Abstract
Prader-Willi syndrome (PWS) is an imprinted neurodevelopmental disease caused by a loss of paternal genes on chromosome 15q11-q13. It is characterized by cognitive impairments, developmental delay, sleep abnormalities, and hyperphagia often leading to obesity. Clinical research has shown that a lack of expression of SNORD116, a paternally expressed imprinted gene cluster that encodes multiple copies of a small nucleolar RNA (snoRNA) in both humans and mice, is most likely responsible for many PWS symptoms seen in humans. The majority of previous research using PWS preclinical models focused on characterization of the hyperphagic and metabolic phenotypes. However, a crucial understudied clinical phenotype is cognitive impairments and thus we investigated the learning and memory abilities using a model of PWS, with a heterozygous deletion in Snord116. We utilized the novel object recognition task, which doesn’t require external motivation, or exhaustive swim training. Automated findings were further confirmed with manual scoring by a highly trained blinded investigator. We discovered deficits in Snord116 +/– mutant mice in the novel object recognition, location memory and tone cue fear conditioning assays when compared to age-, sex- matched, littermate control Snord116 +/+ mice. Further, we confirmed that despite physical neo-natal developmental delays, Snord116 +/− mice had normal exploratory and motor abilities. These results show that the Snord116 + /– deletion murine model is a valuable preclinical model for investigating learning and memory impairments in individuals with PWS without common confounding phenotypes.
Keywords: Behavior, Animal model, Neurodevelopment, Learning and memory, Prader-Willi, Genetics, Snord116, Cognitive
1. Introduction
Prader-Willi syndrome (PWS) is a rare genetic disorder that occurs in approximately 1:15,000 live births. PWS is characterized by neonatal failure to thrive, developmental delay, lack of satiety cues, lack of control of food intake and metabolic abnormalities that lead to obesity. In addition to obesity, PWS affected individuals usually exhibit low to moderate intellectual disability and issues with numerous tasks related to executive function (e.g., response inhibition, set-shifting and planning) (Dykens, Hodapp, Walsh, & Nash, 1992a, 1992b; Dykens, Leckman, & Cassidy, 1996; Dykens et al., 2017; Dykens and Shah, 2003; Martin et al., 1998; Schwartz et al., 2016).
Due to the imprinted status of the 15q11-q13 locus, allele-specific mutations lead to the development of two distinct neurodevelopmental disorders. PWS results from genetic mutations leading to deficient gene expression from the paternal allele of chromosome 15 (15q11-q13), while Angelman Syndrome (AS) results from mutations inherited from the maternal allele of the same locus, emphasizing a critical neurological function of these genes (Jiang, Tsai, Bressler, & Beaudet, 1998). Numerous mutant mouse models were generated by targeting different portions of the orthologous mouse region. The first model deleted the entire PWS/AS locus, similar to the vast majority of PWS and/or AS patients (Gabriel et al., 1999; Stefan, Portis, Longnecker, & Nicholls, 2005). Follow-up model systems highlighted the PWS imprinting center and informed the essential nature of the epigenetic processes (Yang et al., 1998). Several smaller deletion and single gene mutant model systems have been created to delineate the contribution of each gene in the region to aspects of the PWS phenotype. These include models deleting exons in the small nuclear ribonucleoprotein polypeptide N (Snrpn), an RNA-protein complex that forms an RNA spliceosome (Bressler et al., 2001; Dubose, Smith, Yang, Johnstone, & Resnick, 2011; Tsai, Jiang, et al., 1999), necdin (Ndn), which can influence multiple pathways involved in neuronal survival, differentiation and outgrowth and Magel2, a Ndn homolog that is expressed in the hypothalamus, cerebral cortex and spinal cord and key in circadian timekeeping (Kozlov et al., 2007; Lee et al., 2000).
While many genes are likely involved in the entire complicated PWS phenotype (including developmental failure to thrive, metabolic anomalies, obesity, cognitive deficits, obsessive-compulsive behaviors, and other psychiatric co-morbidities), analyses of rare PWS patients have determined that a ~ 200 kb deletion of a cluster of small nucleolar/noncoding RNAs (snoRNAs) within this locus is sufficient to cause PWS (Bieth et al., 2015; de Smith et al., 2009; Duker et al., 2010; Sahoo et al., 2008). Thus, investigation into the loss of this particular group of snoRNA genes, known as the Snord116 cluster, as contributing factors is an essential step toward elucidating the neurobiology of the PWS phenotype.
Ding et al. (2008) derived a novel mouse model for PWS by deleting the paternal copies of Snord116. Mice with paternally inherited Snord116 +/− deletions had growth delays in the first three postnatal weeks, hyperphagia, prolonged mealtime and increased circulating ghrelin but normal pain sensitivity, motor abilities and working memory measured by the spontaneous alternation task (Ding et al., 2008). Lassi, Maggi, et al. (2016) observed significant changes in working-for-food behavioral responses at various timescales in a home cage behavioral system in Snord116 +/− mice (Lassi, Maggi, et al., 2016). It is unknown how or if the missing Snord116 cluster contributes to other components of the intellectual disability behavioral diagnosis in clinical populations since the overwhelming majority of previous research using PWS preclinical models focused on characterization of the hyperphagic and metabolic phenotypes. The following report carefully investigated learning and memory abilities in the heterozygous deletion Snord116 +/− model of PWS. We discovered that Snord116 +/− mice exhibited learning and memory impairments compared to their wildtype littermates on the low stress-standard assays of novel object recognition (NOR) and object location memory (OLM). Second, we illustrated data that were quantified with reasonable signal to noise ratio by automated tracking software, and confirmed and fine-tuned by human manual scoring. We also discovered that the Snord116 +/− group showed deficits in cued but not contextual fear conditioning. Finally, we confirmed our results were not confounded by physical or motor dysfunction.
2. Materials and methods
2.1. Subjects
All animals were housed in a temperature-controlled vivarium maintained on a 12:12 light-dark cycle. All procedures were approved by the Institutional Animal Care and Use Committee at the University of California Davis and were conducted in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals. B6(Cg)-Snord116tm11Uta/J (Snord116 +/– ) mice were obtained from The Jackson Laboratory (Bar Harbor, ME, USA) and fed a standard diet of Teklad global 18% protein rodent diets 2918 (Envigo, Hayward, CA, USA). Heterozygous deletion male mice were bred with C57BL/6J females to generate paternal deletion Snord116 +/− and wildtype (Snord116 +/+) littermates. To identify mice, pups were labelled by paw tattoo over postnatal days (PND) 2–4 using non-toxic animal tattoo ink (Ketchum Manufacturing Inc., Brockville, ON, Canada). Tails of pups were clipped (1–2 mm) for genotyping, following the UC Davis IACUC policy regarding tissue collection. Genotyping was performed with REDExtract-N-Amp (Sigma Aldrich, St. Louis, MO, USA) using primers oIMR7958 AAT CCC CAA CCT ACT TCA AAC AGT C, oIMR7959 TGG ATC TCT CCT TGC TTG TTT TCT C, and oIMR7960 TTT ACG GTA CAT GAC AGC ACT CAA G.
2.2. Behavioral assays
Order of behavioral testing and description of cohorts.
To determine if findings were robust and reproducible 2 cohorts were used for the novel object recognition assessment. The second cohort was also used to assess developmental milestones at PND10, 12 and 14, elevated plus maze, light ↔ dark conflict, open field behavior, balance beam walking, object location memory and fear conditioning. To minimize the carry-over effects from repeated testing, assays were performed in the order of least to the most stressful tasks. Cohort 1 was sampled from 7 litters and was tested in novel object recognition (NOR). Cohort 2 was sampled from 6 litters and was tested in elevated plus-maze, light ↔ dark, open field, beam walking, NOR, object location memory (OLM), and context and cued fear conditioning. All male and female mice were used from every litter. In cohort 1, the order and age of testing was as follows: (1) NOR at 7 weeks of age. In cohort 2, the order of testing was as follows with at least 48-hrs separating tasks: (1) PND 10, 12, 14 developmental milestones, (2) elevated plus-maze at 6 weeks of age, (3) light ↔ dark exploration task at 6 weeks of age, (4) open field locomotion at 7 weeks of age, (5) balance beam walking at 8 weeks of age, (6) NOR at 8 weeks of age, (7) OLM at 10 weeks of age, and (8) fear conditioning at 12 weeks of age. All behavioral assays in all cohorts were performed between 9 am-4 pm PST (ZT2-ZT9).
