Abstract
Nanoflares are intracellular probes consisting of oligonucleotides immobilized on various nanoparticles that can recognize intracellular nucleic acids or other analytes, thus releasing a fluorescent reporter dye. Single-stranded DNA (ssDNA) complementary to mRNA for a target gene is constructed containing a 3′-thiol for binding to gold nanoparticles. The ssDNA “recognition sequence” is prehybridized to a shorter DNA complement containing a fluorescent dye that is quenched. The functionalized gold nanoparticles are easily taken up into cells. When the ssDNA recognizes its complementary target, the fluorescent dye is released inside the cells. Different intracellular targets can be detected by nanoflares, such as mRNAs coding for genes over-expressed in cancer (epithelial-mesenchymal transition, oncogenes, thymidine kinase, telomerase, etc.), intracellular levels of ATP, pH values and inorganic ions can also be measured. Advantages include high transfection efficiency, enzymatic stability, good optical properties, biocompatibility, high selectivity and specificity. Multiplexed assays and FRET-based systems have been designed.
Keywords: Nanoflares, Targeted intracellular fluorescence probes, Nucleic acid hybridization, Cancer cell detection, mRNA detection, ATP detection, Inorganic ion detection
Introduction
Imaging of intracellular processes has remained challenging over the last few decades. The study of intracellular processes has helped scientists to elucidate biological mechanisms, understand gene expression, and explore various enzymatic activities. Over many years, cancer researchers have gained a large body of knowledge about what makes cells become cancerous or malignant. Nevertheless, imaging of intracellular processes within living cells in real time has remained an unsolved challenge. Intracellular physicochemical parameters are difficult to measure without disturbing the cell. A living cell is a complicated system whose intricate workings are ultimately controlled by nucleic acids. Genetic mutations are the main reason for initiation of unwanted and unpredictable processes that result in cancer formation. Therefore, intracellular studies of nucleic acid expression, production and trafficking have attracted much attention. Fluorescent methods relying on external excitation techniques such as those based on Forster resonance energy transfer,1 molecular beacons labeled with fluorescent molecules,2 in situ hybridization and staining with fluorescent antibodies,3 have all been investigated for detection and study of intracellular molecules. In the field of intracellular biomolecular detection, efficient transfection into cells, stability of reagents in the presence of cellular enzymes,4 and fluorescent quenching5 are the main challenges to be overcome. According to many reports, externally delivered fluorescence probes suffer from many unsolved problems, despite having many benefits. Externally delivered probes the advantages of low cost, better availability, and the possibility to be applied for intracellular imaging of a broad range of cells. One of the main disadvantages of these methods however, is an intrinsic limitation on the intensity of fluorescence, and difficulty to generalize the reagents to a variety of target biomolecules inside cells. Internalization of reagents such as antibodies and aptamers into cells is challenging. Moreover, supplementary reagents are often necessary. Unfortunately, these supplementary reagents do not usually have sufficient chemical stability against enzymatic degradation inside cells.6 In addition transfection reagents, such as lipids7 and dendrimers8 can show harmful and toxic side effects. The uptake process of oligonucleotides (which are a critical component of nanoflares) into cells is a significant challenge limiting their use in intracellular imaging processes.
With the discovery of oligonucleotides immobilized onto nanomaterials, many investigators have explored their application to intracellular imaging.9–12 According to recent reports, oligonucleotide-based nanoflares have many advantages that suggest they can play key roles in optical biosensors, for genetic analysis and bio-delivery systems. Generally, nanoflares are formed from nanoparticles with attached oligonucleotides as substrate and probe, respectively. The intrinsic properties of immobilized oligonucleotide-based nanoflares may provide many advantages in imaging of different intracellular species such as DNA, RNA and so on. Different nanoparticulate substrates have been used to immobilize and quench the nanoflares. On one hand, noble metal (Au, Ag, Cu etc.) nanoparticles are good candidates as immobilizing substrates, and as optical quenchers of nanoflares because of their appropriate surface plasmon resonance properties.13 On the other hand, oligonucleotides can possess a highly efficient transfection ability (without any supplementary reagents), have good stability against intracellular enzymatic degradation, and show high sensitivity to detect complementary RNA and DNA sequences. Although the optical properties of nanoflares can be affected by the composition of the cellular milieu, noble metal nanoparticle-based nanoflares exhibit distance-dependent optical properties, with efficient fluorescent emission, high sensitivity for RNA transcripts, and very weak background fluorescence when used for intracellular imaging.14
The understanding of the mechanism of fluorescence in nanoflares plays a key role in the design of optical sensors. So, as a brief introduction to the fluorescence spectroscopy of nanoflares we can mention emission, absorption wavelength and intensity. Typically, many environmental and molecular interactions can reduce the fluorescence intensity, which are called quenching effects. Formation of molecular complexes between fluorophore and other ground state molecules leading to energy transfer between the species leads to quenching. The optical properties of different nanomaterials mean that they can be used both as quenchers or probes in fluorescent-based imaging systems. The nanomaterials that can be used in nanoflares, must have specific optical properties. The crystal phase, size, and band gap of nanoparticles define the absorption and emission properties. Four different types of nanoparticles have been used as substrates and/or probes in nanoflares. These nanoparticles are (a) gold nanoparticles and nanoclusters (AuNPs/NCs); (b) quantum dots (QDs)15–20; (c) polymer-based nanomaterials (conductive polymers, polymer dots etc.)21–25; and (d) rare earth metal-based nanoparticles (upcon-version nanoparticles).26,27
AuNPs have been widely used as imaging agents in cancer diagnosis and tumor detection. AuNPs display the surface plasmon effect. 13,28–30 Interaction between light and AuNPs creates dipoles on the Au surface, and subsequently absorption peaks in the visible wavelength range.31 The high surface electron density characteristic of AuNPs, makes them suitable as powerful quenchers in fluorescence imaging. AuNPs can be used as “on–off;” fluorescence probes in imaging of intracellular biomolecules.32 Although gold nanoclusters (AuNCs) with particle size of less than 2 nm do not have suitable absorption in the visible wavelengths, they have good stability for imaging and detection of cancerous cells and different intracellular biomolecules.33 AuNPs are highly efficient quenchers that can be applied in different nanoflare-based intracellular imaging approaches. The efficient quenching ability of AuNPs makes it possible to use different dyes as molecular beacons in intracellular environments.34 Modified AuNPs can detect a broad range of nucleic acids,35 adenosine triphosphate (ATP),36 cancerous cells37 and several intracellular ions.38 Moreover, different dye molecules can be used as beacons in nanoflare-based intracellular imaging. The optical properties of various organic dyes that have been applied as beacons in intracellular biomolecular detection have recently been reviewed.39 The emission wavelength of fluorescent dyes is a critical factor that should overlap with the surface plasmon resonance of modified AuNPs. The matching of the absorption wavelength of the AuNPs and the emission wavelength of the dye plays a very important role in the “on–off” fluorescence properties of nanoflares.40,41 Currently, many scientific reports have described intracellular detection of a variety of targets using nanoflares, detecting a broad range of RNAs, cations, cancerous cells etc. The great potential of nanoflares can improve the imaging and detection of cancerous cells, and assist in elucidating their unpredictable intercellular pathways. Applications of nanoflares to the detection of RNA, DNA, cancerous cells, intracellular components, and biomolecules will be discussed in the subsequent sections.
Hybrids between AuNPs (13 nm) and oligonucleotides are the basic building block of nanoflares. The dense deposition of oligonucleotides on the AuNP surface makes them suitable for fluorescence-based detection. The fluorophore emission wavelength, the configuration and morphology of the oligonucleotides on the AuNP surface, and the distance dependent plasmonic resonance are all critical features.42–45 Many different types of fluorophore have been tested including, Cy5, Cy3, Texas red, and rhodamine 6G. These dyes are stable against enzymatic degradation, and their emission wavelength can overlap with AuNP surface plasmon resonance absorption peaks.5 The “off–on” generation of fluorescence from nanoprobes has been developed. The specific structural design of the oligonucleotides makes it possible to release fluorophore molecules during the hybridization process between targets and probes. The size of AuNPs bears a direct relation with their biological toxicity. Smaller AuNPs are generally more toxic than large AuNPs. The toxicity was found to be negligible for AuNPs 50 nm in size, but AuNPs with 13 nm size are commonly used in this application.46,47 Moreover, increasing the size of AuNPs increases the specific surface area and hence the number of oligonucleotide molecules that can be immobilized.
From a clinical point of view, the ability of nanoflares be employed in vivo is one of the important issues that will determine their eventual acceptance, There are many factors that are important for in vitro experiments, but more importantly it is necessary to study nanoflares under in vivo conditions. The cytocompatibility, long blood circulation time, specificity to the target and low toxicity to the surrounding tissue are the most important features in designing nanoflares. According to recent reports, the size of metal nanoparticles and the length of the decorated oligonucleotides are the most important structural features. Commonly, metal nanoparticles with larger sizes (up to 50 nm in diameter) and oligonucleotides with shorter length (25–90 bases) show the best results for in vitro conditions. But larger metal nanoparticles have a low specific surface area, and subsequently show lower contrast in imaging applications. Therefore, researchers have proposed that nanoparticles with smaller sizes (13 nm diameter) will improve the contrast in vivo. Moreover, metal nanoparticles with 13 nm size have acceptable performance under in vitro conditions. Unlike short oligonucleotides (25–90 bases) that are suitable for in vitro conditions, longer oligonucleotides are more appropriate for in vivo condition. This issue affects the sensitivity, limit of detection, and background signals from the probes in the intracellular milieu.48,49 Appropriate structural design, chemical stability, suitable size, and low cytotoxicity are desirable properties. So, in summary, the concept of “nanoflares” is relatively new, but is becoming increasingly widely adopted. Nanoflares have some structural features that are crucial to produce photoluminescence used for intracellular detection of a range of important analytes. A nanoflare is a nanocarrier with photoluminescence detection ability that has been constructed from nanoparticles with a plasmon resonance frequency on their surface, and decorated with oligonucleotides as biological recognition elements. In fact, the nanoflares have dual features that make them suitable options for detecting or imaging a range of intracellular physiochemical events, or intracellular analytes. Nanoflares have special optical properties that depend on the plasmon resonance frequency of the nanomaterials such as AuNPs, AgNPs etc., coupled with appropriate dyes or beacons with an emission wavelength that should be matched with the plasmon resonance frequency of the metal nanoparticles. Some useful properties of the nanoflares that make them more attractive than conventional fluorophores for intracellular detection are the nanometer scale of their structure that facilitates their transfection process through the cellular membrane and into the cells; high chemical stability in a harsh enzymatic intracellular milieu coupled with low toxicity and high biocompatibility. According to these explanations, the nanoflares can be selected as promising option for detection of range of range of mutated genes or biomarkers related to serious diseases such as cancer, Alzheimer’s etc. In the next sections, the molecular targets for nanoflare-based imaging and detection inside cells are discussed.
