Abstract
Background:
MicroRNAs (miRNAs or miR-) have been linked to factors associated with aggressive prostate cancer such as biochemical recurrence and metastasis. We investigated whether circulating miRNAs in plasma could be used as diagnostic biomarkers for more aggressive prostate cancer at prostate biopsy.
Methods:
Men, aged ≥ 40 years, newly diagnosed with prostate cancer were categorized into two risk groups, low-grade (Gleason score 6 or 7 (3+4) and serum PSA < 20 ng/mL) and high-grade (Gleason score ≥ 7 (4+3) and serum PSA ≥ 20 ng/mL) prostate cancers. The limma R package was used to compare expression of miRNAs in plasma between the two risk groups, adjusting for age.
Results:
There were 66 men, aged 46-86 years, included: 40 men with low-grade and 26 men with high-grade prostate cancers. There were lower expressions of miR-28, miR-100, miR-942, and miR-28-3p and higher expressions of miR-708, miR-1298, miR-886-3p, miR-374, miR-376c, miR-202, miR-128a, and miR-185 in high-grade compared to low-grade prostate cancer cases at biopsy, after adjusting for age (P-values < 0.05). These differences were no longer statistically significant after adjusting the P-values for multiple comparisons.
Conclusion:
There was no circulating miRNA associated with high-grade prostate cancer at biopsy after adjusting for age and multiple comparisons. Nevertheless, relationships between these circulating miRNAs and high-grade prostate cancer were observed which suggest them as promising prostate cancer biomarkers. Further investigation in a larger cohort may provide insight on their diagnostic potential for aggressive prostate cancer.
Keywords: microRNAs, biomarker, aggressive prostate cancer
Introduction
Even though prostate cancer is the leading cause of cancer death among U.S. men, most men are diagnosed with localized prostate cancer which is curable at a high rate or monitorable through active surveillance for indolent prostate cancer cases. A small percentage of men will have highly aggressive prostate cancer which can lead to metastasis, causing morbidity and possibly prostate cancer-related mortality. Black race, cigarette smoking, and obesity have been linked to aggressive prostate cancer1–3; however, risk factors for this disease remain largely not known. Serum prostate-specific antigen (PSA) and Gleason score have been used as prognostic markers for aggressive prostate cancer; however, serum PSA is not useful in distinguishing between indolent and highly aggressive prostate cancer at biopsy. There are several recently available commercial assays using serum kallikreins and pro-forms of PSA which are associated with higher rates of clinically significant prostate cancer on biopsy. However, there still remains a need for biomarkers for not only aggressive prostate cancer but also for the prediction of worse prostate cancer outcomes.
Several microRNAs (miRNAs), small non-coding RNAs that regulate gene expression post-transcriptionally, have been linked to prostate cancer outcomes such as recurrence4–7 and metastasis4,8–10. However, their diagnostic potential in distinguishing between indolent and aggressive prostate cancer cases in a prostate cancer screening setting remains not known. Because miRNAs have been associated with poor prostate cancer outcomes, it is important to examine whether miRNAs can predict aggressive prostate cancer at early stages of disease. In this cross-sectional study, we examined whether circulating miRNAs in plasma differ between high-grade and low-grade prostate cancer among men screened for prostate cancer, with the purpose of identifying potential miRNAs that may be used to identify aggressive prostate cancer cases at diagnosis. Identifying potential biomarkers for aggressive prostate cancer may have a substantial public health impact of providing on one side a more aggressive treatment for the aggressive prostate cancer cases, while at the same time reducing unnecessary treatment for indolent prostate cancer cases.
Materials and methods
Study population
Men, aged ≥ 40 years, who were scheduled for a prostate biopsy at Northwell Health, New Hyde Park, NY and Penn State Health, Hershey, PA were recruited from February, 2014 to April, 2016; participants from the Emory Biomarker Cohort were included. All participants were part of a study that examined whether miRNAs could be potential biomarkers for the early detection of prostate cancer11. Men were included in this study if they had a newly prostate cancer diagnosis and were excluded if they had factors that may influence miRNA expression such as a previous cancer diagnosis other than non-melanoma skin cancer, a subsequent or current diagnosis of metastatic disease, or currently on hormonal therapy at biopsy.
