Abstract
Brain disorder recognition has becoming a promising area of study. In reality, some disorders share similar features and signs, making the task of diagnosis and treatment challenging. This paper presents a rigorous and robust computer aided diagnosis system for the detection of multiple brain abnormalities which can assist physicians in the diagnosis and treatment of brain diseases. In this system, we used energy of wavelet sub bands, textural features of gray level co-occurrence matrix and intensity feature of MR brain images. These features are ranked using Wilcoxon test. The composite features are classified using back propagation neural network. Bayesian regulation is adopted to find the optimal weights of neural network. The experimentation is carried out on datasets DS-90 and DS-310 of Harvard Medical School. To enhance the generalization capability of the network, fivefold stratified cross validation technique is used. The proposed system yields multi class disease classification accuracy of 100% in differentiating 90 MR brain images into 18 classes and 97.81% in differentiating 310 MR brain images into 6 classes. The experimental results reveal that the composite features along with BPNN classifier create a competent and reliable system for the identification of multiple brain disorders which can be used in clinical applications. The Wilcoxon test outcome demonstrates that standard deviation feature along with energies of approximate and vertical sub bands of level 7 contribute the most in achieving enhanced multi class classification performance results.
Keywords: Magnetic resonance imaging, Discrete wavelet transform, Gray level co-occurrence matrix, Back propagation neural network, Multi class classification
Introduction
Magnetic resonance imaging (MRI) technique uses magnetic field and radio frequency (RF) signals to produce the image of anatomical structure which shows the presence of disease. It is non-invasive technique and does not use harmful radiations. It provides superior contrast for different brain tissues compared to other imaging modalities [1]. These properties of MRI are more useful for brain pathology diagnosis and treatment [2]. However, the amount of MR imaging data is voluminous for manual interpretation, hence computer aided detection (CAD) systems are emerging to reduce the manual workload thus improving the diagnostic accuracy of radiologists [3, 4].
To classify MR brain images, researchers have proposed various useful techniques. El-Dahshan et al. [5] used discrete wavelet transform (DWT) as feature extraction and principal component analysis (PCA) as feature reduction tool for MRI images. They attained classification accuracy of 98% using k-nearest neighbour (k-NN) as classifier. Das et al. [6] utilised combination of ripplet transform (RT) and multi scale geometric analysis (MGA) to extract features from images. Using least square-SVM (LS-SVM) classifier, they achieved average classification accuracy greater than 99%. Saritha et al. [7] proposed a new methodology using spider web plot areas as features and probabilistic neural network as classifier. They achieved 100% binary classification accuracy. El-Dahshan et al. [8] suggested a method using feedback pulse coupled neural network (FPCNN) for MR image segmentation and the discrete wavelet transform (DWT) for features extraction. Yang et al. [9] introduced biogeography based optimization (BBO) method to optimize the weights of the classifier. Using wavelet energy as feature, they achieved binary classification accuracy of 97.78%. Zhang et al. [10] employed entropy of approximation wavelet coefficients as feature. They trained neural network classifier with hybridized BBO and PSO method and obtained significant accuracy. Zhang et al. [11] calculated Shannon entropy (SE) and Tsallis entropy (TE) of extracted wavelet packet coefficients of MR images. They used different combinations of SVM classifier and entropies to test four diagnosis methods. Nayak et al. [12] used probabilistic principal component analysis (PPCA) to reduce approximation wavelet coefficients from MR brain images. Adaboost with random forests (ADBRF) was used for binary classification. Nayak et al. [13] suggested a system using fast discrete curvelet transform (FDCT), principal component analysis (PCA) and least squares SVM (LS-SVM) with three different kernels to perform binary classification. Zhang et al. [14] suggested a novel system employing fourier entropy (FRFE) as feature and multi-layer perceptron (MLP) as classifier. They obtained average classification accuracy of 99.53% using neural network classifier. Gudigar et al. [15] utilised multi-resolution analysis techniques along with SVM classifier to classify MR brain images and they obtained 97.38% classification accuracy using shearlet coefficients. Authors [16], extended the concept of binary classification to multi class disease classification. They employed DWT and PCA as feature extraction and reduction tool respectively. Using different classification models, they perform multi disease classification and achieved 95.70% accuracy. In [17], authors utilised deep neural network to classify multiple brain diseases using DWT coefficients as feature.
The literature review reveals that the most of the research has been undertaken on binary classification. The widely used feature extraction methods include discrete wavelet transform and its variants. The classification features involved are mostly wavelet coefficients, multi resolution analysis techniques’ coefficients, energy and entropy of these coefficients. For classification, most of the systems employed SVM, neural network and their variants. Using these hybrid systems, the researchers achieved promising accuracies in binary classification. However, the research should be explore in the field of multi disease classification, as it is challenging task because of sharing of similar features by many brain disorders. Along with accuracy other performance measures namely sensitivity, specificity of CAD systems should be enhanced as they assist physicians in diagnosis and treatment of MR brain diseases [4]. To address these problems, in this work, instead of using classification features individually, composite features are used, as different features extract different information from image and it results in more competent feature set. The classification of this feature set is carried out using BPNN in which optimization, training functions, stopping criteria are chosen not to over fit the network and to model the training data with 100% accuracy. Also, Wilcoxon test is used to rank the features as per their discriminative capacity in disease classification. The contributions of this work can be outlined as follow.
