Abstract
Over the last few decades, mass spectrometry-based proteomics has become an increasingly powerful tool that is now able to routinely detect and quantify thousands of proteins. A major advance for global protein quantification was the introduction of isobaric tags, which in a single experiment enable the global quantification of proteins across multiple samples. In this review, we refer to these methods as multiplexed proteomics. We discuss the principles, advantages, and drawbacks of various multiplexed proteomics techniques, and compare them to alternative approaches. We discuss how the emerging combination of multiplexing with targeted proteomics might enable the reliable and high-quality quantification of very low-abundance proteins across multiple conditions. Lastly, we suggest that fusing multiplexed proteomics with data-independent acquisition approaches might enable the comparison of hundreds of different samples without missing values while maintaining the superb measurement precision and accuracy obtainable with isobaric tag quantification.
Keywords: Multiplexed proteomics, isobaric tags, mass spectrometry, data independent acquisition, ratio distortion, complementary reporter ions, measurement accuracy, measurement precision, measurement sensitivity, label-free proteomics, heavy isotopes, shotgun proteomics, quantitative proteomics
1. Background on Protein Identification and Quantification in Mass Spectrometry-Based Shotgun Proteomics
Throughout this paper, we will refer to proteomics methods that use isobaric tags to analyze multiple protein samples as multiplexed proteomics. Multiplexed proteomics builds on decades of technological development in proteomics prior to isobaric tags. To put multiplexed experiments in context, we begin this review with an overview of protein identification methods and alternative quantitative approaches. We then cover multiplexed proteomics techniques, which involve the use of isobaric labeling. At the end of this review, we will discuss how merging multiplexed proteomics with other quantification strategies might help to overcome current technical limitations.
Throughout this review, we will only discuss bottom-up proteomics, where proteins are first digested into peptides and the peptides analyzed with the mass spectrometer. An entire field is devoted to the analysis of intact proteins via mass spectrometry, known as top-down proteomics. For these approaches, we refer the reader to excellent reviews published elsewhere.[1]
1.1. Peptide Identification in Shotgun Proteomics
Proteomic analysis is typically performed using liquid chromatography-mass spectrometry (LC-MS).[2] For shotgun proteomics, protein samples derived from cell or tissue lysate are digested into peptides with proteases like trypsin (Fig. 1A).[3] Trypsin cleaves, with fairly high specificity, protein peptide bonds at the C-termini of arginine and lysine residues.[4] To better probe the complexity of the proteolytic mixture, tryptic peptides can be fractionated into multiple samples based on properties such as charge, size, polarity, or hydrophobicity.[5] The peptides in each sample are then separated using liquid chromatography (LC). The peptides elute via a thin opening from the column directly in front of the mass spectrometer (MS).
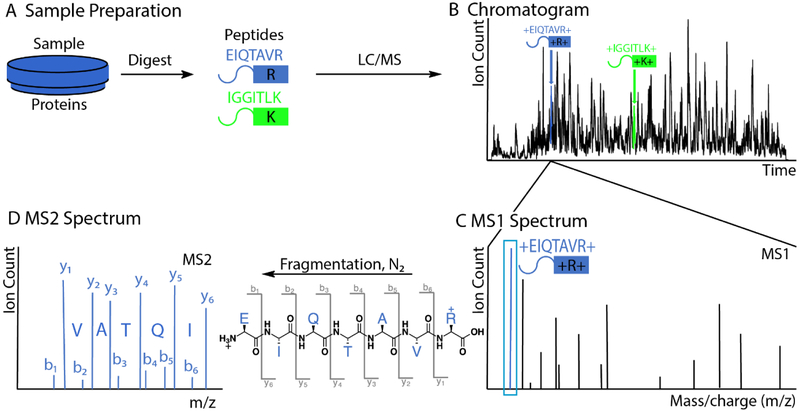
Figure 1: Outline of peptide identification with shotgun proteomics.
A) A sample of proteins is digested by trypsin, which cleaves peptide bonds at the C-terminus of lysine and arginine residues. The example peptide EIQTAVR, which we follow throughout this figure, is shown in blue. To reduce complexity, the peptides are separated by liquid chromatography (LC), ionized via electrospray, and injected into the mass spectrometer (MS). B) Plotted is the chromatogram of the most abundant peak at each retention time. The blue and green peptides elute at different retention times. C) At any given time, e.g., when the blue peptide elutes, multiple different peptides co-elute. The mass spectrometer can typically distinguish them by their mass to charge ratio (m/z). The mass spectrum of the intact peptides is called the MS1 spectrum. The peak corresponding to the peptide EIQTAVR is highlighted in blue. D) In complex mixtures, mass alone is not enough for peptide identification. Inside the mass spectrometer, a peak corresponding to a peptide is isolated and fragmented by collision with inert gas. The fragment ions’ m/z values, derived from the blue peptides, are recorded in the MS2 spectrum. By convention, peptide fragments containing the N-terminus are called b-ions, while fragments with the C-terminus are called y-ions. The characteristic masses of the fragment ions, together with the precursor mass from the MS1-spectrum, are typically sufficient to identify a peptide unambiguously.
Voltage between the column opening and the inlet of the MS leads to a process called electrospray ionization (ESI). The eluting droplets undergo evaporation, concentrating positively charged peptides until coulombic repulsion overwhelms surface tension and the droplets explode, resulting in charged peptides in the gas-phase.[6]
The efficiency of the ionization process can differ by orders of magnitude for different peptides.[7] Additionally, the efficiency of protein conversion into analyzable peptides can also vary drastically due to different digestion efficiencies and/or peptide solubility. Therefore, the number of ions inside the MS is not a direct readout of how many proteins were originally in the sample. Because of this problem, MS is an inherently non-quantitative method and significant additional efforts are required to obtain quantitative information.
Peptide molecules ionize before entering the mass spectrometer where researchers can detect or filter them based on their mass-to-charge (m/z) values. Figure 1B shows the chromatogram of the most abundant ion species collected during a typical ~2-hour experiment. We call a spectrum of all the intact peptides eluting at a given time an MS1 spectrum (Fig. 1C). The height of each peak reflects the number of detected ions.[8] The ~20k human proteins generate ~106 possible tryptic peptides. With the resolving power of current mass analyzers, it is not possible to identify peptides solely based on their intact masses. However, it is possible to fragment peptide ions in a mass spectrometer at the weakest bonds (usually the peptide bond between amino acids) by colliding them with inert gases. The resulting fragment ions are analyzed in an MS2 or MS/MS spectrum, which can be used to identify the amino acid sequence and to detect post-translational modifications such as phosphorylation (Fig. 1D).[9] By convention, the fragment ions containing the peptide’s N-terminus are called b-ions, and the fragments containing the C-terminus are called y-ions.[10]
If all the b- or y-ions were formed and detected, then the differences in m/z values would allow the peptide’s amino acid sequence to be determined de novo, since each genetically-encoded amino acid has a different molecular weight (except leucine and isoleucine). However, the situation is typically not this ideal, and de novo peptide sequencing is not practical for most spectra obtained from actual experiments. Instead, the analyzed samples typically come from organisms for which we know all possible protein sequences and hence all possible peptides. Rather than having to sequence peptides de novo from spectra, we typically only need to find the most likely match to known amino acid sequences. To this end, theoretical MS2 spectra are created for these peptides, based on all the b- and y-ions that can result from fragmentation. By comparing the MS1 mass and the corresponding observed MS2 spectrum to the theoretical spectra of possible peptides, the best match can be found, resulting in the peptide being assigned to the spectrum.[11] Multiple search algorithms are available to automatically perform this analysis.[11–12] Moreover, various machine learning strategies have been developed to confidently assign spectra to peptides and proteins.[13]
1.2. Absolute and Relative Quantification in Proteomics Experiments
Given the intrinsic quantification limitations of mass spectrometry in quantitative proteomics, we distinguish between absolute quantification (determining the absolute concentration of a protein in a sample) and relative quantification (determining the relative ratio of the amounts of a given protein in different samples). As discussed earlier, the efficiencies of turning protein concentrations into MS-signals are nonuniform and currently unpredictable. The signal in the mass spectrometer is therefore only an indirect read-out for the abundance of a peptide in solution.
For the relative quantification of proteins between two or more samples, their peptides must first be relatively quantified from each sample and the data from multiple peptides integrated to get a ratio for the overall protein.[14] The peak size corresponding to a peptide is proportional to the number of peptide ions detected by the instrument. Since the ionization efficiencies of different peptides are different, it is not possible to directly compare the MS signal of different peptides to determine their abundance in a sample. However, it is possible to compare peaks of the same peptide, with the same ionization efficiency, in different samples, which is what relative quantification is based on.[15] All of the methods described below use this as the basis for relative quantification of peptides, and ultimately proteins.
Absolute quantification in proteomics is usually an extension of relative quantification methods that quantify relative to an added spiked-in standard, with known absolute concentration. Due to the high costs of such standards, these experiments are typically limited to studies of a smaller subset of the proteome.[16] While not directly correlated, the total ion-signal of a protein seems to be related to its in vivo abundance via a power law.[17] Using an internal standard, the absolute protein abundance for all proteins detected in a sample can therefore be inferred, but this comes with a wide median ~2-fold error.[17b]
1.3. MS1-Based, Label-free Quantification
Currently, the most widely used form of quantitative proteomics is based on quantifying the MS1 signal of peptides obtained from tryptic digestion. As there is no attempt to covalently modify the peptides, this version of quantitative proteomics is often referred to as label-free. Label-free proteomics involves running different samples consecutively (Fig. 2A).[18] The MS1 signal for a given peptide is integrated over time from all the MS1 spectra in which it can be observed (Fig. 2B, C). The obtained area under the curve is a measure of the total number of ions for a given peptide. While this area is not a good read-out for the absolute amount of peptide in the sample, the corresponding ion-counts (i.e., the area under the curve) for the same peptide in a different sample can be used for relative quantification.[15, 19] These peptide ratios are then combined to give a relative ratio of proteins. This can be done in a variety of ways, such as by using the mean or median of (all or the top n) peptide ratios, taking a weighted average of peptide ratios based on signal intensity, calculating the ratio of total peptide ion counts, or using linear regression to fit a line through the peak intensities for each peptide.[20] If there are more than two samples, pairwise protein ratios can be calculated using any of these methods and a least-squares analysis can be used to interpolate relative protein amounts in each sample.[19] MS2 spectra are required for peptide identification but their signal is typically not used for quantification in these label-free approaches.