(i). Developmental milestones.
Developmental milestones were measured on postnatal days (PND) 10, 12 and 14, similar to those previously described (Fox, 1965; Vogel Ciernia et al., 2017; Yang et al., 2012). All measures were conducted by an experimenter blind to genotype. Body weight, length (nose to edge of tail), and head width were measured using a scale (grams) and a digital caliper (cm). Cliff avoidance was tested by placing each pup near the edge of a cardboard box, gently nudging it towards the edge, and measuring the time for it to turn and back away from the edge. Failures to avoid the cliff was recorded as a maximum score of 30-s. Righting reflex was tested by placing each pup on its back, releasing it, and measuring the time for it to fully flip over onto four paws on each developmental day. Negative geotaxis was tested by placing each pup, facing downwards, on a screen angled at 45° from parallel, and measuring the time for it to completely turn and to climb to the top of the screen. Failures to turn and climb were recorded as a maximum score of 30-s. Circle transverse was tested by placing each pup in the center of a circle with a 5″ (12.5 cm) diameter drawn on a laminated sheet of 8.5″ × 11″ white paper, and measuring the time for it to exit the circle. Failures to exit the circle were recorded as a maximum score of 30-s.
(ii). Novel object recognition (NOR).
The NOR test was conducted in opaque matte white (P95 White, Tap Plastics, Sacramento, CA) open field arenas (41 cm × 41 cm × 30 cm), using methods similar to those previously described (Copping, Christian, et al., 2017; Gompers et al., 2017). The experiment consisted of four sessions: a 30-min exposure to the open field arena the day before the test, a 10-min re-habituation on test day, a 10-min familiarization session and a 5-min recognition test. On day 1, each subject was habituated to a clean, empty open field arena for 30-min. 24-hr later, each subject was returned to the open field arena for another 10- min for the habituation phase. The mouse was then removed from the open field and placed in a clean temporary holding cage for approximately 2-min. Two identical objects were placed in the arena. Each subject was returned to the open field in which it had been habituated and allowed to freely explore for 10-min. After the familiarization session, subjects were returned to their holding cages, which were transferred from the testing room to a nearby holding area. The open field was cleaned with 70% ethanol and let dry. One clean familiar object and one clean novel object were placed in the arena, where the two identical objects had been located during in the familiarization phase. 60-min after the end of the familiarization session, each subject was returned to its open field for a 5-min recognition test, during which time it was allowed to freely explore the familiar object and the novel object. The familiarization session and the recognition test were recorded and scored with Ethovision XT videotracking software (Version 9.0, Noldus Information Technologies, Leesburg, VA). Object investigation was defined as time spent sniffing the object when the nose was oriented toward the object and the nose–object distance was 2-cm or less. Recognition memory was defined as spending significantly more time sniffing the novel object compared to the familiar object via a Student’s paired t-test. Total time spent sniffing both objects was used as a measure of general exploration. Time spent sniffing two identical objects during the familiarization phase confirmed the lack of an innate side bias. Objects used were plastic toys: a small soft plastic orange safety cone and a hard plastic magnetic cone with ribbed sides, as previously described (Gompers et al., 2017; Gulinello et al., 2018).
(iii). Object location memory (OLM).
The OLM test was conducted as previously described (Vogel-Ciernia and Wood, 2014; Yang, Lewis, Sarvi, Foley, & Crawley, 2015), in the same apparatus used for testing NOR. Each animal was habituated to a clean empty arena for 30-min, 24-hrs before the experiment, and again for another 10-min on the day of the experiment. The subject was then exposed to two identical objects (red coral object, ~ 2.5 cm tall, Safari Ltd.) placed 12-cm away from the wall and 18-cm from each other) for a 10-min familiarization session. To facilitate spatial memory, a 2 × 30-cm vertical black stripe was taped on the wall opposite to the start location. To test OLM, one familiar object remained at the original location and the second familiar object was displaced 18-cm from its original location. The relocated object was diagonal to, and 22-cm from, the unmoved object. OLM was tested in a 5-min session, beginning 1-hr after the familiarization session. Object investigation was analyzed with Ethovision XT videotracking software and from the digital recorded videos, by human observers blind to genotype information.
(iv). Open field locomotion.
General exploratory locomotion in a novel open field environment was assayed in an arena sized 40 cm × 40 cm × 30.5 cm, as previously described (Copping, Berg, et al., 2017; Copping, Christian, et al., 2017; Gompers et al., 2017). Open field activity was considered an essential control for effects on physical activity, for example, sedation or hyperactivity which could confound the interpretation of result of interaction with objects, arena and object exploration, and sniffing and investigation times. The testing room was illuminated at ~ 40 lx.
(v). Beam walking.
A beam walking motor task was conducted as previously described, with some modifications as outlined (Carter et al., 2001, chap. 8; Vogel Ciernia et al., 2017). 59 cm long round rods were suspended 68 cm above a cushioned landing pad. A goal box at the end of the beam consisted of a 12 cm diameter cylinder to provide motivation to cross the beam. Each mouse was placed at one end of the beam and the time to cross to the goal box on the other end was measured. Testing sequence moved from largest diameter to smallest diameter rods in order of increased difficulty. On the day prior to testing, all animals were given two practice trials on the largest diameter round beam in order to become accustom to the procedure. On the test day, each animal was sequentially tested on three round rods (35, 18 and 13 mm). Testing sequence was based on presentations of decreasing diameter to present increasing levels of difficulty. Each mouse was given two trials on each beam, separated by approximately 30 min. The time to transverse the beam was recorded and averaged across the two trials for each beam. A maximum time of 60 s was assigned to individuals that failed to cross the beam in that duration. In the small number of cases where mice fell from the beam, a score of 60-s was assigned.
(vi). Elevated plus-maze (EPM).
The assay was performed using a mouse EPM (model ENV-560A) purchased from Med Associates (St. Albans, VT) and performed as previously described (Copping, Christian, et al., 2017; Flannery et al., 2015). The EPM contained two open arms (35.5 cm × 6 cm) and two closed arms (35.5 cm × 6 cm) radiating from a central area (6 cm × 6 cm). The maze was cleaned with 70% ethanol before the beginning of the first test session and after each subject mouse was tested with sufficient time for the ethanol odor to dissipate before the start of the next test session. Room illumination was ~ 30 lx.
(vii). Light ↔ dark conflict.
The light ↔ dark assay was performed in accordance with previously described procedures (Copping, Christian, et al., 2017; Flannery et al., 2015). The test began by placing the mouse in the light side (~ 320 lx; 28 cm × 27.5 cm × 27 cm) of an automated 2-chambered apparatus, in which the enclosed/dark side (~5lx; 28 cm × 27.5 cm × 19 cm) was reached by traversing the small opening of the partition between the two chambers. The mouse was allowed to explore freely for 10-min. Time in the dark side chamber and total number of transitions between the light and dark side chambers were automatically recorded during the 10-min session using Labview 8.5.1 software (National Instruments, Austin, TX).
(viii). Contextual and cued fear conditioning.
Delay contextual and cued fear conditioning was conducted using an automated fear-conditioning chamber (Med Associates, St Albans, VT, USA). The conditioning chamber was interfaced to a PC installed with VideoFreeze software (version 1.12.0.0, Med Associates) and enclosed in a sound-attenuating cubicle. Training consisted of a 2-min acclimation period followed by three tone-shock (CS–US) pairings (80 dB tone, duration 30-s; 0.5 mA footshock, duration 1 s; intershock interval 90-s) and a 2.5-min period, during which no stimuli were presented. The environment was well lit (~ 100 lx), with a stainless steel grid floor and swabbed with almond odor cue (prepared from almond extract; McCormick; 1:100 dilution). A 5-min test of contextual fear conditioning was performed 24-hr after training, in the absence of the tone and footshock, but in the presence of 100 lx overhead lighting, almond odor and chamber cues identical to those used on the training day. Cued fear conditioning, conducted 48-hr after training, was assessed in a novel environment with distinct visual, tactile and orange olfactory cues. Overhead lighting was turned off. The cued test consisted of a 3-min acclimation period followed by a 3-min presentation of the tone CS and a 90-s exploration period. Cumulative time spent freezing in each condition was quantified by VideoFreeze software (Med Associates).