The proposed system has utilised the potential of composite features namely energy of wavelet coefficients, GLCM features, intensity feature to classify multiple brain abnormalities.
The Wilcoxon test is used to find out ranks of features as per their discriminative power in differentiating multiple diseases.
Rigorous experimentations are carried out on BPNN classifier to prove the competency and efficiency of the proposed method.
This paper is organized as follows: Materials are presented in Sect. 2. Section 3 leads to methodology of proposed system. The experimental results with discussion are delineated in Sect. 4. The work is concluded in Sect. 5.
Materials
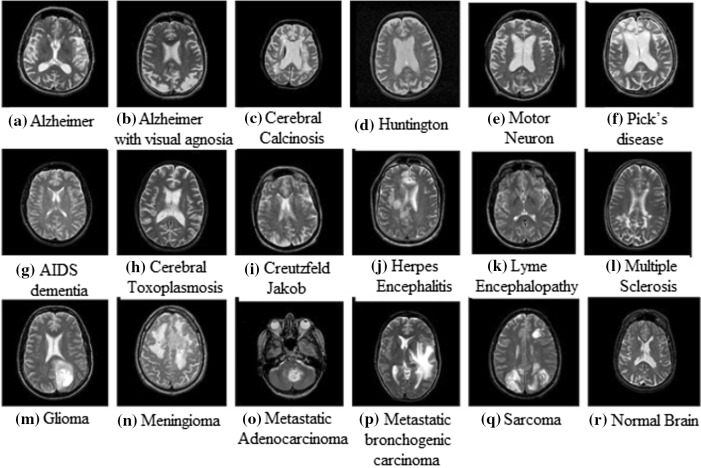
We worked on datasets DS-90 and DS-310 containing 90 and 310 T2-weighted MR brain images respectively obtained from the website of Harvard Medical School [18]. The images are of size in-plane resolution in axial plane. The dataset DS-90 consists of 17 classes of diseased and 1 class of normal brain images. DS-310 consists of 5 classes of diseased and 1 class of normal brain images. Sample images of diseased and normal MR brain are shown in Fig. 1.
Fig. 1.
Sample of brain MRIs: a Alzheimer, b Alzheimer with visual agnosia, c cerebral calcinosis, d Huntington, e motor neuron, f Picks disease, g AIDS dementia, h cerebral toxoplasmosis, i Creutzfeld–Jakob, j herpes encephalitis, k lyme encephalopathy, l multiple sclerosis, m glioma, n meningioma, o metastatic adenocarcinoma, p metastatic bronchogenic carcinoma, q sarcoma, r normal brain
Methodology
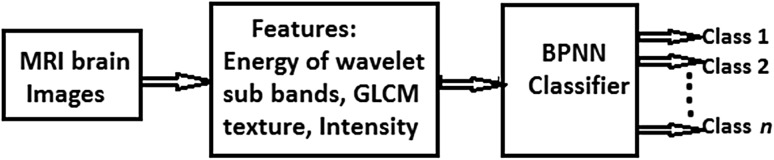
The proposed system consists of major components, feature extraction and classification. Figure 2 depicts the overall block diagram of the proposed system. MR images are decomposed to level-7 using DWT to form 22 sub bands. Wavelet Energy of each sub band, four GLCM features and one intensity feature form feature matrix of MR images. Features are classified using BPNN and their rank is decided by Wilcoxon test. All these components are discussed below.
Fig. 2.
Block diagram of proposed CAD methodology
Feature extraction
The feature set consists of energy of wavelet coefficients, GLCM textural features and intensity feature of MR images. The extracted GLCM textural features are contrast, correlation, energy and homogeneity. Standard Deviation is used as intensity feature.
Energy of wavelet sub bands
Wavelets have emerged as powerful mathematical tools. Wavelet transform provides localized frequency information of data. It can analyses the data at multiple scales [19]. These features of wavelet transform are used by many researchers for feature extraction [20]. Wavelets decomposes image into sub bands. The energy of sub band is computed using following expression.
| 1 |
where is the energy of the kth sub band and k varies from 1 to 7. is the corresponding sub band with dimension .
The energy reveals strength of image information in respective sub band. In this work, MR image is decomposed to level 7 using db4 wavelet resulting into 22 sub bands. Energies of 22 sub bands are considered as features.
Gray level co occurrence matrix (GLCM)
Haralick et al. [21] have introduced GLCM approach in 1973. Researchers have widely used the GLCM approach for texture classification. In this work, four textural features of GLCM namely contrast, correlation, energy and homogeneity are calculated. Let P(i, j) is the GLCM of an image I. Each element (i, j) in GLCM specifies the number of times, the pixel of image with gray level value i occurred horizontally adjacent to a pixel with value j.
In this work, the direction of neighbouring pixel with respect to pixel of reference is set as an angle and the distance is set as 1. Using these parameters, the adjacent pixels get select and local details of abnormalities in image can be detected effectively.