Eligible prostate cancer cases were categorized into two risk groups, low-grade and high-grade prostate cancers. Low-grade prostate cancer was defined as Gleason score 6 or 7 (3+4) at biopsy and serum PSA < 20 ng/mL. High-grade prostate cancer was defined as Gleason score ≥ 7 (4+3) at biopsy and serum PSA ≥ 20 ng/mL. To participate in the study, all study participants had to sign an informed consent form. Blood (15 mL) was collected. A questionnaire that collected demographic information, medical and family history, and lifestyle behaviors (i.e., cigarette smoking) was administered at the time of their prostate biopsy visit. Medical history and clinical information such as Gleason score and serum PSA were extracted from medical records.
All study participants gave a written informed consent to participate in this study. This study received ethical approval by the Institutional Review Boards of Northwell Health, Emory University, and The Pennsylvania State University College of Medicine.
Laboratory methods
Blood collected at prostate biopsy visit were processed into plasma and stored at −80°C. Total RNA from plasma (250 μL) was extracted using miRNeasy Mini kits (Qiagen, Germantown, MD) according to the manufacturer’s protocol. The final elution volume was 30 μL with RNA yield of 5.2-8.0 μg ND-2000 Spectrophotometer, NanoDrop Technologies, Wilmington, DE. All samples had a RNA integrity numbers >9.0 and 260/280 nm absorbance ratios >1.812. TaqMan low density array cards A and B (Applied Biosystem, Foster City, CA) were used to profile 733 miRNAs as laboratory methods are described elsewhere in which the same miRNA profile data were used in the present study13. U6 snRNA was used as the endogenous control to normalize the relative expression level of each candidate miRNA. Cycle threshold (Ct) values were calculated using the SDS2.2.2. Normalized expression was reported as 2−ΔΔCt. Ct value greater than 40 was considered as below the detection threshold of the array.
Statistical analysis
Chi-square or Wilcoxon rank sum test (when appropriate) was used to compare patient characteristics (i.e., demographics, self-reported medical history, and clinical information) between low-grade and high-grade prostate cancer cases. Ct values of miRNA expression levels were log transformed using base 2 before statistical analysis was performed. Only miRNAs (n=197 out of 733 profiled) with 10 non-missing values in both low-grade and high-grade prostate cancer groups were included in the analysis. Wilcoxon rank sum test was used to compare expression of the miRNAs between low-grade and high-grade prostate cancers. The PreprocessCore R package14 was used to perform quantile normalization, then the sva R package15 was used to perform batch correction. Data was normalized and adjusted for batch lab testing days. The limma R package16 was used to compare miRNAs expressions between the two prostate cancer risk groups, adjusting for age. MiRNAs were ranked by the smallest P-values. P-values were then adjusted for multiple comparisons and q-values were computed using the Benjamini-Hochberg procedure. Linear regression was used to model individual miRNA expression level on serum PSA and age in which the partial correlation coefficient (r) between miRNAs and serum PSA was calculated by prostate cancer risk group. The Jonckheere test, as implemented in the clinfun R package, was used to calculate P-value for trend (unadjusted and the Bonferroni-adjusted p-values) between Gleason score (6, 7 [3 +4], 7 [4 +3], 8, and 9-10) and the expression of each miRNA.
The data were analyzed using R 3.4.117 and SAS version 9.4 (Cary, NC). Statistically significance was set at alpha level 0.05.
Results
There were 66 men, aged 46-86 years, with a newly prostate cancer diagnosis included in the study, 40 with low-grade prostate cancer and 26 with high-grade prostate cancer. The majority of the study population was white (66.7%). Patient characteristics did not differ among the three study groups (Northwell Health, Penn State Health, and Emory Biomarker Cohort) (Data not shown). Overall, there was no difference in patient characteristics between low-grade and high-grade prostate cancers, except for serum PSA which was significantly higher in men with high-grade prostate cancer as shown in Table 1 (P-value < 0.0001).
Table 1.
