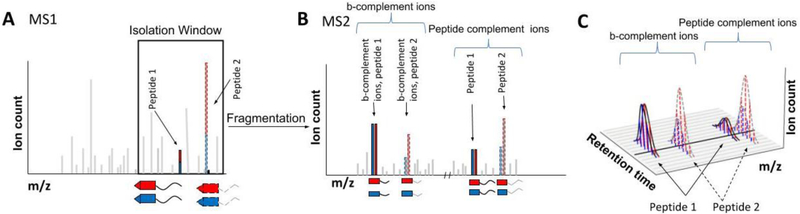
Figure 8: Proposed fusion of DIA with multiplexed proteomics.
A) Peptides would be labeled with isobaric tags similar to a normal multiplexing experiment. For sake of simplicity, we only show two conditions. Like in a normal DIA workflow, all the peaks within a certain wide m/z window in an MS1 scan are co-isolated. This window contains multiple peptides, which will all be simultaneously isolated and fragmented into an MS2 spectrum. For simplicity, only two peptides are shown in detail, depicted with solid and dashed outlines. B) In the MS2 spectrum, the low m/z reporter ions cannot be used for quantification since the reporter ions of all co-isolated peptides will be identical. However, simultaneous quantification is possible via the peptide complement reporter ions. Additionally, complementary b- and y-ions that additionally lost their reporter group can also be used for peptide specific quantification. C) The continuous monitoring of peptide complement reporter ions and b- and y-fragment complement reporter ions allow the relative quantification of multiplexed abundances even between various runs. Additionally, the number of missing values in samples larger than the multiplexing capacity of a single tag should be drastically reduced.