2.3. Statistical analysis
Data were analyzed with Statistica software (Tulsa, OK, USA) and Graphpad Prism. Statistical testing was performed using established assay-specific methods, including Student’s t-test for single parameter comparisons between genotypes, and one-way or two-way repeated-measures ANOVA for comparisons across time points (e.g., development, pre- and post- fear conditioning, open field and beam walking). All significance levels were set at p < 0.05 and all t-tests were two-tailed. Groups sizes were chosen based on past experience and power analyses (Sukoff Rizzo and Silverman, 2016). Significant ANOVAs were followed by Bonferroni-Dunn posthoc testing. Behavioral analysis passed distribution normality tests, was collected using continuous variables and thus was analyzed via parametric analysis in all assays. For all behavioral analyses, variances were similar between groups and data points within 2 standard deviations of the mean were included in analysis.
3. Results
3.1. Snord116+/− mice were delayed in their early physical development and neurological reflexes
Table 1 shows delayed early physical development and neurological reflexes in various parameters in Snord116 +/− compared to Snord116 +/+ littermates. All subjects gained weight and grew in length over time (weight: F2,50 = 120.7, p < 0.0001 and length: F2,50 = 67.51, p < 0.0001). However, Snord116 +/− mice weighed less at all three time points collected (F1,25 = 26.55, p < 0.0001, Bonferroni-Dunn posthoc) and were shorter by total length (body plus tail) at PND 12 and 14 (F1,25 = 13.84, p < 0.002, Bonferroni-Dunn posthoc). Snord116 +/− mice also had smaller head widths at all three time points (F1,25 = 13.84, p < 0.001, Bonferroni-Dunn posthoc). Reflexes including negative geotaxis (F1,25 = 0.074, p > 0.05), cliff aversion (F125 = 0.19, p > 0.05) and the righting reflex (F1,25 = 1.196, p > 0.05) did not differ between genotypes. Onset of ability to walk developed over time by traversing out of the center of a circle (F2,50 = 49.68, p < 0.0001). Snord116 +/− mice had longer times to traverse out of the center of a circle at PND12 (F1,25 = 12.61, p < 0.002, Bonferroni-Dunn posthoc).
Table 1.
Physical characteristics, milestone development and neurological reflexes at postnatal days 10, 12 and 14 in Snord116 + /+ and Snord116 + /- mice.
| Milestones | Domain | Genotype | PND 10 Mean ± SE |
PND 12 Mean ± SE |
PND 14 Mean ± SE |
F value | Significance @ p < 0.05 |
|---|---|---|---|---|---|---|---|
| Weight (g) | Physical | Snord116 +/+ | 5.845 ± 0.106 | 6.818 ± 0.364 | 8.073 ± 0.177 | F(1,25) = 26.55 | Yes, p < 0.0001 |
| Snord116 +/− | 4.525 ± 0.205 | 5.281 ± 0.257 | 5.869 ± 0.256 | ||||
| Length (mm) | Physical | Snord116 +/+ | 69.027 ± 0.800 | 75.318 ± 2.186 | 81.618 ± 1.002 | F(1,25) = 13.84 | Yes, p < 0.001 |
| Snord116 +/− | 65.063 ± 0.830 | 70.938 ± 1.142 | 75.581 ± 1.065 | ||||
| Head width | Physical | Snord116 +/+ | 11.491 ± 0.249 | 11.973 ± 0.288 | 12.318 ± 0.172 | F(1,25) = 27.63 | Yes, p < 0.0001 |
| Snord116 +/− | 10.619 ± 0.232 | 10.706 ± 0.198 | 11.206 ± 0.166 | ||||
| Circle transverse | Reflex | Snord116 +/+ | 22.805 ± 3.096 | 15.791 ± 2.442 | 4.436 ± 1.127 | F(1,25)= 12.61 | Yes, p < 0.002 |
| Snord116 +/− | 27.918 ± 1.313 | 22.751 ± 1.703 | 6.643 ± 1.612 | ||||
| Negative geotaxis | Reflex | Snord116 +/+ | 14.264 ± 3.785 | 6.972 ± 2.368 | 3.195 ± 0.690 | F(1,25) = 0.074 | NS, p > 0.05 |
| Snord116 +/− | 14.201 ± 2.456 | 8.561 ± 1.713 | 3.095 ± 0.639 | ||||
| Righting reflex | Reflex | Snord116 +/+ | 9.257 ± 7.776 | 0.605 ± 0.073 | 0.607 ± 0.060 | F(1,25) = 1.20 | NS, p > 0.05 |
| Snord116 + /– | 1.769 ± 0.177 | 0.939 ± 0.091 | 0.749 ± 0.061 | ||||
| Cliff aversion | Reflex | Snord116 +/+ | 4.735 ± 2.555 | 1.950 ± 0.685 | 0.499 ± 0.073 | F(1,25) = 0.20 | NS, p > 0.05 |
| Snord116 +/− | 3.017 ± 0.884 | 2.617 ± 0.969 | 0.429 ± 0.029 | ||||
3.2. Snord116+/− mice exhibit impaired novel object recognition
Cognitive abilities as measured by the NOR task were tested in two independent cohorts of Snord116+/− compared to Snord116 +/+ mice. In Cohort 1, manual scoring by a highly trained observer blinded to genotype indicated Snord116 +/+ spent more time investigating the novel object versus the familiar object, as expected. In contrast, Snord116 +/− mice did not exhibit typical novel object preference (Fig. 1A: Snord116 +/+ ; t(19) = 4.59, p < 0.002 and Snord116+/−; t(13) = 1.58, p > 0.05). In this cohort, using manual scores, both genotypes explored the two identical objects similarly during the familiarization phase (Fig. 1B: Snord116+/+ ; t(19) = 0.43, p > 0.05 and Snord116 +/–; t(13) = 0.845, p > 0.05). Sexes were combined since there was no sex difference observed on time spent sniffing objects in the novel phase of the test by automated and manual scoring methods in two cohorts (cohort 1 auto: F(1, 50) = 0.000, p > 0.05; cohort 1 manual: F(1,50) = 0.567, p > 0.05; cohort 2 auto: F(1,50) = 0.064, p > 0.05; cohort 2 manual: F(1,50) = 0.003, p > 0.05), consistent with other NOR literature (Gulinello et al., 2018). Snord116 +/+ also displayed normal NOR when quantified by automated tracking software, spending more time in the defined zone with the novel object than in the zone surrounding the familiar object, whereas Snord116 +/− mice did not exhibit typical novel object preference (Fig. 1C: Snord116 +/+ ; t(18) = 2.08, p = 0.065 and Snord116 + /–; t(13) = 0.52, p > 0.05). Both genotypes explored the two identical objects similarly during the automated familiarization phase (Fig. 1D: Snord116+/+ ; t(19) = 0.43, p > 0.05 and Snord116 +/–; t(13) = 0.845, p > 0.05). In automated analysis of the novel phase, data from one Snord116+/+ mouse was removed because of a loss of tracking data.
Fig. 1.