Targets in nanoflare-based imaging
Detection of cancer cells
We know that quenching process and the emission wavelengths of the dyes or beacons are important for intracellular detection of analytes. Nanoflares provide a chance to achieve bioimaging arising from intracellular events or processes. Some diseases have an underlying genetic cause, and the detection of defective cells, and their location within the human body is a complicated and difficult process. Nevertheless, photoluminescence-based methods can reveal altered cells with great precision and accuracy and high specificity. Cancer is one of the most dangerous diseases caused by mutated and defective genes, allowing cells to undergo unbridled divisions, followed by spreading and metastasis throughout the human body. The detection of cancer biomarkers using nanoflare-based methods is a highly potent strategy that may be suitable for commercialization because of high accuracy, low detection time, and an efficient transfection through cell membranes into cells. So, suspicious pathological samples can be tested using lab-on-chip type devices in clinical applications. Moreover, from a technical and spectroscopy point of view, we have been limited by the Hb and HbO2 absorption wavelength which lies between 500 and 600 nm and can affect the contrast in optical bioimaging. Therefore, the ability of nanoflares to match the emission wavelength of the beacons with the plasmon resonance frequency of the nanoparticles is a rare and unique feature. Despite decades of effort, early detection and effective cancer therapy remains a major challenge. Many different biological molecules have been explored as biomarkers of cancer. According to many reports, these biomarkers mostly consist of genes,50–55 ribonucleic acids (RNAs),56 carbohydrates57–59 and glycoproteins.60–66 Many of these biomarkers are present at only low concentrations and their detection is complicated. Moreover, early diagnosis of cancer often requires detection of these biomarkers at low concentrations inside cells, rather than in biological fluids. Nanomaterial-based methods have high sensitivity, accuracy and low limits of detection (LOD) for cancer biomarker detection. QDs and modified QD-based nanomaterials67 fulfill many of the requirements for cancer cell imaging.68 QD-based methods can be used in “lab-on-chip” approaches.69,70 Fluorophore-labeled nanomaterials have been employed for detection of biomarkers related to gastric cancer.71,72 AuNPs have good quenching properties and low levels ofbackground fluorescence. Physical interactions between AuNPs and phosphatase isoenzyme biomarkers can cause the release of absorbed organic quenched beacons to detect the presence of cancer cells (Figure 1).73 (See Table 1.)
Figure 1.
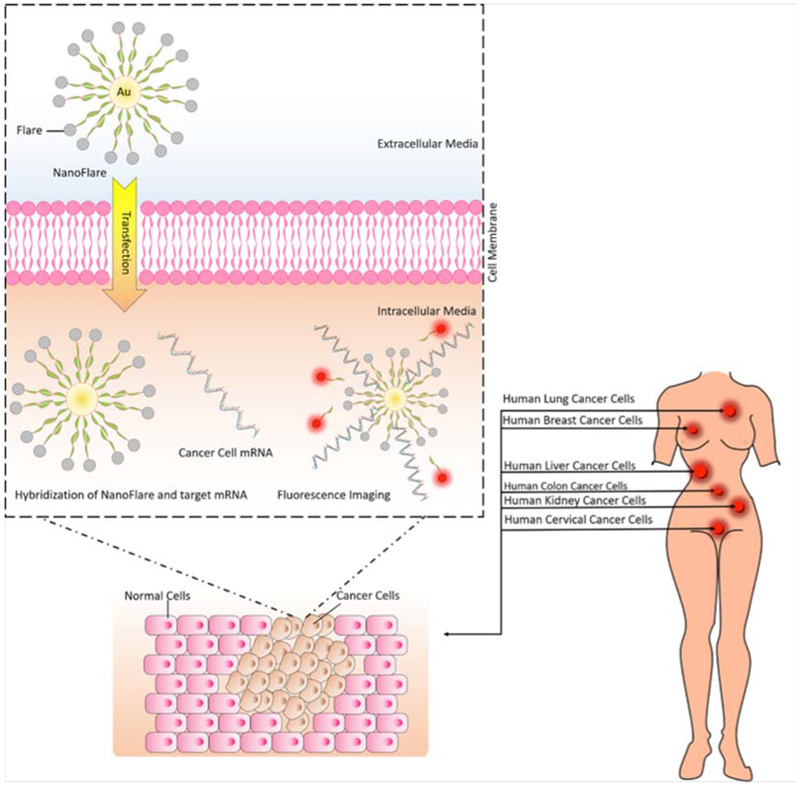
Application of nanoflares for detection of abnormal mRNA in cancer cells.
Table 1.
Recent reports about detection of cancer cells using oligonucleotide.
| Nanomaterials | Target | Design | Cell(s) or virus | Dye | LOD | Linear range | Ref. |
|---|---|---|---|---|---|---|---|
| AuNPs (13 nm) | selenol | Hybridization of AuNPs and Cy5.5 modified peptide chains (Cy5.5-Ser-Asn-Lys-GlyCys) | HepG2a | Cy5.5 | - | - | 104 |
| AuNPs (13 nm) | mRNA | Hybridization of AuNPs and Cy5-modified oligonucleotides | MDA-MB-231b | Cy5 | - | - | 105 |
| AuNPs (13 nm) | mRNA | AuNPs-coupled single-stranded DNA (ssDNA) oligonucleotides | 293Tc, HeLad, U-373 MGe, DU145f, PC3f, A549g, BLMh, RH5i, RH28i, RH30i | Cy3, Cy5 | - | - | 106 |
| AgNCs and AuNPs | miRNA | DNA-modified AgNCs y and AuNPs | MCF-7j, HEK239k | - | 0.4 pM | 1 nM-5 μM | 107 |
| AuNPs (13 nm) | miRNA (miRNA-21, miRNA-141) | AuNPs functionalized with DNA molecules | HeLa, 22Rv1l, SMMC-7721m, LOVE-1n | FAM, Cy5 | 0.9 nM, 1.2 nM | 0–25 nM | 108 |
| AuNPs | miRNA (miRNA-21, let-7d, miRNA-200b) | Hybrid of tetrahedral DNA nanostructure (TDN) and AuNPs (Au-TDNNs) | HepG2, MCF-7, | - | - | - | 109 |
| AuNPs | miRNA-21 | duplex specific nuclease (DSN) and bridge DNA-AuNPs | - | - | 6.8 aM | 10−17 -10−11 M | 110 |
| AuNPs | mRNA | AuNPs functionalized with oligonucleotides | C166o, SKBR3p | Cy5 | - | - | 111 |
| AuNPs | Human telomerase mRNA | AuNPs and DNA hybridization for construction of “sticky-fare” probe | KBq, A549 | Cy5 | - | - | 112 |
| AuNPs | mRNA | Commercial spherical nucleic acid modified AuNPs | HeLa | Cy3, Cy5 | - | - | 113 |
| Core-shell Ag@SiO2 | mRNA, ATP, | Core-shell Ag@SiO2 and cDNA hybridization | Human blood cells | Cy5 | 8 μM | 0–0.5 mM | 114 |
| AuNPs | mRNA | self-assembly of DNA flares hybridization on AuNPs | HeLa | TAMRAr | - | - | 115 |
| AuNPs (13 nm) | DNA, cocaine, | DNA-modified AuNPs | - | OliGreen, | - | - | 116 |
| AuNPs (17 nm) | HIV-DNA | dendrite-modified AuNPs | HIV virus | - | 73 pM | 150 pM-6 nM | 117 |
| AuNPs | Vimentin mRNA | DNA-coated AuNPs | 16-HBEs, MRC-5t, MEFsu | Cy5 | - | - | 118 |
| AuNPs | ssDNA, dsDNA | DNA probes labeled with various fluorophores | - | FAMv, ROXw | 330 pM | 1.0 nM–500 nM | 119 |
| gold nanodots (AuNDs) | Sulfide ions | MTTx capped AuNDs | - | - | 2 nM | 870 nM-16 μM | 120 |
Human liver carcinoma cells.
Human breast cancer cells.
Human embryonic kidney cells.
Human cervical cancer cells.
Human astrocytoma cells).
Human prostate cancer cells.
Human lung cancer cells.
Human melanoma cells.
Human rhabdomyosarcoma cells.
Human breast cancer cell.
Normal human embryonic kidney cell.
Human prostate cancer cells line.
Human hepatocyte cell.
Human colon cancer cell.
Mouse endothelial cells.
Human breast cancer cells.
Keratin-forming tumor cell of HeLa.
5-Carboxytetramethylrhodamine.
16-Human bronchial epithelial cells.
Human fetal lung fibroblast.
Primary mouse embryonic fibroblasts.
6-Carboxyfluorescein.
Carboxy-X-rhodamine.
6-Mercapto-s-triazolo(4,3-b)-s-tetrazine.
Ag nanoclusters.
Detection of cancer cells can involve cancer metastasis or circulating cancer cells,74–76 proteins expressed on the cell surface,77–80 or gene expression-based methodology.81–83 Nanoflares are an important and highly efficient method of detecting changes in gene expression. Detection of different RNAs has been achieved using AuNPs with attached oligonucleotides. Prigodich et al. developed hybrid-based nanoflares consisting of AuNPs with fluorophore-labeled oligonucleotide (DNA-LNA chimeras) as theranostic agents for detection of HeLa (human cervical cancer) and C166 (mouse endothelial) cells. The marker that was targeted was messenger ribonucleic acid (mRNA) encoding for endogenous survivin, an anti-apoptotic protein over-expressed in many cancers. A schematic illustration of the use of AuNP-oligonucleotide nanoflares for detection of survivin mRNA in HeLa and C166 cancer cells is shown in Figure 2. The hybridization process between survivin mRNA and nanoflares resulted in an increase in the intensity of Cy5 dye fluorescence.84
Figure 2.
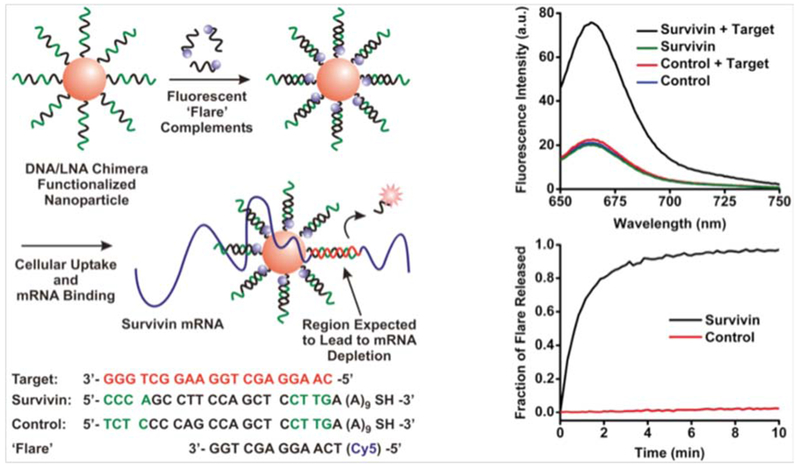
Surface-modified AuNP nanoflares and their fluorescence intensity change (upper plot). Transfection of the nanoflare inside cells, interaction between survivin mRNA and the nanoflare, and released flare was monitored over time (lower plot)84 with permission.
The detection of abnormal RNAs within cells located in different parts of the human body is a challenge. The importance of mRNA expression levels for cancer detection has led researchers to concentrate on multiple RNA detection methods.37 Cellular metabolism and progression,85,86 activation,87–90 and ganglioside pathways18,91 are all controlled by different mRNAs, and disruption in their levels can be related to the presence of cancer. Li et al. devised a detection method for multiple tumor-related mRNAs based on nanoflares with multicolor optical emissions. They used AuNP-oligonucleotide hybrids as nanoflares for simultaneous detection of mRNAs coding for TK1 (human thymidine kinase), c-myc (oncogene) and GalNAc-T (N-acetylgalactosaminyltransferase) in breast cancer cells. TK1, c-myc and GalNAc-T all play key roles in tumor cell progression. The nanoprobes used the following fuorochromes: green (Rh110), yellow (Cy3) and red (Cy5) colors. The response of the combined nanoflares was up to five times higher than the response of single ones. The background fluorescence signals did not change in the presence of irrelevant intracellular biomolecules. The LOD of these nanoprobes was 1.6 nM, 1.4 nM and 1.2 nM for GalNAc-T, TK1 and c-myc mRNAs, respectively. The nanoprobes had high stability against nuclease, deoxyribonuclease I, and endonuclease enzymes (Figure 3).83
Figure 3.