Contrast: This property of image is measure of intensity contrast between pixel and its neighbour over whole image. It is calculated as,
| 2 |
Correlation: Correlation indicates how pixel is correlated with its neighbour over the whole image. It is given by equation,
| 3 |
where, indicates mean and indicates standard deviation.
Energy: Energy of image is the sum of squared elements of GLCM.
| 4 |
Homogeneity: Homogeneity measures the closeness of the distribution of elements in the GLCM to the GLCM diagonal. It shows uniformity in intensity values.
| 5 |
Intensity based feature
Standard Deviation: Standard deviation is measure of spread of gray level around the mean. It is given by following equation.
| 6 |
where, I(i, j) is image matrix, indicates mean and N is total number of pixels in image.
The feature matrix consists of 27 features of all images which is given as input to BPNN classifier.
Wilcoxon test
The Wilcoxon test is a non parametric test for independent samples [22, 23]. Let, X and Y are independent samples with different sample sizes and respectively. Then the rank sum of sample is calculated using Wilcoxon test. The Wilcoxon test is equivalent to the mann whitney U-test. U is the number of times y precedes x in an ordered arrangement of the elements in samples X and Y. In the following way, mann whitney U-test is related to the Wilcoxon rank sum statistic,
| 7 |
where , is sum of ranks for sample X with size ; , is U value based on sample size .
The U statistic of Wilcoxon test is used to rank classification features. Out of 27 classification features used in this work, extracted feature with highest discriminative power has highest U value and highest feature rank. Feature ranking decides the discriminative capacity of feature.
Classification based on back propagation neural network (BPNN)
Neural networks (NNs) are biologically inspired [24]. NNs can learn from previous experience and generalize using past examples to new ones. They have achieved considerable success when applied to solve difficult, multivariate, non linear, diverse problems and noisy domains [24, 25] . Ojha et al. [26] has given excellent review on feed forward neural network.
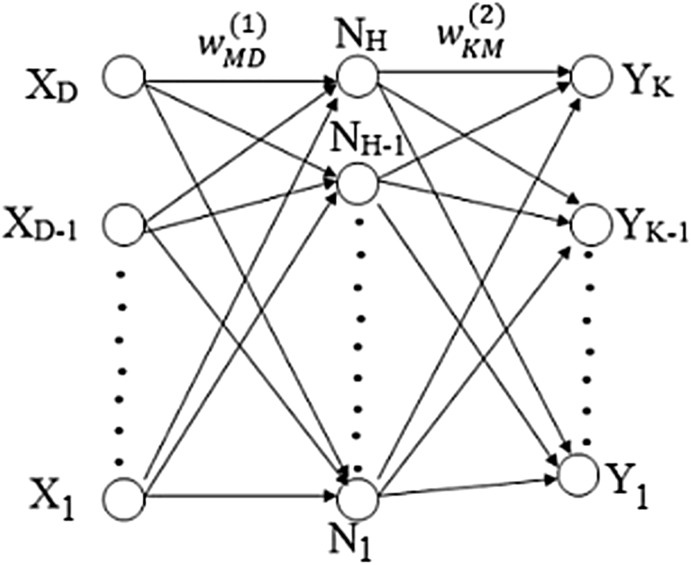
BPNN with single hidden layer and sufficient number of hidden neurons can approximate virtually any function to any degree of accuracy [27, 28]. The selection of optimal number of neurons in hidden layer is an important issue in the design of BPNN. A network with few neurons may not capture the input and output patterns accurately. If network is having excess of hidden neurons then it can lead to poor generalization and over fit the training data. To overcome over fitting issue of NN, bayesian regularization algorithm is used in this work. This training function provides a measure of how many weights and biases out of total network parameters are being effectively used by the network, thus avoiding the over fitting issue of network [27]. In this work, hyperbolic tangent sigmoid transfer function is used in hidden layer and output layer as activation function. It ranges from − 1 to 1 so, it can prevent training difficulties caused by saturation of the sigmoid functions [27]. It is used in NN where speed is important [22]. The BPNN architecture details are tabulated in Table 1. The structure of two layer network with input neurons, hidden neurons and output neurons is depicted in Fig. 3. For simplicity biases are not shown in the figure. The overall network function is given by following equation,
| 8 |
where is input variable and ; , is output variable and K is total number of outputs; and ] represents weight parameters of first and second layer of network respectively; M, is total number of parameters of network; and , are activation functions of first and second layer of network respectively.
Table 1.
BPNN parameters
| Parameters | BPNN |
|---|---|
| Output layer activation function | Hyperbolic tangent sigmoid |
| Hidden layer activation function | Hyperbolic tangent sigmoid |
| Performance function | Mean squared normalized error |
| Optimization method | Levenberg–Marquardt algorithm |
| Training function | Bayesian regulation backpropagation |
| Minimum performance gradient | |
| Stopping criteria | Minimum gradient |
Fig. 3.