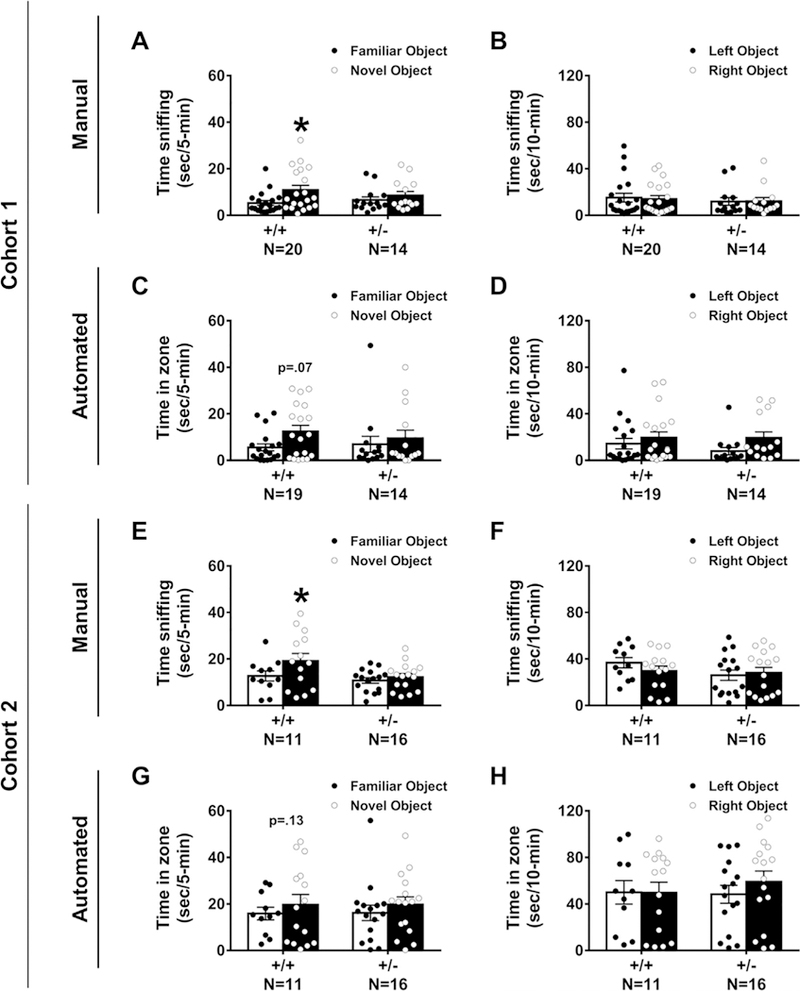
Snord116 +/– displayed robust impairments on the novel object recognition (NOR) assay in two independent cohorts by manual and automated quantification measures. This version of NOR started with animal habituation to the open field testing arena for 30-min. After 24-hr, the mouse received another 10-min habituation session. Next, the subjects were given a 10-min familiarization session during which time spent sniffing each object was recorded. The objects were then cleaned and after a 1-hr inter-trial interval, the mouse was placed back into the arena with one familiar object and one novel object. (A) Snord116 +/– mice do not spend more time sniffing the novel object as compared to the familiar object. Snord116 +/+ litter- mates spend more time sniffing the novel object as compared to familiar object. (B) During the familiarization phase, manual scoring of objects revealed no left-right side bias in either Snord116 + /+ or Snord116 +/–. (C) Automated scoring via Noldus Ethovision 9.0XT illustrated Snord116 +/– mice do not spend more time in the zone defined around the novel object versus the zone defined around the familiar object, while Snord116 +/+ exhibit this typical object preference (p = 0.07) during the 5-min testing phase. (D) Similar to manual data, in the familiarization phase, automated tracings confirmed the absence of a left-right side bias in Snord116 +/+ and Snord116 +/– mice. (E) Manual scoring of the 5-min testing phase in Cohort 2 confirmed that Snord116 +/– mice do not spend more time sniffing the novel object as compared to the familiar object. Once again, Snord116 +/+ littermates spend more time sniffing the novel object when compared to familiar object. (F) Reproducing Cohort 1, during the familiarization phase, manual scoring of objects revealed no left-right side bias in Snord116+/+ and Snord116 +/–. (G) Similar to manual data and Cohort 1, automated scoring via Noldus Ethovision 9.0XT illustrated Snord116 + /– mice do not spend more time in the zone defined around the novel object versus the zone defined around the familiar object, while Snord116 +/+ exhibited a trend to toward novel object preference (p = 0.13) during the 5-min testing phase. (H) In the familiarization phase, automated tracings repeatedly confirmed the absence of a left-right side bias in either Snord116 +/+ or Snord116 +/– mice. *p < 0.05, novel object versus familiar object, paired t-test within genotype.
In Cohort 2, manual and automated scoring indicated Snord116+/+ spent more time investigating the novel object versus the familiar object whereas Snord116+/− mice did not exhibit typical novel object preference (Fig. 1E: Snord116 +/+ ; t(10) = 3.16, p < 0.02 and Snord116 +/–; t(15) = 1.74, p > 0.05 and Fig. 1G: Snord116 +/+ ; t(10) = 2.186, p < 0.05 and Snord116 +/–; t(15) = 1.034, p > 0.05). Once again, manual and automated scoring illustrated that both genotypes explored the two identical objects similarly during the familiarization phase that tested for side bias and object salience (Fig. 1F: Snord116 +/+ ; t(10) = 0.439, p > 0.05 and Snord116 +/− ; t(15) = 0.324, p > 0.05 and Fig. 1H: Snord116 +/+ ; t(10) = 0.203, p > 0.05 and Snord116+/− ; t(14) = 0.007, p > 0.05). During automated analysis of side bias, data from one Snord116+/− mouse was removed because of loss of video tracking data. These results indicated that the NOR deficit in Snord116 +/− mice is replicable in different cohorts with differing laboratory personnel and when scored either from videos by human observer or by automated video tracking methods.
3.3. Snord116 +/− mice exhibit impaired object location memory
OLM was defined as spending significantly more time sniffing the relocated object than sniffing the object in the original location. Manual scoring of object investigation by sniffing indicated Snord116+/+ spent more time investigating an object displaced to a novel location versus the familiar location whereas Snord116+/– mice did not exhibit this location memory (Fig. 2A: Snord116+/+ ; t(10) = 2.254, p < 0.05 and Snord116 +/– ;t(15) = 0.203, p > 0.05). Automated video tracking corroborated the manual methods confirming that Snord116 +/+ spent more time investigating an object displaced to a novel location versus the familiar location whereas Snord116+/– mice did not exhibit this location memory (Fig. 2B: Snord116 +/+ ; t(10) = 4.169, p < 0.02 and Snord116+/– ;t(15) = 1.654, p > 0.05). Sexes were combined since there was no sex difference observed on time spent sniffing objects in the novel phase of the test by automated and manual scoring methods (auto: F(1, 50) = 2.211, p > 0.05; manual: F(1, 50) = 1.175, p > 0.05).
Fig. 2.
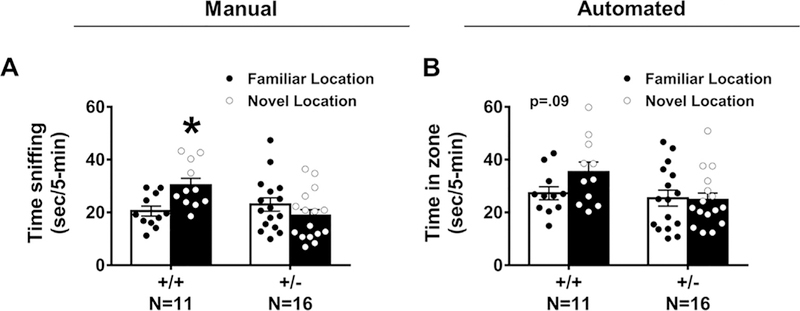
Snord116 +/− displayed impairments on the object location memory (OLM) task by manual and automated quantification measures. This version of the OLM used the same apparatus that was used for testing NOR and used the same habituation and familiarization procedures. However, the subject was exposed to different objects. Two identical objects were placed 12-cm away from the wall and 18-cm from each other) for a 10-min familiarization session. (A) Snord116 +/− mice do not spend more time sniffing the displaced object in the new location as compared to the object in the original location. Snord116 +/+ littermates spend more time sniffing the displaced object in the new location when compared to familiar location. (B) Automated scoring via Noldus Ethovision 9.0XT illustrated Snord116 +/− mice do not spend more time in the zone defined around the displaced object versus the zone defined around the familiar location, while Snord116 +/+ exhibit this object location memory (p = 0.09) during the 5-min testing phase.
3.4. Snord116 +/− mice have deficits in tone cued fear conditioning
Learning and memory was further evaluated using two measures of Pavlovian fear conditioning with a 24-hr contextual component and a tone cued fear conditioning. High levels of freezing were observed, subsequent to the conditioned stimulus (CS) – unconditioned stimulus (UCS) pairings, on the training day, in both genotypes (Fig. 3A: main effect of training, F(1, 25) = 31.62, p < 0.0001; no genotype difference in training freeze scores F(1, 25) = 0.919, p > 0.05), indicating no confounds and no deficits in the learning of the associations between the context stimuli and tone cues. No genotype difference was observed 24-hrs following CS-UCS training between Snord116 +/– and Snord116 +/+ freezing (Fig. 3B: t(25) = 0.176, p > 0.05), when placed in the context chamber from conditioning training with identical stimulus cues. No sex difference was observed in context freezing (F(1, 50) = 0.671, p > 0.05).
Fig. 3.