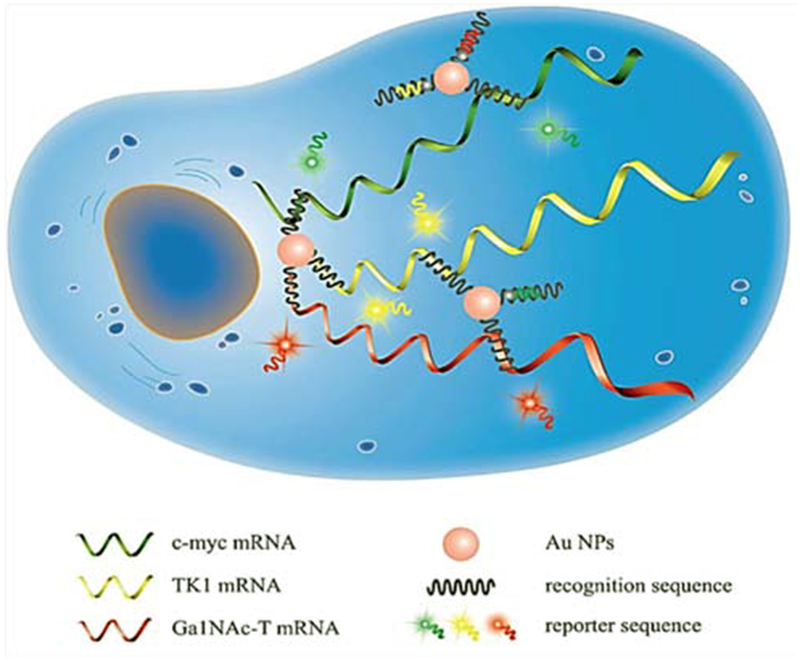
Illustration of simultaneous detection of c-myc, TK1 and Ga1NAc mRNAs in human breast cancer cells (MCF-7, MCF-10A) and HepG2, HL-7702 cells using AuNP-oligonucleotide nanoflares83 with permission.
During the metastatic process, cancer cells can be distributed all over the body before the appearance of discrete lesions. The transition from the epithelial to mesenchymal cell phenotype (EMT) governs the spreading of cancer cells to distant organs. Typically, reduced expression of adhesion molecules (such as, E-cadherin, fibronectin and vimentin) leads to loss of intercellular contacts92 leading to cell migration to other parts of the body.93 Halo et al. devised a nanoflare-based approach to detect mRNAs related to EMT in metastatic cancer cells. The mRNAs coded for the mesenchymal markers, twist, vimentin, and fibronectin, and the epithelial marker E-cadherin. The nanoflares consisted of a spherical AuNP densely functionalized with a monolayer of ssDNA attached via a 3′ thiol group that was complementary to mRNAs for the target genes. The ssDNA “recognition sequence” was prehybridized to a shorter DNA complement containing a fluorescent reporter (the “reporter flare”) whose fluorescence was quenched based on its proximity to the AuNP. When target mRNA binds the recognition sequence, the reporter flare strand was displaced, liberating the fluorescent Cy5 dye.94
Prigodich et al. reported multiplex nanoflares that could detect HeLa, Jurkat, and MCF-7 human cancer cells using commercial siRNAs (small interfering RNAs). Their results indicated that the nanoflares showed a high ability to detect HeLa, Jurkat, and MCF-7 cell mRNAs using actin and survivin as complementary sequences. They used Dharma-FECT 1 transfection reagent in the imaging process. The multiplexed nanoflares and their different detector sequences showed independent behavior in buffer media. A schematic representation of simultaneous and independent intracellular imaging of mRNAs in human cancer cells using multiplex nanoflares is shown in Figure 4.95
Figure 4.
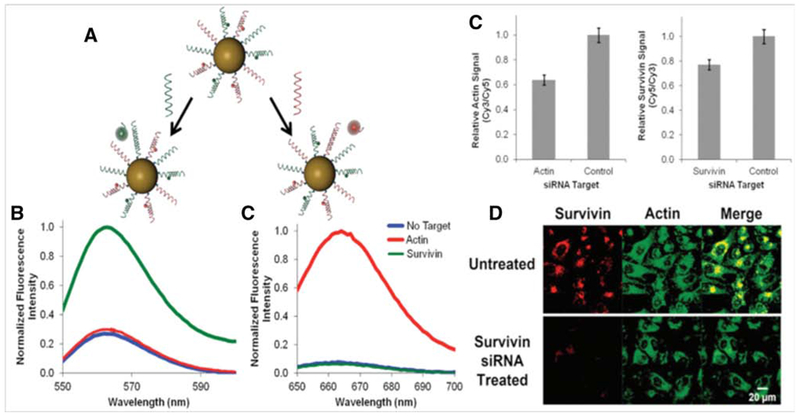
(A) Spherical AuNP-based nanoflares and detection process of different cancer cell mRNAs, (B) Fluorescence spectrum of actin and survivin based nanoflares with independent fluorescence wavelengths, (C) Actin and survivin relative fluorescence signals using Cy3 and Cy5 dyes, and (D) fluorescence confocal microscopy (FCM) images of survivin and actin mRNAs, and the combination, for intracellular imaging in human cancer cells95 with permission.
Pan and coworkers developed AuNP-based nanoflares with four color probes for simultaneous detection of mRNAs. The intracellular detection of TK1, survivin, c-myc and GalNAc-T related mRNAs were possible. They used HepG2, MCF-7, MCF-10A and HL-7702 cancer cells. A schematic of the four color-based nanoflares is shown in Figure 5.96
Figure 5.
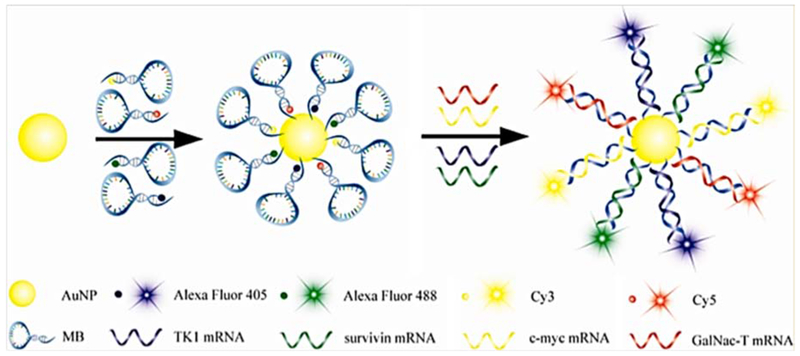
Molecular mechanism of multiplex four color AuNP-based nanoflares for detection of TK1, survivin, c-myc and GalNAc-T mRNAs in cancer cells96 with permission.
It is important to minimize the thermodynamic fluctuation in order to remove unwanted false positive signals. Yang et al. fabricated AuNP-based nanoflares with acceptor-donor pairs that could detect mRNAs in HepG2, MCF-7 and L02 cancer cells using a fluorescence resonance energy transfer (FRET) system (Figure 6). They utilized the TK1 mRNA as the main target in cells. They found that intracellular DNAse I had an important influence on the optical properties of nanoflares by cleaving the nucleotide sequences of both target and flare. DNAse I had a remarkable effect on the fluorescence background optical signal.97
Figure 6.
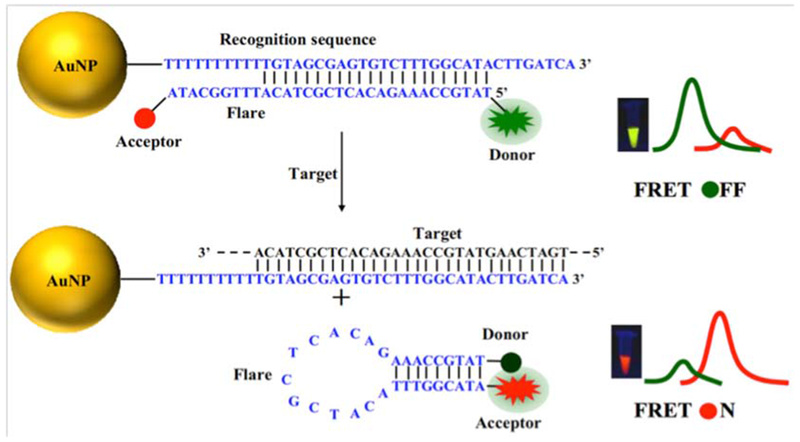
AuNPs-based nanoflares with acceptor-donor flare beacon97 with permission.
An abnormal level of telomerase enzyme activity is another marker of cancer that can distinguish malignant cells from normal ones. The telomerase enzyme is responsible for adding telomere units onto the end of chromosomes. Telomerase controls proliferation and cell division and effectively makes cells “immortal”.98 Hong et al. described AuNP-graphene oxide (GO) hybrid-based nanoflares for in situ detection of telomerase activity in cancer and normal cells. They used this novel nanoflare-based method to analyze HeLa, A549, MCF-7, Caco-2, HBL-100 and HL-7702 Cells. The high transfection efficiency, enzyme-free detection, and the amplification process were claimed as the main advantages of this novel nanoflare-based method. A schematic illustration of AuNPs-GO hybrid-based nanoflares is shown in Figure 7.99
Figure 7.
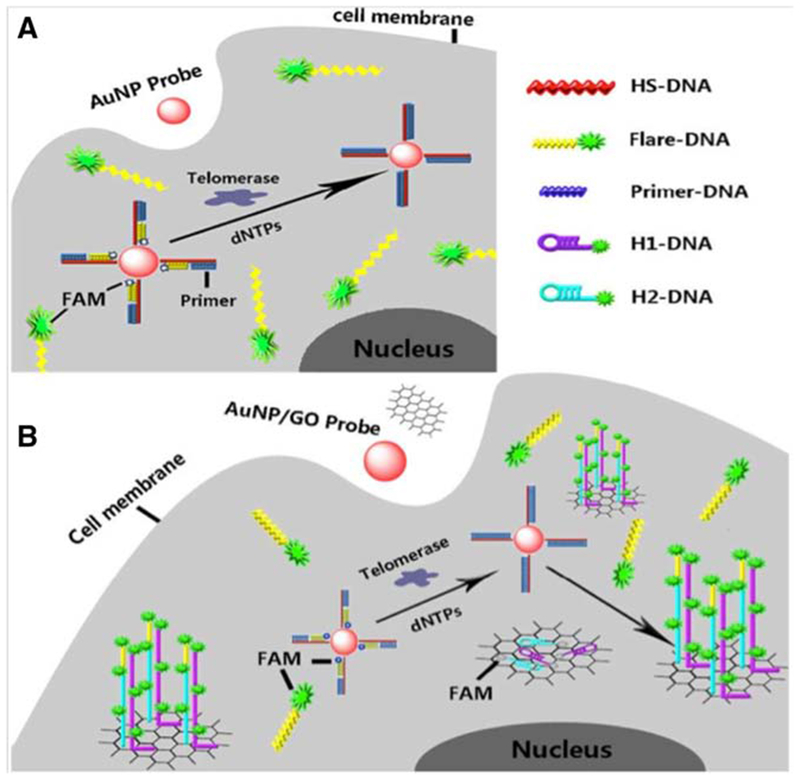
Schematic representation of AuNP-GO hybrid-based nanoflares99 with permission.
In another study, He and co-workers, developed a ratiometric fluorescent-based biosensor for detection of cancer cells based on differences in telomerase expression levels. They prepared modified AuNP-based nanoflares with oligonucleotides to detect telomerase mRNA in cancer (HeLa, HepG2, A549, and 293 T) and normal (QSG) cells. Their results indicated that the ratiometric nanoflare probes could detect telomerase levels via the complementary mRNA sequences. A schematic illustration is shown in Figure 8.100
Figure 8.
