Abstract
To understand the role of RAS-signaling networks in the pathogenesis of renal cell carcisnoma, we clarified the relationship between miR-143 and RAS. The expression of miR-143 was extremely downregulated in tumor tissues from renal cell carcinoma patients compared with that in the adjacent normal tissues and Caki-1 cells. We developed a synthetic miR-143#12, and we found that the ectopic expression of it inhibited cell growth with autophagy in Caki-1 cells. Also, the expression level of c-Myc was markedly decreased, resulting in the perturbation of cancer-specific energy metabolism by negatively modulating the expression of GLUT1 and the PTBP1/PKMs axis. A partial metabolic shift from glycolysis to oxidative phosphorylation induced autophagy through increasing the intracellular level of reactive oxygen species (ROS). In an in vivo study, the potent anti-tumor activity of polyion complex (PIC)-loaded miR-143#12 (miR-143#12/PIC) was shown by systemic administration of it to Caki-1 cell-xenografted mice. Higher levels of miR-143 were found in both blood and tumor tissues after the systemic administration with miR-143#12/PIC compared to those with lipoplexes in the xenografted mice. These findings indicated that this synthetic miR-143#12 induced a marked growth inhibition by impairing K-RAS-signaling networks in vitro and in vivo.
Keywords: synthetic miR-143, K-RAS, renal cell cancer, cancer specific metabolism, autophagy
Synthetic miR-143 loaded with a PIC nanocarrier can be delivered specifically to tumor tissue. Also, the miR-143 can induce marked growth inhibition, cell-cycle arrest, and autophagy through impairing K-RAS-signaling networks.
Introduction
Renal cell carcinoma (RCC) is a frequent malignancy affecting nearly 300,000 individuals worldwide, and clear cell RCC (ccRCC) is one of the most frequent RCC subtypes.1, 2 Approximately 25%–30% of RCC patients are initially diagnosed as having metastatic RCC (mRCC).3 The prognosis for patients with mRCC is very poor, showing a 5-year survival rate of only approximately 5%–10%.4 Sunitinib (Sutent), which is now the first-line therapy for mRCC, is a small-molecule inhibitor of the receptor tyrosine kinases of the vascular endothelial growth factor (VEGF),5 which is activated by loss of the von Hippel-Lindau (VHL) function.6 Other VEGF receptor (VEGFR) inhibitors, such as sorafenib,7 axitinib,8 and pazopanib,9 were all reported to be active against ccRCC. However, approximately 20%–30% of the patients do not respond to these inhibitors, and nearly all become resistant to them.10
Our strategy is developing microRNA (miRNA) medicine for RNAi, particularly by using miR-143.11, 12 miRNAs are small noncoding RNAs that can bind to the 3′ UTR of the targeted mRNA, thus inhibiting translation or promoting RNA degradation. Our previous study revealed that most of the human cancer cell lines tested exhibited extremely low expression levels of miR-143, whereas their originating normal tissues showed a good expression of miR-143.11, 12 Also, miR-143 acts as an anti-oncomiR and has a function of growth inhibition in various kinds of cancers.12, 13, 14, 15, 16 An extremely low expression level of miR-143 has been detected and contributes to tumor development, differentiation, proliferation, invasion, and metastasis, because miR-143 silences the Kirsten rat sarcoma viral oncogene homolog (K-RAS) or ERK5.
K-RAS is one of the RAS gene family members and encodes a small guanosine triphosphatase.17, 18 K-RAS performs its essential function by participating in more than 10 signaling pathways, and it is promoted mainly by receptor tyrosine kinases for epidermal growth factor (EGF), transforming growth factor α (TGF-α), and VEGF. However, the overexpression of K-RAS with a mutation or not has crucial functions in various biological processes, including cellular proliferation, invasion, metastasis, and angiogenesis. Once guanosine diphosphate (GDP)-K-RAS is converted to guanosine triphosphate (GTP)-K-RAS, this K-RAS activates its growth-related effector-signaling pathways, such as mitogen-activated protein kinase (MAPK)/ERK and PI3K/AKT. In addition, K-RAS can induce the expression of c-Myc via its effector signaling pathways.19
Also, GLUT1 has been found to be aberrantly expressed in K-RAS-overexpressing cells;20, 21, 22, 23 and RAS can promote glycolysis,24, 25, 26 which would maintain cancer-specific energy metabolism. RAS-signaling networks promote glucose uptake by increasing the expression of the glucose transporter GLUT1, which in turn promotes glycolytic activity and increases lactate production. This phenomenon is known as the Warburg effect, which is regulated by the expression profiles of pyruvate kinase muscle (PKM) isoforms.27, 28 RAS upregulates the GLUT1 glucose transporter, thereby contributing to the Warburg effect in cancer cells through the c-Myc/PTBP1/PKMs axis. Therefore, the ectopic expression of miR-143 in RCC may be a potential therapeutic approach for suppressing the action of K-RAS. However, there are well-known barriers to overcome, such as degradation by RNase; therefore, the development of a novel drug delivery system is essential for the establishment of effective RNA medicine. To further enhance the anti-tumor effect of miR-143 and to make it resistant to RNase, we developed a novel synthetic miR-143.
Recent studies on RNA delivery vehicles for use in drug delivery systems have been reported, such as polymers,29, 30 lipids,31 and inorganic nanoparticles,32 all of which have sought to prolong blood circulation time and to enhance tumor selectivity. Among them, we have developed a novel efficient polyion complex (PIC)-based nanocarrier for systemic delivery of RNA medicine.33, 34 This PIC was engineered to provide the RNA medicine with enhanced colloidal stability and biocompatibility due to the poly(ethylene glycol) (PEG) palisade surrounding the PIC core of the nanocarrier loaded with RNA medicine. Furthermore, the PIC nanocarrier enables preferential tumor accumulation and appears to be safe, because there are no significant changes in hematological and biochemical parameters in mice treated with these nanocarriers.35 In the current study, treatment by RNAi using synthetic miR-143 loaded in the PIC nanocarrier exhibited a great anti-cancer effect when administered systemically.
Results
Expression of miR-143 Was Extremely Downregulated in Tumor Samples from Clear Cell Renal Cancer Patients and in the RCC Caki-1 Cell Line Used in This Study
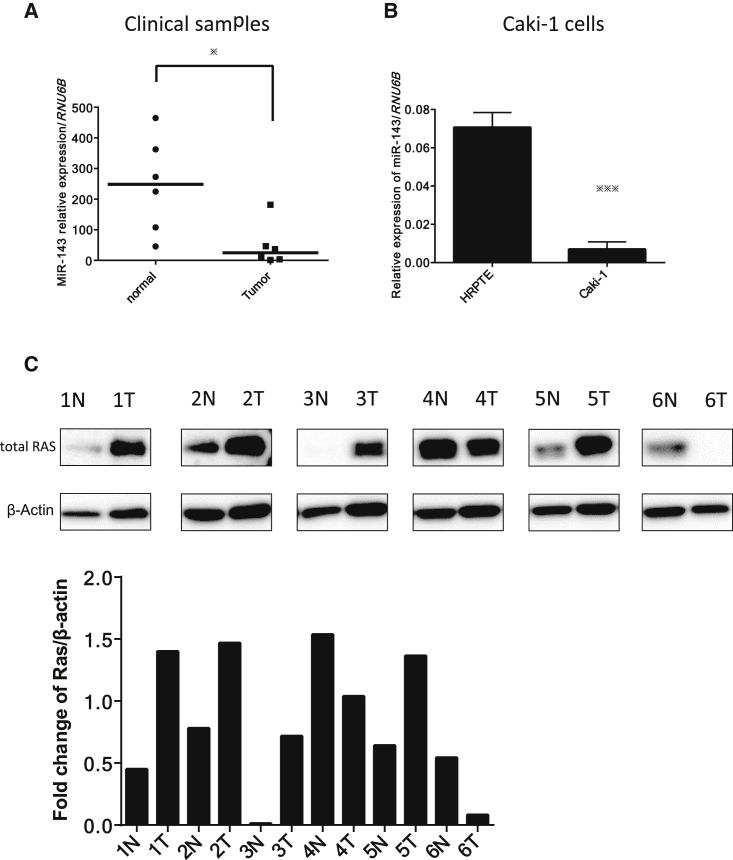
We first examined the expression levels of miR-143 in clinical tumor samples of RCC and in samples of the adjacent normal tissue in the same patients, as well as that in the RCC Caki-1 cell line used in this study. The expression levels of miR-143 in RCC samples examined by RT-PCR using a real-time PCR were extremely downregulated compared with those in the normal tissue samples (Figure 1A), as was the case for the human RCC Caki-1 cell line compared with human renal proximal tubule epithelial (HRPTE) cells (Figure 1B). Furthermore, we investigated whether Ras in clinical RCC samples was expressed; we examined the paired samples from 6 RCC patients by western blot analysis (Figure 1C). As shown in Figure 1C, Ras expression was upregulated in more than 50% of patients.