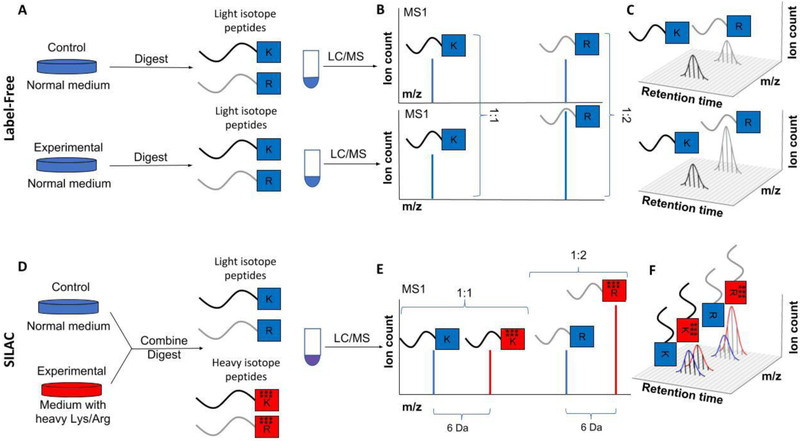
Figure 2: Outline of label-free and SILAC quantification.
A-C) Principles of label-free quantification. A) In label-free quantification, multiple protein samples are digested with trypsin (which cleaves after K or R). The resulting peptides are separated via liquid chromatography (LC) and ionized before entering the mass spectrometer (MS). Shown throughout are two peptides. The one ending in K has equal concentration in the two analyzed samples, and the one ending in R is concentrated 2-fold higher in the experimental sample compared to the control. B) The MS1 spectrum records the number of ions for various m/z values of the intact peptide eluting at a given time. C) The elution time of a peptide takes ~20 seconds. During this time, ~10 MS1 spectra are collected, each showing the peptide at potentially different intensities. The integration of this intensity over time approximates the total number of ions ionizing into the mass spectrometer. D-F) Principles of MS1 based quantification via heavy isotope labeling (e.g., SILAC). D) In SILAC, cell samples are grown either in media with amino acids with naturally occurring isotopes (light) or media where amino acids (K and R) contain heavy isotopes (here 6). Importantly, the heavy isotopes do not alter the chemical properties of the peptides. Cells are lysed and combined. The proteins are digested together, and the resulting peptides are simultaneously separated via LC and ionized before entering the mass spectrometer. E) Peptides in the heavy sample are shifted to the right on the MS1 spectrum compared to those from the light sample. Ratios between peak sizes within one spectrum can thus be used for relative quantification. F) To utilize all available information, typically the ion intensity is integrated over the entire elution profile.
Compared to quantitation methods involving tags or labels, a label-free method avoids additional expense and sample preparation steps. Furthermore, label-free quantification is feasible on hundreds or even thousands of samples. On the other hand, there are some major limitations to a label-free approach. A major limitation is the requirement for multiple runs, which reduces throughput. Another drawback is the comparatively poor measurement precision, with the median protein coefficients of variation (CVs) between replicates typically being ~20%.[19] Many less abundant proteins typically exhibit even larger variability, though this is also tends to be a problem for other quantification methods. This comparatively poor reproducibility comes from each sample being run separately and data acquisition varying when MS1 and MS2 spectra are obtained. Another major limitation of the label-free approach is that, even in replicates, a significant fraction of peptides will not be detected in every sample. This is known as the missing value problem.[21] One can decide to concentrate on the proteins identified in every sample, but this will quickly reduce the number of quantified proteins to only the most abundant proteins in all samples. However, some statistical methods have been developed to tackle the missing value problem, and multiple papers have discussed the effective imputation of missing values.[21–22] Jürgen Cox, Matthias Mann, and colleagues have developed a system of tools, known as MaxLFQ, which imputes missing values by matching retention times and m/z values between different samples.[19] The continuous improvement in MS-technology enables faster and faster collection of MS2 spectra, which cover more and more peaks from the MS1 spectrum.[23] These advances might help to overcome the missing value problem in label-free quantification.
1.4. MS1-Based Quantification with Heavy Isotope Labeling
In contrast to label-free quantification, multiple methods label peptides with heavy, non-radioactive, isotopes. Peptides can be either labeled in vivo e.g., by the addition of heavy amino acids to tissue culture medium (SILAC, i.e., Stable Isotope Labeling with Amino acids in Cell culture),[24] or in vitro, e.g., by performing chemical modifications after proteolytic digestion.[25] Heavy isotopes, with the exception of deuterium, have essentially identical chemistry and elution patterns as their light equivalents, but the mass spectrometer can easily distinguish between their different mass-to-charge ratios. The main advantage of this approach is that samples can be labeled separately with different isotopes and then combined before injection into the mass spectrometer. The samples can therefore be co-analyzed (Fig. 2D, E) and the relative quantification occurs within a single experiment rather than between runs (Fig. 2E, F). This inherently leads to much higher reproducibility (i.e., higher measurement precision) and avoids the missing value problem of label-free approaches, provided the number of samples does not exceed the maximum number of labelling combinations.[26] If there is no signal for a peptide it is known to be below the detection limit rather than not being picked up by chance, which can be the case for label-free approaches.
The major limitation of using the MS1 signal to quantify isotopically labeled peptides is that the complexity of the MS1 spectrum increases with the number of samples, as each sample is isotopically labeled with a different mass. In practice, this limits the number of samples that can be compared in a single experiment to 2 or 3.[26] A recent clever extension of SILAC can avoid this limitation by using labels whose masses only differ by a few mDa.[27] However, these experiments require current mass analyzers engineered to exceptionally high standards, which hinders the technology’s wider application. Another limitation of MS1-based quantification is that the number of ions that can be accumulated in the most commonly used high resolution analyzer, the Orbitrap, is limited. The number of ions for low-abundance peptides can therefore be very small if some very high abundant peptides co-elute at the same time in the MS1 spectrum, resulting in less precise quantification due to poor ion statistics. This limitation has been somewhat alleviated by ion-mobility separation or BoxCar.[28]
1.5. Data-Independent Acquisition
One feature common to the standard implementations of label-free and many other quantitative proteomics methods is the Data-Dependent Acquisition (DDA) of MS2 spectra (Figure 3A-D). Based on the MS1 spectrum, the instrument successively chooses the largest peaks for acquisition of MS2 spectra and peptide identification (Fig. 3A).[29] Intuitively, this makes sense, as the goal is to spend the limited number of MS2 spectra on the peaks in the MS1 spectrum, which can most likely be successfully identified and quantified (Fig. 3 B-E). However, there are usually more peaks available than the MS can isolate for fragmentation and which peaks are chosen is an inherently stochastic process. Which MS2 spectra are acquired and at what time during the elution profile will differ from run to run even if the exact same sample is reanalyzed.
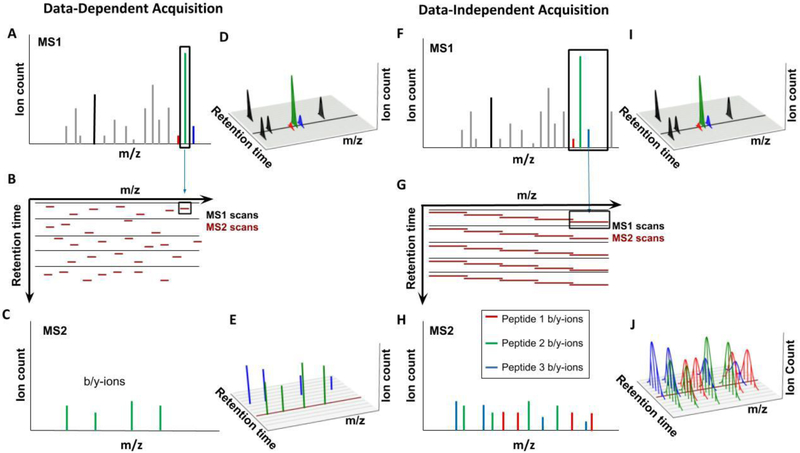
Figure 3: Comparison of Data-Dependent and Data-Independent Acquisition approaches.
