hippo and TOR pathways. Curr Biol 22, R368–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tumaneng K, Schlegelmilch K, Russell RC, Yimlamai D, Basnet H, Mahadevan N, Fitamant J, Bardeesy N, Camargo FD, Guan KL, 2012b. YAP mediates crosstalk between the Hippo and PI(3)K-TOR pathways by suppressing PTEN via miR-29. Nat Cell Biol 14, 1322–1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver M, Krasnow MA, 2008. Dual origin of tissue-specific progenitor cells in Drosophila tracheal remodeling. Science 321, 1496–1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Stallock JP, Ng JC, Reinhard C, Neufeld TP, 2000. Regulation of cellular growth by the Drosophila target of rapamycin dTOR. Genes Dev 14, 2712–2724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, Pei C, Wang X, Xiang J, Sun BF, Cheng Y, Qi X, Marchetti M, Xu JW, Sun YP, Edgar BA, Yuan Z, 2017. A Balance of Yki/Sd Activator and E2F1/Sd Repressor Complexes Controls Cell Survival and Affects Organ Size. Dev Cell 43, 603–617 e605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou F, Qiang KM, Beckingham KM, 2016. Failure to Burrow and Tunnel Reveals Roles for jim lovell in the Growth and Endoreplication of the Drosophila Larval Tracheae. PLoS One 11, e0160233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zielke N, Kim KJ, Tran V, Shibutani ST, Bravo MJ, Nagarajan S, van Straaten M, Woods B, von Dassow G, Rottig C, Lehner CF, Grewal SS, Duronio RJ, Edgar BA, 2011. Control of Drosophila endocycles by E2F and CRL4(CDT2). Nature 480, 123–127. [DOI] [PMC free article] [PubMed] [Google Scholar]