Figure 1.
Expression of miR-143 in Clinical Clear Renal Cell Carcinoma Samples and in the Renal Carcinoma Caki-1 Cell Line
(A) Relative expression levels of miR-143 in clinical clear renal cell carcinoma samples and normal tissue samples from the same patients. (B) Relative expression levels of miR-143 in normal renal cell HRPTE and Caki-1 cells. (C) Ras and β-actin expression in 6 renal cell carcinoma patients as determined by western blot analysis and fold change Ras/β-actin. *p < 0.05, **p < 0.01, ***p < 0.001. Error bars, means + SD.
Ectopic Expression of miR-143 Exhibited Growth Inhibition by Silencing K-RAS
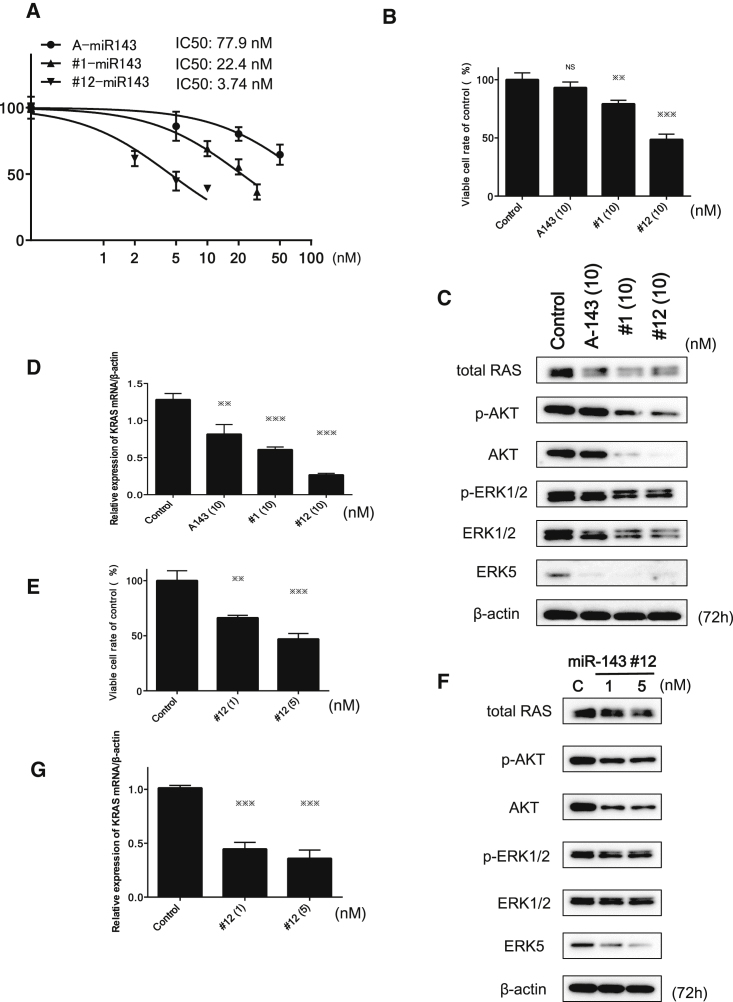
To enhance the nuclease resistance of miR-143, we developed more than 100 kinds of chemically modified miR-143s, and we examined the growth inhibition after the transfection with each miR-143 derivative. Among them, we chose to use the synthetic miR-143 (#1-miR-143 [#1]) and chemically modified synthetic miR-143 (#12-miR-143 [#12]) in view of anti-proliferative activity (Figure 2A). The #12 form was chemically modified with fluorine, methoxy group, phosphorylation, deoxythymidine, and phosphorothioate in the antisense strand. The seed sequences of #1 and #12 were identical (Table 1).
Figure 2.
Effects of Ectopic Expression of A143, miR-143#1, and miR-143#12 on Cell Growth and Protein and mRNA Expression Levels of Target Genes of miR-143s in Caki-1 Cells
(A) Dose-response effect of miR-143s on growth inhibition. Cell counts were performed at 72 h after the transfection. p values were determined for differences between the cells transfected with control miRNA and those transfected with A143, #1, or #12. (B) Comparison of the effect on cell growth inhibition among different miR-143s tested at the same concentration. Cell counts were performed at 72 h after the transfection with A143, #1, or #12 (10 nM). (C) Effects of transfection with A143, #1, or #12 (10 nM) on the protein expression levels of target genes of miR-143, as estimated by western blot analysis. (D) mRNA expression of K-RAS in Caki-1 cells at 72 h after transfection with A143, #1, or #12 miR-143 (10 nM). (E) Cell growth inhibition at 72 h after transfection with #12 (1 and 5 nM) in the cells. (F and G) The protein expression of target genes of miR-143 (F) and mRNA expression of K-RAS (G) in Caki-1 cells, as examined by western blot analysis and real-time PCR at 72 h after transfection with #12 at a concentration of 1 or 5 nM. *p < 0.05, **p < 0.01, ***p < 0.001. Error bars, means + SD.
Table 1.
Sequences of Synthetic Wild-Type miR-143 (#1) and Chemically Modified miR-143 (#12)
The sequences of wild type miR-143(#1) and chemically modified miR-143(#12) are shown. The antisense strand of #12 was modified with fluorine, methoxy group, phosphorylation, deoxythymidine, and phosphorothioate. SS, sense sequence; AS, antisense sequence; MW, molecular weight.
Next, we examined the effects of these miR-143s on cell growth, and we calculated the half-maximum inhibitory concentration (IC50), comparing their anti-proliferative activity with that of the mature type of miR-143 (A-miR-143 or A143, commercially available from Ambion, Foster City, CA, USA) as a standard. The IC50 values of A143, #1, and #12 were 77.9, 22.4, and 3.74 nM, respectively (Figure 2A). Expectedly, #12 had the greatest growth-inhibitory activity among them. At the same dose of 10 nM, the activity was #12 > #1 > A143 (Figure 2B).
Western bolt analysis indicated that #1 and #12 strikingly reduced the expression levels of RAS, ERK5, AKT, and ERK1/2 more than A143 did, except in the case of ERK5 (Figure 2C; Figure S1A). Furthermore, the introduction of #1 or #12 caused a marked reduction in the level of KRAS mRNA compared with the level obtained with A143 (Figure 2D), which results almost paralleled those for the growth inhibition activity. The #12 had a greater growth-inhibitory activity than the other miR-143s, whereas #12 as well as #1 exhibited similar RAS protein expression profiles of target genes to that of A143 (Figure 2C; Figure S1A).
The ectopic expression of #12 or #1 at the concentration of around their IC50 values also inhibited the cell growth and significantly downregulated the protein expression levels of AKT and ERK as well as total RAS in Caki-1 cells (Figures S2A and 2B). Also, a marked reduction of K-RAS mRNA level was found following the introduction of any of the miR-143s (Figure S2C). Thus, #12 and #1 silenced not only K-RAS but also AKT and ERKs, leading to the suppression of both K-RAS effector PI3K/AKT- and MAPK/ERK-signaling pathways in Caki-1 cells. As shown in Figures 2E–2G, the introduction of #12 caused concentration-dependent growth inhibition (Figure 2E); downregulated expression of proteins such as RAS, AKT, ERK5, and ERK1/2 (Figure 2F; Figure S1B); and a decrease in the K-RAS mRNA expression level (Figure 2G).