A-E) Data-Dependent Acquisition (DDA). A) The goal of DDA is to identify as many peptides as possible, one at a time. The highest peaks in the MS1 spectrum are selected for isolation, with an isolation window of ~1 Th. Peptides in this window are isolated and fragmented for readout in the MS2 spectra. B) Shown is the sequence of MS1 spectra (black) and the data-dependent MS2 isolation windows (dark red), centered on the highest abundant peaks. Each MS1 is followed by multiple MS2 spectra - with current instrumentation and duty cycles of ~2 seconds this would be ~30 MS2 spectra following each MS1 spectrum. C) The MS2 spectrum resulting after isolation and fragmentation consists mainly of b- and y-ions from the target peptide, which allows for comparatively simple peptide identification. D) For quantification, e.g., with label-free approaches, the peptides in the MS1 spectrum are continuously monitored via the peak intensities in the MS1 spectra. Shown are the retention profiles for various peptides - the area under this curve is typically used for peptide quantification. The black line represents the single MS1 scan shown in B. E) Shown here are peaks for the b- and y-ions for the green and blue peptide. A peptide is typically only isolated once for MS2 analysis, the peak height cannot be used for quantification. Not all peaks in the MS1 will trigger the collection of an MS2 spectrum (peaks for the red peptide are missing). Additionally, low-abundance peaks might be below the detection limit in the MS1 spectrum and thus cannot trigger MS2 spectra. The dark red line represents the single MS2 scan shown in C. F-J) Data-Independent Acquisition (DIA). F) The goal of DIA is to continuously collect fragment ion intensities for all eluting peptides. To make this approach compatible with current MS speed requires significantly wider isolation windows (~10 Th) compared to the DDA approach (~1 Th). All the ions within this comparatively wide isolation window are isolated and simultaneously fragmented. G) Shown is the schedule of MS1 spectra (black) and the isolation windows of MS2 spectra (red). H) The simultaneous isolation and fragmentation of multiple peptides results in a complex MS2 spectrum consisting of b- and y-ions from all isolated peptides. I) Similarly to DDA, MS1 intensities of peptides are collected and can be used for quantification. The black line indicates the time for the MS1 spectrum in G. J) Unlike in the DDA equivalent, ion intensity information for b- and y-ions are available throughout the entire elution profile for each peptide. This makes it possible to use fragment ion intensities for quantification. Because the entire m/z space is continuously covered, information for more peptides than with the DDA approach is available. Here, the red peptide’s abundance can be quantified. The dark red line represents the single MS2 scan shown in H
Data-Independent Acquisition (DIA) was envisioned to overcome this limitation by continuously and methodically collecting MS2 spectra covering the entire MS1 spectrum so that for each m/z value information in the MS1 level and the MS2 level are available (Fig. 3F-J).[30] Current instruments are not fast enough to collect enough MS2 spectra with the typically ~1 Th (Dalton/elementary charge) isolation window. Therefore, wider windows are chosen to reduce the number of MS2 spectra needed to cover the total m/z range, and so the resulting MS2 spectra typically contain fragment ion series from multiple precursors (Fig. 3F). Another alternative is to simultaneously isolate multiple small MS2 windows.[31] This results in a very complex series of MS2 spectra which are more difficult to analyze than in DDA methods (Fig. 3H). The Aebersold group introduced an approach, known as SWATH-MS to analyze these complex spectra, using prior knowledge of peptides’ chromatographic and mass spectrometric behavior.[30b] This approach was recently reviewed by Ludwig et al., who described improvements to DIA and how SWATH-MS can be used to analyze both total cell lysates and protein samples enriched for post-translational modifications.[32] The recent drastic improvements of DIA measurements are mostly due to computational advances.[33] The major advantage of DIA is its coverage: every peptide is fragmented multiple times. Due to this, DIA does not have as severe of a missing value problem as label-free DDA approaches. DIA seems particularly attractive when comparing many samples. For quantification via DIA, either MS1 or MS2 spectra can be chosen, though MS2 quantification is predominantly used (Fig. 3I, J).[30b,30c] While currently DDA and DIA acquisition strategies are mutually exclusive, the rapid advance of instrument speed will likely result in the two different approaches merging.[23] Once the instrument is fast enough to continuously cover the entire precursor space with the small (~1 Th) windows commonly used for DDA strategies, the DDA and DIA methods may start to become identical in terms of window size and breadth of coverage.
2. Multiplexed Proteomics with Isobaric Labeling
The methods discussed so far have key limitations. Label-free quantification provides comparatively poor measurement precision. Additionally, missing values of peptides that are only identified in some samples are hard to interpret even qualitatively. While MS1-based isotope labeling offers exquisite quantification for more abundant peptides, it suffers from a lack of multiplexing capability, because, as the number of samples increases, so does the complexity of the MS1 spectrum. DIA mostly overcomes the missing value problem of a label-free approach but samples are still analyzed one at a time, limiting measurement precision and requiring lots of instrument time.
2.1. Principles of Quantitative Multiplexed Proteomics
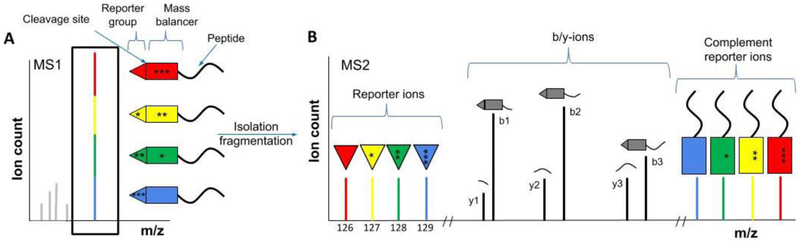
Multiplexed proteomics, based on isobaric mass tags, promises to overcome or at least mitigate these limitations.[34] The most commonly used isobaric tags are TMT[35] and iTRAQ[36], which are both commercially available, but there are other isobaric tags we will describe later. Isobaric tags are reagents used to covalently modify peptides, using the heavy isotope distribution in the tag to encode the different conditions, and are generally added after digestion. Unlike the isotopic labeling methods such as SILAC that were discussed earlier (Fig. 2D-F), each variant of an isobaric tag set has an identical total mass. The only difference is how the heavy isotopes are distributed among the tag, since each tag contains a site that fragments in the MS2 spectrum, resulting in reporter ions with different masses depending on which sample the peptide originated from. In addition to the reactive group, which reacts with the peptide, each tag contains a reporter group with differential number of heavy isotopes. To keep the total mass of the tag constant, the number of heavy isotopes on the mass balancer group is adjusted accordingly (Fig. 4A). Identical peptides from different samples elute at the same time and therefore appear as a single peak in the MS1 spectrum. This is a major advantage, since the complexity of the MS1 spectra does not increase significantly with the number of samples. Complexity only increases with the addition of new proteins in the new samples. This is in contrast to SILAC-like experiments, where even comparing replicates will double the number of peaks in the MS1 spectra. Therefore, the number of conditions that can be compared with isobaric tag experiments in a single experiment is higher (currently up to 11) than with SILAC-like methods. Quantification occurs after isolation and fragmentation of these labeled peptides in the MS2 spectrum (Fig. 4B). Usually, the amount of energy added for fragmentation is only enough for one bond to break. This could either be a peptide bond on the backbone or the intended breakage point in the isobaric tag. Each tag has several heavy isotopes that are distributed differently relative to this fragile bond. Upon breakage, the isobaric tag produces low m/z reporter ions that contain different masses depending on the condition they come from and can therefore be used for relative quantification (Fig. 4B). Additionally, the breakage of the isobaric tag leads to the formation of complementary reporter ions, which contain the balancing part of the isobaric tag and the intact peptide{Wuhr, 2012 #8} or fragment ions, which result from breakage in the peptide backbone and the isobaric tag.[37] The balancing group of the isobaric tag also encodes the experimental conditions and the complementary reporter ions can therefore be similarly used for quantification (Fig. 4B).[38] We will discuss later the key advantages and disadvantages of the utilization of the different reporter ions for quantification. The complementary reporter ions were noticed, e.g., by the Mechtler group, but were not initially used for quantification.[39]. Instead, they removed these peaks to increase peptide identification success-rate. Using the complementary reporter ions for quantification is similar to an approach by Yan et al., who labeled peptides differentially on their N- and C-termini with heavy isotopes to generate isobaric peptides. The authors used the fragment ions for quantification.[40]
Figure 4: Outline of multiplexed proteomics with isobaric tags.
A) Isobaric tags have the same total mass, but differing distributions of heavy isotopes between the reporter group and mass balancer. Heavy isotopes are shown as asterisks. Peptides from 4 different samples are labeled with tags of the same mass, resulting in a single MS1 peak which can be isolated. With more tags (conditions), the complexity of the MS1 spectrum does not increase. This makes isobaric tags compatible with higher multiplexing (currently up to 11) compared to e.g., SILAC (see Fig. 2). B) After a peptide is isolated based on the MS1 spectrum, fragmentation will either cleave off the reporter ions, or lead to fragmentation of the peptide backbone. The reporter ions show different masses in the MS2 spectrum and can be used for relative quantification. Similarly, the intact peptide with the balancing groups, i.e., the complementary reporter ions, can also be used for quantification. The b- and y-ions are used for peptide identification (see Fig. 1).
Although the labeling step after protein digestion could introduce some variability, and though there is a limit to the number of samples that can be labeled by an isobaric tag system, there are many advantages of isobaric tags that make up for these. The ability to analyze many samples at once mostly circumvents the missing value problem. If no signal is detected for a peptide in a particular condition it can be inferred that the peptide is indeed much less abundant than it is in the other conditions. It is still possible for peptides to be excluded from MS2 fragmentation, but since all samples have peptides eluting under the same peak, all labeled versions of the same peptide with either be isolated together or none at all. Another major advantage is the inherent high reproducibility between samples due to the samples being combined after labeling and co-analyzed. Compared to MS1 quantification methods like SILAC, data quality is even further improved because each analysis heavily enriches the peptides of interest in the MS2 spectrum, where quantification occurs, resulting in peptide ion statistics even for low-abundance peptides.[41] Multiplexed proteomics therefore demonstrates very high reproducibility with CVs of ~5% and very few peptides with CVs above 10%.[42] The last major advantage, at least compared to label-free quantification, is that throughput is markedly increased, since all samples can be shot in one run. This limits expense and makes it compatible with the analysis of pre-fractionated samples[43]. For equivalent amounts of machine time, this results in significantly more proteins that can be quantified compared to label-free approaches.[42,44] Together, these benefits make multiplexed proteomics a very attractive option for relative quantification.