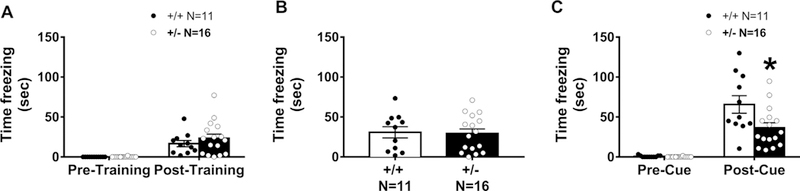
Snord116 +/− displayed impairments in learning and memory in cued but not contextual fear conditioning. Learning and memory was evaluated by Pavlovian fear conditioning using two components, contextual cues and an auditory tone, with assessments of freezing time before and after the presentation of three tone-shock pairings. (A) Normal levels of freezing post-training indicate associations between tone and shock were made in all genotypes. In addition, both genotypes showed typical basal levels of freezing suggesting no confounds of deficits in sensory reactivity or pain threshold. (B) The 24-hr contextual component illustrated typical levels freezing, indicating no abnormal contextual learning and memory in either Snord116 + /+ or Snord116 +/− mice. (C) Cued conditioning freeze time 48-hr after initial training, before and after, the presentation of three tone-cues is illustrated. Snord116 +/− mice had lower freezing scores compared to Snord116 +/+ following the auditory reminder tone cue. *p < 0.05 versus Snord116 +/+ by student’s unpaired t-tests.
Levels of freezing, between the pre- and post-cue presentation 48-hrs after training, revealed significant main effects of cued training (main effect of cue, F(1, 25) = 75.86, p < 0.001). Snord116 +/– froze less than Snord116 +/+ when the tone was presented in the novel contextual arena (Fig. 3C: t(25) = 2.49, p < 0.0199). No sex difference was observed in pre or post cue freezing (pre cued: F(1, 50) = 3.228, p > 0.05 and post-cued: F(1, 50) = 0.387, p > 0.05).
3.5. Snord116 +/− mice have normal motor abilities in open field locomotion and balance beam walking
Normal motor function in Snord116 +/− mice was confirmed by lack of genotype effect in the open field exploratory locomotion task across a 30-min session. The time course for total distance traversed in the novel open field, was significant, representing normal habituation to the novelty of the open field (Fig. 4A: F(5,125) = 63.81, p < 0.0001). No sex difference was observed in activity metrics (F (1, 50) = 0.649, p > 0.05). No genotype effects were observed in total distances traversed (Fig. 4A; F(1, 25) = 0.187, p > 0.05), horizontal activity scores (Fig. 4B: F(1, 25) = 2.479, p > 0.05), vertical activity counts (Fig. 4C: F(1, 25) = 1.094, p > 0.05), nor time spent in the center of the arena (Fig. 4D: F(1, 25) = 0.0439, p > 0.05) between Snord116+/− and Snord116 +/+ mice. No genotype effect was observed when the activity over the 30-min session was summed (Fig. 4E: t (25) = 0.433, p > 0.05). Both groups showed longer latencies to cross the rod shaped beams as they became thinner and more difficult to traverse, as expected across the task (Fig. 4F: F(2, 50) = 35.98, p < 0.0001). No genotype effects were observed in time to traverse any of the various widths of beams F(1, 25) = 0.342, p > 0.05). Sexes were combined since there was no sex difference observed on time to cross the beams (F(1, 50) = 2.53, p > 0.05).
Fig. 4.
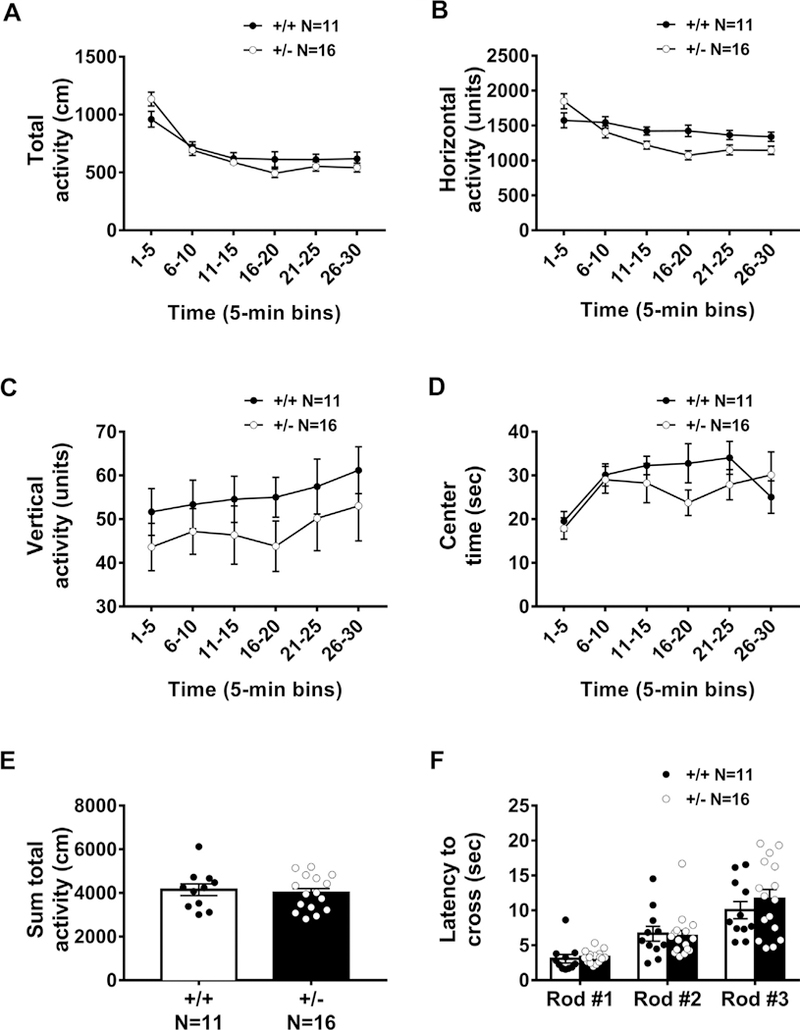
General motor ability and coordination in a novel open field and a balance beam assay were normal in Snord116 +/− mice. Activity and motor abilities are essential controls because hypolocomotion could confound the interpretation of result of interaction with objects, arena and object exploration, and sniffing and investigation times. Motor abilities were measured by (A) total distance traversed, (B) horizontal activity, (C) vertical activity and (D) time spent in the center of an arena over the course of a 30-min trial. Data are shown in 5-min time bins. (E) Summed activity over the course of the 30-min session highlights similar exploratory levels in Snord116 +/+ and Snord116 +/− mice. (F) Both groups showed longer latencies to cross the rod-shaped beams as they became thinner and more difficult to traverse. However, no genotype effects were observed in latency to traverse any of the various widths of beams, regardless of difficulty.
3.6. Snord116+/− mice showed inconsistent and/or mild anxiety-like phenotypes
Anxiety-like behavior was assessed with the elevated plus-maze and the light ↔ dark conflict assay. As compared to Snord116 +/+ , Snord116 +/− mice spent less time on the open arm of the maze (Fig. 5A: t(25) = 3.056, p < 0.006). Snord116 +/− also made fewer total arm entries calculated by open arm entries + closed arm entries (Fig. 5B: t(25) = 2.142, p > 0.05), suggesting less overall navigational motion during this short task in the mutants. No sex differences were observed in the percent time spent on the open arm (F(1,50) = 2.8733, p > 0.05), or the total entries (F(1>50) = 1.50, p > 0.05). In the light« dark conflict assay, no genotype effects were observed on the amount of time spent in the dark chamber (Fig. 5C: t(25) = 0.124, p > 0.05) nor the number of transitions between chambers (Fig. 5D: t(25) = 0.948, p > 0.05), suggesting that anxiety-like phenotypes are either subtle or absent in our examination. No sex differences were observed in the parameters of light↔dark conflict (F(1,50) = 1.75, p > 0.05), or the total transitions (F(1,50) = 2.55, p > 0.05).
Fig. 5.