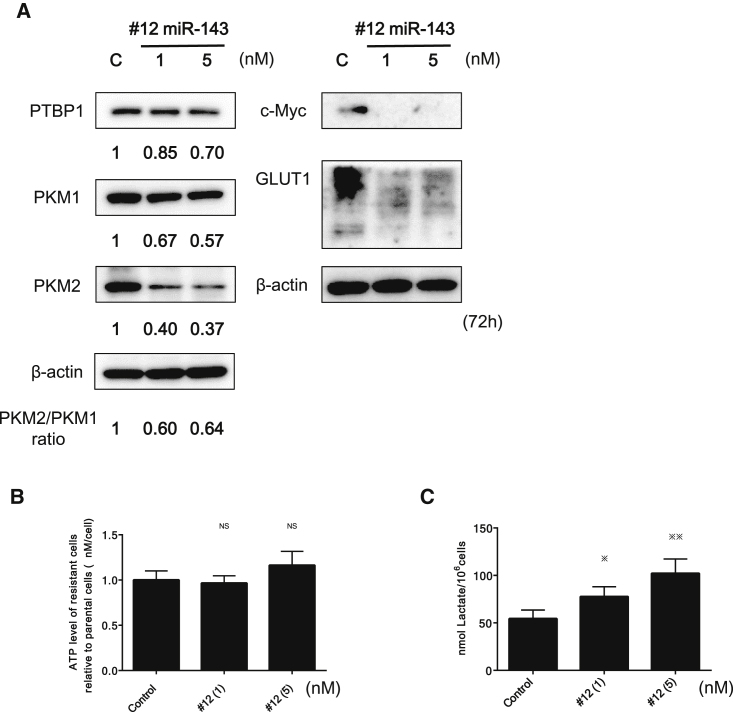
Ectopic Expression of #12 Perturbed Cancer-Specific Energy Metabolism through RAS/GLUT1 and c-Myc/PTBP1/PKMs Pathways
PTBP1 is a chief splicer of the PKM gene, which encodes a rate-limiting enzyme of glycolysis. Our previous study revealed that the overexpression of PTBP1, which promotes the expression of PKM2 over that of PKM1, contributes to the maintenance of cancer-specific energy metabolism.36, 37 It was reported that K-RAS positively regulates the expression of GLUT1.28 To elucidate the effect of #12 on cancer-specific energy metabolism through the downregulation of K-RAS, we performed ectopic expression of #12. As a result, a significant downregulation of GLUT1 and reversed PKM2:PKM1 ratio was induced, probably through the downregulation of c-Myc and its downstream PTBP1 (Figure 3A). The intracellular lactate and ATP levels tended to increase in a concentration-dependent manner (Figures 3B and 3C). We considered that the metabolic shift from glycolysis to oxidative phosphorylation (OXPHOS) occurred partly due to the decreased ratio of PKM2:PKM1, which would result in increased intracellular reactive oxygen species (ROS) levels associated with the tricarboxylic acid (TCA) cycle.
Figure 3.
miR-143 Induced a Significant Downregulation of GLUT1 and Decreased the PKM2:PKM1 Ratio by Downregulating c-Myc and PTBP1
(A) Protein expression of GLUT1 and glycolysis-related genes, including c-Myc, PTBP1, PKM1, and PKM2, was examined by western blot analysis. The numbers under the panels indicate values relative to the control taken as “1,” as determined by densitometry. (B and C) Intracellular ATP (B) and lactic acid (C) levels were measured at 72 h after transfection of Caki-1 cells with #12 (1 or 5 nM). *p < 0.05, **p < 0.01, ***p < 0.001. Error bars, means + SD.
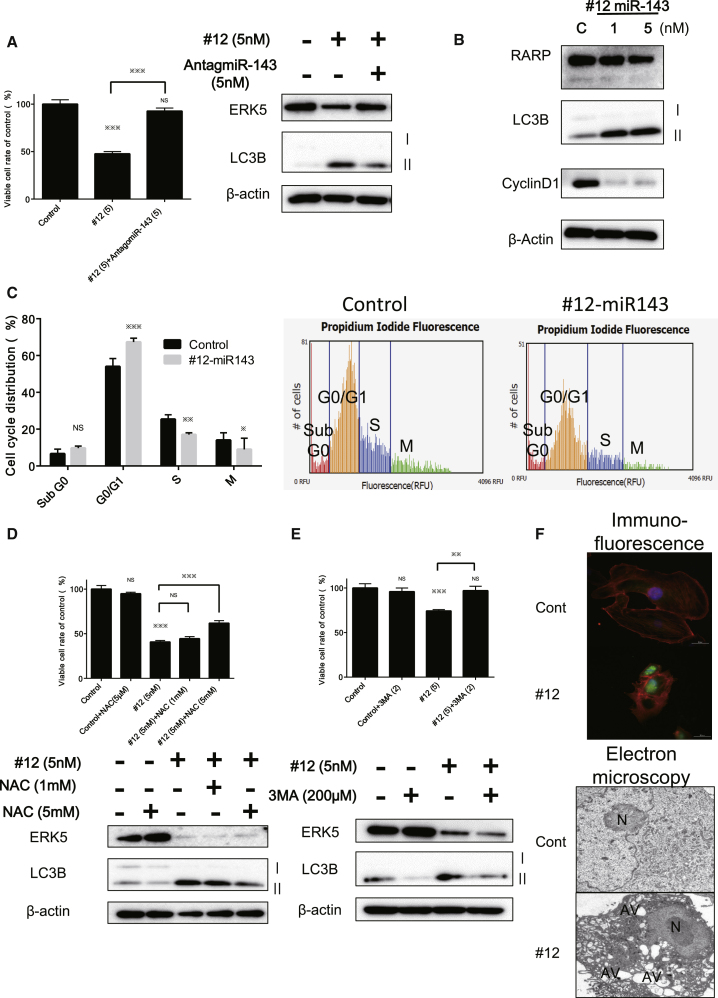
Furthermore, western blot analysis indicated the downregulation of cyclin D1 and the transition of the LC3BI to LC3BII; and, to the contrary, treatment with antagomiR-143 reversed the growth inhibition and the decreased level of LC3BII elicited by #12 (Figures 4A and 4B). Expectedly, #12 induced autophagy along with G0/G1 cell-cycle arrest in Caki-1 cells (Figure 4C). The free radical scavenger N-acetyl-L-cysteine (NAC) partially canceled the autophagy induced by the transfection with #12, a finding that was validated by the reduced transition of LC3BI to II and recovery of the viable cell rate (Figure 4D). Also, we validated the occurrence of autophagy by using an inhibitor of autophagy, 3-methyladenine (3-MA). The treatment with 3-MA (200 μM) reversed the growth inhibition and the decrease in the transition from the LC3BI to LC3BII elicited by #12 (Figure 4E).
Figure 4.