2.2. Main Problem of Multiplexed Proteomics: Interference/Ratio Distortion
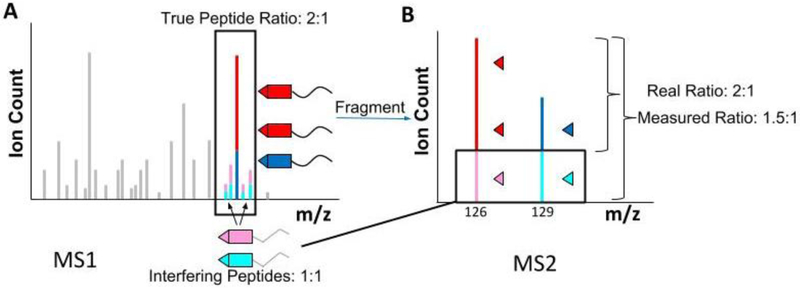
In the previous section we discussed the principles and promises of multiplexed proteomics. However, in its initial implementation, the method came with a major measurement artifact, ratio distortion. In Figure 4, we pretended that it is possible to specifically isolate one peptide of interest. For technical reasons, however, the smallest possible isolation window currently achievable with a mass filter (e.g., a quadrupole) is approximately 0.5 Th.[45] In complex mixtures, like tryptic digests derived from cell lysates, whenever a peptide is isolated in the MS1 spectrum for MS2 analysis, other peptides with similar m/z values will nearly always be co-isolated (Fig. 5A).[46] Since both the target peptide and the contaminating peptides carry the same reporter groups, after MS2 fragmentation the reporter ion signal for that particular isolation will be a combination of reporter ions stemming from the peptide of interest and from all other contaminating peptide ions (Fig. 5B). Nearly all measurements are therefore distorted, often to a significant extent. In general, these contaminating ions tend to bias the relative ratios between different conditions towards a 1:1 ratio.[46c] This distortion tends to be more significant for low-abundance peptides, where the interfering signal is relatively greater. However, it is also possible that a 1:1 peptide is distorted by a changing contaminant resulting in unsubstantiated measurements of changes.[47] Despite these problems, many groups successfully use multiplexed methods which are vulnerable to interference.[48] For some studies, the qualitative knowledge on which proteins change are sufficient. However, if one is interested in the quantitative change of protein expression levels, addressing interference is essential.[46b,46c,49] Recently, multiple statistical methods have been suggested to bioinformatically correct for this distortion.[50] Nevertheless, the best sample quantification can currently be obtained by applying experimental remedies for this major problem, which we will discuss in the following section.
Figure 5: The problem of multiplexed proteomics: ratio distortion.
A) Even when using the smallest technically possible isolation window centered on a peptide of interest (red and dark blue), in a real experiment, other peptides with similar m/z and retention time will be co-isolated (pink and light blue). These interfering peptides will also be labeled with identical isobaric tags. B) Upon co-isolation and co-fragmentation, in the MS2 spectrum the low m/z reporter ions are identical, regardless of origin, and distort the quantification. Most background peptides tend to not change, showing a 1:1 ratio between control and experiment. The observed ratio for a peptide of interest, which changes 2-fold between control and experiment, will typically be compressed towards a 1:1 ratio.
2.3. Overcoming Interference with further gas-phase purification (Quantmode, MS3)
One of the earliest methods to reduce interference from contaminating ions is an approach known as QuantMode.[46b] This method reduces the charge of all peptides by one. After isolation of the new desired m/z window, interfering peptides with similar mass but different charge than the targeted peptide are removed. QuantMode was thus able to significantly reduce interfering ions, resulting in more accurate quantification. The main drawback of the method is that interfering ions of the same charge as the target ion can still be co-isolated. Additionally, the proton transfer process, which alters the ion charge, is comparatively slow, resulting in fewer collected spectra and a shallower assaying of the sample.
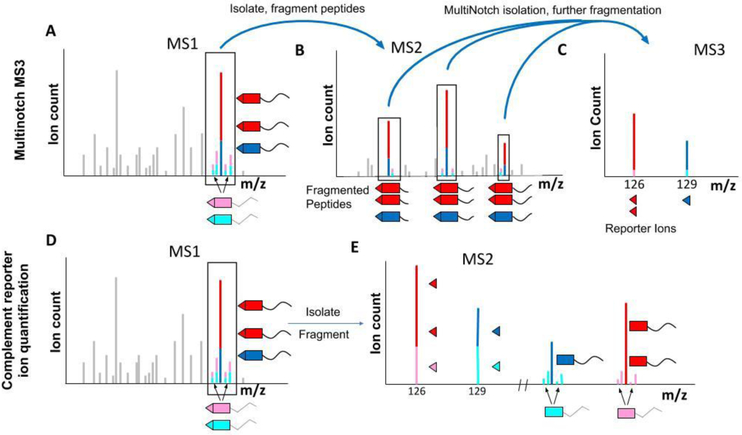
Currently, the most widely used approach to counteract ratio distortion involves a further fragmentation and isolation step to produce an MS3 spectrum.[46c] An MS3 spectrum results from the isolation of ions in the MS2 spectrum and their further fragmentation (Fig. 6A-C). This filters out the interfering peptides, allowing the target peptide to be quantified more accurately. The original version of the MS3 method only isolated a single isobaric-tag-labeled fragment ion from each MS2 spectrum, which greatly reduced the sensitivity of the quantification.[46c] This drawback was overcome by a more advanced method called MultiNotch MS3.[47] The use of isolation waveforms with multiple frequency notches enables the simultaneous precursor selection (SPS) in the linear ion trap.[51]
Figure 6: Strategies to overcome ratio distortion.
A-C) Overview of the MultiNotch MS3 approach A) The MultiNotch MS3 method acquires an MS2 spectrum similar to the standard approach by isolating a target peptide (red and dark blue), along with interfering peptides (pink and light blue). This spectrum is used for peptide identification. B) Instead of quantifying the reporter ions in the MS2 spectrum, the highest abundant peaks, which typically are b- and y-ions from the peptide of interest, are simultaneously isolated and further fragmented for an MS3 spectrum. C) Reporter ions in the MS3 spectrum are used for quantification. The additional gas-phase purification typically leads to removal of most interfering signal. While not perfect, the measured ratios are typically significantly more accurate than with a standard MS2 approach. D-E) Overview of the complementary reporter ion quantification strategy. D) For the complementary reporter ion method, a standard MS2 spectrum is acquired, which will co-isolate and co-fragment the peptide of interest and interfering species. E) The low m/z reporter ions show interference, as discussed in Figure 5. However, this method involves analyzing the complement reporter ions in the MS2 spectrum, where the peptide is still attached to the mass balancer group. Since the target peptide and interfering peptides typically have slightly different masses, this allows them to be distinguished with a high resolution mass analyzer like an Orbitrap. The results are significantly more accurate quantification compared to MS2 and even the MS3 approach. While interference still will occasionally lead to ratio distortion, to our knowledge this method currently generates the most accurate data.
With this approach multiple fragment ions from each MS2 spectrum are simultaneously isolated, resulting in greater sensitivity (Fig. 6A-C). Thermo Fisher Scientific commercialized this approach on the Orbitrap Fusion and Lumos mass spectrometers. As a result, MultiNotch MS3 is now widely used and currently considered state-of-the-art, able to detect ~10% changes in protein abundance with high confidence.[52]
Despite this, there are a number of limitations to the MS3 approach. Perhaps the most significant disadvantage is the requirement for additional MS scans. This results in a loss of ions, and comparatively slow cycle times. Furthermore, the MS3-based methods require instrumentation that is more complex and expensive. Finally, even MultiNotch MS3 fails to completely remove interference, especially for peptides with low abundance, since interfering ions in the MS2 spectrum are still co-isolated into the MS3 spectrum (Fig. 6C).[42, 52a]. It is likely that MultiNotch MS3 data-quality can be further improved by setting the notches in a peptide specific manner. For shotgun approaches, this would require the ability to identify MS2 spectra immediately after their acquisition and before the corresponding MS3 scan. [53]
Another approach to reduce interference is ion mobility spectrometry. [28b, 54] This separation method, which is orthogonal to the LC and the m/z analyzers, promises to reduce ratio distortion, since interfering ions will be separated from the peptide of interest.
2.4. Overcoming Interference with the Complement Reporter Ion-Based Approach (TMTc).
An alternative method to overcome the ratio distortion problem is based on the complementary fragment ions in the MS2 spectrum.[38] When an isobaric tag (e.g., TMT) breaks it produces a low m/z reporter ions but also the intact peptide with the balancing group of the isobaric tag still attached (Fig. 4B). Due to their complementary nature, these were named complementary reporter ions, or TMTc ions when the experiment is performed with TMT tags. Although TMTc-based methods have not been widely used outside of our lab, we think that they provide a viable alternative to the more prevalent MS3-based methods.
TMTc ions containing the same peptide differ in mass depending on the experimental condition, just as the low m/z reporter ions do. These TMTc ions can therefore also be used for multiplexed quantification (Figure 6D-E). The key advantage of using TMTc over low m/z reporter ions for quantification is that any interfering peptides typically will have slightly different masses compared to the target peptide. The ability to distinguish different TMTc masses in the Orbitrap is ~100-fold higher than the lowest feasible resolving power of the quadrupole ion isolation. TMTc is therefore much more robust to interfering ions than the standard MS2 approach. TMTc is even able to outperform MS3-based methods in terms of measurement accuracy.{Sonnett, 2018 #129} Furthermore, compared to Quantmode or MultiNotch MS3, TMTc does not require an additional fragmentation step, saving time and in principle increasing sensitivity. Because no higher order scans are required, the complement reporter ion approach can be performed on comparatively simple instruments like quadrupole Orbitraps or QTOFs.