Network diagram for two layer Neural Network
The training phase of the network progresses by adaptively adjusting the values of weights and biases of the network based on the performance function, given by following formula,
| 9 |
where , is the output produced by network; , is the target output; K, is total number of input-target pairs; and , are performance function parameters; N, is total number of network weights and biases; , is network weights and biases.
The performance function causes the network to have smaller weights and biases and this forces the network response to be smoother and less likely to over fit, improving generalization ability of network. The key point is optimal selection of parameter and . This is achieved using bayesian regularization. When the value of E gets minimized, the training phase of network stops and the network represents optimal values of weights and biases which are further used for testing phase. In this work, BPNN with single hidden layer and 26 hidden neurons are used in case of dataset DS-90. Dataset DS-310 is having more images resulting in large number of training equations so, two hidden layers with 25 and 10 hidden neurons in first and second layer respectively are used to train BPNN model. The number of hidden neurons are decided by performing experimentation.
Experimental results with discussion
To validate the proposed scheme, experimentation has been carried out using combination of wavelet and neural network toolboxes of MATLAB 2013a, running under windows 7 operating system with 1.60 GHz, dual core processor and 2 GB RAM.
Cross validation setting
Stratified cross validation (SCV) is commonly used for rearranging the dataset into different folds so that each fold fully represents the whole dataset. It helps classifier to be more generalize [27]. Running SCV more times gives more accurate results.
In this work, we have used 5-fold stratified cross validation technique to arrange MR brain images into 5 folds. One of the folds used for testing and remaining 4 folds used for training. The process is repeated until each fold is used for testing. The setting of training and test images is shown in Table 2. The 5-fold SCV has repeated for 75 runs with the aim of improving generalization ability of the network [12, 13, 15] which results in reliable and robust system.
Table 2.
Training and test image setting
| Dataset | Total images | Total diseases | Total classes | k-fold SCV | Total training images | Total testing images |
|---|---|---|---|---|---|---|
| DS-90 | 90 | 17 | 18 | 5 | 72 | 18 |
| DS-310 | 310 | 5 | 6 | 5 | 248 | 62 |
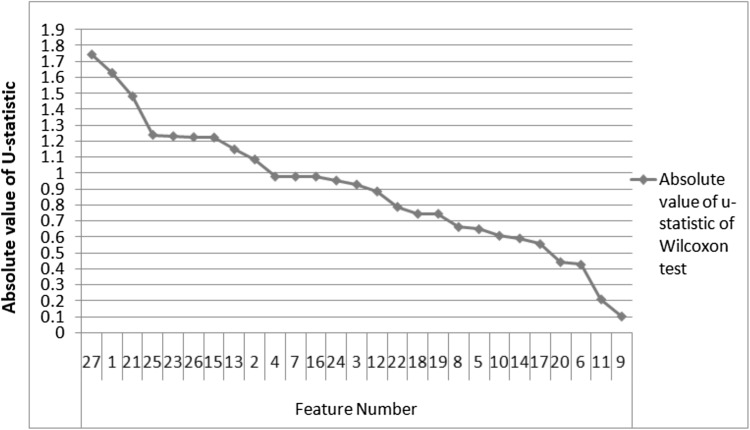
Feature ranking using Wilcoxon test
The absolute value of U-statistic of Wilcoxon test is used to rank the features. The values are plotted as shown in Fig.4. Feature 1 is the energy of approximation wavelet coefficients of level 7. Features 2 to 22 represent energies of horizontal, vertical and diagonal sub bands of levels 1 to 7 respectively. Features 23 to 26 are GLCM extracted four features namely correlation, homogeneity, contrast and energy. Feature 27 represents intensity feature namely standard deviation. The graph demonstrates that the standard deviation is the most significant feature for classification. Energies of approximate and vertical sub bands of level-7 are the 2nd and 3rd ranked discriminative features. The textural features contrast, correlation and energy have major role in classification. In summary, the combination of wavelet, GLCM texture and intensity feature of MR images lead to enhance the diagnostic accuracy of brain diseases.
Fig. 4.