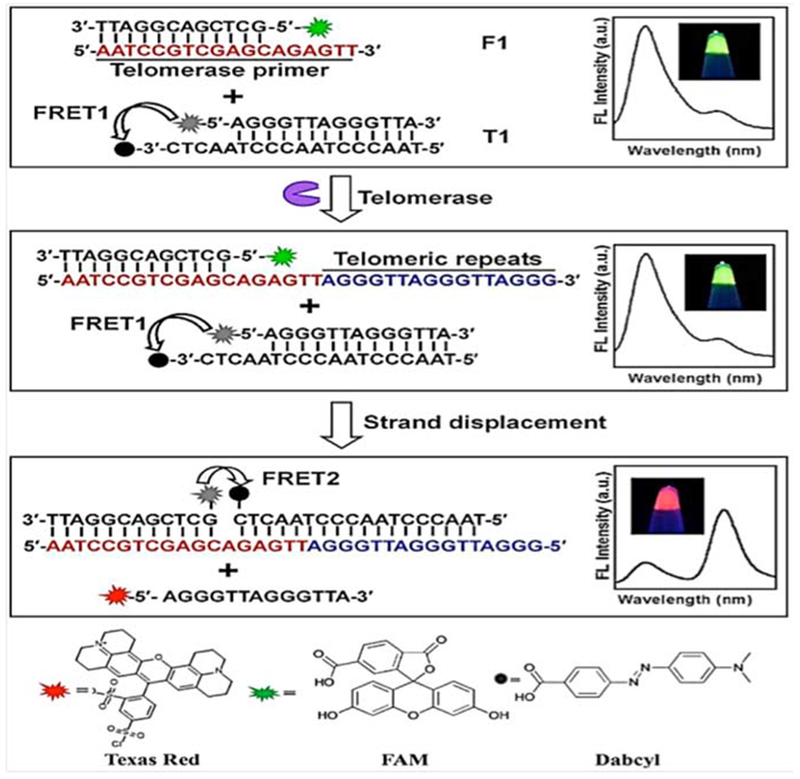
Telomerase activity detection using ratiometric nanoflares100 with permission.
MicroRNAs (miRNAs) are biomarkers that have been used as for cancer cell detection. miRNAs are related to the malignant properties ofcells such as a disruption in the transcription process, uncontrolled epigenetic processes, and defects in protein synthesis.101 Hong et al. prepared nanoflares using a hybrid between AuNPs and GO with oliogonucleotides for detection of mature/pre-miRNA-21 inside cells. Their results suggested that nanoprobes had an excellent LOD and a linear range of activity. Expression levels of pre-miRNAs were unchanged, but levels of mature miRNAs expression showed a significant change in cancer cells. Whether the expression levels were increased or decreased depended on the nature of the miRNA. Mature miRNA-21 was upregulated and mature let-7a was down-regulated.102
Nanoflares based on Au-Se bonds have also been used in various biosensing systems. They are particularly useful in intracellular environments, in which high concentrations of free thiols (such as reduced glutathione) would destroy Au-S bonds, and lead to high background signals. Gao et al. developed surface-modified AuNPs using selenol peptides as linking agents, for the detection of the matrix metalloproteinase 2 (MMP-2) enzyme with remarkable stability and low chemical anti-interference. Comparison between Au-S and Au-Se nanoflares indicated that the Au-Se bonded one performed better than the nanoflare with Au-S bonds due to higher stability in the presence of glutathione.103
Cancer therapy
Nanoflares can have applications in cancer therapy as well as in cancer detection. Combining drug delivery methods with nanoflare-based systems can provide novel clinical approaches to cancer treatment. Kyriazi and co-workers developed a multiplex nanoflare-based drug delivery system using hybrids between DNA and AuNPs prepared by a self-assembly method. Two anticancer drugs were tested, mitoxantrone (MTX) and doxorubicin (DOX). Two separate AuNPS were linked together via DNA linkers, with attached sense strands of ssDNA that recognized mRNAs in living cells, and flare strands that were released upon binding to the target. The high surface area of oligonucleotide-modified AuNPs made it possible to load large amounts of drug, and specifically release it upon binding to the target, with the nanoflare as a fluorescent reporter. This multiplex nanoflare system is shown in Figure 9.121
Figure 9.
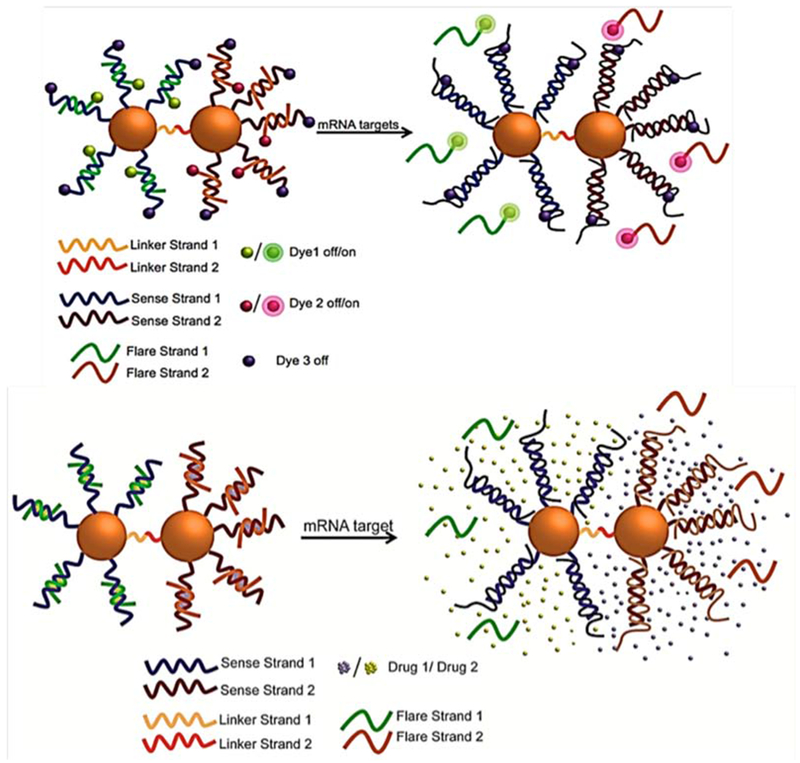
Multiplex dimeric AuNPs and DNAs hybrid-based nanoflares with fluorescent dyes Cy3, Cy5 and FAM have been used in flares. Simultaneous drug loading (MTX and DOX) on the nanoflare and delivery inside cells for anticancer therapy121 with permission.
The human mutT homologue (MTH1) is an oxidized purine nucleoside triphosphatase enzyme, which has been explored as a cancer marker. Simultaneous detection and inhibition of MTH1 activity could be beneficial in cancer therapy. Gao et al. developed AuNPs bound to mesoporous silica nanoparticles (MSNPs)-as nanoflares for detection and inhibition of MTH1 inside living cells. They used S-crizotinib (an aminopyridine drug) as a MTH1 inhibitor that was loaded onto the capped hybrid MSNP-AuNP nanoflare-based system. Release of S-crizotinib inside live cells was triggered by the hybridization process with the MTH1 mRNA. The rate of S-crizotinib release depended on the MSNPs pores that opened when the nanoflares hybridized to their target. They tested this novel method for different cancer types such as kidney, liver, and lung using both in vitro cultured cells (HeLa, HepG2 and HL7702 cancer cells), and in vivo in xenografted mouse tumors. Significant reductions in tumor size were observed. These MSNPs-AuNPs-based nanoflares are shown in Figure 10.122
Figure 10.
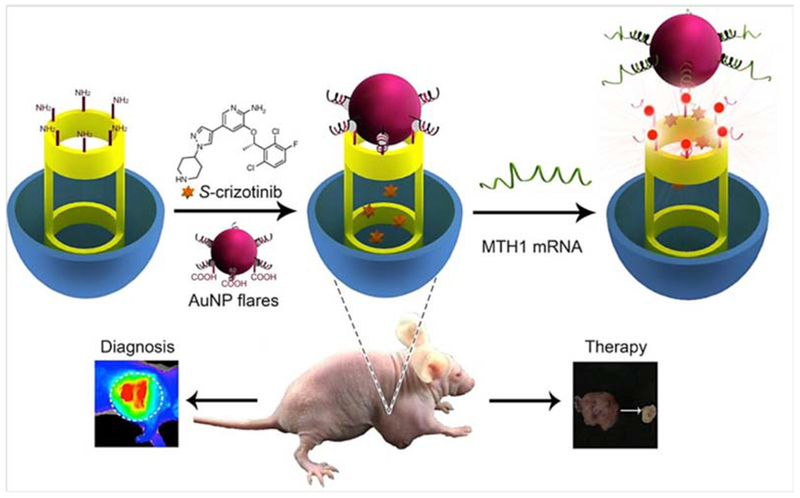
Schematic illustration of capped MSNP-AuNP nanoflares for MTH1 detection and inhibition in cancer therapy122 with permission.
The shapes and sizes of different nanomaterials,123,124 and their interfacial properties125 have large effects on their interactions with different tissues and organs. AuNP-DNA hybrids can take advantage of nucleic interactions to tailor the shape and size of the nanostructures, and have been termed “satellite-like”.126,127 Raeesi et al. reported satellite-like nanoflares using AuNPs modified with DNAs for efficient drug release. Intelligent design of these satellitelike nanoflares made it possible to manage the interactions between drug molecules and their carrier. The preparation and design of different parts of the satellite-like AuNP-based nanoflares is illustrated in Figure 11.128
Figure 11.
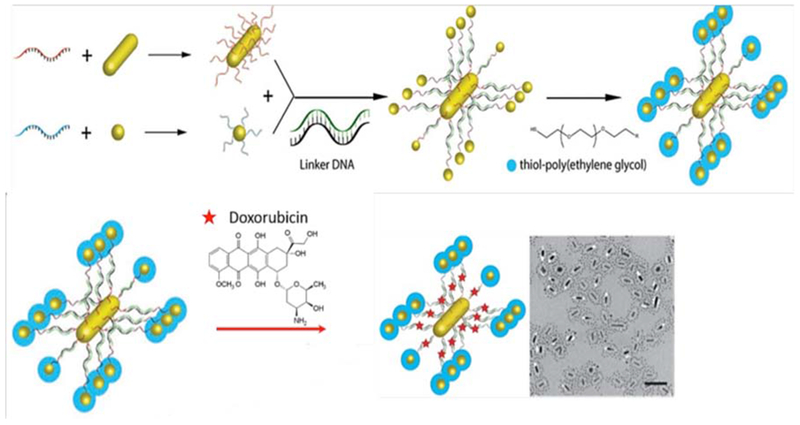
Gold nanorods (AuNRs) and AuNPs have been used in satellite-like nanoflares designed for loading with doxorubicin anticancer drug (upper panel), transmission electron microscopy (lower panel)128 with permission.
Adenosine triphosphate (ATP) detection
ATP is an important biomolecule with a key role in energy metabolism, intracellular biological pathways, and cellular activity. The natural range of ATP concentrations inside cells varies from 0.1–3.0 mM. Zheng et al. modified AuNPs with an RNA aptamer sequence to specifically detect ATP inside lysosomes.129 Jin et al. developed a nanoflare-based sensor with subcellular detection capability. According to the different ATP concentrations inside cells (<0.4 mM), and in the extracellular environment (1–10 mM),130 their AuNP-modified nanoflares had the appropriate sensitivity and activity for detection. The presence of other ATP-like biomolecules such as: uridine triphosphate, guanidine triphosphate and thymidine triphosphate did not interfere with the nanoflare-based sensor.131
pH Sensing
The pH levels inside cells can control different biological pathways. pH levels have been implicated in drug resistance, transportation of different ions, regulation of biological systems, membrane dynamics, and cell division and proliferation.132–135 Moreover correlations have been established between intracellular pH and myocardial ischemia,136 Alzheimer’s disease137 and many forms of cancer.138 “i-motif” biosensors have been constructed from ssDNA that have an intrinsic ability for detection of intracellular pH.139 At high pH, the fluorescent dye is hybridized and quenched, but at low pH the fluorescent probe sequence is released. Huang et al. modified ssDNA-AuNP-based nanoflares that could sense pH inside living cells. Changing the structural conformation of the i-motifs, made it possible to measure pH values ranging from 5.0–7.0. The pH-independent properties of the fluorophores, ensured that pH changes from 7.5–5.0 had no effect on the fluorescence intensity. Figure 12 shows a schematic illustrations of the i-motif for pH measurement.140
Figure 12.