Figure 6 heavily simplifies the actual picture by portraying a single peak corresponding to each peptide. In reality, peptides elute as an isotopic envelope of multiple peaks, spaced apart by 1 Th due to the natural frequency of 13C, 15N, 18O, and other heavy isotopes, in biological molecules. When the entire isotopic envelope of a peptide is isolated, the complement reporter ion cluster has to be deconvolved from the isotopic envelope of the precursor peptide.[38] This deconvolution process results in a loss of quantitative precision. In order to combat this shortcoming, we have developed a refinement of the TMTc deconvolution method known as TMTc+.[42] The TMTc+ method uses a narrow isolation window. In the extreme case, where only one precursor peak is chosen, the deconvolution becomes similar to the simplified cases represented in Figure 6, and only isotopic impurities have to be accounted for. The resulting data comes with a drastic improvement of measurement precision while still preserving the superb measurement accuracy.
Despite the promises of the complement reporter ion approach, several key limitations remain: At high m/z values, mass spectrometers cannot distinguish the extra neutron in heavy nitrogen or carbon in commercial isobaric tags with currently feasible resolving power. Also, with MS2 or MS3 approaches, up to 11 conditions can currently be compared, but only 5 TMT channels are currently distinguishable with the complement reporter ion strategy (Fig. S1). This lowered multiplexing capacity is a major drawback to the TMTc method, but future isobaric tags should be able to address this limitation. Furthermore, emerging super-resolution approaches are, at least in principle, able to further increase multiplexing capacity by providing the resolving power to distinguish the extra neutron in different elements even at high m/z regions.[55] Another major hurdle is the poor formation of the complement reporter ions. Commercially available tags were not intended for this purpose and the complement reporter ions form inefficiently. Recently, two tags (the SO-tag and EASI-tag, further discussed below)[37,56] were designed specifically for the formation of the complement reporter ions. But while the complement forms efficiently, they come with the drawback of making identification of peptides difficult, since breakages of both the tag and the peptide backbone can occur, leading to many additional peaks, which are not classical b- or y-ions and are not recognized by standard search algorithms. The advantageous combination of measurement sensitivity, precision, and accuracy of TMTc+ make it our current method of choice for most experiments in our laboratory. Nevertheless, many shortcomings remain, and considerable extra effort will be required to exploit the method’s full potential.
2.5. Overview of different Isobaric Tags
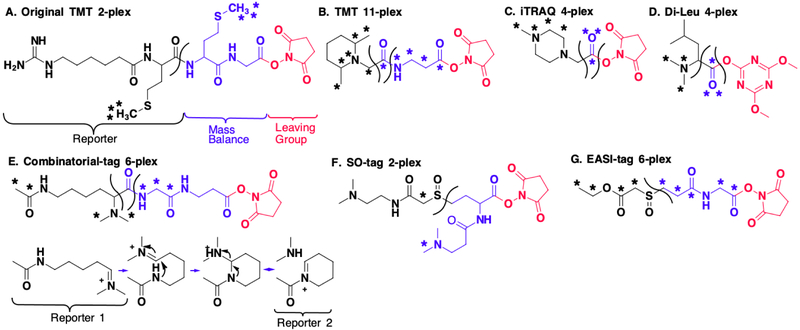
All isobaric tags contain a functional group that enables covalent attachment to peptides. Typically, this group reacts with primary amines on a peptide’s N-terminus or lysine side-chains, as is the case here. However, some tags react with carbonyl or sulfhydryl groups.[57] In addition, all isobaric tags contain a reporter group and a mass balancer group (Fig. 7A). The total number of heavy isotopes in the tag is constant, making them isobaric in the MS1 spectrum. However, the distribution of the heavy isotopes between the reporter and balancing group differs for the different conditions (Fig. S1).
Figure 7: Overview of isobaric tags.
In each structure, asterisks denote heavy isotopes. The black part of the structure indicates the reporter part, the balancing group is blue, and the leaving group, which is removed after the tag reacts with the peptides, is red A) The original 2-plex TMT from Thompson et al. B) Current commercial TMT, which can encode up to 11 different conditions (See Fig. S1 for heavy isotope distribution). C) In the iTRAQ structure, the marked oxygen can be either 16O or 18O. D) The DiLeu-tag is a 4-plex tag developed by Xiang et. al. E) Braun et al. developed combinatorial tags. These tags generate multiple reporter ions, which allows for high multiplexing capacity for a given number of heavy isotopes. After fragmentation at the shown cleavage site reporter 1 forms. However, this further fragments into reporter 2 and a neutral loss. F) Stadlmeier et al. developed the sulfoxide-based tag, which is optimized for complement reporter ion formation due to fragmentation of the sulfoxide bond at lower energies. The two tertiary amines result in higher charge states of peptides after ionization and further facilitate fragmentation. G) The EASI-tag developed by Winter et al. similarly fragments comparatively easily. The “reporter” part of the EASI-tag is a neutral loss. Therefore, quantification with the EASI-tag is only possible via the complement reporter ions.
Isobaric tags were first introduced by Thompson et al. for the relative quantification of peptides.[34] The original tag was a 2-plex called Tandem Mass Tag (TMT) (Fig. 7A). While this original TMT tag was used to prove an important new concept, the structure was comparatively bulky, which led to additional unintentional fragmentation patterns, poor ionization properties, and poor identification success rates. Thompson and colleagues further developed the tandem mass tag into the significantly smaller version currently available commercially from Pierce (Fig. 7B).[35] This TMT tag is currently able to encode up to 11 different conditions (Fig. S1). To obtain this high multiplexing capacity, a mass analyzer is required that can distinguish between the additional neutron masses in 13C versus 14N, which differ by 6 mDa (Fig. S1).[58]
An alternative commercially available tag from AB Sciex is the isobaric Tag for Relative and Absolute Quantification (iTRAQ) (Fig. 7C). The structure, shown in Figure 7C, can encode up to four different conditions.[36] An 8-plex iTRAQ is also commercially available, but to our knowledge, its structure has not been published.[59] Pichler et al. have found that 4-plex iTRAQ has a higher peptide identification rate than 8-plex iTRAQ or 6-plex TMT.[39] The authors conclude that the 8-plex iTRAQ may suffer due to the appearance of fragment ions from the larger tag in the MS2 spectrum, which they suggest hinders peptide identification. This indicates that isobaric tags should be designed to be as small as possible while allowing sufficient multiplexing capacity.
An alternative to the commercial tags are the N,N-Dimethyl Leucine (DiLeu) tags (Fig. 7D).[60] These tags contain a reporter group consisting of a dimethyl amine connected to a leucine side chain, and a mass balancing group consisting of the CO-atoms of the carboxyl group. These DiLeu tags were originally 4-plex. Using deuterium isotopes as labels, Frost et al. reported an upgraded version of DiLeu that increased its multiplexing capacity to 12.[61] Nevertheless, deuterium-labeled peptides typically show different elution profiles compared to unlabeled peptides. For MS1 based quantification this can be acceptable, as the entire elution profile can be integrated (Fig. 2F).[24] However, for multiplexed proteomics typically only one MS2/MS3 spectrum is acquired per peptide. Differential elution profiles in different channels could therefore lead to serious quantification artifacts.
A clever set of isobaric tags was showcased by Braun et. al. (Fig 7E).[62] Known as Combinatorial isobaric Mass Tags (CMT), the fragmentation of these molecules results in multiple reporter ions. Because of this, their multiplexing capacities are larger compared to conventional tags of comparable size and number of heavy isotopes. Nevertheless, the quantification depends on a deconvolution approach, which comes at the cost of measurement precision, particularly if interfering ions alter the true peptide ratios. The CMT publication demonstrates a 6-plex version by using two different reporter ions. Furthermore, by taking into account a 3rd reporter ion that was also detected, the chemical structure of the tag allows for 28-plex tags when utilizing the 6 mDa spacing between heavy carbon and nitrogen. These high values are achieved with just 5 heavy isotopes on each tag.
Motivated by the inefficient formation of the complementary reporter ions (Fig. 6B) from the commercial isobaric tags, Stadlmeier et al. developed a sulfoxise-based tag, known as the SulfOxide Tag (SOT) (Fig. 7F).[37] In this tag, the reporter and balancer groups are linked by a sulfoxide group. This allows the tag to be fragmented at low energies, increasing the yield of complementary reporter ions. Indeed, the SO tag is much more favorable to fragmentation compared to the peptide backbone, and so typically plenty of signal is available for quantification. An interesting idea put forward with this paper was quantification using the complementary b- and y-ions. These are the fragment ions that develop when both the isobaric tag and the peptide backbone break. Like the complementary reporter ions that result from only the breakage of the isobaric tag, these ions also encode the different sample conditions.
Another sulfoxide-based was recently developed by Virreira Winter et al., known as EASI-tag (Easily Abstractable Sulfoxide-based Isobaric tag) (Fig. 7G).[56] The EASI-tag also contains an asymmetric sulfoxide bond that is cleaved at relatively low energy. The authors used a shifted isolation window of 0.4 Th to isolate a single MS1 peak in the isotopic envelope, making deconvolution easier when the pseudo-monoisotopic peak is isolated. Another interesting novelty of the EASI-tag is that the low m/z reporter ion equivalent is a neutral loss, which makes quantification only possible via the complement reporter ions.
Both sulfoxide-based tags, SOT and EASI-tag, seem to suffer from comparatively poor success-rates in identifying peptide spectra (Fig. 1D). This is because the tags fragment much easier than the peptide backbone. Typically, the b- and y-ions additionally lose the low m/z reporter part of the isobaric tag, resulting in peaks that standard search algorithms will not consider for identification. Improved search algorithms that consider these ions might mitigate this problem. However, the spectra are much more complex, and it is not clear how much adapted search algorithms will be able to overcome this major limitation, which results in comparatively few quantified peptides and ultimately proteins. We believe the most promising way forward for complement reporter ion quantification is the development of new chemical structures that balance the formation of complement reporter ions with the ability to reliably and efficiently identify peptides.