Ectopic Expression of miR-143 Induced Autophagy and G0/G1 Cell-Cycle Arrest in Caki-1 Cells
(A) Treatment with antagomiR-143 reversed the growth inhibition, increased the transition from LC3BI to II, and decreased the protein level of ERK5 elicited by miR-143. (B) Western blot analysis was performed for the validation of autophagy and cell-cycle arrest. #12 caused the transition from LC3BI to II and suppressed the expression of Cyclin D1 without appearance of the cleaved form of PARP. (C) Cell cycle distribution analyzed by cytometry at 72 h after transfection with control miRNA or #12 (5 nM). (D) Effects of NAC on the growth and cell death of Caki-1 cells after transfection with #12. Caki-1 cells were treated with NAC (1 and 5 mM) at 12 h after transfection with #12 (5 nM). Cell viability and results of western blot analysis at 72 h after the transfection of Caki-1 cells with #12 are shown. (E) Inhibition of autophagy in Caki-1 cells by 3-MA. The cells were pretreated with 3-MA (200 μM) and then transfected with #12 (5 nM) for 72 h. *p < 0.05, **p < 0.01, ***p < 0.001. Error bars, means + SD. (F) Top: immunofluorescence of LC3B at 72 h after transfection of Caki-1 cells with #12 (5 nM). LC3B, green; cytoskeleton, red; nucleus, blue. Bottom: morphological study using electron microscopy. Caki-1 cells were treated with control miRNA or #12 (5 nM) for 72 h. N, fragmented nuclei; AV, autophagosomes.
Morphologically, we performed immunofluorescence (IF) and electron microscopic studies on Caki-1 cells transfected with #12 (5 nM). As a result, immunostaining for LC3B showed a significant increase in signal intensity in the treated cells at the single-cell level, and typical autophagic features such as the formation of autolysosomes were also observed by electron microscopy (Figure 4F). These results strongly suggested that #12 induced autophagy through impairing the K-RAS/c-MYC/PTBP1/PKMs network, downregulation of GLUT1, and finally inducing a partial metabolic shift from glycolysis to OXPHOS due to the decrease in the PKM2:PKM1 ratio. Moreover, #12 suppressed various growth-signaling pathways, including PI3K/AKT and MAPK/ERK.
The Stability of #12 against RNase in Medium Containing Mouse Serum and in Caki-1 Cell-Xenografted Tumor
To acquire a significant resistance to RNase, #12 was chemically modified in the antisense strand, as given in Table 1. We estimated the resistance to RNase of #12 compared with that of A143 and #1 by incubating these miRNAs in 30% mouse serum-containing medium. The amount of #12 and A143 remaining at 60 min after the start of incubation was approximately 50% and 20%, respectively, whereas most of #1 had decayed within 30 min after the incubation (Figure S3A). In addition, the decay of miR-143 in the Caki-1 cell-xenografted tumor was also examined. As shown in Figure S3B, the amount of miR-143 remaining in the tumors at 24 h after the injection of #12 was significantly higher than that for the tumors injected with A143 or #1. These results suggested that miR-143 chemically modified with fluorine, methoxy group, deoxythymidine, and phosphrothioate in the antisense strand could acquire a strong resistance to RNase and be stable in the blood.
Inhibition of Tumor Growth in Caki-1 Cell-Xenografted Mouse by General Infusion of the Synthetic miR-143 Carried by PIC
The PIC can be prepared through the spontaneous assembly of cationic copolymers with oppositely charged miRNAs (Figure S4). The PIC micelle is approximately 20 nm in diameter and has a PEG-coated surface, which is expected to protect miRNA from RNase and accumulate in the blood and tumors by an enhanced permeability and retention (EPR) effect.38, 39 On the other hand, the size distribution of LipoTrust (Lipo) is more than 100 nm.40, 41
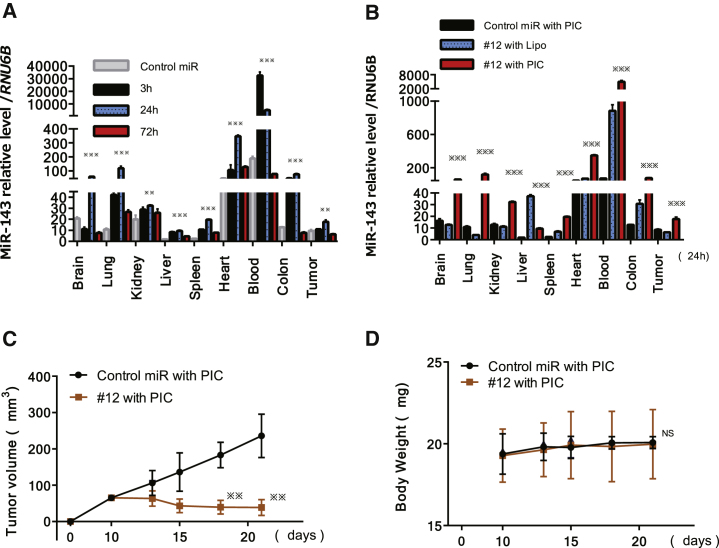
To validate the tissue distribution of miR-143 after the treatment with PIC-loaded miR-143#12 (#12/PIC), we examined the levels of miR-143 in the blood, xenografted tumors, and other organs by performing qRT-PCR. First, the time course of the miR-143 level in various organs, including tumor tissue after the last injection with #12/PIC, was examined. The miR-143 levels in the tumor and each organ were the highest at 24 h, excluding the level in the blood at 3 h, after the last injection (Figure 5A). Next, we compared the tissue distribution of miR-143 between #12/Lipo and #12/PIC at 24 h after the last injection (Figure 5B). Notably, the levels of miR-143 in blood and tumor tissues of the #12/PIC group were significantly higher than those of the #12/Lipo group. Based on the results of blood and tumor miR-143 levels, we designed the experiments to validate the anti-tumor effect of #12. We compared the effect between control miR/PIC and #12/PIC with the same drug delivery system. After the last treatment of #12/PIC or control miR/PIC, the mice were sacrificed, and the tumor volumes were then measured. As a result, #12/PIC exhibited a marked tumor growth inhibition compared with the control mice, even at the dose of 210 μg/kg/injection (Figure 5C). As shown in Figure 5D, there was no difference in body weight between the groups.
Figure 5.
Comparison between PIC Carrier and Lipofection for Delivery in Tissue Distribution of miR-143 after General Administration and Tumor-Suppressive Effects of miR-143#12/PIC on Caki-1 Cell-Xenografted Mice
(A) Time course of miR-143 relative to the level of RNU6B in each indicated organ at 3, 24, and 72 h after the last intravenous injection of #12/PIC. The levels of miR-143 in control miR/PIC are shown at 3 h after the last administration. (B) miR-143 relative to the level of RNU6B in each indicated organ at 24 h after the last intravenous injection with control miR/PIC, #12/Lipo, or #12/PIC. (C) Time course of tumor size in mice injected intravenously with control miR/PIC or #12/PIC. p values were determined for differences between the tumors injected with control miR/PIC and #12/PIC. (D) Time course of body weight after injection of each formulation in (C). *p < 0.05, **p < 0.01, ***p < 0.001. Error bars, means + SD.
Discussion
So far, we have been studying the roles of miRNAs in carcinogenesis and the pathogenesis of cancer.37, 42 In the progression of our research, we focused on miRs-143 and -145, which are from the same primary miRNA at chromosomal location 5q33 and, acting as tumor suppressor miRNAs, are extremely downregulated commonly in most kinds of cancer cells.11, 12 It is considerably important that miR-143 silences K-RAS by binding to 2 different regions in the 3′ UTR43 and that miR-143 silences the effector signal molecules AKT and ERK, because they are downstream of PI3K and Raf, which are frequently mutated. However, the commercially available miR-143 (A-miR-143) from Ambion does not show sufficient growth inhibition in human K-RAS-driven colon cancer cells or gastric cells, because A-miR-143 can’t silence K-RAS expression significantly.
In contrast, the chemically modified miR-143 (#12), which was markedly resistant to RNase, significantly silenced K-RAS. Consequently, we could gain a better understanding of most of the networks associated with K-RAS by using miR-143#12. miR-143 silenced other oncogenic genes, including those encoding ERK1/2, AKT, and ERK5, which are signal molecules of RAS effector-signaling pathways. We consider that the downregulation of miR-143 caused gain of functions of K-RAS and activation of its signaling networks during carcinogenesis. Although VHL is well known as an anti-oncogene that degrades HIF-1α in RCC, any other onco-related genes of RCC have not been identified yet. K-RAS is a well-known oncogene in various types of cancers. We certified the K-RAS gene product as being the main isomer of RAS in Caki-1 cells by PCR and nucleotide-sequencing analyses. However, no mutation at the hotspots was observed.