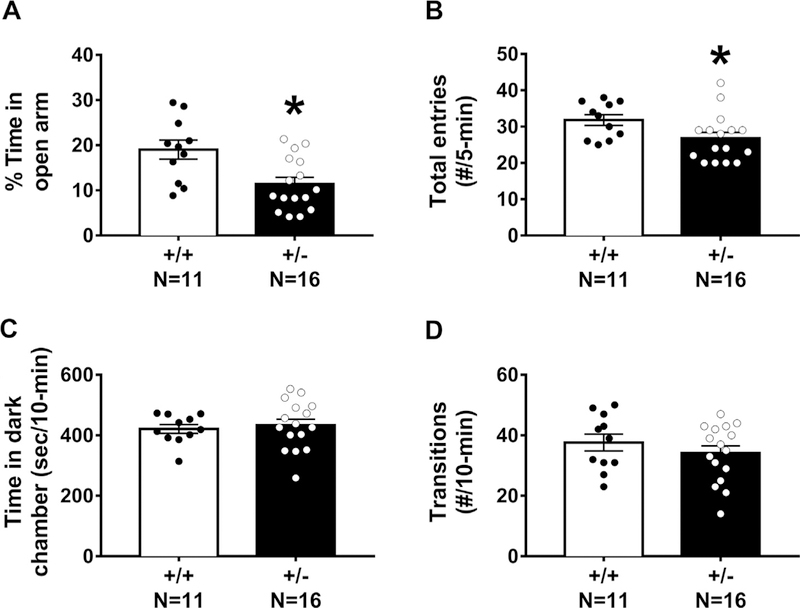
Anxiety-like behavioral testing illustrated inconsistent and confounded behavioral phenotypes in Snord116+/− mice. Anxiety-like behaviors were assessed in two gold standard assays: elevated plus-maze and light↔dark conflict. (A) During the elevated plus-maze, percent time spent on the open arm and (B) number of total arm entries (open+closed) were fewer in Snord116+/− mice as compared to Snord116+/ +. (C) In the light↔dark conflict assay, time spent in the dark chamber and (D) transitions between chambers did not differ between Snord116+/+ and Snord116+/− mice. *p < 0.05 versus Snord116+/+ by student’s unpaired t-test.
4. Discussion
Basic research to discover highly specific intervention targets requires well-controlled in vivo studies in model organisms with high construct validity. PWS arises by paternal 15q11–13 deletions, maternal uniparental disomy, or imprinting errors which lead to aberrant methylation and downregulation of paternally expressed transcripts. Genetic mapping data from unique “microdeletion” patients suggest SNORD116, is a critical mediator of the neurodevelopmental phenotypes. Therefore, we evaluated a mouse model of paternally inherited Snord116 deletion on physical characteristics and functional outcome measures. For the first time, we reliably identified the clinically relevant phenotype of cognitive deficits due to the loss of Snord116 using several standard behavioral metrics of learning and memory. Further, we corroborated earlier reports of postnatal failure to thrive. Our data highlight a functional cognitive contribution of this critical snoRNA complex and suggest a broader role for Snord116/SNORD116, beyond known cellular processes.
We discovered that Snord116 +/− mice exhibited robust learning and memory impairments in the low stress standard assay of NOR using optimized methods of our laboratory and the MIND Institute’s Intellectual and Developmental Disabilities Research Center (IDDRC) Rodent Behavioral Core, recently highlighted as a collaborative effort by numerous prominent neuroscientists (Gulinello et al., 2018). The outcome parameter of time spent in zone of close proximity using Noldus 9.0XT automated tracking software was corroborated by an expert technician blinded to genotype that scored time spent sniffing the objects manually. Both methods produced quantifiable data with reasonable signal to noise ratio. However, we discovered that only manual observation was able to eliminate misdetections in the automated tracking such as subjects knocking over objects, subjects sitting on objects, subjects grooming next to objects, and/or when the nose and tail detection indicator points switch amongst one another.
Our study also discovered deficits of the Snord116+/− mutants in learning and memory by OLM on which rodents are presented with two identical, familiar objects, one of which is in its previous location while the other is in a new location. Rodents spend more time exploring the object in the novel location (Ennaceur & Meliani, 1992). The NOR and OLM impairments were detected in the typical short-term range by a 1-hr time interval between familiarization and testing. This window is thought to occur within the consolidation phase of memory formation and may not be dependent on protein synthesis. Snord116+/− mutants showed impairments in cued but not contextual fear conditioning. The cued conditioning deficits but intact responses in context learning and memory were detected 48-hr post-training and therefore were transcriptionally and translationally dependent learning and memory as previously shown by this Pavlovian paradigm (Schafe and LeDoux, 2000; Schafe et al., 2000), and as further suggested by studies performed using a latent inhibition cued conditioning protocol (Lewis & Gould, 2007). Our detection of impairments in the short and longer term intervals implicate the multiple biochemical mechanisms that underlie learning and memory.
These cognitive behavioral deficiencies are intriguing and lend suggestions toward the role of the well-studied underlying neural circuitry. OLM and NOR both heavily rely on the spontaneous exploratory behavior of rodents towards objects (Bevins and Besheer, 2006; Ennaceur and Delacour, 1988; Vogel-Ciernia and Wood, 2014). Broadly, neuroanatomical substrates implicated in these functional outcomes include the hippocampus and its supporting limbic regions, such as the medial prefrontal, anterior cingulate, and retrosplenial cortices. In fact, an array of lesions in limbic sites connected with the hippocampus did not disrupt performance on a 15-min retention test of NOR while similar studies in OLM revealed impairments from lesions of the fornix and the cingulate cortex but not the medial prefrontal cortex (Ennaceur, Neave, & Aggleton, 1997). The amygdala is important for associating objects or spatial relationships with positive or aversive outcomes and appears to mediate fear responses to novel stimuli or neophobia (Ennaceur, Michalikova, & Chazot, 2009). For example, rodents with amygdala lesions show attenuated neophobia for novel food (Burns, Annett, Kelley, Everitt, & Robbins, 1996). Moreover, a rich literature of lesion work indicated that specific but different brain regions appear to regulate cued versus contextual fear conditioning. Although the responses of behavioral freezing elicited by contextual and cued fear conditioning are identical, the processing demands underlying the two forms of fear conditioning are different. The preponderance of the evidence supports the hypothesis that primary sensory information from the auditory thalamus into the lateral amygdala mediate cued fear conditioning (Phillips and LeDoux, 1992; Schafe and LeDoux, 2000) while contextual fear conditioning require both the amygdala and the hippocampus (Phillips and LeDoux, 1995; Maren and Holt, 2000). Given our reported impairments in Snord116+/− mice using novelty-based tasks and cued fear conditioning, we hypothesize a broad range of neural substrates underlying the dysfunction. A study by Lassi, Priano et al. (2016) also supports our suggestion of subcortical and limbic anomalies with evidence of morphological changes measured by structal MRI. Moreover, they discovered a major reduction in the size of the hippocampus in Snord116 +/− mice and PWS patients (Lassi, Priano, et al., 2016).
We corroborated the previously reported physical neo-natal developmental delays in weight and growth in Snord116 +/− (Ding et al., 2008; Powell et al., 2013). We extended these phenotypic outcomes by also quantifying smaller body lengths and smaller head circumferences. Development of walking measured by the animals’ skills to traverse outward of a circle disk was delayed in the Snord116 +/− mice. Interestingly, other standard neonatal neurological reflexes such as negative geotaxis, righting reflex and cliff aversion remained intact. We confirmed that the cognitive deficits observed in Snord116 +/− mice were not the result of confounding motor hypo activity, despite the substantial growth delays in the first 2 weeks of postnatal life. In extension of earlier work, we also examined anxiety-related phenotypes. Earlier characterization studies had reported an increased anxiety-like phenotype measured by increased ratio of time spent in the closed arm by the Snord116 +/− mice (Ding et al., 2008). We corroborated this finding by the standard measure of anxiety-like behavior on the plus- maze of percent time spent on the open arm. While a mild anxiety-like phenotype was observed on the elevated plus-maze, it was not supported by data from the light-dark conflict task nor the center time in the open field, which suggested to us a very mild anxiety-like phenotype which probably did not confound learning and memory results.
Earlier research using the same Snord116 +/− mutation mice reported disrupted circadian rhythms, sleep-wake cycles, frontal cortical transcriptional and epigenetic regulation, as well as reduced forebrain neuronal and cerebellar cellular sizes, which could mechanistically contribute to the behavioral phenotypes reported (Burnett et al., 2017; Coulson et al., 2018; Lassi, Priano, et al., 2016). Other in vivo model systems of PWS deleted the entire critical imprinting center and reported low locomotor abilities and impairments in the frontal cortical dependent five choice serial reaction time task (Relkovic et al., 2010). Given the number of genes in the critical imprinting region, other models were generated that focused on mutations of single paternally expressed genes. Necdin (Ndn) null mutants were the first to be generated and characterized since Necdin protein is postulated to govern the permanent arrest of cell growth of post-mitotic neurons during development. Ndn mutant mouse models were generated from multiple laboratories and displayed highly variable functional phenotypes, that ranged from no abnormality to respiratory distress and lethality, depending on the background strain of mouse (Gerard, Hernandez, Wevrick, & Stewart, 1999; Kuwako et al., 2005; Muscatelli et al., 2000; Tsai, Armstrong, et al., 1999). The other gene in the region that is highly homologous to Ndn, paternally expressed and has multiple key cell cycle regulatory functions is Magel2. Magel2 mutant mice have disrupted circadian rhythm and reduced brain volumes however relatively normal motor and learning abilities (Fountain, Tao, Chen, Yin, & Schaaf, 2017; Kozlov et al., 2007; Mercer et al., 2009) in contrast to the learning and memory deficits reported herein.