The absolute value of U-statistic of Wilcoxon test of features
Comparison with existing methods
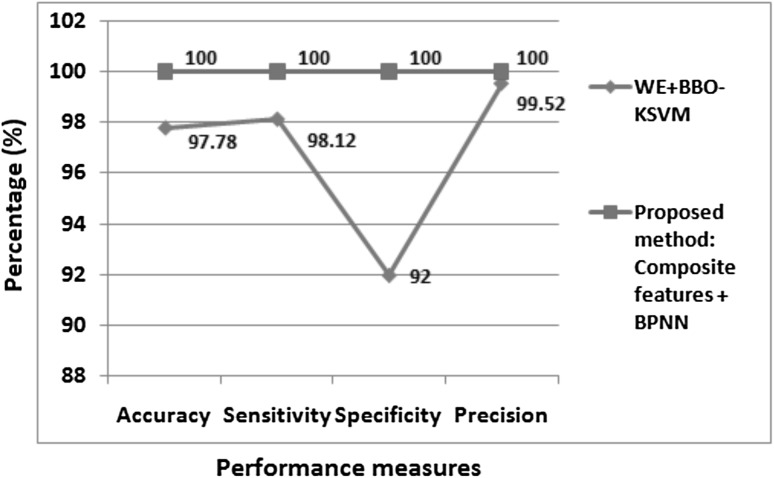
To evaluate the performance of proposed multi class disease classifier, different classification methods are compared on Harvard Medical School brain dataset. Table 3 presents the comparative results of multi class disease classification. Authors [16], used various methods to differentiate 310 images into 6 brain image classes . Initially, (DWT + PCA + kNN), (RT + PCA + LS-SVM (RBF)), (DWPT + GEPSVM), these methods were proposed by authors [5, 6, 11] respectively for binary classification. Authors [16] used these methods along with their proposed method (DWT + PCA + RF) for multi class classification and attained highest average accuracy of 95.70% with their proposed approach. Authors [17], attained 96.97% classification accuracy over 4 brain image classes of 66 images. On dataset DS-310, our proposed method achieved higher accuracy of 97.81% than existing approaches. Table 3, reveals that classification accuracy of 100% is yielded by our proposed method in differentiating 90 images into 18 brain image classes. Also, in binary classification, our proposed method achieved higher classification performances metrics of 100% than existing method [9] as shown in Fig. 5.
Table 3.
Multi class disease classification accuracy of existing methods on Harvard Medical School brain dataset
| Paper | Method | Number of images | Number of brain diseases | Number of classes to distinguish | Accuracy (%) |
|---|---|---|---|---|---|
| [16] | DWT + PCA + kNN | 310 | 5 | 6 | 83.87 |
| [16] | RT + PCA + LS-SVM (RBF) | 310 | 5 | 6 | 86.02 |
| [16] | DWPT + GEPSVM | 310 | 5 | 6 | 88.92 |
| [16] | DWT + PCA + RF | 310 | 5 | 6 | 95.70 |
| [17] | DWT + PCA + Deep NN | 66 | 3 | 4 | 96.97 |
| Proposed method | Composite features + BPNN | 90 | 17 | 18 | 100 |
| 310 | 5 | 6 | 97.81 |
Fig. 5.
Performance measures of existing binary classification methods on Harvard Medical School brain dataset DS-90
Back propagation neural network (BPNN) classifier
Experimentation on dataset DS-90
Following two types of experiments have carried out on dataset DS-90:
The BPNN is retrained for 375 folds (75 runs 5 fold = 375 folds), with different initial weights-biases, and training, test sets. However, these different conditions can lead to different solutions for the same problem [22]. But, under these conditions, we achieved 100% accuracy over training and test sets in each fold. It shows the robustness of system. Randomly 10 runs are selected whose performance results are tabulated in Table 4. It provides information about training performance error, testing performance error, network performance error, number of epochs (iterations), number of effective parameters (weights-biases) and approximate neurons used by network model and accuracy of network. Thus under diverse initial conditions of network, the performance errors are minimal and we can observe 100% classification accuracy. Also, in each fold the approximate number of neurons i.e. number of neurons used by network is almost same. It indicates the system is reliable and accurate.
The BPNN is retrained with different initial weight-biases, and training—test sets are kept same. With neural network such tests are generally carried out if satisfactory results are not achieved. By changing initial weight -biases, the network might produce different results [22]. It means, for same diagnosis problem, system can produce different results. It imply, with same training, test sets and varying initial weight-biases, if a network can produce same results continuously, the network is reliable and robust. We have performed this experiment 14 times and attained 100% accuracy in multiple class disease detection. The performance results of these 14 cases are tabulated in Table 5. The performance errors are minimal and in each case the number of neurons used by network is approximately same. It indicates the system is robust and reliable.
Table 4.