Description of Study Population
| Low-grade Prostate Cancer (n=40) | High-grade Prostate Cancer (n=26) | P-value | |
|---|---|---|---|
| Median age, years (n; range) | 63.5 (40; 46-79) | 65 (26; 47-86) | 0.16 |
| Median BMI, kg/m2 (n; 25-75 percentile) | 28.4 (39; 25.8-34.2) | 27.9 (25; 25.8-30.5) | 0.44 |
| Race % (n) | -------------- | ||
| White | 62.5 (25/40) | 73.1 (19/26) | |
| Black | 20 (8/40) | 15.4 (4/26) | |
| Asian | 7.5 (3/40) | 7.7 (2/26) | |
| Other | 10 (4/40) | 3.9 (1/26) | |
| Ethnicity % (n) | 0.39 | ||
| Non-Hispanic | 87.5 (35/40) | 96.2 (25/26) | |
| Hispanic | 12.5 (5/40) | 3.9(1/26) | |
| Education % (n) | 1.00 | ||
| ≤ 11 years | 7.9 (3/38) | 4 (1/25) | |
| ≥ 12 years | 92.1 (35/38) | 96 (24/25) | |
| Ever smoked cigarettes % (n) | 0.94 | ||
| Yes | 62.5 (35/40) | 61.5 (16/26) | |
| No | 37.5 (15/40) | 38.5 (10/26) | |
| BPH % (n) | 1.00 | ||
| Yes | 7.5 (3/40) | 4 (1/25) | |
| No | 92.5 (37/40) | 96.0 (24/25) | |
| Prostatitis % (n) | 0.20 | ||
| Yes | 5.0 (2/40) | 15.4 (4/26) | |
| No | 95.0 (38/40) | 84.6 (22/26) | |
| ASAP/atypia % (n) | 0.16 | ||
| Yes | 27.6 (8/29) | 9.1 (2/22) | |
| No | 72.4(21/29) | 90.9 (20/22) | |
| HGPIN % (n) | 0.30 | ||
| Yes | 3.5 (1/29) | 13.6 (3/22) | |
| No | 96.6 (28/29) | 86.4 (19/22) | |
| Family history of prostate cancer % (n) | 0.29 | ||
| Yes | 20.6 (7/34) | 8.7 (2/23) | |