In this study, the ectopic expression of miR-143#12 induced autophagic cell death through the downregulation of K-RAS and inactivation of both MAPK/AKT- and PI3K/ERK-signaling pathways in Caki-1 cells. Furthermore, it impaired the PTBP1/PKM2 axis, causing a shift away from cancer-specific energy metabolism. These results demonstrated that the impairment of K-RAS networks by miR-143#12 led to perturbation of the Warburg effect. Furthermore, we successfully addressed the issue of miRNA stability in the bloodstream by confirming the resistance to RNase of miR-143#12. We developed more than 100 kinds of synthetic miR-143 derivatives, #1 being the wild-type of miR-143 and #12 being #1 chemically modified with fluorine, methoxy group, phosphorylation, deoxythymidine, and phosphorothioate in its antisense strand (Table 1). Among the chemically modified miR-143 derivatives, the miR-143 derivatives that fully modified only in the antisense strand exhibited a growth-inhibitory effect. Among them, #12 had the most potent growth-inhibitory activity and acquired the higher RNase resistance (Figure S3). It is possible that suitable chemical modification depends on the structure of double-stranded miRNA that has mismatched regions. The target genes of #1 and #12 were similar to those of miR-143, including A143, because both miRNAs targeted K-RAS, AKT, ERK1/2, and ERK5.
In this study, we used a PIC nanocarrier as an RNA delivery system. The PIC nanocarrier more efficiently delivered miR-143 to tumor tissues to a relatively greater degree compared with Lipofection (Figures 5A and 5B). One of the reasons why miR-143 was delivered more effectively by PIC is that the #12/PIC was considerably stable, remaining almost 90%, even at 180 min after exposure in the presence of serum in vitro (data not shown). Second, the diameter of PIC is approximately 20 nm, which was characterized to accumulate in tumor tissue rather than Lipo (Figure 5B; Figure S4). PIC could protect synthesized miR-143s in the blood. In Caki-1 cell-xenografted mice, #12/PIC exhibited a marked anti-tumor effect, as observed in in vitro experiments.
Nair et al.44 reported that more than 50% of an injected dose of nucleic acid is delivered to the liver in vivo. Nucleic acids carried by liposomes accumulate well in the liver, and so the toxicity caused by liposomes and the on-target effects of miRNAs delivered via liposomes would occur more in the liver tissue than elsewhere. PIC micelles can be prepared through the spontaneous assembly of cationic block copolymers with oppositely charged miRNA (Figure S4). Their core-shell architectures offer a delivery platform for vulnerable miRNA, improving their biological activities for medicinal applications such as tumor-targeted therapy. The surface of the complex is covered with PEG, which causes it to reduce the renal clearance and immune surveillance. As a result, the level of blood miR-143/PIC was increased, however, the hepatotoxicity was not observed in our model mice (data not shown). It was considered that PIC is really safe and suitable for the delivery of nucleic acid to tumor tissue. These findings suggest that novel miR-143#12 carried by PIC nanocarriers, which can deliver nucleic acids specifically to tumor tissue, has the potential to be a new strategy as an RNA medicine for the treatment of various cancers, including RCC.
Materials and Methods
Cell Culture and Cell Viability
Caki-1 cells were obtained from the JCRB (Japanese Collection of Research Bioresources) Cell Bank. The cells were cultured in Eagle’s minimum essential medium (EMEM) supplemented with 10% (v/v) heat-inactivated fetal bovine serum (FBS) (Sigma-Aldrich, St. Louis, MO, USA) and 2 mM L-glutamine under an atmosphere of 95% air and 5% CO2 at 37°C. The number of viable cells was determined by performing the trypan blue dye exclusion test.
Patients and Samples
All human samples were obtained from patients who had undergone biopsy or surgery for resection at Osaka Medical College Hospital (Takatsuki, Osaka, Japan). Informed consent in writing was obtained from each patient. The consent and this study were reviewed and approved by the University Hospital Medical Information Network Center (approval R000027312 on April 8, 2016), in accordance with the tenets of the Declaration of Helsinki. Six patients with previously untreated (or recently diagnosed) RCC were selected. The distribution according to other clinical parameters is shown in Table S1. Under a pathologist’s supervision, all tissue sample pairs were collected from surgically resected tissues, with these paired samples being from the primary tumor and its adjacent non-tumor tissue in the same patient. These paired samples were examined by western blotting analysis and real-time RT-PCR.
Transfection Experiments
Caki-1 cells were seeded in 6-well plates at a concentration of 0.35 × 105/well (10%–30% confluence) on the day before the transfection. The mature type of miR-143, A143 (mirVana miRNA mimic; Ambion, Foster City, CA, USA); synthetic wild-type miR-143, #1 (Shionogi, Osaka, Japan); and chemically modified miR-143, #12 (Shionogi, Osaka, Japan) were used for the transfection of the cells, which was achieved by using cationic liposomes, Lipofectamine RNAiMAX (Invitrogen), according to the manufacturer’s Lipofection protocol. The nonspecific control miRNA (Hokkaido System Science [HSS], Hokkaido, Japan) sequence was 5′-GUA GGA GUA GUG AAA GGCC-3′, which was used as a control miRNA for nonspecific effects.13 The nonspecific control miRNA was designed to target the firefly (Lampyris pyralis) luciferase mRNA and, thus, have less interaction with human mRNA. Then, the sequences were slightly modified to have fewer similarities to human mRNA sequences by using BLAST (NCBI, MD, USA). Microarray analysis confirmed that the control miRNA has minimal effects in several types of cancer cell lines as well. We used it in in vitro and in vivo experiments. The sequences of the synthetic miR-143 and chemically modified miR-143 are shown in Table 1. The effects manifested by the introduction of each miR-143 into the cells were assessed at 72 h after the transfection.
Western Blotting
Protein extraction and western blotting analysis were performed in triplicate as described in our previous reports.45, 46 The following primary antibodies were used: antibodies against c-Myc, PTBP1, p-AKT, AKT, p-ERK1/2, ERK1/2, PARP, LC3B, ERK5, CyclinD1, and GLUT1 (Cell Signaling Technology, Danvers, MA, USA); PKM1 and PKM2 (Novus Biologicals, USA); RAS (Abcam, Cambridge, UK); and β-actin antibody (Sigma-Aldrich). Horseradish peroxidase (HRP)-conjugated goat anti-rabbit and horse anti-mouse immunoglobulin G (IgG) antibodies (Cell Signaling Technology) were used as secondary antibodies. β-actin served as an internal control.
Real-Time RT-PCR
Total RNA was isolated from cultured cells or tumor tissues by using a NucleoSpin miRNA isolation kit (TaKaRa, Otsu, Japan). Total RNA of HRPTE cells was available from TOYOBO Life Science (Tokyo, Japan). RNA concentrations and purity were assessed by UV spectrophotometry. RNA integrity was checked by formaldehyde gel electrophoresis. To determine the expression levels of miR-143, we conducted qRT-PCR by using TaqMan MicroRNA Assays (Applied Biosystems) and THUNDERBIRD Probe qPCR Mix (TOYOBO,Osaka, Japan), according to the manufacturer’s protocol. RNU6B was used as an internal control.
For determination of the expression levels of K-RAS and β-actin (β-actin) mRNAs, total RNA was reverse transcribed with a PrimeScriptH RT reagent Kit (TaKaRa). RT-PCR was then performed in triplicate with primers specific for them by using THUNDERBIRD SYBR qPCR Mix (TOYOBO). The primers for K-RAS and β-actin were the following: K-RAS sense, 5′-CCT GCT CCA TGC AGA CTG TTA-3′, and K-RAS antisense, 5′-TGG GGA GAG TGA CCA GACT-3′; and β-actin sense, 5′-TGA CGG GGT CAC CCA CAC TGT GCC CAT CTA-3′, and β-actin antisense, 5′-CTA GAA GCA TTT GCG GTG GAC GAT GGA GGG-3′. β-actin was used as an internal control. The relative expression levels were calculated by the ΔΔCt method.