Training and test results of 10 runs out of 75 runs of dataset DS-90 (training, test sets and weight-biases are different in each fold)
| Run | Fold | Training performance error | Testing performance error | Network performance error | Epochs | Effective parameters | Actual neurons | Approximate neurons | Classification accuracy (%) |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 1 | 28 | 258 | 5.22 | 6 | 100 | |||
| 2 | 25 | 275 | 5.59 | 6 | 100 | ||||
| 3 | 30 | 275 | 5.59 | 6 | 100 | ||||
| 4 | 35 | 256 | 5.17 | 6 | 100 | ||||
| 5 | 29 | 260 | 5.26 | 6 | 100 | ||||
| 2 | 1 | 25 | 275 | 5.59 | 6 | 100 | |||
| 2 | 25 | 273 | 5.54 | 6 | 100 | ||||
| 3 | 29 | 273 | 5.54 | 6 | 100 | ||||
| 4 | 30 | 266 | 5.39 | 6 | 100 | ||||
| 5 | 32 | 255 | 5.15 | 6 | 100 | ||||
| 3 | 1 | 28 | 268 | 5.43 | 6 | 100 | |||
| 2 | 30 | 268 | 5.43 | 6 | 100 | ||||
| 3 | 32 | 254 | 5.13 | 6 | 100 | ||||
| 4 | 29 | 270 | 5.48 | 6 | 100 | ||||
| 5 | 30 | 265 | 5.37 | 6 | 100 | ||||
| 4 | 1 | 29 | 255 | 5.15 | 6 | 100 | |||
| 2 | 27 | 248 | 5.00 | 5 | 100 | ||||
| 3 | 28 | 269 | 5.46 | 6 | 100 | ||||
| 4 | 39 | 261 | 5.28 | 6 | 100 | ||||
| 5 | 25 | 286 | 5.83 | 6 | 100 | ||||
| 5 | 1 | 25 | 263 | 5.33 | 6 | 100 | |||
| 2 | 38 | 259 | 5.24 | 6 | 100 | ||||
| 3 | 29 | 257 | 5.20 | 6 | 100 | ||||
| 4 | 32 | 271 | 5.50 | 6 | 100 | ||||
| 5 | 26 | 271 | 5.50 | 6 | 100 | ||||
| 6 | 1 | 26 | 283 | 5.76 | 6 | 100 | |||
| 2 | 26 | 272 | 5.52 | 6 | 100 | ||||
| 3 | 30 | 270 | 5.48 | 6 | 100 | ||||
| 4 | 29 | 247 | 4.98 | 5 | 100 | ||||
| 5 | 25 | 264 | 5.35 | 6 | 100 | ||||
| 7 | 1 | 36 | 259 | 5.24 | 6 | 100 | |||
| 2 | 31 | 262 | 5.30 | 6 | 100 | ||||
| 3 | 23 | 280 | 5.70 | 6 | 100 | ||||
| 4 | 26 | 284 | 5.78 | 6 | 100 | ||||
| 5 | 26 | 271 | 5.50 | 6 | 100 | ||||
| 8 | 1 | 29 | 270 | 5.48 | 6 | 100 | |||
| 2 | 29 | 256 | 5.17 | 6 | 100 | ||||
| 3 | 31 | 248 | 5.00 | 5 | 100 | ||||
| 4 | 29 | 254 | 5.13 | 6 | 100 | ||||
| 5 | 26 | 262 | 5.30 | 6 | 100 | ||||
| 9 | 1 | 29 | 265 | 5.37 | 6 | 100 | |||
| 2 | 38 | 267 | 5.41 | 6 | 100 | ||||
| 3 | 31 | 274 | 5.57 | 6 | 100 | ||||
| 4 | 25 | 273 | 5.54 | 6 | 100 | ||||
| 5 | 29 | 266 | 5.39 | 6 | 100 | ||||
| 10 | 1 | 27 | 287 | 5.85 | 6 | 100 | |||
| 2 | 28 | 264 | 5.35 | 6 | 100 | ||||
| 3 | 22 | 270 | 5.48 | 6 | 100 | ||||
| 4 | 36 | 266 | 5.39 | 6 | 100 | ||||
| 5 | 31 | 252 | 5.09 | 6 | 100 |
Table 5.
Network performance results of 14 cases of dataset DS-90 (training, test sets are same and weight-biases are different in each case)
| Case | Training performance error | Testing performance error | Network performance error | Epochs | Effective parameters | Actual neurons | Approximate neurons | Classification accuracy (%) |
|---|---|---|---|---|---|---|---|---|
| 1 | 24 | 307 | 6.28 | 7 | 100 | |||
| 2 | 27 | 262 | 5.30 | 6 | 100 | |||
| 3 | 27 | 273 | 5.54 | 6 | 100 | |||
| 4 | 27 | 267 | 5.41 | 6 | 100 | |||
| 5 | 25 | 270 | 5.48 | 6 | 100 | |||
| 6 | 26 | 275 | 5.59 | 6 | 100 | |||
| 7 | 26 | 273 | 5.54 | 6 | 100 | |||
| 8 | 47 | 266 | 5.39 | 6 | 100 | |||
| 9 | 28 | 260 | 5.26 | 6 | 100 | |||
| 10 | 29 | 271 | 5.50 | 6 | 100 | |||
| 11 | 26 | 279 | 5.67 | 6 | 100 | |||
| 12 | 27 | 296 | 6.04 | 7 | 100 | |||
| 13 | 31 | 272 | 5.52 | 6 | 100 | |||
| 14 | 28 | 276 | 5.61 | 6 | 100 |
Experimentation on dataset DS-310
The BPNN is retrained for 75 runs, with different initial weights-biases and training, test sets. The results of 10 runs out of 75 runs are tabulated in Table 6. Table 6 provides information about training performance error, testing performance error, network performance error, training classification accuracy and testing classification accuracy. The performance errors are minimal. The average classification accuracy in training phase is 100% and in testing phase is 97.81%.
Table 6.