| No | 79.4 (27/34) | 91.3 (21/23) | |
| Median serum PSA, ng/mL (n; 25-75 percentile) | 5 (39; 4.2-7.13) | 10.0 (26; 6.6-20.5) | <0.01 |
Note: BMI, body mass index; BPH, benign prostatic hyperplasia; ASAP, atypical small acinar proliferation; HGPIN, high-grade prostatic intraepithelial neoplasia; PSA, prostate-specific antigen; statistically significance between low- and high-grade prostate cancer at P-value ≤ 0.05.
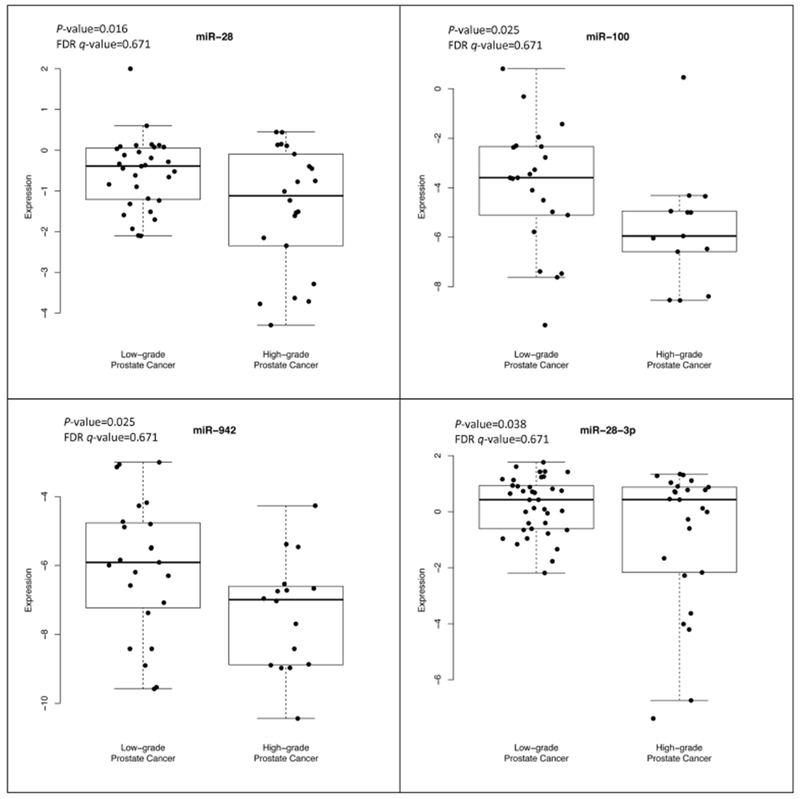
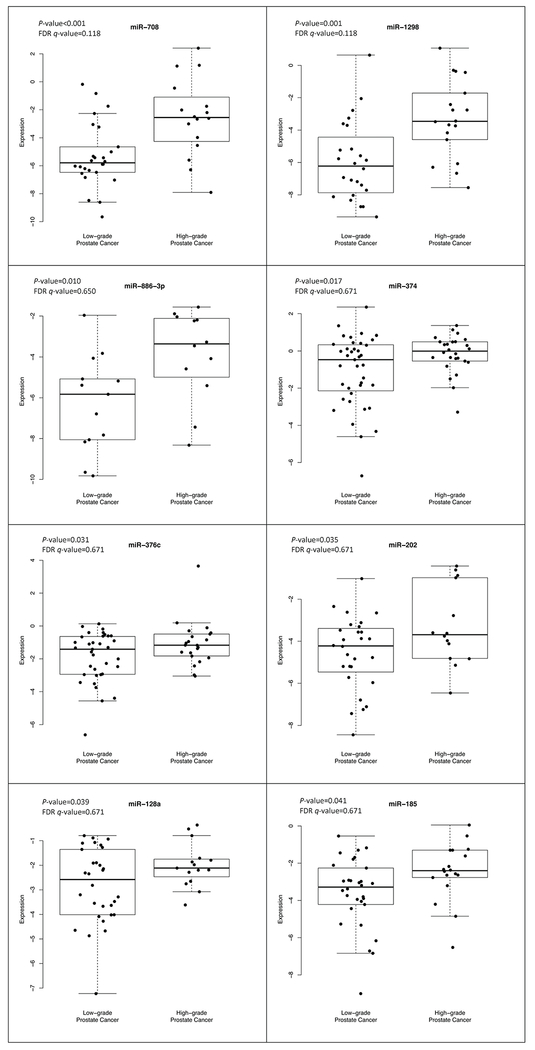
Of the 197 miRNAs evaluated in plasma, 12 had a statistically significant association with high-grade prostate cancer, after adjusting for age; of these, four miRNA expressions (miR-28, miR-100, miR-942, and miR-28-3p) were statistically significantly lower in high-grade compared to low-grade prostate cancer cases as shown in Figure 1 (P-values < 0.04). There were eight miRNA expressions (miR-708, miR-1298, miR-886-3p, miR-374, miR-376c, miR-202, miR-128a, and miR-185) that were statistically significantly higher in high-grade compared to low-grade prostate cancer cases as shown in Figure 2 (P-values < 0.05). After adjusting for age and the P-values for multiple comparisons, these miRNAs were no longer statistically significant.
Figure 1.
Lower expression of microRNAs in high-grade compared to low-grade prostate cancer identified at biopsy
Figure 2.