3. Emerging Multiplexed Proteomics Technologies
Multiplexed proteomics in its current form is highly attractive and well suited for many studies. However, significant shortcomings remain. Among them are the difficulties in detecting low-abundance proteins. These are often some of the most interesting proteins, such as transcription factors or signaling molecules. To overcome these limitations, we discuss the emerging fusion of targeted proteomics with multiplexing technologies to reliably reach low-abundance proteins. Another major limitation of multiplexed proteomics is the maximal multiplexing capacity. The current limit, with TMT tags, is 11-plex. But for many studies, it is desirable to compare hundreds or even thousands of different samples. In principle, these can be split into several 11-plex experiments, but then similar to label-free approaches, some peptides will only be analyzed in a subset of the experiments. The interpretation of these missing values is difficult. Additionally, quantification between 11-plexes is challenging. Here, we suggest the fusion of the complement reporter ion quantification strategy with DIA approaches to enable the comparison of hundreds of samples with few missing values and high measurement quality.
3.1. Targeted Multiplexed Proteomics
So far, the methods we described involve analyzing protein samples globally, with the aim of identifying and quantifying as many proteins as possible. However, it is also possible to sacrifice global coverage and focus the limited ion injection times on peptides of a few (~100) proteins of particular interest. Such approaches are called targeted proteomics.[63] By predefining the data acquisition towards specific ions that elute at specific times, targeting, at least in principle, enables detection and quantification of much less abundant peptides that could otherwise be overlooked. Although this approach requires a significant amount of setup, it can be used to analyze low-abundance peptides which would be missed by a shotgun approach.
The simple combination of targeted proteomics with an isobaric MS2 approach is not attractive. The interference problem is especially problematic for low-abundance peptides and quantification would be very unreliable and likely severely distorted (Fig. 5). However, the reduction of ratio distortion using MS3-based methods made targeted multiplexing of proteolyzed cell lysates more feasible (Fig 6A). As a result, Erickson et al. developed a targeted multiplexing method known as TOMAHAQ.[64] In this method, samples were spiked with trigger peptides labeled with TMT0, which is the standard TMT structure without any heavy isotopes. This resulted in the trigger peptides eluting simultaneously at a known m/z offset away from the target sample peptides in the MS1 spectrum, which were labeled with standard TMT 10-plex labels. The spiked-in trigger peptides were sufficiently abundant to be consistently observed in the MS1 scan. The instrument was programmed to isolate and fragment the sample peptides at the known mass offset, even if there were no detectable peaks from the target peptides at that m/z. The notches for the MS3 scan were preprogrammed to fit the peptide of interest and to be specific for fragment ions containing an intact TMT tag. Furthermore, this approach was refined by selecting in real time from the MS2 spectra only those b- and y-ions that had minimal interfering ions. This allowed the researchers to obtain accurate quantifications of even dilute peptides, which suffer from significant interference even in the standard SPS-MS3 method.
While this MS3-based method has proved critical to targeted multiplexed proteomics, the complement reporter ion approach is, in our opinion, particularly attractive for this purpose (Fig. 6B). The complement reporter ion strategy is superbly able to distinguish signal from interfering background noise, and the lack of an additional gas-phase isolation makes it, at least in principle, more sensitive. Thus, we believe that for a targeted approach, the use of complementary b- and y-ions could be particularly attractive as this would provide an additional layer of distinction (besides the precursor mass) and allow the separation of signal even for isobaric peptides with nearly identical elution times.
3.2. Fusing Multiplexed Proteomics with Data-Independent Acquisition
Currently, multiplexed proteomics is very attractive when comparing up to 11 samples, the maximal number that can be analyzed in a single experiment. However, many studies require the comparison of hundreds or even thousands of different conditions. In such scenarios, multiplexed experiments suffer from the missing value problem, similar to label-free approaches. Additionally, it is hard to compare proteins quantified in one 11-plex with another 11-plex. Often, so-called bridging channels are used, which are analyzed in all experiments.[65] However, it is possible that these do not contain a subset of proteins or contain it at a significantly different concentration than the other analyzed samples. In such cases, the quantification between two 11-plexes relies on the ratios of unreliable ratios. Currently, the best remedy for quantitative comparison of many samples might be DIA approaches. However, these come with comparatively low measurement precision and inefficient instrument time usage. A fusion of DIA with isobaric labeling approaches might be able to combine the advantages from both methods.
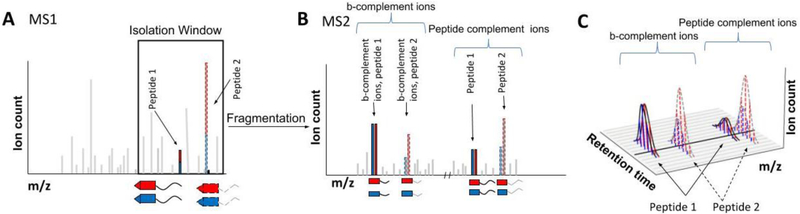
Since DIA involves fragmenting all the MS1 ions in a certain mass range (Fig. 3F-G), it is incompatible with MS2 or MS3 methods using the low m/z reporter ions (Fig. 5, 6A-C). However, for multiplexed methods based on the complement reporter ion approach the reporter ion signal is precursor specific (Fig. 6D-E). Complement ions from different peptides would have different masses and would be distinguishable (Fig. 8). In a proof of principle, we have already demonstrated the quantification of two different peptides in one spectrum.[38] Particularly attractive might be the use of b- or y-ions that additionally have a broken isobaric tag, thereby forming complementary fragment ions which can also be used for quantification.[37] Regardless, several hurdles have to be overcome to make this approach feasible. Simultaneously fragmenting all of the precursor ions within an m/z range will give a very complex MS2 spectrum, both due to the large number of isolated peptide species and due to the differing masses of the complement reporter ions, which will likely make the analysis quite challenging. Although interference will lead to some ratio distortion, the combination of multiple quantification events over multiple spectra might provide enough data to overcome such challenges. Additionally, the very wide isolation windows will require deconvolution of the isotopic envelopes. Isobaric tags with multiple Dalton spacing might make this approach more feasible with high measurement precision. Despite these challenges, a successful fusion of DIA with multiplexing could be a highly attractive method.
Figure 8: Proposed fusion of DIA with multiplexed proteomics.
A) Peptides would be labeled with isobaric tags similar to a normal multiplexing experiment. For sake of simplicity, we only show two conditions. Like in a normal DIA workflow, all the peaks within a certain wide m/z window in an MS1 scan are co-isolated. This window contains multiple peptides, which will all be simultaneously isolated and fragmented into an MS2 spectrum. For simplicity, only two peptides are shown in detail, depicted with solid and dashed outlines. B) In the MS2 spectrum, the low m/z reporter ions cannot be used for quantification since the reporter ions of all co-isolated peptides will be identical. However, simultaneous quantification is possible via the peptide complement reporter ions. Additionally, complementary b- and y-ions that additionally lost their reporter group can also be used for peptide specific quantification. C) The continuous monitoring of peptide complement reporter ions and b- and y-fragment complement reporter ions allow the relative quantification of multiplexed abundances even between various runs. Additionally, the number of missing values in samples larger than the multiplexing capacity of a single tag should be drastically reduced.
4. Conclusion
Multiplexed proteomics in its current form is highly attractive and often the best suited quantitative proteomics option for many studies. The higher throughput enabled by multiplexing has made it possible to analyze hundreds of samples with reasonable depth [66]. Over the last several years, remarkable technological progress has been made, particularly in addressing ratio distortion, the major shortcoming of multiplexed proteomics. Currently, isobaric tag-based multiplexed proteomics can accurately, precisely, and sensitively quantify thousands of proteins simultaneously across up to 11 samples. With the resulting data, changes of less than ~10% can be detected with high confidence. Despite these advantages, major limitations remain. One major remaining hurdle is the reliable quantification of low-abundance proteins. Emerging methods for targeted multiplexing promise to overcome the problem of quantifying low-abundance proteins across multiple conditions. Another major remaining challenge is how to quantify protein abundances among hundreds of samples while limiting missing values. In this review, we suggested that the fusion of DIA with the complement reporter ion approach might be able to unite the best of both worlds.
Supplementary Material
Acknowledgements
We would like to thank members of the Wühr Lab for helpful suggestions and discussions. We thank Michael Stadlmeier for comments on the manuscript. N.P. was supported by NIH grant T32GM007388. This work was supported by NIH grant 1R35GM128813 and the U.S. Department of Energy, Office of Science, Office of Biological and Environmental Research under award numbers DE-SC0018420 and DE-SC0018260.