Training and test results of 10 runs out of 75 runs of dataset DS-310 (training, test sets and weight-biases are different in each fold)
| Run | Fold | Training performance error | Testing performance error | Network performance error | Training classification accuracy (%) | Testing classification accuracy (%) |
|---|---|---|---|---|---|---|
| 1 | 1 | 100 | 96.77 | |||
| 2 | 100 | 98.39 | ||||
| 3 | 100 | 96.77 | ||||
| 4 | 100 | 98.39 | ||||
| 5 | 100 | 100.00 | ||||
| 2 | 1 | 100 | 100.00 | |||
| 2 | 100 | 98.39 | ||||
| 3 | 100 | 100.00 | ||||
| 4 | 100 | 96.77 | ||||
| 5 | 100 | 96.77 | ||||
| 3 | 1 | 100 | 98.39 | |||
| 2 | 100 | 96.77 | ||||
| 3 | 100 | 98.39 | ||||
| 4 | 100 | 96.77 | ||||
| 5 | 100 | 98.39 | ||||
| 4 | 1 | 100 | 96.77 | |||
| 2 | 100 | 100.00 | ||||
| 3 | 100 | 96.77 | ||||
| 4 | 100 | 100.00 | ||||
| 5 | 100 | 96.77 | ||||
| 5 | 1 | 100 | 100.00 | |||
| 2 | 100 | 98.39 | ||||
| 3 | 100 | 98.39 | ||||
| 4 | 100 | 96.77 | ||||
| 5 | 100 | 96.77 | ||||
| 6 | 1 | 100 | 96.77 | |||
| 2 | 100 | 98.39 | ||||
| 3 | 100 | 96.77 | ||||
| 4 | 100 | 95.16 | ||||
| 5 | 100 | 100.00 | ||||
| 7 | 1 | 100 | 96.77 | |||
| 2 | 100 | 98.39 | ||||
| 3 | 100 | 98.39 | ||||
| 4 | 100 | 96.77 | ||||
| 5 | 100 | 98.39 | ||||
| 8 | 1 | 100 | 98.39 | |||
| 2 | 100 | 100.00 | ||||
| 3 | 100 | 95.16 | ||||
| 4 | 100 | 96.77 | ||||
| 5 | 100 | 100.00 | ||||
| 9 | 1 | 100 | 98.39 | |||
| 2 | 100 | 96.77 | ||||
| 3 | 100 | 98.39 | ||||
| 4 | 100 | 96.77 | ||||
| 5 | 100 | 95.16 | ||||
| 10 | 1 | 100 | 98.39 | |||
| 2 | 100 | 96.77 | ||||
| 3 | 100 | 98.39 | ||||
| 4 | 100 | 96.77 | ||||
| 5 | 100 | 96.77 |
The average computation time for each image is 0.9428 s. Table 3 and Fig. 5 explain the efficiency of our proposed method over state-of-the-art methods. Tables 4, 5 and 6 reveal competency of proposed method in classifying different brain disorders.
Conclusion
In this paper, we have proposed a method utilising wavelet, GLCM extracted textural features and intensity feature of MR images and BPNN as classifier for the diagnosis of different MR brain diseases. The experimental results show that the composite features could achieve 100% multi class disease classification accuracy on dataset DS-90 and 97.81% on dataset DS-310 which are higher than the existing approaches. The standard deviation, energies of approximate and vertical sub bands of level 7 are proved to be first three dominant features for classification as per Wilcoxon test. Also, GLCM textural features contrast, correlation and energy are significant classification features. The rigorous experimentation using BPNN classifier reveals the robustness and effectiveness of proposed method in diagnosis of multi class brain disorders and it can be applied for practical use.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Vandana V. Kale, Email: vandanav_kale@yahoo.co.in
Satish T. Hamde, Email: sthamde@sggs.ac.in
Raghunath S. Holambe, Email: rsholambe@sggs.ac.in
References
- 1.Sprawls P. Magnetic resonance imaging: principles, methods and techniques. Montreat: Sprawls Education Foundation; 2000. [Google Scholar]
- 2.Chaplot S, Patnaik LM, Jagannathan NR. Classification of magnetic resonance brain images using wavelets as input to support vector machine and neural network. Biomed Signal Process Control. 2006;1:86–92. doi: 10.1016/j.bspc.2006.05.002. [DOI] [Google Scholar]
- 3.Fujita H, Uchiyama Y, Nakagawa T, Fukuoka D, Hatanaka Y, Hara T, Lee GN, Hayashi Y, Ikedo Y, Gao X, Zhou X. Computer-aided diagnosis: The emerging of three CAD systems induced by Japanese health care needs. Comput Methods Programs Biomed. 2008;92(3):238–248. doi: 10.1016/j.cmpb.2008.04.003. [DOI] [PubMed] [Google Scholar]
- 4.Doi K. Computer-aided diagnosis in medical imaging: historical review, current status and future potential. Comput Med Imaging Gr. 2007;31(4):198–211. doi: 10.1016/j.compmedimag.2007.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.El-Dahshan E-SA, Hosny T, Salem A-BM. Hybrid intelligent techniques for MRI brain images classification. Digit Signal Process. 2010;20(2):433–441. doi: 10.1016/j.dsp.2009.07.002. [DOI] [Google Scholar]
- 6.Das S, Chowdhury M, Kundu MK. Brain MR image classification using multiscale geometric analysis of ripplet. Prog Electromagn Res. 2013;137:1–17. doi: 10.2528/PIER13010105. [DOI] [Google Scholar]