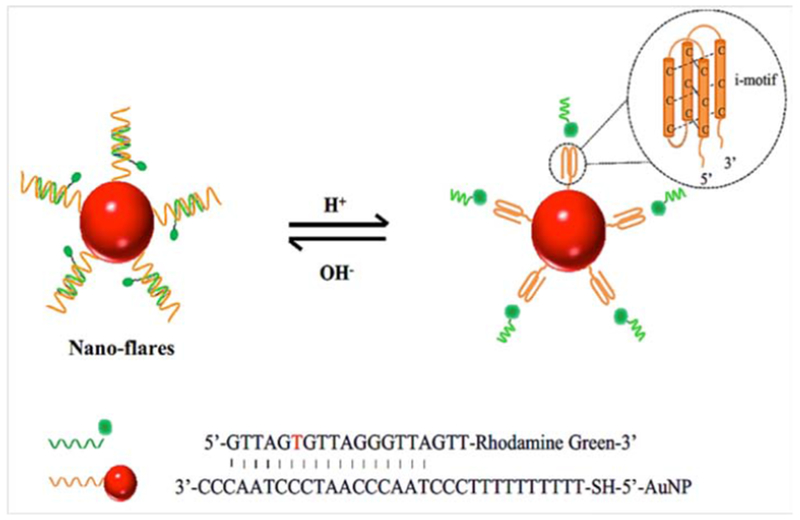
Schematic illustration of pH dependent i-motif-based nanoflares as pH biosensors140 with permission.
Inorganic ion detection
The intracellular levels of inorganic ions can affect numerous disease conditions.141 Zn2+ ions are involved in gene transcription and the transmission of neural signals.142,143 Cu2+ ions can regulate and control the mitochondrial respiratory function within cells, by modifying enzyme activities, and can modulate the immune system.144,145 Excessively high levels of Cu2+ can produce harmful effects such as disruption of neural systems, kidney and liver failure,146,147 and even contribute to prion-based diseases.148 Simultaneous detection of Zn2+ and Cu2+ inside cells could therefore be attractive. Nanoflare-based biosensors can be a powerful tool for the early-detection of biomarkers of Alzheimer’s disease and prion-based diseases in the human body. These biomarkers are often related to unbalanced levels of metal ions like Zn2+ and Cu2+. Li and co-workers designed a AuNP-DNAzyme nanoflare-based method for measurement of Zn2+ and Cu2+ ions in living cells. DNAzymes are specific for Zn2+ and Cu2+ ions, comprising substrate strands labeled with fluorophores at the 5′ end and quenchers at the 3′ end. The fluorescence of the fluorophores is quenched both by the gold nanoparticle and the quencher. After the nanoprobes are transferred into the cells, the substrate strands would be cleaved in the presence of the Zn2+ and Cu2+ target ions, resulting in disassociation of the shorter DNA fragments containing fluorophores. These multiplex nanoflares had high enzymatic stability and the ability to monitor and image Zn2+ and Cu2+ ions. They showed a response at Zn2+ and Cu2+ concentrations from 0 to 5.0 μM. The linear ranges were for Zn2+ (1.0–30.0 nM) and for Cu2+ (1.0–20.0 nM). The LOD was 0.47 nM and 0.45 nM for Zn2+ and Cu2+ ions, respectively. Other ions (Mn2+, Cd2+, Mg2+, Ni2+, K2+) did not show undue interferences because the nanoflares showed high optical signals for their target Zn2+ and Cu2+ ions. The use of AuNP-DNAzyme nanoflares for Zn2+ and Cu2+ ion detection is shown in Figure 13.149
Figure 13.
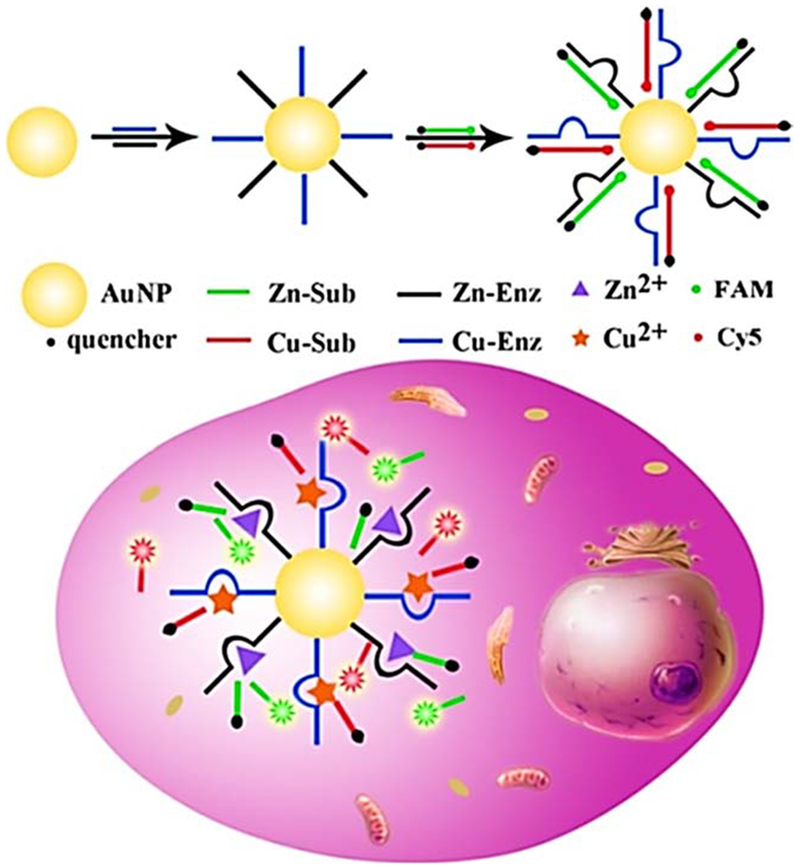
Detection of intracellular Zn2+ and Cu2+ ions based on AuNPs-DNAzyme nanoflares149 with permission.
K+ ions play a unique role biological in the metabolism of the heart and in hyperpolarization of cells in the nervous system.150,151 Yang et al. developed a FRET-based nanoflare system composed of AuNPs modified with G-quadruplex oligonucleotides for intracellular K+ ion detection. The K+ ions can interact with the G-quadruplex oligonucleotides which are dual-fluorophore-labeled so that FAM acts as a donor and TAMRA as an acceptor. The complementary oligonucleotides are designed to bind with the G-quadruplex sequences (flares) and immobilized on the AuNP surface via an Au–S bond. In the absence of target K+ ions, the G-quadruplex sequences bind to the complementary strands, separating the donor (FAM) and acceptor (TAMRA), and destroying the FRET coupling. In this open state, only the fluorescence of the donor dyes can be detected. However, in the presence of target K+, the flares are gradually displaced from the complementary strands, subsequently forming G-quadruplex structures that bring the donor and acceptor into close proximity and result in a high FRET coupling efficiency. In this closed state, the fluorescence of the acceptor can be detected when the donor is excited. Thus, the fluorescence emission ratio of acceptor to donor (A/D) can be used as a signal for measurement of target K+. Sodium ions (Na+) could interfere with assays for K+ ions, but for the modified AuNP-aptamer nanoflares the sensitivity for K+ ions was 4.5-fold higher than other ions. A schematic description of the AuNP-aptamer nanoflare system is shown in Figure 14.152
Figure 14.
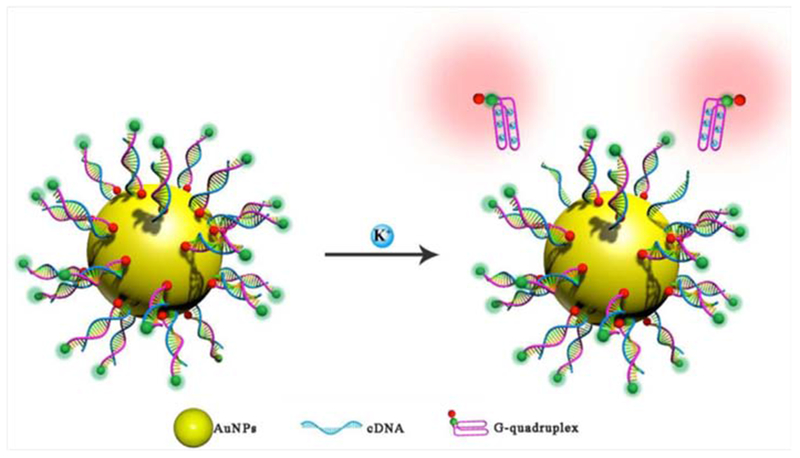
Interaction of K+ ions with AuNP-based G-quadrnplex oligonucleotide FRET nanoflares152 with permission.
Outlook and future perspectives
Nanoflares have become an exciting new alternative to more traditional intracellular fluorescence probes. They are able to enter cells relatively easily and detect the target genes (or other analytes) with high sensitivity and selectivity. Future work should be undertaken to explore the wider possibilities of using nanoflares for the detection and analysis of many genetic-based diseases in addition to cancer. Furthermore, we anticipate that nanoflares will have applications for investigating RNA functions inside cells, and its dysregulation in diseases. Nevertheless, knowledge about the intracellular trafficking of nanoflares at the molecular and tissue levels is still relatively sparse. More studies on the subcellular localization, transport from cell to cell within living systems, and penetration into tissues and organs are required.
Conclusion
Optical nanoflare-based imaging systems are becoming an efficient detection approach for many intracellular biomolecules. AuNPs modified with oligonucleotides can function as nanoflares and fluorescent biosensors with a high transfection capability, good enzymatic stability, appropriate surface plasmon resonance absorption peaks, and ability to match the emission wavelength. Their high selectivity and specificity could overcome weaknesses of traditional detection methods. The main topic of this review focused on the range of different targets that the nanoflares could detect inside cells. The ability to detect intracellular biomolecules is important for early-diagnosis of a range of diseases especially cancers, and for increased understanding of cancer cell signaling pathways. Nanoflares can be used to monitor intracellular drug delivery methods, and even to increase the efficiency of drug delivery. The structural and optical features of nanoflares are just starting to be fully understood, and their properties must be optimized for a diverse range of applications. These applications have so far have included the detection of a wide range of mRNAs (many of which are related to cancer such as EMT and telomerase), DNA expression, ATP levels, and levels of various inorganic ions inside cells. The unique design of nanoflares makes it possible to detect ultra-low concentrations of biomarkers in the complicated intracellular milieu. Interference between the target and other similar biomolecules was an unsolved challenge for many years, while nanoflares have helped to solve this problem. We predict that nanoflare-based fluorescence methods will continue to play a remarkable role in medical and biological sciences in the future.
Acknowledgments
Funding information: MRH was supported by US NIH grants R01AI050875 and R21AI121700.
Footnotes
Conflicts of interest: Dr. Hamblin is on the following Scientific Advisory Boards: Transdermal Cap Inc., Cleveland, OH. Photothera Inc., Carlsbad, CA. BeWell Global Inc., Wan Chai, Hong Kong. Hologenix Inc. Santa Monica, CA. LumiThera Inc., Poulsbo, WA. Vielight, Toronto, Canada. Bright Photomedicine, Sao Paulo, Brazil. Quantum Dynamics LLC, Cambridge, MA. Global Photon Inc., Bee Cave, TX. Medical Coherence, Boston, MA. NeuroThera, Newark, DE. JOOVV Inc., Minneapolis-St. Paul, MN. AIRx Medical, Pleasanton, CA. FIR Industries, Inc. Ramsey, NJ. UVLRx Therapeutics, Oldsmar, FL. Ultralux UV Inc., Lansing, MI. Illumiheal & Petthera, Shoreline, WA. MB Lasertherapy, Houston, TX. Dr. Hamblin has been a consultant for:. Lexington Int, Boca Raton, FL. USHIO Corp, Japan. Merck KGaA, Darmstadt, Germany. Philips Electronics Nederland B.V. Johnson & Johnson Inc., Philadelphia, PA. Sanofi-Aventis Deutschland GmbH, Frankfurt am Main, Germany. Dr. Hamblin is a stockholder of:. Global Photon Inc., Bee Cave, TX. Mitonix, Newark, DE.