References
- [1] a.Catherman AD, Skinner OS, Kelleher NL, Biochem Biophys Res Commun 2014, 445, 683–693; [DOI] [PMC free article] [PubMed] [Google Scholar]; b Toby TK, Fornelli L, Kelleher NL, Annu Rev Anal Chem (Palo Alto Calif) 2016, 9, 499–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Ardrey RE, Liquid chromatography-mass spectrometry: an introduction, J. Wiley, New York, 2003. [Google Scholar]
- [3].Gundry RL, White MY, Murray CI, Kane LA, Fu Q, Stanley BA, Van Eyk JE, Curr Protoc Mol Biol 2009, Chapter 10, Unit10 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Olsen JV, Ong SE, Mann M, Mol Cell Proteomics 2004, 3, 608–614. [DOI] [PubMed] [Google Scholar]
- [5] a.Manadas B, Mendes VM, English J, Dunn MJ, Expert Rev Proteomics 2010, 7, 655–663; [DOI] [PubMed] [Google Scholar]; b Lee HJ, Kim HJ, Liebler DC, J Proteome Res 2016, 15, 2346–2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Fenn JB, Mann M, Meng CK, Wong SF, Whitehouse CM, Science 1989, 246, 64–71. [DOI] [PubMed] [Google Scholar]
- [7].Muntel J, Boswell SA, Tang S, Ahmed S, Wapinski I, Foley G, Steen H, Springer M, Mol Cell Proteomics 2015, 14, 430–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Chelius D, Bondarenko PV, J Proteome Res 2002, 1, 317–323. [DOI] [PubMed] [Google Scholar]
- [9].Hunt DF, Yates JR 3rd, Shabanowitz J, Winston S, Hauer CR, Proc Natl Acad Sci U S A 1986, 83, 6233–6237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Roepstorff P, Fohlman J, Biomed Mass Spectrom 1984, 11, 601. [DOI] [PubMed] [Google Scholar]
- [11].Eng JK, McCormack AL, Yates JR, J Am Soc Mass Spectrom 1994, 5, 976–989. [DOI] [PubMed] [Google Scholar]
- [12] a.Perkins DN, Pappin DJ, Creasy DM, Cottrell JS, Electrophoresis 1999, 20, 3551–3567; [DOI] [PubMed] [Google Scholar]; b Cox J, Neuhauser N, Michalski A, Scheltema RA, Olsen JV, Mann M, J Proteome Res 2011, 10, 1794–1805; [DOI] [PubMed] [Google Scholar]; c Eng JK, Jahan TA, Hoopmann MR, Proteomics 2013, 13, 22–24. [DOI] [PubMed] [Google Scholar]
- [13].Nesvizhskii AI, Keller A, Kolker E, Aebersold R, Anal Chem 2003, 75, 4646–4658. [DOI] [PubMed] [Google Scholar]
- [14] a.Bantscheff M, Schirle M, Sweetman G, Rick J, Kuster B, Anal Bioanal Chem 2007, 389, 1017–1031; [DOI] [PubMed] [Google Scholar]; b Bantscheff M, Lemeer S, Savitski MM, Kuster B, Anal Bioanal Chem 2012, 404, 939–965. [DOI] [PubMed] [Google Scholar]
- [15] a.Bondarenko PV, Chelius D, Shaler TA, Anal Chem 2002, 74, 4741–4749; [DOI] [PubMed] [Google Scholar]; b Wang W, Zhou H, Lin H, Roy S, Shaler TA, Hill LR, Norton S, Kumar P, Anderle M, Becker CH, Anal Chem 2003, 75, 4818–4826. [DOI] [PubMed] [Google Scholar]
- [16].Gerber SA, Rush J, Stemman O, Kirschner MW, Gygi SP, Proc Natl Acad Sci U S A 2003, 100, 6940–6945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17] a.Ludwig C, Claassen M, Schmidt A, Aebersold R, Mol Cell Proteomics 2012, 11, M111 013987; [DOI] [PMC free article] [PubMed] [Google Scholar]; b Wuhr M, Freeman RM Jr., Presler M, Horb ME, Peshkin L, Gygi S, Kirschner MW, Curr Biol 2014, 24, 1467–1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18] a.Ong SE, Mann M, Nat Chem Biol 2005, 1, 252–262; [DOI] [PubMed] [Google Scholar]; b Andersen JS, Wilkinson CJ, Mayor T, Mortensen P, Nigg EA, Mann M, Nature 2003, 426, 570–574. [DOI] [PubMed] [Google Scholar]
- [19].Cox J, Hein MY, Luber CA, Paron I, Nagaraj N, Mann M, Mol Cell Proteomics 2014, 13, 2513–2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20] a.Hultin-Rosenberg L, Forshed J, Branca RM, Lehtio J, Johansson HJ, Mol Cell Proteomics 2013, 12, 2021–2031; [DOI] [PMC free article] [PubMed] [Google Scholar]; b Carrillo B, Yanofsky C, Laboissiere S, Nadon R, Kearney RE, Bioinformatics 2010, 26, 98–103. [DOI] [PubMed] [Google Scholar]
- [21].Webb-Robertson BJ, Wiberg HK, Matzke MM, Brown JN, Wang J, McDermott JE, Smith RD, Rodland KD, Metz TO, Pounds JG, Waters KM, J Proteome Res 2015, 14, 1993–2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22] a.Lazar C, Gatto L, Ferro M, Bruley C, Burger T, J Proteome Res 2016, 15, 1116–1125; [DOI] [PubMed] [Google Scholar]; b Wei R, Wang J, Su M, Jia E, Chen S, Chen T, Ni Y, Sci Rep 2018, 8, 663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23] a.Meier F, Brunner AD, Koch S, Koch H, Lubeck M, Krause M, Goedecke N, Decker J, Kosinski T, Park MA, Bache N, Hoerning O, Cox J, Rather O, Mann M, Mol Cell Proteomics 2018, 17, 2534–2545; [DOI] [PMC free article] [PubMed] [Google Scholar]; b Espadas G, Borras E, Chiva C, Sabido E, Proteomics 2017, 17. [DOI] [PubMed] [Google Scholar]
- [24].Ong SE, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A, Mann M, Mol Cell Proteomics 2002, 1, 376–386. [DOI] [PubMed] [Google Scholar]
- [25] a.Gygi SP, Rist B, Gerber SA, Turecek F, Gelb MH, Aebersold R, Nat Biotechnol 1999, 17, 994–999; [DOI] [PubMed] [Google Scholar]; b Tolonen AC, Haas W, J Vis Exp 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Li Z, Adams RM, Chourey K, Hurst GB, Hettich RL, Pan C, J Proteome Res 2012, 11, 1582–1590. [DOI] [PubMed] [Google Scholar]
- [27] a.Merrill AE, Hebert AS, MacGilvray ME, Rose CM, Bailey DJ, Bradley JC, Wood WW, El Masri M, Westphall MS, Gasch AP, Coon JJ, Mol Cell Proteomics 2014, 13, 2503–2512; [DOI] [PMC free article] [PubMed] [Google Scholar]; b Overmyer KA, Tyanova S, Hebert AS, Westphall MS, Cox J, Coon JJ, Nat Protoc 2018, 13, 293–306; [DOI] [PMC free article] [PubMed] [Google Scholar]; c Hebert AS, Merrill AE, Bailey DJ, Still AJ, Westphall MS, Strieter ER, Pagliarini DJ, Coon JJ, Nat Methods 2013, 10, 332–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28] a.Yi EC, Marelli M, Lee H, Purvine SO, Aebersold R, Aitchison JD, Goodlett DR, Electrophoresis 2002, 23, 3205–3216; [DOI] [PubMed] [Google Scholar]; b Pfammatter S, Bonneil E, McManus FP, Prasad S, Bailey DJ, Belford M, Dunyach JJ, Thibault P, Mol Cell Proteomics 2018, 17, 2051–2067; [DOI] [PMC free article] [PubMed] [Google Scholar]; c Meier F, Geyer PE, Virreira Winter S, Cox J, Mann M, Nat Methods 2018, 15, 440–448. [DOI] [PubMed] [Google Scholar]
- [29].Stahl DC, Swiderek KM, Davis MT, Lee TD, J Am Soc Mass Spectrom 1996, 7, 532–540. [DOI] [PubMed] [Google Scholar]
- [30] a.Venable JD, Dong MQ, Wohlschlegel J, Dillin A, Yates JR, Nat Methods 2004, 1, 39–45; [DOI] [PubMed] [Google Scholar]; b Gillet LC, Navarro P, Tate S, Rost H, Selevsek N, Reiter L, Bonner R, Aebersold R, Mol Cell Proteomics 2012, 11, O111 016717; [DOI] [PMC free article] [PubMed] [Google Scholar]; c Rardin MJ, Schilling B, Cheng LY, MacLean BX, Sorensen DJ, Sahu AK, MacCoss MJ, Vitek O, Gibson BW, Mol Cell Proteomics 2015, 14, 2405–2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Egertson JD, Kuehn A, Merrihew GE, Bateman NW, MacLean BX, Ting YS, Canterbury JD, Marsh DM, Kellmann M, Zabrouskov V, Wu CC, MacCoss MJ, Nat Methods 2013, 10, 744–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Ludwig C, Gillet L, Rosenberger G, Amon S, Collins BC, Aebersold R, Mol Syst Biol 2018, 14, e8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33] a.Searle BC, Pino LK, Egertson JD, Ting YS, Lawrence RT, MacLean BX, Villen J, MacCoss MJ, Nat Commun 2018, 9, 5128; [DOI] [PMC free article] [PubMed] [Google Scholar]; b Tsou CC, Avtonomov D, Larsen B, Tucholska M, Choi H, Gingras AC, Nesvizhskii AI, Nat Methods 2015, 12, 258–264, 257 p following 264; [DOI] [PMC free article] [PubMed] [Google Scholar]; c Rost HL, Rosenberger G, Navarro P, Gillet L, Miladinovic SM, Schubert OT, Wolski W, Collins BC, Malmstrom J, Malmstrom L, Aebersold R, Nat Biotechnol 2014, 32, 219–223; [DOI] [PubMed] [Google Scholar]; d Bruderer R, Bernhardt OM, Gandhi T, Miladinovic SM, Cheng LY, Messner S, Ehrenberger T, Zanotelli V, Butscheid Y, Escher C, Vitek O, Rinner O, Reiter L, Mol Cell Proteomics 2015, 14, 1400–1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Thompson A, Schafer J, Kuhn K, Kienle S, Schwarz J, Schmidt G, Neumann T, Johnstone R, Mohammed AK, Hamon C, Anal Chem 2003, 75, 1895–1904. [DOI] [PubMed] [Google Scholar]