Cell Cycle Analysis
Quantification of cellular DNA content at 72 h after transfection with control miRNA or #12 (5 nM) was determined by using a cytometer. Briefly, the cells were harvested and fixed with 70% cold ethanol at −20°C overnight. The fixed cells were then washed twice with PBS and resuspended in 100 μL PBS-based propidium iodide solution containing 0.1% Triton X-100 (Wako, Osaka, Japan), 0.2 mg/mL RNase A (Life Technologies), and 20 μg/mL propidium iodide (Life Technologies). Thereafter, they were incubated for 30 min at room temperature protected from the light. The DNA content in the cells was analyzed with a cytometer (The Tail Image-Based Cytometer, Life Technologies).
Inhibitor Agents
We used the pan-caspase inhibitor 3-MA, from Calbiochem (San Diego, CA, USA), and the free radical scavenger NAC, from Sigma-Aldrich (St. Louis, MO, USA). Caki-1 cells were pretreated with 3-MA (0.3 mM) before the transfection with miR-143. Caki-1 cells were treated with NAC (1 and 5 mM) 12 h after transfection with each miR-143.
Lactate Assay
Cells were incubated with #12 for 72 h. Intracellular L-lactate was extracted by using an L-Lactate Assay kit (Cayman Chemical, Ann Arbor, MI, USA). L-lactate production was measured with a Lactate Colorimetric/Fluorometric Assay kit (Biovision, Milpitas, CA, USA), according to the manufacturer’s instructions. Lactate production was normalized to cell numbers.
ATP Assay
To measure the ATP levels before the cells were committed to programmed cell death, we incubated them with #12 for 72 h. ATP production was measured with an ATP Determination Kit (A22066; Invitrogen), according to the manufacturer’s instructions. ATP production was normalized to cell numbers.
Immunofluorescence Study
We transfected Caki-1 cells with control miRNA or #12 (5 nM). At 72 h after the transfection, the cells were fixed for 15 min with 4% paraformaldehyde. They were then transferred to PBS and kept in it for 15 min, followed by exposure to 1% Block Ace (Dainippon Sumitomo Pharma, Tokyo, Japan) for 20 min to block nonspecific antibody binding. Next, the cells were incubated with primary antibody against LC3B (Cell Signaling Technology, Danvers, MA, USA) for 2 h at room temperature (RT). After a rinse in PBS for 15 min, the cells were then incubated with biotin anti-rabbit antibody (Dako, Carpenteria, CA, USA) for 60 min, followed by streptavidin-conjugated Alexa Fluor 555 (Molecular Probes, Eugene, OR, USA) for 30 min at RT. Finally, the samples were counterstained with DAPI for 15 min and observed under a fluorescence microscope.
Electron Microscopic Study
Caki-1 cells treated with control miRNA or #12 (5 nM) were harvested and rinsed with PBS. The cells were then fixed for 2 h with 2% paraformaldehyde and 2.5% glutaraldehyde in 0.2 M phosphate buffer (PB [pH 7.4]), rinsed in PB, and postfixed in 2% osmium tetraoxide for 2 h. After having been washed with PB, the cells were progressively dehydrated by passage through a 10% graded series of 30%–100% ethanol and then cleared in QY-1 (Nissin EM, Tokyo, Japan). Thereafter, they were embedded in Epon 812 resin (TAAB Laboratories Equipment, Reading, UK), and thin sections (70-nm thickness) were prepared. Finally, the sections were stained with uranyl acetate and lead citrate and examined by transmission electron microscopy with a Hitachi-7650 (Hitachi, Tokyo, Japan), operating at 80 kV.
Assay for the Stability of miRNA In Vitro
To evaluate the stability of miRNA in vitro, we incubated 20 pmol of each miR-143 (A143, #1, and #12) without Lipofectamine RNAiMAX in 30% fresh mouse serum with 90 μL EMEM at 37°C for 0, 5, 30, or 60 min. Then, total RNA was isolated, and real-time RT-PCR using TaqMan MicroRNA Assay was thereafter performed to quantify the level of miR-143 remaining, which was calculated by the ΔΔCt method.
In Vivo Xenograft Model
BALB/cSLC-nu/nu (nude) mice were obtained from Japan SLC (Hamamatsu, Japan). Human RCC Caki-1 cells were inoculated at 3.0 × 106 cells/100 μL/site into the back of each mouse. The inoculation day was set as day 0. First, to decide the frequency of miRNA administration, at 14 days after inoculation of the tumor cells, A143, #1, or #12 (0.1 nmol) in 50 μL Opti-MEM (Invitrogen) was mixed with 2 μL cationic liposomes (LipoTrust, HSS, Sapporo); the mixture was injected into the tumors at one time. At 0 or 24 h after the injection, the mice were sacrificed, and the whole tissues of transplanted tumors were harvested for total RNA extraction and subsequent evaluation of their miR-143 expression levels.
To investigate the effect of drug delivery systems, we compared the effect between Lipo and PIC nanocarrier. For the treatment with the #12 carried by Lipo (#12/Lipo) group, #12 (210 μg/kg/1 administration) in 100 μL Opti-MEM was incubated with 2 μL Lipo. For the treatments with control miRNA and #12 carried by PIC nanocarrier (#12/PIC) groups, control miRNA or #12 (210 μg/kg/1 administration) in 100 μL liquid (0.5 M NaCl and 1 mM HEPES) was loaded into the nanocarrier (250 μg/1 administration) by mixing with PEG-based block copolymer.34, 47 We administered each formulation to mice intravenously every 72 h from 14 to 23 days.
At 3, 24, and 72 h after the last administration, mice were killed to obtain various organs, including tumors for examination of the tissue distribution of miR-143. The tissue distribution of miR-143 in the certain groups was examined by qRT-PCR. For evaluation of the antitumor effect of #12/PIC in vivo, we confirmed the engraftment of the tumors at 14 days after inoculation. Before each treatment and after the last treatment, the tumor volume was calculated by the formula 0.5236 L1 (L2)2, where L1 is the long axis and L2 is the short axis of the tumor. Animal experiments in this study were performed in compliance with the guidelines of the Institute for Laboratory Animal Research of Gifu University (approval 11021) and with the UKCCCR Guidelines for the Welfare of Animals Used for Experimental Neoplasia. Y.A. was named in the approved experiment.
Statistics
Each examination was performed in triplicate. Statistical differences between clinicopathologic parameters and the miR-143 level of tumor samples were evaluated by using Pearson’s χ2 test or Fisher’s exact test, unless otherwise specified. For in vitro and in vivo experiments, statistical significances of differences were evaluated by performing the two-sided Student’s t test. The values were presented as the mean ± SD. A p value < 0.05 was considered to be statistically significant.
Author Contributions
Conceptualization of Ideas, Y.A.; Methodology, Y.A.; Validation, T. Takai, T. Tsujino, K. Heishima, and Y.Y.; Investigation, T. Takai; Resources, T. Takai, T. Tsujino, Y.Y., T.I., N.S., Y.K., K. Hayashi, K.M., and H.A.; Data Curation, T. Takai and T.S.; Writing – Original Draft, T. Takai; Writing – Review & Editing, K.M., K.K., and Y.A.; Visualization, T. Takai and T.S.; Supervision, Y.A.; Project Administration, Y.A.; Funding Acquisition, Y.A.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
This work was supported by the grant Project for Cancer Research and Therapeutic Evolution from the Japan Agency for Medical Research and Development (AMED: 16cm0106202h001) This work was also performed with the support of SHIONOGI, japan.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.ymthe.2019.03.004.