Higher expression of microRNAs in high-grade compared to low-grade prostate cancer identified at biopsy
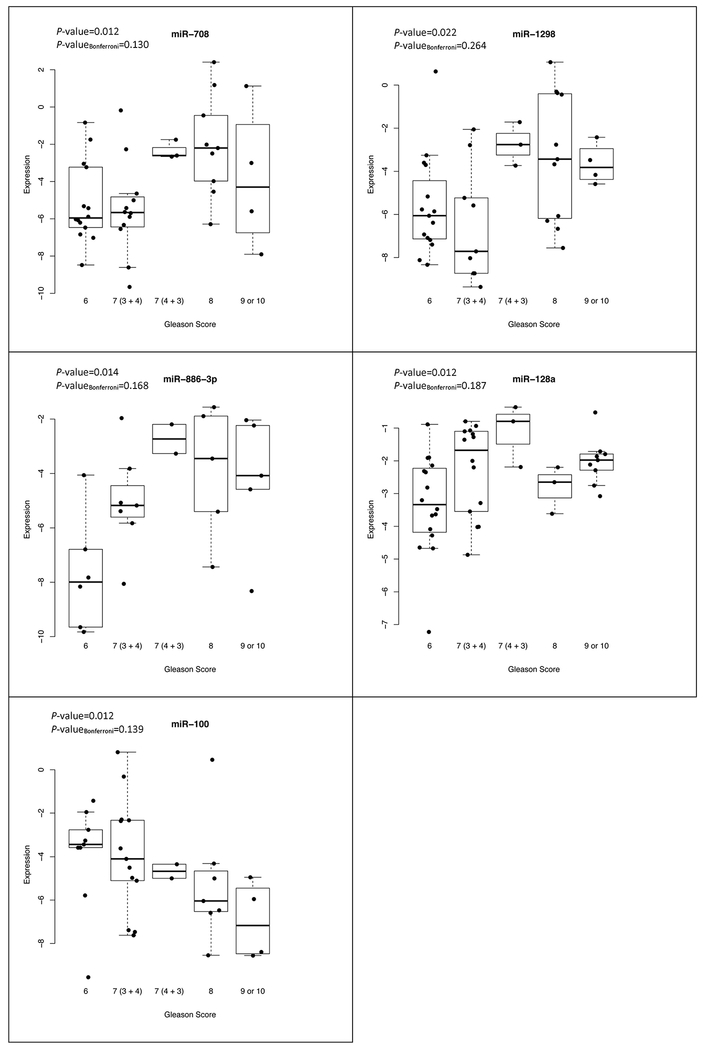
Among the 12 miRNAs, there were five miRNAs (miR-708, −1298, −886-3p, 128a, and −100) that displayed a statistically significant trend of expression with increasing Gleason scores as shown in Figure 3 (P-values < 0.03). After applying a Bonferroni correction, this trend was no longer statistically significant. There was no statistically significant correlation between serum PSA and the miRNA expression levels, after adjusting for age, except for miR-128a among men with low-grade prostate cancer (r = 0.36; P-value = 0.05).
Figure 3.
Association between microRNA expression and Gleason score among prostate cancer cases identified at biopsy
Discussion
Circulating miRNAs such as in plasma and serum are noted for their stability in blood and being non-invasive biomarkers18,19. However, identifying miRNAs as potential cancer biomarkers has been challenging due to inconsistent study findings in which different miRNAs associated with prostate cancer have been reported20–22. These inconsistences may be explain by differences in study population compared, samples collected and how they were processed (i.e., serum, plasma, or isolated exosomes from serum/plasma), laboratory methods used for detection, and data analysis performed on miRNA data (i.e., data normalization and analysis with or without adjustment for multiple comparisons). In addition, their cellular origin remains not known.
In the present study, there were 12 circulating miRNAs that were differentially expressed in plasma between men with low-grade and high-grade prostate cancer at biopsy, after adjusting for age. There were four miRNAs that had a lower expression and eight miRNAs that had higher expression in men with high-grade prostate cancer compared to low-grade prostate cancer. None of these miRNAs were associated with high-grade prostate cancer at biopsy, after adjusting for age and multiple comparisons, possibly due to the small study sample size. Nevertheless, relationships between these circulating miRNAs in plasma and high-grade prostate cancer were observed which suggest them as promising prostate cancer biomarkers. In addition, previous studies have reported relationships between these miRNAs and poor prostate cancer outcomes, which suggest their potential as biomarkers for aggressive prostate cancer.