References
- 1.Bratu DP, Cha B-J, Mhlanga MM, Kramer FR, Tyagi S. Visualizing the distribution and transport of mRNAs in living cells. Proc Natl Acad Sci 2003;100:13308–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Peng X-H, Cao Z-H, Xia J-T, Carlson GW, Lewis MM, Wood WC, et al. Real-time detection of gene expression in cancer cells using molecular beacon imaging: new strategies for cancer research. Cancer Res 2005;65:1909–17. [DOI] [PubMed] [Google Scholar]
- 3.Kloosterman WP, Wienholds E, de Bruijn E, Kauppinen S, Plasterk RH. In situ detection of miRNAs in animal embryos using LNA-modified oligonucleotide probes. Nat Methods 2006;3:27. [DOI] [PubMed] [Google Scholar]
- 4.Rosi NL, Giljohann DA, Thaxton CS, Lytton-Jean AK, Han MS, Mirkin CA. Oligonucleotide-modified gold nanoparticles for intracellular gene regulation. Science 2006;312:1027–30. [DOI] [PubMed] [Google Scholar]
- 5.Dubertret B, Calame M, Libchaber AJ. Single-mismatch detection using gold-quenched fluorescent oligonucleotides. Nat Biotechnol 2001;19:365. [DOI] [PubMed] [Google Scholar]
- 6.Opalinska JB, Gewirtz AM. Nucleic-acid therapeutics: basic principles and recent applications. Nat Rev Drug Discov 2002;1:503. [DOI] [PubMed] [Google Scholar]
- 7.Zabner J, Fasbender AJ, Moninger T, Poellinger KA, Welsh MJ. Cellular and molecular barriers to gene transfer by a cationic lipid. J BiolChem 1995;270:18997–9007. [DOI] [PubMed] [Google Scholar]
- 8.Kukowska-Latallo JF, Bielinska AU, Johnson J, Spindler R, Tomalia DA, Baker JR. Efficient transfer of genetic material into mammalian cells using starburst polyamidoamine dendrimers. Proc Natl Acad Sci 1996;93:4897–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mirkin CA, Letsinger RL, Mucic RC, Storhoff JJ. A DNA-based method for rationally assembling nanoparticles into macroscopic materials. Nature 1996;382:607. [DOI] [PubMed] [Google Scholar]
- 10.Elghanian R, Storhoff JJ, Mucic RC, Letsinger RL, Mirkin CA. Selective colorimetric detection of polynucleotides based on the distance-dependent optical properties of gold nanoparticles. Science 1997;277:1078–81. [DOI] [PubMed] [Google Scholar]
- 11.Storhoff JJ, Elghanian R, Mucic RC, Mirkin CA, Letsinger RL. One-pot colorimetric differentiation of polynucleotides with single base imperfections using gold nanoparticle probes. J Am Chem Soc 1998;120:1959–64. [Google Scholar]
- 12.Chen C-C, Chang T-W, Chen F-M, Hou M-F, Hung S-Y, Chong I-W, et al. Combination of multiple mRNA markers (PTTG1, Survivin, UbcH10 and TK1) in the diagnosis of Taiwanese patients with breast cancer by membrane array. Oncology 2006;70:438–46. [DOI] [PubMed] [Google Scholar]
- 13.Chhabra R, Sharma J, Wang H, Zou S, Lin S, Yan H, et al. Distance-dependent interactions between gold nanoparticles and fluorescent molecules with DNA as tunable spacers. Nanotechnology 2009;20:485201. [DOI] [PubMed] [Google Scholar]
- 14.Altieri DC. Survivin, versatile modulation of cell division and apoptosis in cancer. Oncogene 2003;22:8581. [DOI] [PubMed] [Google Scholar]
- 15.Chan WC, Maxwell DJ, Gao X, Bailey RE, Han M, Nie S. Luminescent quantum dots for multiplexed biological detection and imaging. Curr Opin Biotechnol 2002;13:40–6. [DOI] [PubMed] [Google Scholar]
- 16.Medintz IL, Uyeda HT, Goldman ER, Mattoussi H. Quantum dot bioconjugates for imaging, labelling and sensing. Nat Mater 2005;4:435. [DOI] [PubMed] [Google Scholar]
- 17.Michalet X, Pinaud F, Bentolila L, Tsay J, Doose S, Li J, et al. Quantum dots for live cells, in vivo imaging, and diagnostics. Science 2005;307:538–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Taback B, Chan AD, Kuo CT, Bostick PJ, Wang H-J, Giuliano AE, et al. Detection of occult metastatic breast cancer cells in blood by a multimolecular marker assay: correlation with clinical stage of disease. Cancer Res 2001;61:8845–50. [PubMed] [Google Scholar]
- 19.Resch-Genger U, Grabolle M, Cavaliere-Jaricot S, Nitschke R, Nann T. Quantum dots versus organic dyes as fluorescent labels. Nat Methods 2008;5:763. [DOI] [PubMed] [Google Scholar]
- 20.Yu WW, Qu L, Guo W, Peng X. Experimental determination of the extinction coefficient of CdTe, CdSe, and CdS nanocrystals. Chem Mater 2003;15:2854–60. [Google Scholar]
- 21.Wu C, Chiu DT. Highly fluorescent semiconducting polymer dots for biology and medicine. Angew Chem Int Ed 2013;52:3086–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu C, Peng H, Jiang Y, McNeill J. Energy transfer mediated fluorescence from blended conjugated polymer nanoparticles. J Phys Chem B 2006;110:14148–54. [DOI] [PubMed] [Google Scholar]
- 23.Wu C, Bull B, Szymanski C, Christensen K, McNeill J. Multicolor conjugated polymer dots for biological fluorescence imaging. ACS Nano 2008;2:2415–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu C, Schneider T, Zeigler M, Yu J, Schiro PG, Burnham DR, et al. Bioconjugation of ultrabright semiconducting polymer dots for specific cellular targeting. J Am Chem Soc 2010;132:15410–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim S, Lim C-K, Na J, Lee Y-D, Kim K, Choi K, et al. Conjugated polymer nanoparticles for biomedical in vivo imaging. Chem Commun 2010;46:1617–9. [DOI] [PubMed] [Google Scholar]
- 26.Wang F, Banerjee D, Liu Y, Chen X, Liu X. Upconversion nanoparticles in biological labeling, imaging, and therapy. Analyst 2010;135:1839–54. [DOI] [PubMed] [Google Scholar]
- 27.Chen G, Qiu H, Prasad PN, Chen X. Upconversion nanoparticles: design, nanochemistry, and applications in theranostics. Chem Rev 2014;114:5161–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Anker JN, Hall WP, Lyandres O, Shah NC, Zhao J, Van Duyne RP. Biosensing with plasmonic nanosensors. Nat Mater 2008;7:442. [DOI] [PubMed] [Google Scholar]
- 29.Homola J Surface plasmon resonance sensors for detection of chemical and biological species. Chem Rev 2008;108:462–93. [DOI] [PubMed] [Google Scholar]
- 30.Hutter E, Fendler JH. Exploitation of localized surface plasmon resonance. Adv Mater 2004;16:1685–706. [Google Scholar]
- 31.Willets KA, Van Duyne RP. Localized surface plasmon resonance spectroscopy and sensing. Annu Rev Phys Chem 2007;58:267–97. [DOI] [PubMed] [Google Scholar]
- 32.Kelly KL , Coronado E, Zhao LL, Schatz GC. The Optical Properties of Metal Nanoparticles: The Influence of Size, Shape, and Dielectric Environment. ACS Publications; 2003. [Google Scholar]
- 33.Qian H, Zhu M, Wu Z, Jin R. Quantum sized gold nanoclusters with atomic precision. Acc Chem Res 2012;45:1470–9. [DOI] [PubMed] [Google Scholar]
- 34.Torres VA, Tapia JC, Rodriguez DA, Parraga M, Lisboa P, Montoya M, et al. Caveolin-1 controls cell proliferation and cell death by suppressing expression of the inhibitor of apoptosis protein survivin. J Cell Sci 2006;119:1812–23. [DOI] [PubMed] [Google Scholar]
- 35.Crooke ST. Progress in antisense technology. Annu Rev Med 2004;55:61–95. [DOI] [PubMed] [Google Scholar]
- 36.Huisman W, Bouma J, Gruber M. Facilitation of lysosome disruption by ATP at low pH. Nature 1974;250:428. [DOI] [PubMed] [Google Scholar]
- 37.Sidransky D Nucleic acid-based methods for the detection of cancer. Science 1997;278:1054–8. [DOI] [PubMed] [Google Scholar]
- 38.Blacker TS, Mann ZF, Gale JE, Ziegler M, Bain AJ, Szabadkai G, et al. Separating NADH and NADPH fluorescence in live cells and tissues using FLIM. Nat Commun 2014;5:3936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Berezin MY, Achilefu S. Fluorescence lifetime measurements and biological imaging. Chem Rev 2010;110:2641–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Peterson JE, Zurakowski D, Italiano JE, Michel LV, Connors S, Oenick M, et al. VEGF, PF4 and PDGF are elevated in platelets of colorectal cancer patients. Angiogenesis 2012;15:265–73. [DOI] [PubMed] [Google Scholar]
- 41.Liu J, Liu C, Qiu L, Li J, Zhang P, Sun Y. Overexpression of both platelet-derived growth factor-BB and vascular endothelial growth factor-C and its association with lymphangiogenesis in primary human non-small cell lung cancer. Diagn Pathol 2014;9:128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Maxwell DJ, Taylor JR, Nie S. Self-assembled nanoparticle probes for recognition and detection of biomolecules. J Am Chem Soc 2002;124:9606–12. [DOI] [PubMed] [Google Scholar]
- 43.Kang KA, Wang J, Jasinski JB, Achilefu S. Fluorescence manipulation by gold nanoparticles: from complete quenching to extensive enhancement. J Nanobiotechnol 2011;9:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dulkeith E, Morteani A, Niedereichholz T, Klar T, Feldmann J, Levi S, et al. Fluorescence quenching of dye molecules near gold nanoparticles: radiative and nonradiative effects. Phys Rev Lett 2002;89:203002. [DOI] [PubMed] [Google Scholar]
- 45.Dulkeith E, Ringler M, Klar T, Feldmann J, Munoz Javier A, Parak W. Gold nanoparticles quench fluorescence by phase induced radiative rate suppression. Nano Lett 2005;5:585–9. [DOI] [PubMed] [Google Scholar]
- 46.Chithrani BD, Ghazani AA, Chan WC. Determining the size and shape dependence of gold nanoparticle uptake into mammalian cells. Nano Lett 2006;6:662–8. [DOI] [PubMed] [Google Scholar]
- 47.Alkilany AM, Murphy CJ. Toxicity and cellular uptake of gold nanoparticles: what we have learned so far? J Nanopart Res 2010;12:2313–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Giljohann DA, Seferos DS, Patel PC, Millstone JE, Rosi NL, Mirkin CA. Oligonucleotide loading determines cellular uptake of DNA-modified gold nanoparticles. Nano Lett 2007;7:3818–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cederquist KB, Keating CD. Curvature effects in DNA: au nanoparticle conjugates. ACS Nano 2009;3:256–60. [DOI] [PubMed] [Google Scholar]
- 50.Cao YC, Jin R, Thaxton CS, Mirkin CA. A two-color-change, nanoparticle-based method for DNA detection. Talanta 2005;67:449–55. [DOI] [PubMed] [Google Scholar]
- 51.Jin R, Cao YC, Thaxton CS, Mirkin CA. Glass-bead-based parallel detection of DNA using composite Raman labels. Small 2006;2:375–80. [DOI] [PubMed] [Google Scholar]
- 52.Stoeva SI, Lee JS, Thaxton CS, Mirkin CA. Multiplexed DNA detection with biobarcoded nanoparticle probes. Angew Chem 2006;118:3381–4. [DOI] [PubMed] [Google Scholar]