Supplemental Information
References
- 1.Zhang L., Xul B., Chen S., Lu K., Liu C., Wang Y., Zhao Y., Zhang X., Liu D., Chen M. The complex roles of microRNAs in the metastasis of renal cell carcinoma. J. Nanosci. Nanotechnol. 2013;13:3195–3203. doi: 10.1166/jnn.2013.6712. [DOI] [PubMed] [Google Scholar]
- 2.Frees S., Kamal M.M., Knoechlein L., Bell R., Ziesel C., Neisius A., Thomas C., Brenner W., Jäger W., Thüroff J.W., Roos F.C. Differences in Overall and Cancer-specific Survival of Patients Presenting With Chromophobe Versus Clear Cell Renal Cell Carcinoma: A Propensity Score Matched Analysis. Urology. 2016;98:81–87. doi: 10.1016/j.urology.2016.05.048. [DOI] [PubMed] [Google Scholar]
- 3.Crispen P.L., Breau R.H., Allmer C., Lohse C.M., Cheville J.C., Leibovich B.C., Blute M.L. Lymph node dissection at the time of radical nephrectomy for high-risk clear cell renal cell carcinoma: indications and recommendations for surgical templates. Eur. Urol. 2011;59:18–23. doi: 10.1016/j.eururo.2010.08.042. [DOI] [PubMed] [Google Scholar]
- 4.Hadoux J., Vignot S., De La Motte Rouge T. Renal cell carcinoma: focus on safety and efficacy of temsirolimus. Clin. Med. Insights Oncol. 2010;4:143–154. doi: 10.4137/CMO.S4482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marangoni E., Vincent-Salomon A., Auger N., Degeorges A., Assayag F., de Cremoux P., de Plater L., Guyader C., De Pinieux G., Judde J.G. A new model of patient tumor-derived breast cancer xenografts for preclinical assays. Clin. Cancer Res. 2007;13:3989–3998. doi: 10.1158/1078-0432.CCR-07-0078. [DOI] [PubMed] [Google Scholar]
- 6.Patel P.H., Chaganti R.S., Motzer R.J. Targeted therapy for metastatic renal cell carcinoma. Br. J. Cancer. 2006;94:614–619. doi: 10.1038/sj.bjc.6602978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Escudier B., Eisen T., Stadler W.M., Szczylik C., Oudard S., Siebels M., Negrier S., Chevreau C., Solska E., Desai A.A., TARGET Study Group Sorafenib in advanced clear-cell renal-cell carcinoma. N. Engl. J. Med. 2007;356:125–134. doi: 10.1056/NEJMoa060655. [DOI] [PubMed] [Google Scholar]
- 8.Sonpavde G., Hutson T.E., Rini B.I. Axitinib for renal cell carcinoma. Expert Opin. Investig. Drugs. 2008;17:741–748. doi: 10.1517/13543784.17.5.741. [DOI] [PubMed] [Google Scholar]
- 9.Bukowski R.M. Pazopanib: a multikinase inhibitor with activity in advanced renal cell carcinoma. Expert Rev. Anticancer Ther. 2010;10:635–645. doi: 10.1586/era.10.38. [DOI] [PubMed] [Google Scholar]
- 10.Rini B.I., Atkins M.B. Resistance to targeted therapy in renal-cell carcinoma. Lancet Oncol. 2009;10:992–1000. doi: 10.1016/S1470-2045(09)70240-2. [DOI] [PubMed] [Google Scholar]
- 11.Akao Y., Nakagawa Y., Naoe T. MicroRNA-143 and -145 in colon cancer. DNA Cell Biol. 2007;26:311–320. doi: 10.1089/dna.2006.0550. [DOI] [PubMed] [Google Scholar]
- 12.Akao Y., Nakagawa Y., Naoe T. MicroRNAs 143 and 145 are possible common onco-microRNAs in human cancers. Oncol. Rep. 2006;16:845–850. [PubMed] [Google Scholar]
- 13.Akao Y., Nakagawa Y., Hirata I., Iio A., Itoh T., Kojima K., Nakashima R., Kitade Y., Naoe T. Role of anti-oncomirs miR-143 and -145 in human colorectal tumors. Cancer Gene Ther. 2010;17:398–408. doi: 10.1038/cgt.2009.88. [DOI] [PubMed] [Google Scholar]
- 14.Akao Y., Nakagawa Y., Iio A., Naoe T. Role of microRNA-143 in Fas-mediated apoptosis in human T-cell leukemia Jurkat cells. Leuk. Res. 2009;33:1530–1538. doi: 10.1016/j.leukres.2009.04.019. [DOI] [PubMed] [Google Scholar]
- 15.Akao Y., Nakagawa Y., Kitade Y., Kinoshita T., Naoe T. Downregulation of microRNAs-143 and -145 in B-cell malignancies. Cancer Sci. 2007;98:1914–1920. doi: 10.1111/j.1349-7006.2007.00618.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin T., Dong W., Huang J., Pan Q., Fan X., Zhang C., Huang L. MicroRNA-143 as a tumor suppressor for bladder cancer. J. Urol. 2009;181:1372–1380. doi: 10.1016/j.juro.2008.10.149. [DOI] [PubMed] [Google Scholar]
- 17.Ahmadian M.R., Zor T., Vogt D., Kabsch W., Selinger Z., Wittinghofer A., Scheffzek K. Guanosine triphosphatase stimulation of oncogenic Ras mutants. Proc. Natl. Acad. Sci. USA. 1999;96:7065–7070. doi: 10.1073/pnas.96.12.7065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sasaki A.T., Carracedo A., Locasale J.W., Anastasiou D., Takeuchi K., Kahoud E.R., Haviv S., Asara J.M., Pandolfi P.P., Cantley L.C. Ubiquitination of K-Ras enhances activation and facilitates binding to select downstream effectors. Sci. Signal. 2011;4:ra13. doi: 10.1126/scisignal.2001518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang W., Zheng Y., Xia Y., Ji H., Chen X., Guo F., Lyssiotis C.A., Aldape K., Cantley L.C., Lu Z. ERK1/2-dependent phosphorylation and nuclear translocation of PKM2 promotes the Warburg effect. Nat. Cell Biol. 2012;14:1295–1304. doi: 10.1038/ncb2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.de Geus-Oei L.F., van Krieken J.H., Aliredjo R.P., Krabbe P.F., Frielink C., Verhagen A.F., Boerman O.C., Oyen W.J. Biological correlates of FDG uptake in non-small cell lung cancer. Lung Cancer. 2007;55:79–87. doi: 10.1016/j.lungcan.2006.08.018. [DOI] [PubMed] [Google Scholar]
- 21.Ganapathy V., Thangaraju M., Prasad P.D. Nutrient transporters in cancer: relevance to Warburg hypothesis and beyond. Pharmacol. Ther. 2009;121:29–40. doi: 10.1016/j.pharmthera.2008.09.005. [DOI] [PubMed] [Google Scholar]
- 22.Sasaki H., Shitara M., Yokota K., Hikosaka Y., Moriyama S., Yano M., Fujii Y. Overexpression of GLUT1 correlates with Kras mutations in lung carcinomas. Mol. Med. Rep. 2012;5:599–602. doi: 10.3892/mmr.2011.736. [DOI] [PubMed] [Google Scholar]
- 23.Szablewski L. Expression of glucose transporters in cancers. Biochim. Biophys. Acta. 2013;1835:164–169. doi: 10.1016/j.bbcan.2012.12.004. [DOI] [PubMed] [Google Scholar]
- 24.Chiaradonna F., Sacco E., Manzoni R., Giorgio M., Vanoni M., Alberghina L. Ras-dependent carbon metabolism and transformation in mouse fibroblasts. Oncogene. 2006;25:5391–5404. doi: 10.1038/sj.onc.1209528. [DOI] [PubMed] [Google Scholar]
- 25.Racker E., Resnick R.J., Feldman R. Glycolysis and methylaminoisobutyrate uptake in rat-1 cells transfected with ras or myc oncogenes. Proc. Natl. Acad. Sci. USA. 1985;82:3535–3538. doi: 10.1073/pnas.82.11.3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vizan P., Boros L.G., Figueras A., Capella G., Mangues R., Bassilian S., Lim S., Lee W.N., Cascante M. K-ras codon-specific mutations produce distinctive metabolic phenotypes in NIH3T3 mice [corrected] fibroblasts. Cancer Res. 2005;65:5512–5515. doi: 10.1158/0008-5472.CAN-05-0074. [DOI] [PubMed] [Google Scholar]