Among the circulating miRNAs in plasma that had a lower expression in high-grade compared to low-grade prostate cancer cases in the present study, miR-100 has shown to be downregulated in various cancers including prostate cancer, primarily in prostate tissues 5,23–25. Studies suggest that miR-100 can serve as a tumor suppressor or oncogene depending on the tumor type, cell type, or mRNA it targets. For prostate cancer, miR-100 has been shown to be a tumor suppressor whose expression has been downregulated in more advanced stages of disease. A lower expression in prostate cancer tissue compared to normal prostate tissue and lower expression in bone metastatic prostate cancer tissue compared to primary prostate cancer tissue have been reported23,24 . Studies have also reported downregulation of miR-100 during prostate cancer progression24. This decreased in miR-100 expression with more advanced prostate cancer is consistent with our study finding of lower expression levels among high-grade prostate cancer cases compared to low-grade prostate cancer cases. However, higher expression of miR-100 in prostate tissues among localized prostate cancer cases who had a biochemical recurrence after radical prostatectomy was reported in a previous study26. In the present study, circulating miR-100 in plasma demonstrated a trend of lower expression with higher Gleason scores which may suggest its relationship with more advanced prostate cancer as shown in previous studies that evaluated miRNAs in prostate tissues. The present study demonstrates possible relationships between circulating miR-100 in plasma and high-grade prostate cancer. Further evaluation of circulating miR-100 as well as other miRNAs identified in the present study among men at high-risk for aggressive prostate cancer at prostate cancer diagnosis and during disease progression will provide more insight on their influence on aggressive disease.
Among the circulating miRNAs in plasma that had a higher expression in high-grade compared to low-grade prostate cancer cases in the present study, miR-708 and −376c have been linked to factors associated with aggressive prostate cancer in tissues or prostate cancer cell lines27,28. MiR-708 has been previously associated with biochemical recurrence and prostate cancer metastasis; however, the reported association with these outcomes was in both directions. For example, Guo et al. reported upregulation of miR-708 in prostate tumor samples from primary prostate cancer cases to metastasis prostate cancer cases27, which supports our study findings of higher expression of miR-708 in high-grade compared to low-grade prostate cancer cases and the positive correlation observed between miR-708 levels and Gleason score. However, Watahiki et al. found miR-708 statistically significantly downregulated from primary prostate cancer cell lines to metastatic prostate cancer cell lines, where it reached undetectable levels10. As for miR-376c, downregulation of this miRNA has been linked to prostate cancer progression directly or indirectly through its targets such as via matrix metalloproteases and Thrombospondin-2, factors that contribute to the development of metastatic disease29. In addition, miR-376c has been shown to target UDP-glucuronosyltransferases conjugating enzymes UGT2B15 and UGT2B17 which control bioavailability of androgen, a steroid known to play a role in prostate cancer progression30. Studies have shown miR-376c downregulated in metastatic prostate cancer cell lines or tissues compared to normal prostate epithelial cell lines or tissues28,30. However, higher expression of miR-376c in metastatic prostate tissues has been found to be correlated with higher serum PSA (r=0.577; P-value=0.039)30. In the present study, no statistically significant correlation was observed between the expression of circulating miR-376 in plasma and serum PSA levels by prostate cancer grade status. Previous studies have linked the expression of miR-708 and −376c in prostate tissue or prostate cancer cell lines to more aggressive disease. As for their relationship as circulating miRNAs with aggressive prostate cancer, further investigation is warranted in a larger cohort of men with prostate cancer.
A strength to the present study was that several miRNAs were examined to determine their diagnostic potential for aggressive prostate cancer among men being screened for prostate cancer. However, there were study limitations. Study participants came from three different study populations; however, there were no difference in patient characteristics. We profiled miRNA expression in a cohort of men screened for prostate cancer in which there were more people who had low-grade prostate cancer compared to high-grade prostate cancer, possibly due to early screening. In addition, our study was underpowered (< 80%) because of the small sample size, in particular, men with high-grade prostate cancer. Another limitation was that there was only 27% of the miRNAs evaluated in the analysis out of 733 miRNAs profiled due to the minimum requirement of 10 men in the low-grade and high-grade prostate cancer groups. Therefore, miRNAs that may be biologically relevant to aggressive prostate cancer may have been missed due to the number of miRNAs detected in plasma among the study participants in the present study. Finally, we did not examine miRNAs in tumor tissue to confirm their tumor specificity. These men were identified at prostate biopsy; therefore, surgical specimens were not obtained for miRNA profiling.