5. Conclusion
Prader-Willi syndrome (PWS) is a complex neurodevelopmental disease caused by a loss of imprinted paternal genes on chromosome 15q11-q13 and is characterized by cognitive impairments, developmental delay, hyperphagia, and obesity. The majority of previous research using PWS preclinical models focused on characterization of the hyperphagic and metabolic phenotypes. This work reports the clinically relevant phenotype of cognitive impairments on three different learning and memory behavioral assays in the Snord116+/− model of PWS. These deficits in the Snord116 +/− mouse model highlight the critical role of Snord116 +/− in PWS and extend validity of the model as a valuable preclinical tool for investigating learning and memory impairments in PWS.
Acknowledgements
We are grateful for the support of Ms. Heather Boyle of the Stem Cell and MIND Institute vivarium teams for maintaining the mouse colonies. This work was supported by the NIH R01NS097808 (JLS), and the MIND Institute’s Intellectual and Developmental Disabilities Research Center (IDDRC) Grant HD079125 (PI Abbeduto).
References
- Bevins RA, & Besheer J (2006). Object recognition in rats and mice: A one-trial non-matching-to-sample learning task to study ‘recognition memory’. Nature Protocols, 1, 1306–1311. [DOI] [PubMed] [Google Scholar]
- Bieth E, Eddiry S, Gaston V, Lorenzini F, Buffet A, Conte Auriol F, … Tauber M (2015). Highly restricted deletion of the SNORD116 region is implicated in Prader- Willi syndrome. European Journal of Human Genetics: EJHG, 23, 252–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bressler J, Tsai TF, Wu MY, Tsai SF, Ramirez MA, Armstrong D, & Beaudet AL (2001). The SNRPN promoter is not required for genomic imprinting of the Prader- Willi/Angelman domain in mice. Nature Genetics, 28, 232–240. [DOI] [PubMed] [Google Scholar]
- Burnett LC, Hubner G, LeDuc CA, Morabito MV, Carli JFM, & Leibel RL (2017). Loss of the imprinted, non-coding Snord116 gene cluster in the interval deleted in the Prader Willi syndrome results in murine neuronal and endocrine pancreatic developmental phenotypes. Human Molecular Genetics, 26, 4606–4616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns LH, Annett L, Kelley AE, Everitt BJ, & Robbins TW (1996). Effects of lesions to amygdala, ventral subiculum, medial prefrontal cortex, and nucleus accumbens on the reaction to novelty: Implication for limbic-striatal interactions. Behavioral Neuroscience, 110, 60–73. [DOI] [PubMed] [Google Scholar]
- Carter RJ, Morton J, & Dunnett SB (2001). Motor coordination and balance in rodents. Current Protocols in Neuroscience, 8–12. [DOI] [PubMed] [Google Scholar]
- Copping NA, Berg EL, Foley GM, Schaffler MD, Onaga BL, Buscher N, … Yang M (2017). Touchscreen learning deficits and normal social approach behavior in the Shank3B model of Phelan-McDermid Syndrome and autism. Neuroscience, 345, 155–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copping NA, Christian SGB, Ritter DJ, Islam MS, Buscher N, Zolkowska D,… Dindot SV (2017). Neuronal overexpression of Ube3a isoform 2 causes behavioral impairments and neuroanatomical pathology relevant to 15q11.2-q13.3 duplication syndrome. Human Molecular Genetics, 26, 3995–4010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulson R, Yasui D, Dunaway K, Laufer B, Vogel-Ciernia A, Mordaunt C, … LaSalle J (2018). Snord116-dependent diurnal rhythm of DNA methylation in mouse cortex. Nature Communications, 9(1), 1616 10.1101/184788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Smith AJ, Purmann C, Walters RG, Ellis RJ, Holder SE, Van Haelst MM, … Blakemore AI (2009). A deletion of the HBII-85 class of small nucleolar RNAs (snoRNAs) is associated with hyperphagia, obesity and hypogonadism. Human Molecular Genetics. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding F, Li HH, Zhang S, Solomon NM, Camper SA, Cohen P, & Francke U (2008). SnoRNA Snord116 (Pwcr1/MBII-85) deletion causes growth deficiency and hyperphagia in mice. PloS One, 3, e1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubose AJ, Smith EY, Yang TP, Johnstone KA, & Resnick JL (2011). A new deletion refines the boundaries of the murine Prader-Willi syndrome imprinting center. Human Molecular Genetics, 20, 3461–3466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duker AL, Ballif BC, Bawle EV, Person RE, Mahadevan S, Alliman S, … Sahoo T (2010). Paternally inherited microdeletion at 15q11.2 confirms a significant role for the SNORD116 C/D box snoRNA cluster in Prader-Willi syndrome. European Journal of Human Genetics, 18(11), 1196–1201. 10.1038/ejhg.2010.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dykens EM, Hodapp RM, Walsh K, & Nash LJ (1992a). Adaptive and maladaptive behavior in Prader-Willi syndrome. Journal of the American Academy of Child & Adolescent Psychiatry, 31, 1131–1136. [DOI] [PubMed] [Google Scholar]
- Dykens EM, Hodapp RM, Walsh K, & Nash LJ (1992b). Profiles, correlates, and trajectories of intelligence in Prader-Willi syndrome. Journal of the American Academy of Child & Adolescent Psychiatry, 31, 1125–1130. [DOI] [PubMed] [Google Scholar]
- Dykens EM, Leckman JF, & Cassidy SB (1996). Obsessions and compulsions in Prader-Willi syndrome. Journal of Child Psychology and Psychiatry, 37, 995–1002. [DOI] [PubMed] [Google Scholar]
- Dykens EM, Roof E, Hunt-Hawkins H, Dankner N, Lee EB, Shivers CM, … Kim SJ (2017). Diagnoses and characteristics of autism spectrum disorders in children with Prader-Willi syndrome. J Neurodev Disord, 9, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dykens E, & Shah B (2003). Psychiatric disorders in Prader-Willi syndrome: Epidemiology and management. CNS Drugs, 17, 167–178. [DOI] [PubMed] [Google Scholar]
- Ennaceur A, & Delacour J (1988). A new one-trial test for neurobiological studies of memory in rats. 1: Behavioral data. Behavioural Brain Research, 31, 47–59. [DOI] [PubMed] [Google Scholar]
- Ennaceur A, & Meliani K (1992). A new one-trial test for neurobiological studies of memory in rats. III. Spatial vs. non-spatial working memory. Behavioural Brain Research, 51(1), 83–92. [DOI] [PubMed] [Google Scholar]
- Ennaceur A, Michalikova S, & Chazot PL (2009). Do rats really express neophobia towards novel objects? Experimental evidence from exposure to novelty and to an object recognition task in an open space and an enclosed space. Behavioural Brain Research, 197, 417–434. [DOI] [PubMed] [Google Scholar]
- Ennaceur A, Neave N, & Aggleton JP (1997). Spontaneous object recognition and object location memory in rats: The effects of lesions in the cingulate cortices, the medial prefrontal cortex, the cingulum bundle and the fornix. Experimental Brain Research, 113, 509–519. [DOI] [PubMed] [Google Scholar]
- Flannery BM, Silverman JL, Bruun DA, Puhger KR, McCoy MR, Hammock BD, Lein PJ (2015). Behavioral assessment of NIH Swiss mice acutely intoxicated with tetramethylenedisulfotetramine. Neurotoxicology and Teratology, 47, 36–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fountain MD, Tao H, Chen CA, Yin J, & Schaaf CP (2017). Magel2 knockout mice manifest altered social phenotypes and a deficit in preference for social novelty. Genes, Brain and Behavior, 16, 592–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox WM (1965). Reflex-ontogeny and behavioural development of the mouse. Animal Behaviour, 13, 234–241. [DOI] [PubMed] [Google Scholar]
- Gabriel JM, Merchant M, Ohta T, Ji Y, Caldwell RG, Ramsey MJ, … Nicholls RD (1999). A transgene insertion creating a heritable chromosome deletion mouse model of Prader-Willi and angelman syndromes. Proceedings of the National Academy of Sciences of the United States of America, 96, 9258–9263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerard M, Hernandez L, Wevrick R, & Stewart CL (1999). Disruption of the mouse necdin gene results in early post-natal lethality. Nature Genetics, 23, 199–202. [DOI] [PubMed] [Google Scholar]