- 7.Saritha M, Joseph KP, Mathew AT. Classification of MRI brain images using combined wavelet entropy based spider web plots and probabilistic neural network. Pattern Recognit Lett. 2013;34:2151–2156. doi: 10.1016/j.patrec.2013.08.017. [DOI] [Google Scholar]
- 8.El-Dahshan E-SA, Mohsen HM, Revett K, Salem A-BM. Computer-aided diagnosis of human brain tumor through MRI: a survey and a new algorithm. Expert Syst Appl. 2014;41(11):5526–5545. doi: 10.1016/j.eswa.2014.01.021. [DOI] [Google Scholar]
- 9.Yang G, Zhang Y, Yang J, Ji G, Dong Z, Wang S, Feng C, Wang Q. Automated classification of brain images using wavelet-energy and biogeography-based optimization. Multimed Tools Appl. 2015;75:1560115617. [Google Scholar]
- 10.Zhang Y, Wang S, Dong Z, Phillip P, Ji G, Yang J. Pathological brain detection in magnetic resonance imaging scanning by wavelet entropy and hybridization of biogeography-based optimization and particle swarm optimization. Prog Electromagn Res. 2015;152:4158. [Google Scholar]
- 11.Zhang Yudong, Dong Zhengchao, Wang Shuihua, Ji Genlin, Yang Jiquan. Preclinical diagnosis of magnetic resonance (MR) brain images via discrete wavelet packet transform with Tsallis entropy and generalized eigenvalue proximal support vector machine (GEPSVM), Entropy, 17, 4, 2015;1795–1813, Multidisciplinary Digital Publishing Institute
- 12.Nayak DR, Dash R, Majhi B. Brain MR image classification using two-dimensional discrete wavelet transform and AdaBoost with random forests. Neurocomputing. 2016;177(Supplement C):188–197. doi: 10.1016/j.neucom.2015.11.034. [DOI] [Google Scholar]
- 13.Nayak DR, Dash R, Majhi B. Pathological brain detection using curvelet features and least squares SVM. Multimed Tools Appl. 2016 [Google Scholar]
- 14.Zhang Y, Sun Y, Phillips P, Liu G, Zhou X, Wang S. A multilayer perceptron based smart pathological brain detection system by fractional Fourier entropy. J Med Syst. 2016;40:1–11. doi: 10.1007/s10916-015-0365-5. [DOI] [PubMed] [Google Scholar]
- 15.Gudigar A, Raghavendra U, San TR, Ciaccio EJ, Acharya UR. Application of multiresolution analysis for automated detection of brain abnormality using MR images: a comparative study. Future Gen Comput Syst. 2019;90:359–367. doi: 10.1016/j.future.2018.08.008. [DOI] [Google Scholar]
- 16.Siddiqui MF, Mujtaba G, Reza AW, Shuib L. Multi-class disease classification in brain mris using a computer-aided diagnostic system. Symmetry. 2017;9(3):37. doi: 10.3390/sym9030037. [DOI] [Google Scholar]
- 17.Mohsen H, El-Dahshan E-SA, El-Horbaty E-SM, Salem A-BM. Classification using deep learning neural networks for brain tumors. Future Comput Inform J. 2018;3(1):68–71. doi: 10.1016/j.fcij.2017.12.001. [DOI] [Google Scholar]
- 18.Harvard Medical School: Database. http://www.med.harvard.edu/AANLIB. Accessed 2016.
- 19.Vetterli M, Herley C. Wavelets and filter banks: theory and design. IEEE Trans Signal Process. 1992;40:2207–2232. doi: 10.1109/78.157221. [DOI] [Google Scholar]
- 20.Mallat SG. A theory for multiresolution signal decomposition: the wavelet representation. IEEE Trans Pattern Anal Mach Intell. 1989;11:674–693. doi: 10.1109/34.192463. [DOI] [Google Scholar]
- 21.Haralick RM, Shanmugam K, Dinstein I. Textural features for image classification. IEEE Trans Syst Man Cybern. 1973;SMC–3:610–621. doi: 10.1109/TSMC.1973.4309314. [DOI] [Google Scholar]
- 22.Mark HB, Hagan MT, Demuth, HB. Neural network toolbox™ user’s guide. The Mathworks Inc; 1992.
- 23.Lowry R. Concepts and applications of inferential statistics, VassarStats website
- 24.Haykin S. Neural networks: a comprehensive foundation. Upper Saddle River: Prentice Hall PTR; 1994. [Google Scholar]
- 25.Bishop CM. Neural networks for pattern recognition. Oxford: Oxford University Press; 1995. [Google Scholar]
- 26.Ojha VK, Abraham A, Snášel V. Metaheuristic design of feedforward neural networks: a review of two decades of research. Eng Appl Artif Intell. 2017;60:97–116. doi: 10.1016/j.engappai.2017.01.013. [DOI] [Google Scholar]
- 27.Hagan M, Demuth HB, Beale MH, De Jess O. Neural network design. 2. Boston: PWS Publishing; 1996. [Google Scholar]
- 28.Okut H. Bayesian regularized neural networks for small n big p data. In: Artificial neural networks-models and applications. London: InTech; 2016.