Conclusions
In conclusion, there were miRNAs that were differentially expressed between low-grade and high-grade prostate cancer, after adjusting for age, in the present study. However, these miRNAs were not associated with high-grade prostate cancer, after adjusting for multiple comparisons. Nevertheless, relationships between these circulating miRNAs in plasma and high-grade prostate cancer were observed. In addition, higher expression of miR-100 and lower expression of miR-708 in men with more advanced disease compared to early stage disease are consistent with previous studies23,24,27. Because circulating miRNAs are stable in blood, a non-invasive biomarker, and differentially expressed between low-grade and high-grade prostate cancer cases, they still remain promising prostate cancer diagnostic biomarkers. Further evaluation of these circulating miRNAs in a larger cohort of men at different stages of prostate cancer may provide more insight on their diagnostic potential for aggressive prostate cancer.
Acknowledgements:
Special thanks to research assistants Alexia Watson from Northwell Health, and Kathleen Lehman and Bhavyata Pandya from The Pennsylvania State University College of Medicine for data collection and administrative support for this study.
Grant Support: This study was supported by funding received from the NIH grant (R21CA178864; A.C. McDonald - PI), NIH grant (U01 CA113913; – M. Sanda -PI), and the Pennsylvania State University College of Medicine.
Footnotes
Institution at which the work was performed: Pennsylvania State University College of Medicine, Department of Public Health Sciences, Hershey, PA 17033
Disclosure Statement: There is no conflict of interest to disclose.
References
- 1.Wood HM, Reuther AM, Gilligan TD, Kupelian PA, Modlin CS Jr., Klein EA Rates of biochemical remission remain higher in black men compared to white men after radical prostatectomy despite similar trends in prostate specific antigen induced stage migration. Journal of Urology . 2007;178(4 Pt 1):1271–1276. [DOI] [PubMed] [Google Scholar]
- 2.Zapata DF, Howard LE, Aronson WJ, et al. Smoking is a predictor of adverse pathological features at radical prostatectomy: Results from the Shared Equal Access Regional Cancer Hospital database. Int J Urol . 2015;22(7):658–662. [DOI] [PubMed] [Google Scholar]
- 3.Vidal AC, Freedland SJ. Obesity and Prostate Cancer: A Focused Update on Active Surveillance, Race, and Molecular Subtyping. Eur Urol . 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Spahn M, Kneitz S, Scholz C-J, et al. Expression of microRNA-221 is progressively reduced in aggressive prostate cancer and metastasis and predicts clinical recurrence. International Journal of Cancer . 2010;127(2):394–403. [DOI] [PubMed] [Google Scholar]
- 5.Tong AW, Fulgham P, Jay C, et al. MicroRNA profile analysis of human prostate cancers. Cancer Gene Therapy . 2009;16(3):206–216. [DOI] [PubMed] [Google Scholar]
- 6.Zheng Q, Peskoe SB, Ribas J, et al. Investigation of miR-21, miR-141, and miR-221 expression levels in prostate adenocarcinoma for associated risk of recurrence after radical prostatectomy. Prostate . 2014;74(16):1655–1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Selth LA, Townley SL, Bert AG, et al. Circulating microRNAs predict biochemical recurrence in prostate cancer patients. Br J Cancer . 2013;109(3):641–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Leite KRM, Sousa-Canavez JM, Reis ST, et al. Change in expression of miR-let7c, miR-100, and miR-218 from high grade localized prostate cancer to metastasis. Urologic Oncology . 2011;29(3):265–269. [DOI] [PubMed] [Google Scholar]
- 9.Brase JC, Johannes M, Schlomm T, et al. Circulating miRNAs are correlated with tumor progression in prostate cancer. International Journal of Cancer . 2011;128(3):608–616. [DOI] [PubMed] [Google Scholar]
- 10.Watahiki A, Wang Y, Morris J, et al. MicroRNAs associated with metastatic prostate cancer. PLoS One . 2011;6(9):30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McDonald AC, Vira M, Shen J, et al. Circulating microRNAs in plasma as potential biomarkers for the early detection of prostate cancer. Prostate . 2018. [DOI] [PubMed] [Google Scholar]
- 12.Ghandhi SA, Yaghoubian B, Amundson SA. Global gene expression analyses of bystander and alpha particle irradiated normal human lung fibroblasts: synchronous and differential responses. BMC Med Genomics . 2008;1:63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McDonald AC, Vira MA, Vidal AC, Gan W, Freedland SJ, Taioli E. Association between systemic inflammatory markers and serum prostate-specific antigen in men without prostatic disease - the 2001-2008 National Health and Nutrition Examination Survey. Prostate . 2014;74(5):561–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.PreprocessCore: A collection of pre-processing functions [computer program]. Version 1.44.0: R package 2018.