- Gompers AL, Su-Feher L, Ellegood J, Copping NA, Riyadh MA, Stradleigh TW, … Zdilar, I. (2017). Germline Chd8 haploinsufficiency alters brain development in mouse. Nature Neuroscience, 20, 1062–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulinello M, Mitchell HA, Chang Q, Timothy O’Brien W, Zhou Z, Abel T, … Crawley JN (2018). Rigor and reproducibility in rodent behavioral research. Neurobiology of Learning and Memory. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Tsai TF, Bressler J, & Beaudet AL (1998). Imprinting in Angelman and Prader-Willi syndromes. Current Opinion in Genetics & Development, 8, 334–342. [DOI] [PubMed] [Google Scholar]
- Kozlov SV, Bogenpohl JW, Howell MP, Wevrick R, Panda S, Hogenesch JB, … Stewart CL (2007). The imprinted gene Magel2 regulates normal circadian output. Nature Genetics, 39, 1266–1272. [DOI] [PubMed] [Google Scholar]
- Kuwako K, Hosokawa A, Nishimura I, Uetsuki T, Yamada M, Nada S, … Yoshikawa K (2005). Disruption of the paternal necdin gene diminishes TrkA signaling for sensory neuron survival. The Journal of Neuroscience: The Official Journal of the Society for Neuroscience, 25, 7090–7099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassi G, Maggi S, Balzani E, Cosentini I, Garcia-Garcia C, & Tucci V (2016). Working-for-food behaviors: a preclinical study in Prader-Willi Mutant mice. Genetics, 204, 1129–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassi G, Priano L, Maggi S, Garcia-Garcia C, Balzani E, El-Assawy N, … Tucci V (2016). Deletion of the Snord116/SNORD116 alters sleep in mice and patients with Prader-Willi Syndrome. Sleep, 39, 637–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Kozlov S, Hernandez L, Chamberlain SJ, Brannan CI, Stewart CL, & Wevrick R (2000). Expression and imprinting of MAGEL2 suggest a role in Prader- willi syndrome and the homologous murine imprinting phenotype. Human Molecular Genetics, 9, 1813–1819. [DOI] [PubMed] [Google Scholar]
- Lewis MC, & Gould TJ (2007). Signal transduction mechanisms within the entorhinal cortex that support latent inhibition of cued fear conditioning. Neurobiology of Learning and Memory, 88(3), 359–368. [DOI] [PubMed] [Google Scholar]
- Maren S, & Holt W (2000). The hippocampus and contextual memory retrieval in Pavlovian conditioning. Behavioural Brain Research, 110, 97–108. [DOI] [PubMed] [Google Scholar]
- Martin A, State M, Koenig K, Schultz R, Dykens EM, Cassidy SB, & Leckman JF (1998). Prader-Willi syndrome. American Journal of Psychiatry, 155, 1265–1273. [DOI] [PubMed] [Google Scholar]
- Mercer RE, Kwolek EM, Bischof JM, van Eede M, Henkelman RM, & Wevrick R (2009). Regionally reduced brain volume, altered serotonin neurochemistry, and abnormal behavior in mice null for the circadian rhythm output gene Magel2. American Journal of Medical Genetics Part B: Neuropsychiatric Genetics, 150B, 1085–1099. [DOI] [PubMed] [Google Scholar]
- Muscatelli F, Abrous DN, Massacrier A, Boccaccio I, Le Moal M, Cau P, & Cremer H (2000). Disruption of the mouse Necdin gene results in hypothalamic and behavioral alterations reminiscent of the human Prader-Willi syndrome. Human Molecular Genetics, 9, 3101–3110. [DOI] [PubMed] [Google Scholar]
- Phillips RG, & LeDoux JE (1992). Differential contribution of amygdala and hippocampus to cued and contextual fear conditioning. Behavioral Neuroscience, 106, 274–285. [DOI] [PubMed] [Google Scholar]
- Phillips RG, & LeDoux JE (1995). Lesions of the fornix but not the entorhinal or perirhinal cortex interfere with contextual fear conditioning. The Journal of Neuroscience: The Official Journal of the Society for Neuroscience, 15, 5308–5315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell WT, Coulson RL, Crary FK, Wong SS, Ach RA, Tsang P, … Lasalle JM (2013). A Prader-Willi locus lncRNA cloud modulates diurnal genes and energy expenditure. Human Molecular Genetics, 22, 4318–4328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Relkovic D, Doe CM, Humby T, Johnstone KA, Resnick JL, Holland AJ, … Isles AR (2010). Behavioural and cognitive abnormalities in an imprinting centre deletion mouse model for Prader-Willi syndrome. European Journal of Neuroscience, 31, 156–164. [DOI] [PubMed] [Google Scholar]
- Sahoo T, del Gaudio D, German JR, Shinawi M, Peters SU, Person RE, … Beaudet AL (2008). Prader-Willi phenotype caused by paternal deficiency for the HBII-85 C/D box small nucleolar RNA cluster. Nature Genetics, 40, 719–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafe GE, Atkins CM, Swank MW, Bauer EP, Sweatt JD, & LeDoux JE (2000). Activation of ERK/MAP kinase in the amygdala is required for memory consolidation of pavlovian fear conditioning. The Journal of Neuroscience: The Official Journal of the Society for Neuroscience, 20, 8177–8187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafe GE, & LeDoux JE (2000). Memory consolidation of auditory pavlovian fear conditioning requires protein synthesis and protein kinase A in the amygdala. The Journal of Neuroscience: The Official Journal of the Society for Neuroscience, 20, RC96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz L, Holland A, Dykens E, Strong T, Roof E, & Bohonowych J (2016). Prader-Willi syndrome mental health research strategy workshop proceedings: The state of the science and future directions. Orphanet Journal of Rare Diseases, 11, 131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefan M, Portis T, Longnecker R, & Nicholls RD (2005). A nonimprinted Prader-Willi Syndrome (PWS)-region gene regulates a different chromosomal domain in trans but the imprinted pws loci do not alter genome-wide mRNA levels. Genomics, 85, 630–640. [DOI] [PubMed] [Google Scholar]
- Sukoff Rizzo SJ, & Silverman JL (2016). Methodological considerations for optimizing and validating behavioral assays. Current Protocols in Mouse Biology, 6, 364–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai TF, Armstrong D, & Beaudet AL (1999). Necdin-deficient mice do not show lethality or the obesity and infertility of Prader-Willi syndrome. Nature Genetics, 22, 15–16. [DOI] [PubMed] [Google Scholar]
- Tsai TF, Jiang YH, Bressler J, Armstrong D, & Beaudet AL (1999). Paternal deletion from Snrpn to Ube3a in the mouse causes hypotonia, growth retardation and partial lethality and provides evidence for a gene contributing to Prader-Willi syndrome. Human Molecular Genetics, 8, 1357–1364. [DOI] [PubMed] [Google Scholar]
- Vogel Ciernia A, Pride MC, Durbin-Johnson B, Noronha A, Chang A, Yasui DH, … LaSalle JM (2017). Early motor phenotype detection in a female mouse model of Rett syndrome is improved by cross-fostering. Human Molecular Genetics, 26, 1839–1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel-Ciernia A, & Wood MA (2014). Examining object location and object recognition memory in mice. Current Protocols in Neuroscience, 69, 8–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang T, Adamson TE, Resnick JL, Leff S, Wevrick R, Francke U, … Brannan CI (1998). A mouse model for Prader-Willi syndrome imprinting-centre mutations. Nature Genetics, 19, 25–31. [DOI] [PubMed] [Google Scholar]
- Yang M, Bozdagi O, Scattoni ML, Wohr M, Roullet FI, Katz AM, … Crawley JN (2012). Reduced excitatory neurotransmission and mild autism-relevant phenotypes in adolescent Shank3 null mutant mice. The Journal of Neuroscience: The Official Journal of the Society for Neuroscience, 32, 6525–6541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M, Lewis FC, Sarvi MS, Foley GM, & Crawley JN (2015). 16p11.2 Deletion mice display cognitive deficits in touchscreen learning and novelty recognition tasks. Learning & Memory, 22, 622–632. [DOI] [PMC free article] [PubMed] [Google Scholar]