- 53.Hill HD, Vega RA, Mirkin CA. Nonenzymatic detection of bacterial genomic DNA using the bio bar code assay. Anal Chem 2007;79:9218–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xu X, Georganopoulou DG, Hill HD, Mirkin CA. Homogeneous detection of nucleic acids based upon the light scattering properties of silver-coated nanoparticle probes. Anal Chem 2007;79:6650–4. [DOI] [PubMed] [Google Scholar]
- 55.Schwarzenbach H, Hoon DS, Pantel K. Cell-free nucleic acids as biomarkers in cancer patients. Nat Rev Cancer 2011;11:426. [DOI] [PubMed] [Google Scholar]
- 56.Alhasan AH, Kim DY, Daniel WL, Watson E, Meeks JJ, Thaxton CS, et al. Scanometric microRNA array profiling of prostate cancer markers using spherical nucleic acid–gold nanoparticle conjugates. Anal Chem 2012;84:4153–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ueda K Glycoproteomic strategies: from discovery to clinical application of cancer carbohydrate biomarkers. Proteomics Clin Appl 2013;7:607–17. [DOI] [PubMed] [Google Scholar]
- 58.Füzéry AK, Levin J, Chan MM, Chan DW. Translation of proteomic biomarkers into FDA approved cancer diagnostics: issues and challenges. Clin Proteomics 2013;10:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hakomori S-i. Tumor-Associated Carbohydrate Antigens Defining Tumor Malignancy: Basis for Development of Anti-Cancer Vaccines The molecular immunology of complex carbohydrates—2. Springer; 2001. p. 369–402. [DOI] [PubMed] [Google Scholar]
- 60.Nam J-M, Thaxton CS, Mirkin CA. Nanoparticle-based bio-bar codes for the ultrasensitive detection of proteins. Science 2003;301:1884–6. [DOI] [PubMed] [Google Scholar]
- 61.Hill HD, Mirkin CA. The bio-barcode assay for the detection of protein and nucleic acid targets using DTT-induced ligand exchange. Nat Protoc 2006;1:324. [DOI] [PubMed] [Google Scholar]
- 62.Stoeva SI, Lee J-S, Smith JE, Rosen ST, Mirkin CA. Multiplexed detection of protein cancer markers with biobarcoded nanoparticle probes. J Am Chem Soc 2006;128:8378–9. [DOI] [PubMed] [Google Scholar]
- 63.Kim E-Y, Stanton J, Korber BT, Krebs K, Bogdan D, Kunstman K, et al. Detection of HIV-1 p24 Gag in Plasma by a Nanoparticle-Based Bio-Barcode-Amplification Method; 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lee J-S, Ulmann PA, Han MS, Mirkin CA. A DNA– gold nanoparticle-based colorimetric competition assay for the detection of cysteine. Nano Lett 2008;8:529–33. [DOI] [PubMed] [Google Scholar]
- 65.Findeisen P, Neumaier M. Functional protease profiling for diagnosis of malignant disease. Proteomics Clin Appl 2012;6:60–78. [DOI] [PubMed] [Google Scholar]
- 66.Surinova S, Schiess R, Huttenhain R, Cerciello F, Wollscheid B, Aebersold R. On the development of plasma protein biomarkers. J Proteome Res 2010;10:5–16. [DOI] [PubMed] [Google Scholar]
- 67.Shitrit D, Zingerman B, Shitrit AB-G, Shlomi D, Kramer MR. Diagnostic value of CYFRA 21-1, CEA, CA 19-9, CA 15-3, and CA 125 assays in pleural effusions: analysis of 116 cases and review of the literature. Oncologist 2005;10:501–7. [DOI] [PubMed] [Google Scholar]
- 68.Alivisatos AP. Semiconductor clusters, nanocrystals, and quantum dots. Science 1996;271:933–7. [Google Scholar]
- 69.Ying L, Wang Q. Microfluidic chip-based technologies: emerging platforms for cancer diagnosis. BMC Biotechnol 2013;13:76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Saylor RA, Lunte SM. A review of microdialysis coupled to microchip electrophoresis for monitoring biological events. J Chromatogr A 2015;1382:48–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hsu P-I, Chen C-H, Hsieh C-S, Chang W-C, Lai K-H, Lo G-H, et al. α1-antitrypsin precursor in gastric juice is a novel biomarker for gastric cancer and ulcer. Clin Cancer Res 2007;13:876–83. [DOI] [PubMed] [Google Scholar]
- 72.Khazanov E, Yavin E, Pascal A, Nissan A, Kohl Y, Reimann-Zawadzki M, et al. Detecting a secreted gastric cancer biomarker molecule by targeted nanoparticles for real-time diagnostics. Pharm Res 2012;29:983–93. [DOI] [PubMed] [Google Scholar]
- 73.Keshaviah A, Dellapasqua S, Rotmensz N, Lindtner J, Crivellari D, Collins J, et al. CA15-3 and alkaline phosphatase as predictors for breast cancer recurrence: a combined analysis of seven international breast Cancer study group trials. Ann Oncol 2007;18:701–8. [DOI] [PubMed] [Google Scholar]
- 74.Budd GT, Cristofanilli M, Ellis MJ, Stopeck A, Borden E, Miller MC, et al. Circulating tumor cells versus imaging—predicting overall survival in metastatic breast cancer. Clin Cancer Res 2006;12:6403–9. [DOI] [PubMed] [Google Scholar]
- 75.Hayes DF, Cristofanilli M, Budd GT, Ellis MJ, Stopeck A, Miller MC, et al. Circulating tumor cells at each follow-up time point during therapy of metastatic breast cancer patients predict progression-free and overall survival. Clin Cancer Res 2006;12:4218–24. [DOI] [PubMed] [Google Scholar]
- 76.Pantel K, Brakenhoff RH, Brandt B. Detection, clinical relevance and specific biological properties of disseminating tumour cells. Nat Rev Cancer 2008;8:329. [DOI] [PubMed] [Google Scholar]
- 77.He H, Xie C, Ren J. Nonbleaching fluorescence of gold nanoparticles and its applications in cancer cell imaging. Anal Chem 2008;80:5951–7. [DOI] [PubMed] [Google Scholar]
- 78.Haun JB, Devaraj NK, Hilderbrand SA, Lee H, Weissleder R. Bioorthogonal chemistry amplifies nanoparticle binding and enhances the sensitivity of cell detection. Nat Nanotechnol 2010;5:660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Liu M, Zhao H, Chen S, Yu H, Zhang Y, Quan X. A “turn-on” fluorescent copper biosensor based on DNA cleavage-dependent graphene-quenched DNAzyme. Biosens Bioelectron 2011;26:4111–6. [DOI] [PubMed] [Google Scholar]
- 80.Song E-Q, Hu J, Wen C-Y, Tian Z-Q, Yu X, Zhang Z-L, et al. Fluorescent-magnetic-biotargeting multifunctional nanobioprobes for detecting and isolating multiple types of tumor cells. ACS Nano 2011;5:761–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Choi CHJ, Hao L, Narayan SP, Auyeung E, Mirkin CA. Mechanism for the endocytosis of spherical nucleic acid nanoparticle conjugates. Proc Natl Acad Sci 2013;110:7625–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Massich MD, Giljohann DA, Schmucker AL, Patel PC, Mirkin CA. Cellular response of polyvalent oligonucleotide – gold nanoparticle conjugates. ACS Nano 2010;4:5641–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Li N, Chang C, Pan W, Tang B. A multicolor nanoprobe for detection and imaging of tumor-related mRNAs in living cells. Angew Chem Int Ed 2012;51:7426–30. [DOI] [PubMed] [Google Scholar]
- 84.Prigodich AE, Seferos DS, Massich MD, Giljohann DA, Lane BC, Mirkin CA. Nano-flares for mRNA regulation and detection. ACS Nano 2009;3:2147–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Robertson J, O’Neill K, Thomas M, McKenna P, Blamey R. Thymidine kinase in breast cancer. Cancer 1990;62:663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Broet P, Romain S, Daver A, Ricolleau G, Quillien V, Rallet A, et al. Thymidine kinase as a proliferative marker: clinical relevance in 1,692 primary breast cancer patients. J Clin Oncol 2001;19:2778–87. [DOI] [PubMed] [Google Scholar]
- 87.Meyer N, Penn LZ. Reflecting on 25 years with MYC. Nat Rev Cancer 2008;8:976. [DOI] [PubMed] [Google Scholar]
- 88.Liao D, Dickson R. c-Myc in breast cancer. Endocr Relat Cancer 2000;7:143–64. [DOI] [PubMed] [Google Scholar]
- 89.Xu J, Chen Y, Olopade OI. MYC and breast cancer. Genes Cancer 2010;1:629–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Efstratiadis A, Szabolcs M, Klinakis A. Notch, Myc and breast cancer. Cell Cycle 2007;6:418–29. [DOI] [PubMed] [Google Scholar]
- 91.Kuo CT, Bostick PJ, Ire RF, Morton DL, Conrad AJ, Hoon D. Assessment of messenger RNA of beta 1–> 4-N-acetylgalactosaminyl-transferase as a molecular marker for metastatic melanoma. Clin Cancer Res 1998;4:411–8. [PubMed] [Google Scholar]
- 92.Radisky DC. Epithelial-mesenchymal transition. J Cell Sci 2005;118:4325–6. [DOI] [PubMed] [Google Scholar]
- 93.Gunasinghe ND, Wells A, Thompson EW, Hugo HJ. Mesenchymal–epithelial transition (MET) as a mechanism for metastatic colonisation in breast cancer. Cancer Metastasis Rev 2012;31:469–78. [DOI] [PubMed] [Google Scholar]
- 94.Halo TL, McMahon KM, Angeloni NL, Xu Y, Wang W, Chinen AB, et al. NanoFlares for the detection, isolation, and culture of live tumor cells from human blood. Proc Natl Acad Sci 2014;111:17104–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Prigodich AE, Randeria PS, Briley WE, Kim NJ, Daniel WL, Giljohann DA, et al. Multiplexed nanoflares: mRNA detection in live cells. Anal Chem 2012;84:2062–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Pan W, Zhang T, Yang H, Diao W, Li N, Tang B. Multiplexed detection and imaging of intracellular mRNAs using a four-color nanoprobe. Anal Chem 2013;85:10581–8. [DOI] [PubMed] [Google Scholar]
- 97.Yang Y, Huang J, Yang X, Quan K, Wang H, Ying L, et al. FRET nanoflares for intracellular mRNA detection: avoiding false positive signals and minimizing effects of system fluctuations. J Am Chem Soc 2015;137:8340–3. [DOI] [PubMed] [Google Scholar]
- 98.Zhou X, Xing D. Assays for human telomerase activity: progress and prospects. Chem Soc Rev 2012;41:4643–56. [DOI] [PubMed] [Google Scholar]
- 99.Hong M, Xu L, Xue Q, Li L, Tang B. Fluorescence imaging of intracellular telomerase activity using enzyme-free signal amplification. Anal Chem 2016;88:12177–82. [DOI] [PubMed] [Google Scholar]
- 100.He C, Liu Z, Wu Q, Zhao J, Liu R, Liu B, et al. Ratiometric fluorescent biosensor for visual discrimination of cancer cells with different telomerase expression levels. ACS Sensors 2018;3:757–62. [DOI] [PubMed] [Google Scholar]
- 101.Peng Y, Croce CM. The role of MicroRNAs in human cancer. Signal Transduct Targeted Ther 2016;1:15004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hong M, Sun H, Xu L, Yue Q, Shen G, Li M, et al. In situ monitoring of cytoplasmic precursor and mature microRNA using gold nanoparticle and graphene oxide composite probes. Anal Chim Acta 2018;1021:129–39. [DOI] [PubMed] [Google Scholar]