- [35].Dayon L, Hainard A, Licker V, Turck N, Kuhn K, Hochstrasser DF, Burkhard PR, Sanchez JC, Anal Chem 2008, 80, 2921–2931. [DOI] [PubMed] [Google Scholar]
- [36].Ross PL, Huang YN, Marchese JN, Williamson B, Parker K, Hattan S, Khainovski N, Pillai S, Dey S, Daniels S, Purkayastha S, Juhasz P, Martin S, Bartlet-Jones M, He F, Jacobson A, Pappin DJ, Mol Cell Proteomics 2004, 3, 1154–1169. [DOI] [PubMed] [Google Scholar]
- [37].Stadlmeier M, Bogena J, Wallner M, Wuhr M, Carell T, Angew Chem Int Ed Engl 2018, 57, 2958–2962. [DOI] [PubMed] [Google Scholar]
- [38].Wuhr M, Haas W, McAlister GC, Peshkin L, Rad R, Kirschner MW, Gygi SP, Anal Chem 2012, 84, 9214–9221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Pichler P, Kocher T, Holzmann J, Mazanek M, Taus T, Ammerer G, Mechtler K, Anal Chem 2010, 82, 6549–6558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Yan W, Luo J, Robinson M, Eng J, Aebersold R, Ranish J, Mol Cell Proteomics 2011, 10, M110 005611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Mertins P, Udeshi ND, Clauser KR, Mani DR, Patel J, Ong SE, Jaffe JD, Carr SA, Mol Cell Proteomics 2012, 11, M111 014423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Sonnett M, Yeung E, Wuhr M, Anal Chem 2018, 90, 5032–5039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43] a.Ruprecht B, Zecha J, Zolg DP, Kuster B, Methods Mol Biol 2017, 1550, 83–98; [DOI] [PubMed] [Google Scholar]; b Edwards A, Haas W, Methods Mol Biol 2016, 1394, 1–13. [DOI] [PubMed] [Google Scholar]
- [44].O'Connell JD, Paulo JA, O'Brien JJ, Gygi SP, J Proteome Res 2018, 17, 1934–1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Scheltema RA, Hauschild JP, Lange O, Hornburg D, Denisov E, Damoc E, Kuehn A, Makarov A, Mann M, Mol Cell Proteomics 2014, 13, 3698–3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46] a.Karp NA, Huber W, Sadowski PG, Charles PD, Hester SV, Lilley KS, Mol Cell Proteomics 2010, 9, 1885–1897; [DOI] [PMC free article] [PubMed] [Google Scholar]; b Wenger CD, Lee MV, Hebert AS, McAlister GC, Phanstiel DH, Westphall MS, Coon JJ, Nat Methods 2011, 8, 933–935; [DOI] [PMC free article] [PubMed] [Google Scholar]; c Ting L, Rad R, Gygi SP, Haas W, Nat Methods 2011, 8, 937–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].McAlister GC, Nusinow DP, Jedrychowski MP, Wuhr M, Huttlin EL, Erickson BK, Rad R, Haas W, Gygi SP, Anal Chem 2014, 86, 7150–7158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48] a.Savitski MM, Reinhard FB, Franken H, Werner T, Savitski MF, Eberhard D, Martinez Molina D, Jafari R, Dovega RB, Klaeger S, Kuster B, Nordlund P, Bantscheff M, Drewes G, Science 2014, 346, 1255784; [DOI] [PubMed] [Google Scholar]; b Keshishian H, Burgess MW, Specht H, Wallace L, Clauser KR, Gillette MA, Carr SA, Nat Protoc 2017, 12, 1683–1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Ow SY, Salim M, Noirel J, Evans C, Rehman I, Wright PC, J Proteome Res 2009, 8, 5347–5355. [DOI] [PubMed] [Google Scholar]
- [50] a.O'Brien JJ, O'Connell JD, Paulo JA, Thakurta S, Rose CM, Weekes MP, Huttlin DL, Gygi SP, J Proteome Res 2018, 17, 590–599; [DOI] [PMC free article] [PubMed] [Google Scholar]; b Bai B, Tan H, Pagala VR, High AA, Ichhaporia VP, Hendershot L, Peng J, Methods Enzymol 2017, 585, 377–395; [DOI] [PMC free article] [PubMed] [Google Scholar]; c Ahrne E, Glatter T, Vigano C, Schubert C, Nigg EA, Schmidt A, J Proteome Res 2016, 15, 2537–2547. [DOI] [PubMed] [Google Scholar]
- [51] a.Marshall AG, Wang TCL, Ricca TL, Journal of the American Chemical Society 1985, 107, 7893–7897; [Google Scholar]; b McLafferty FW, Stauffer DB, Loh SY, Williams ER, Analytical Chemistry 1987, 59, 2212–2213; [Google Scholar]; c Chen L, Marshall AG, International Journal of Mass Spectrometry and Ion Processes 1987, 79, 115–125; [Google Scholar]; d Schwartz JC, Senko MW, Syka JE, J Am Soc Mass Spectrom 2002, 13, 659–669. [DOI] [PubMed] [Google Scholar]
- [52] a.Wuhr M, Guttler T, Peshkin L, McAlister GC, Sonnett M, Ishihara K, Groen AC, Presler M, Erickson BK, Mitchison TJ, Kirschner MW, Gygi SP, Curr Biol 2015, 25, 2663–2671; [DOI] [PMC free article] [PubMed] [Google Scholar]; b Peshkin L, Ryazanova L, Wuhr M, bioRxiv 2017. [Google Scholar]
- [53].Graumann J, Scheltema RA, Zhang Y, Cox J, Mann M, Mol Cell Proteomics 2012. 11, M111 013185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Swearingen KE, Moritz RL, Expert Rev Proteomics 2012, 9, 505–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Kozhinov AN, Tsybin YO, Anal Chem 2012, 84, 2850–2856. [DOI] [PubMed] [Google Scholar]
- [56].Virreira Winter S, Meier F, Wichmann C, Cox J, Mann M, Meissner F, Nat Methods 2018, 15, 527–530. [DOI] [PubMed] [Google Scholar]
- [57] a.Palmese A, De Rosa C, Chiappetta G, Marino G, Amoresano A, Anal Bioanal Chem 2012, 404, 1631–1635; [DOI] [PubMed] [Google Scholar]; b Pan KT, Chen YY, Pu TH, Chao YS, Yang CY, Bomgarden RD, Rogers JC, Meng TC, Khoo KH, Antioxid Redox Signal 2014, 20, 1365–1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58] a.Werner T, Becher I, Sweetman G, Doce C, Savitski MM, Bantscheff M, Anal Chem 2012, 84, 7188–7194; [DOI] [PubMed] [Google Scholar]; b McAlister GC, Huttlin EL, Haas W, Ting L, Jedrychowski MP, Rogers JC, Kuhn K, Pike I, Grothe RA, Blethrow JD, Gygi SP, Anal Chem 2012, 84, 7469–7478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Pierce A, Unwin RD, Evans CA, Griffiths S, Carney L, Zhang L, Jaworska E, Lee CF, Blinco D, Okoniewski MJ, Miller CJ, Bitton DA, Spooncer E, Whetton AD, Mol Cell Proteomics 2008, 7, 853–863. [DOI] [PubMed] [Google Scholar]
- [60].Xiang F, Ye H, Chen R, Fu Q, Li L, Anal Chem 2010, 82, 2817–2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Frost DC, Greer T, Li L, Anal Chem 2015, 87, 1646–1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Braun CR, Bird GH, Wuhr M, Erickson BK, Rad R, Walensky LD, Gygi SP, Haas W, Anal Chem 2015, 87, 9855–9863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Lange V, Picotti P, Domon B, Aebersold R, Mol Syst Biol 2008, 4, 222; [DOI] [PMC free article] [PubMed] [Google Scholar]; a Addona TA, Abbatiello SE, Schilling B, Skates SJ, Mani DR, Bunk DM, Spiegelman CH, Zimmerman LJ, Ham AJ, Keshishian H, Hall SC, Allen S, Blackman RK, Borchers CH, Buck C, Cardasis HL, Cusack MP, Dodder NG, Gibson BW, Held JM, Hiltke T, Jackson A, Johansen EB, Kinsinger CR, Li J, Mesri M, Neubert TA, Niles RK, Pulsipher TC, Ransohoff D, Rodriguez H, Rudnick PA, Smith D, Tabb DL, Tegeler TJ, Variyath AM, Vega-Montoto LJ, Wahlander A, Waldemarson S, Wang M, Whiteaker JR, Zhao L, Anderson NL, Fisher SJ, Liebler DC, Paulovich AG, Regnier FE, Tempst P, Carr SA, Nat Biotechnol 2009, 27, 633–641; [DOI] [PMC free article] [PubMed] [Google Scholar]; b Peterson AC, Russell JD, Bailey DJ, Westphall MS, Coon JJ, Mol Cell Proteomics 2012, 11, 1475–1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Erickson BK, Rose CM, Braun CR, Erickson AR, Knott J, McAlister GC, Wuhr M, Paulo JA, Everley RA, Gygi SP, Mol Cell 2017, 65, 361–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65] a.Paulo JA, McAllister FE, Everley RA, Beausoleil SA, Banks AS, Gygi SP, Proteomics 2015, 15, 462–473; [DOI] [PMC free article] [PubMed] [Google Scholar]; b Paulo JA, Gygi SP, Proteomics 2015, 15, 474–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66] a.Chick JM, Munger SC, Simecek P, Huttlin EL, Choi K, Gatti DM, Raghupathy N, Svenson KL, Churchill GA, Gygi SP, Nature 2016, 534, 500–505; [DOI] [PMC free article] [PubMed] [Google Scholar]; b Lapek JD Jr., Greninger P, Morris R, Amzallag A, Pruteanu-Malinici I, Benes CH, Haas W, Nat Biotechnol 2017, 35, 983–989; [DOI] [PMC free article] [PubMed] [Google Scholar]; c Rudnick PA, Markey SP, Roth J, Mirokhin Y, Yan X, Tchekhovskoi DV, Edwards NJ, Thangudu RR, Ketchum KA, Kinsinger CR, Mesri M, Rodriguez H, Stein SE, J Proteome Res 2016, 15, 1023–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.