- 27.Gaglio D., Metallo C.M., Gameiro P.A., Hiller K., Danna L.S., Balestrieri C., Alberghina L., Stephanopoulos G., Chiaradonna F. Oncogenic K-Ras decouples glucose and glutamine metabolism to support cancer cell growth. Mol. Syst. Biol. 2011;7:523. doi: 10.1038/msb.2011.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yun J., Rago C., Cheong I., Pagliarini R., Angenendt P., Rajagopalan H., Schmidt K., Willson J.K., Markowitz S., Zhou S. Glucose deprivation contributes to the development of KRAS pathway mutations in tumor cells. Science. 2009;325:1555–1559. doi: 10.1126/science.1174229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jhaveri A.M., Torchilin V.P. Multifunctional polymeric micelles for delivery of drugs and siRNA. Front. Pharmacol. 2014;5:77. doi: 10.3389/fphar.2014.00077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miyata K., Nishiyama N., Kataoka K. Rational design of smart supramolecular assemblies for gene delivery: chemical challenges in the creation of artificial viruses. Chem. Soc. Rev. 2012;41:2562–2574. doi: 10.1039/c1cs15258k. [DOI] [PubMed] [Google Scholar]
- 31.Kanasty R., Dorkin J.R., Vegas A., Anderson D. Delivery materials for siRNA therapeutics. Nat. Mater. 2013;12:967–977. doi: 10.1038/nmat3765. [DOI] [PubMed] [Google Scholar]
- 32.Pittella F., Cabral H., Maeda Y., Mi P., Watanabe S., Takemoto H., Kim H.J., Nishiyama N., Miyata K., Kataoka K. Systemic siRNA delivery to a spontaneous pancreatic tumor model in transgenic mice by PEGylated calcium phosphate hybrid micelles. J. Control. Release. 2014;178:18–24. doi: 10.1016/j.jconrel.2014.01.008. [DOI] [PubMed] [Google Scholar]
- 33.Matsumoto S., Christie R.J., Nishiyama N., Miyata K., Ishii A., Oba M., Koyama H., Yamasaki Y., Kataoka K. Environment-responsive block copolymer micelles with a disulfide cross-linked core for enhanced siRNA delivery. Biomacromolecules. 2009;10:119–127. doi: 10.1021/bm800985e. [DOI] [PubMed] [Google Scholar]
- 34.Shimizu H., Hori Y., Kaname S., Yamada K., Nishiyama N., Matsumoto S., Miyata K., Oba M., Yamada A., Kataoka K., Fujita T. siRNA-based therapy ameliorates glomerulonephritis. J. Am. Soc. Nephrol. 2010;21:622–633. doi: 10.1681/ASN.2009030295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Christie R.J., Matsumoto Y., Miyata K., Nomoto T., Fukushima S., Osada K., Halnaut J., Pittella F., Kim H.J., Nishiyama N., Kataoka K. Targeted polymeric micelles for siRNA treatment of experimental cancer by intravenous injection. ACS Nano. 2012;6:5174–5189. doi: 10.1021/nn300942b. [DOI] [PubMed] [Google Scholar]
- 36.Taniguchi K., Sugito N., Kumazaki M., Shinohara H., Yamada N., Nakagawa Y., Ito Y., Otsuki Y., Uno B., Uchiyama K., Akao Y. MicroRNA-124 inhibits cancer cell growth through PTB1/PKM1/PKM2 feedback cascade in colorectal cancer. Cancer Lett. 2015;363:17–27. doi: 10.1016/j.canlet.2015.03.026. [DOI] [PubMed] [Google Scholar]
- 37.Takai T., Yoshikawa Y., Inamoto T., Minami K., Taniguchi K., Sugito N., Kuranaga Y., Shinohara H., Kumazaki M., Tsujino T. A Novel Combination RNAi toward Warburg Effect by Replacement with miR-145 and Silencing of PTBP1 Induces Apoptotic Cell Death in Bladder Cancer Cells. Int. J. Mol. Sci. 2017;18:E179. doi: 10.3390/ijms18010179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Matsumura Y., Maeda H. A new concept for macromolecular therapeutics in cancer chemotherapy: mechanism of tumoritropic accumulation of proteins and the antitumor agent smancs. Cancer Res. 1986;46:6387–6392. [PubMed] [Google Scholar]
- 39.Maeda H., Wu J., Sawa T., Matsumura Y., Hori K. Tumor vascular permeability and the EPR effect in macromolecular therapeutics: a review. J. Control. Release. 2000;65:271–284. doi: 10.1016/s0168-3659(99)00248-5. [DOI] [PubMed] [Google Scholar]
- 40.Xu Y., Hui S.W., Frederik P., Szoka F.C., Jr. Physicochemical characterization and purification of cationic lipoplexes. Biophys. J. 1999;77:341–353. doi: 10.1016/S0006-3495(99)76894-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Furst T., Dakwar G.R., Zagato E., Lechanteur A., Remaut K., Evrard B., Braeckmans K., Piel G. Freeze-dried mucoadhesive polymeric system containing pegylated lipoplexes: Towards a vaginal sustained released system for siRNA. J. Control. Release. 2016;236:68–78. doi: 10.1016/j.jconrel.2016.06.028. [DOI] [PubMed] [Google Scholar]
- 42.Taniguchi K., Sakai M., Sugito N., Kumazaki M., Shinohara H., Yamada N., Nakayama T., Ueda H., Nakagawa Y., Ito Y. PTBP1-associated microRNA-1 and -133b suppress the Warburg effect in colorectal tumors. Oncotarget. 2016;7:18940–18952. doi: 10.18632/oncotarget.8005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen X., Guo X., Zhang H., Xiang Y., Chen J., Yin Y., Cai X., Wang K., Wang G., Ba Y. Role of miR-143 targeting KRAS in colorectal tumorigenesis. Oncogene. 2009;28:1385–1392. doi: 10.1038/onc.2008.474. [DOI] [PubMed] [Google Scholar]
- 44.Nair J.K., Willoughby J.L., Chan A., Charisse K., Alam M.R., Wang Q., Hoekstra M., Kandasamy P., Kel’in A.V., Milstein S. Multivalent N-acetylgalactosamine-conjugated siRNA localizes in hepatocytes and elicits robust RNAi-mediated gene silencing. J. Am. Chem. Soc. 2014;136:16958–16961. doi: 10.1021/ja505986a. [DOI] [PubMed] [Google Scholar]
- 45.Noguchi S., Iwasaki J., Kumazaki M., Mori T., Maruo K., Sakai H., Yamada N., Shimada K., Naoe T., Kitade Y., Akao Y. Chemically modified synthetic microRNA-205 inhibits the growth of melanoma cells in vitro and in vivo. Mol. Ther. 2013;21:1204–1211. doi: 10.1038/mt.2013.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yamada N., Tsujimura N., Kumazaki M., Shinohara H., Taniguchi K., Nakagawa Y., Naoe T., Akao Y. Colorectal cancer cell-derived microvesicles containing microRNA-1246 promote angiogenesis by activating Smad 1/5/8 signaling elicited by PML down-regulation in endothelial cells. Biochim. Biophys. Acta. 2014;1839:1256–1272. doi: 10.1016/j.bbagrm.2014.09.002. [DOI] [PubMed] [Google Scholar]
- 47.Moriya C., Taniguchi H., Miyata K., Nishiyama N., Kataoka K., Imai K. Inhibition of PRDM14 expression in pancreatic cancer suppresses cancer stem-like properties and liver metastasis in mice. Carcinogenesis. 2017;38:638–648. doi: 10.1093/carcin/bgx040. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.