- 103.Gao X, Jiang L, Hu B, Kong F, Liu X, Xu K, et al. Au–se-bond-based Nanoprobe for imaging MMP-2 in tumor cells under a high-thiol environment. Anal Chem 2018;90:4719–24. [DOI] [PubMed] [Google Scholar]
- 104.Hu B, Cheng R, Liu X, Pan X, Kong F, Gao W, et al. A nanosensor for in vivo selenol imaging based on the formation of AuSe bonds. Biomaterials 2016;92:81–9. [DOI] [PubMed] [Google Scholar]
- 105.Shi J, Zhou M, Gong A, Li Q, Wu Q, Cheng GJ, et al. Fluorescence lifetime imaging of Nanoflares for mRNA detection in living cells. Anal Chem 2016;88:1979–83. [DOI] [PubMed] [Google Scholar]
- 106.Czarnek M, Bereta J. SmartFlares fail to reflect their target transcripts levels. Sci Rep 2017;7:11682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Borghei Y-S, Hosseini M, Ganjali MR, Ju H. Colorimetric and energy transfer based fluorometric turn-on method for determination of microRNA using silver nanoclusters and gold nanoparticles. Microchim Acta 2018;185:286. [DOI] [PubMed] [Google Scholar]
- 108.Li J, Huang J, Yang X, Yang Y, Quan K, Xie N, et al. Two-color-based nanoflares for multiplexed MicroRNAs imaging in live cells. Nanotheranostics 2018;2:96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Bai S, Xu B, Guo Y, Qiu J, Yu W, Xie G. High-discrimination factor nanosensor based on tetrahedral DNA nanostructures and gold nanoparticles for detection of MiRNA-21 in live cells. Theranostics 2018;8:2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Bo B, Zhang T, Jiang Y, Cui H, Miao P. Triple signal amplification strategy for ultrasensitive determination of miRNA based on duplex specific nuclease and bridge DNA–gold nanoparticles. Anal Chem 2018;90:2395–400. [DOI] [PubMed] [Google Scholar]
- 111.Seferos DS, Giljohann DA, Hill HD, Prigodich AE, Mirkin CA. Nanoflares: probes for transfection and mRNA detection in living cells. J Am Chem Soc 2007;129:15477–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Wu Q, Liu Z, Su L, Han G, Liu R, Zhao J, et al. Sticky-flares for in situ monitoring of human telomerase RNA in living cells. Nanoscale 2018;10:9386–92. [DOI] [PubMed] [Google Scholar]
- 113.Levy R, Held M, Mason D, Comenge J, Carolan G. The spherical nucleic acids mRNA detection paradox. Science Open Res 2015. [Google Scholar]
- 114.Lu L, Qian Y, Wang L, Ma K, Zhang Y. Metal-enhanced fluorescence-based core–shell ag@ SiO2 nanoflares for affinity biosensing via target-induced structure switching of aptamer. ACS Appl Mater Interfaces 2014;6:1944–50. [DOI] [PubMed] [Google Scholar]
- 115.Yang B, Zhang X-B, Kang L-P, Huang Z-M, Shen G-L, Yu R-Q, et al. Intelligent layered nanoflare:“lab-on-a-nanoparticle” for multiple DNA logic gate operations and efficient intracellular delivery. Nanoscale 2014;6:8990–6. [DOI] [PubMed] [Google Scholar]
- 116.Liang P, Canoura J, Yu H, Alkhamis O, Xiao Y. Dithiothreitol-regulated coverage of oligonucleotide-modified gold nanoparticles to achieve optimized biosensor performance. ACS Appl Mater Interfaces 2018;10:4233–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Liang S, He G, Tian J, Zhao Y, Zhao S. Fluorescence polarization gene assay for HIV-DNA based on the use of dendrite-modified gold nanoparticles acting as signal amplifiers. Microchim Acta 2018;185:119. [DOI] [PubMed] [Google Scholar]
- 118.Vilela P, Heuer-Jungemann A, El-Sagheer A, Brown T, Muskens OL, Smyth NR, et al. Sensing of vimentin mRNA in 2D and 3D models of wounded skin using DNA-coated gold nanoparticles. Small 2018;14:1703489. [DOI] [PubMed] [Google Scholar]
- 119.Ma H, Li Z, Xue N, Cheng Z, Miao X. A gold nanoparticle based fluorescent probe for simultaneous recognition of single-stranded DNA and double-stranded DNA. Microchim Acta 2018;185:93. [DOI] [PubMed] [Google Scholar]
- 120.Vasimalai N, Fernández-Argüelles MaT, Espiña B. Detection of sulfide using mercapto tetrazine-protected fluorescent gold nanodots: preparation of paper-based testing kit for on-site monitoring. ACS Appl Mater Interfaces 2018;10:1634–45. [DOI] [PubMed] [Google Scholar]
- 121.Kyriazi M-E, Giust D, El-Sagheer AH, Lackie PM, Muskens OL, Brown T, et al. Multiplexed mRNA sensing and combinatorial-targeted drug delivery using DNA-gold nanoparticle dimers. ACS Nano 2018;12:3333–40. [DOI] [PubMed] [Google Scholar]
- 122.Gao W, Cao W, Sun Y, Wei X, Xu K, Zhang H, et al. AuNP flares-capped mesoporous silica nanoplatform for MTH1 detection and inhibition. Biomaterials 2015;69:212–21. [DOI] [PubMed] [Google Scholar]
- 123.Florez L, Herrmann C, Cramer JM, Hauser CP, Koynov K, Landfester K, et al. How shape influences uptake: interactions of anisotropic polymer nanoparticles and human mesenchymal stem cells. Small 2012;8:2222–30. [DOI] [PubMed] [Google Scholar]
- 124.Jokerst JV, Lobovkina T, Zare RN, Gambhir SS. Nanoparticle PEGylation for imaging and therapy. Nanomedicine 2011;6:715–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Yang Z, Wang J, Lu Q, Xu J, Kobayashi Y, Takakura T, et al. PEGylation confers greatly extended half-life and attenuated immunogenicity to recombinant methioninase in primates. Cancer Res 2004;64:6673–8. [DOI] [PubMed] [Google Scholar]
- 126.Chou LY, Zagorovsky K, Chan WC. DNA assembly of nanoparticle superstructures for controlled biological delivery and elimination. Nat Nanotechnol 2014;9:148–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Ohta S, Glancy D, Chan WC. DNA-controlled dynamic colloidal nanoparticle systems for mediating cellular interaction. Science 2016;351:841–5. [DOI] [PubMed] [Google Scholar]
- 128.Raeesi V, Chou LY, Chan WC. Tuning the drug loading and release of DNA-assembled gold-Nanorod superstructures. Adv Mater 2016;28:8511–8. [DOI] [PubMed] [Google Scholar]
- 129.Zheng D, Seferos DS, Giljohann DA, Patel PC, Mirkin CA. Aptamer nano-flares for molecular detection in living cells. Nano Lett 2009;9:3258–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Mo R, Jiang T, DiSanto R, Tai W, Gu Z. ATP-triggered anticancer drug delivery. Nat Commun 2014;5:3364. [DOI] [PubMed] [Google Scholar]
- 131.Jin F, Zheng J, Liu C, Yang S, Li Y, Li J, et al. Dual-stimuli responsive i-motif/nanoflares for sensing ATP in lysosomes. Analyst 2014;139:3714–7. [DOI] [PubMed] [Google Scholar]
- 132.Li J, Eastman A. Apoptosis in an interleukin-2-dependent cytotoxic T lymphocyte cell line is associated with intracellular acidification role of the Na/H-antiport. J Biol Chem 1995;270:3203–11. [DOI] [PubMed] [Google Scholar]
- 133.Caceres-Cortes J, Rajotte D, Dumouchel J, Haddad P, Hoang T. Product of the steel locus suppresses apoptosis in hemopoietic cells. Comparison with pathways activated by granulocyte macrophage colony-stimulating factor. J Biol Chem 1994;269:12084–91. [PubMed] [Google Scholar]
- 134.Barry MA, Reynolds JE, Eastman A. Etoposide-induced apoptosis in human HL-60 cells is associated with intracellular acidification. Cancer Res 1993;53:2349–57. [PubMed] [Google Scholar]
- 135.Busa WB. Mechanisms and consequences of pH-mediated cell regulation. Annu Rev Physiol 1986;48:389–402. [DOI] [PubMed] [Google Scholar]
- 136.Liu R, Liu L, Liang J, Wang Y, Wei Y, Gao F, et al. Detection of pH change in cytoplasm of live myocardial ischemia cells via the ssDNA-SWCNTs nanoprobes. Anal Chem 2014;86:3048–52. [DOI] [PubMed] [Google Scholar]
- 137.Davies T, Fine R, Johnson R, Levesque C, Rathbun W, Seetoo K, et al. Non-age related differences in thrombin responses by platelets from male patients with advanced Alzheimer’ s disease. Biochem Biophys Res Commun 1993;194:537–43. [DOI] [PubMed] [Google Scholar]
- 138.Izumi H, Torigoe T, Ishiguchi H, Uramoto H, Yoshida Y, Tanabe M, et al. Cellular pH regulators: potentially promising molecular targets for cancer chemotherapy. Cancer Treat Rev 2003;29:541–9. [DOI] [PubMed] [Google Scholar]
- 139.Wang W, Yang Y, Cheng E, Zhao M, Meng H, Liu D, et al. A pH-driven, reconfigurable DNA nanotriangle. Chem Commun 2009:824–6. [DOI] [PubMed] [Google Scholar]
- 140.Huang J, He Y, Yang X, Wang K, Ying L, Quan K, et al. I-motif-based nano-flares for sensing pH changes in live cells. Chem Commun 2014;50:15768–71. [DOI] [PubMed] [Google Scholar]
- 141.Zhang L, Zhang Y-M, Liang R-P, Qiu J-D. Colorimetric logic gates based on ion-dependent DNAzymes. J Phys Chem C 2013;117:12352–7. [Google Scholar]
- 142.Nies DH. Microbial heavy-metal resistance. Appl Microbiol Biotechnol 1999;51:730–50. [DOI] [PubMed] [Google Scholar]
- 143.Carol P, Sreejith S, Ajayaghosh A. Ratiometric and near-infrared molecular probes for the detection and imaging of zinc ions. Chem Asian J 2007;2:338–48. [DOI] [PubMed] [Google Scholar]
- 144.Şengör SS, Gikas P, Moberly JG, Peyton BM, Ginn TR. Comparison of single and joint effects of Zn and cu in continuous flow and batch reactors. J Chem Technol Biotechnol 2012;87:374–80. [Google Scholar]
- 145.Balch PA. Prescription for Nutritional Healing. Penguin; 2006. [Google Scholar]
- 146.Georgopoulos PG, A R, Yonone-Lioy MJ, Opiekun RE, Lioy PJ. Environmental copper: its dynamics and human exposure issues. Crit Rev 2001;4:341–94. [DOI] [PubMed] [Google Scholar]
- 147.Zietz BP, Dieter HH, Lakomek M, Schneider H, Keßler-Gaedtke B, Dunkelberg H. Epidemiological investigation on chronic copper toxicity to children exposed via the public drinking water supply. Sci Total Environ 2003;302:127–44. [DOI] [PubMed] [Google Scholar]
- 148.Brown DR, Kozlowski H. Biological inorganic and bioinorganic chemistry of neurodegeneration based on prion and Alzheimer diseases. Dalton Trans 2004:1907–17. [DOI] [PubMed] [Google Scholar]
- 149.Li L, Feng J, Fan Y, Tang B. Simultaneous imaging of Zn2+ and Cu2+ in living cells based on DNAzyme modified gold nanoparticle. Anal Chem 2015;87:4829–35. [DOI] [PubMed] [Google Scholar]
- 150.Kofuji P, Newman E. Potassium buffering in the central nervous system. Neuroscience 2004;129:1043–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Malinska D, Mirandola SR, Kunz WS. Mitochondrial potassium channels and reactive oxygen species. FEBS Lett 2010;584:2043–8. [DOI] [PubMed] [Google Scholar]
- 152.Yang Y, Huang J, Yang X, Quan K, Xie N, Ou M, et al. Aptamer-based FRET nanoflares for imaging potassium ions in living cells. Chem Commun 2016;52:11386–9. [DOI] [PubMed] [Google Scholar]