- 15.Sva: Surrogate Variable Analysis [computer program]. Version 3.30.0: R package 2018.
- 16.Ritchie ME, Phipson B, Wu D, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic acids research . 2015;43(7):e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.R: A language and environment for statistical computing [computer program]. Vienna, Austria: R Foundation for Statistical Computing; 2017. [Google Scholar]
- 18.Mitchell PS, Parkin RK, Kroh EM, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proceedings of the National Academy of Sciences of the United States of America . 2008;105(30):10513–10518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reid G, Kirschner MB, van Zandwijk N. Circulating microRNAs: Association with disease and potential use as biomarkers. Critical Reviews in Oncology-Hematology . 2011;80(2):193–208. [DOI] [PubMed] [Google Scholar]
- 20.Luu HN, Lin HY, Sorensen KD, et al. miRNAs associated with prostate cancer risk and progression. BMC Urol . 2017;17(1):18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cannistraci A, Di Pace AL, De Maria R, et al. MicroRNA as New Tools for Prostate Cancer Risk Assessment and Therapeutic Intervention: Results from Clinical Data Set and Patients’ Samples. BioMed Research International . 2014;2014:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Alhasan AH, Scott AW, Wu JJ, et al. Circulating microRNA signature for the diagnosis of very high-risk prostate cancer. Proceedings of the National Academy of Sciences of the United States of America . 2016;113(38):10655–10660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peng X, Guo W, Liu T, et al. Identification of miRs-143 and −145 that is associated with bone metastasis of prostate cancer and involved in the regulation of EMT. PLoS One . 2011;6(5):27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang M, Ren D, Guo W, et al. Loss of miR-100 enhances migration, invasion, epithelial-mesenchymal transition and stemness properties in prostate cancer cells through targeting Argonaute 2. Int J Oncol . 2014;45(1):362–372. [DOI] [PubMed] [Google Scholar]
- 25.Giangreco AA, Vaishnav A, Wagner D, et al. Tumor suppressor microRNAs, miR-100 and −125b, are regulated by 1,25-dihydroxyvitamin D in primary prostate cells and in patient tissue. Cancer Prev Res (Phila) . 2013;6(5):483–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leite KR, Tomiyama A, Reis ST, et al. MicroRNA-100 expression is independently related to biochemical recurrence of prostate cancer. J Urol . 2011;185(3):1118–1122. [DOI] [PubMed] [Google Scholar]
- 27.Guo K, Liang Z, Li F, Wang H. Comparison of miRNA and gene expression profiles between metastatic and primary prostate cancer. Oncol Lett . 2017;14(5):6085–6090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Formosa A, Markert EK, Lena AM, et al. MicroRNAs, miR-154, miR-299-5p, miR-376a, miR-376c, miR-377, miR-381, miR-487b, miR-485-3p, miR-495 and miR-654-3p, mapped to the 14q32.31 locus, regulate proliferation, apoptosis, migration and invasion in metastatic prostate cancer cells. Oncogene . 2014;33(44):5173–5182. [DOI] [PubMed] [Google Scholar]
- 29.Chen P-C, Tang C-H, Lin L-W, et al. Thrombospondin-2 promotes prostate cancer bone metastasis by the up-regulation of matrix metalloproteinase-2 through down-regulating miR-376c expression. Journal of hematology & oncology . 2017;10(1):33–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Margaillan G, Levesque E, Guillemette C. Epigenetic regulation of steroid inactivating UDP-glucuronosyltransferases by microRNAs in prostate cancer. J Steroid Biochem Mol Biol . 2016;155(Pt A):85–93. [DOI] [PubMed] [Google Scholar]