Understanding functional capacities of gut microbiomes is important to improve our understanding of symbiotic associations. Here, we use peptide-based functional annotation to show that the gut microbiomes of fungus-farming termites code for a wealth of enzymes that likely target the fungal diet the termites eat. Comparisons to other termites showed that fungus-growing termite guts have relatively more fungal cell wall-degrading enzyme genes, whereas wood-feeding termite gut communities have relatively more plant cell wall-degrading enzyme genes. Across termites with different diets, the dominant biomass-degrading enzymes are predominantly coded for by the most abundant bacterial taxa, suggesting tight links between diet and gut community compositions.
KEYWORDS: HiSeq, HotPep, carbohydrate-active enzymes, cellulase, chitinase, metagenomics, peptide-based functional predictions
ABSTRACT
Termites forage on a range of substrates, and it has been suggested that diet shapes the composition and function of termite gut bacterial communities. Through comparative analyses of gut metagenomes in nine termite species with distinct diets, we characterize bacterial community compositions and use peptide-based functional annotation method to determine biomass-degrading enzymes and the bacterial taxa that encode them. We find that fungus-growing termite guts have relatively more fungal cell wall-degrading enzyme genes, while wood-feeding termite gut communities have relatively more plant cell wall-degrading enzyme genes. Interestingly, wood-feeding termite gut bacterial genes code for abundant chitinolytic enzymes, suggesting that fungal biomass within the decaying wood likely contributes to gut bacterial or termite host nutrition. Across diets, the dominant biomass-degrading enzymes are predominantly coded for by the most abundant bacterial taxa, suggesting tight links between diet and gut community composition, with the most marked difference being the communities coding for the mycolytic capacity of the fungus-growing termite gut.
IMPORTANCE Understanding functional capacities of gut microbiomes is important to improve our understanding of symbiotic associations. Here, we use peptide-based functional annotation to show that the gut microbiomes of fungus-farming termites code for a wealth of enzymes that likely target the fungal diet the termites eat. Comparisons to other termites showed that fungus-growing termite guts have relatively more fungal cell wall-degrading enzyme genes, whereas wood-feeding termite gut communities have relatively more plant cell wall-degrading enzyme genes. Across termites with different diets, the dominant biomass-degrading enzymes are predominantly coded for by the most abundant bacterial taxa, suggesting tight links between diet and gut community compositions.
INTRODUCTION
Termites are widespread in tropical, subtropical, and warm temperate regions (1) and form a diverse group of more than 3,000 described species in 281 genera and seven families (2–5). They have major impacts on their environments (1), and this success has been attributed to their capacity to use nutritionally imbalanced, recalcitrant food sources, allowing for colonization of otherwise inaccessible niches (6). Different termites forage on distinct substrates, including soil, wood, dung, and fungus (7, 8), decomposed through intricate interactions with complex gut microbial communities (6, 9). In most termites, the main role of gut microbiota is believed to be the digestion of lignocellulose (10, 11), but gut microbes also play key roles in nitrogen fixation (12–14), microbial defense (see, for example, reference 15), and immune regulation (16, 17), which have major importance for the evolutionary history of the symbioses.
Approximately 30 million years ago, the basal higher termite subfamily Macrotermitinae engaged in a mutualistic association with Termitomyces fungi (18, 19) and have a distinct composition of the gut microbiota (20–22). Termitomyces decomposes plant material within external fungus gardens (combs) (23, 24), but the gut still remains central in the association because plant substrate is macerated and mixed with asexual Termitomyces spores in a first gut passage prior to comb deposition (25). After Termitomyces breaks down the plant substrate, the termites ingest mature parts of the comb in a second gut passage (25), where gut microbes may contribute enzymes for final digestion of any remaining plant components (24). This division of labor is consistent with gut bacteria being of importance mainly when the comb material passes through the termite gut in a second passage (cf. references 23, 24, and 26), but recent work has suggested that partial lignin breakdown may also be accomplished during this first gut passage in Odontotermes formosanus (27).
A set of microbes distinct from the gut microbiota of other termites persists in the fungus-growing termite guts, but limited work has examined functional implications of these differences (8, 24). It has been hypothesized that it was associated with the more protein-rich fungal diet (20) and/or to break down chitin and other fungal cell wall components (8, 24, 28). Termitomyces domestication exposed fungus-growing termite gut communities to large quantities of fungal cell wall glucans (composed of d-glucose monomers), chitin (glucosamine polymer), and glycoproteins (see, for example, reference 29). Their breakdown requires a combination of carbohydrate-active enzymes (CAZymes; www.cazy.org) (30, 31) and fungus-growing termite gut bacteria indeed encode glycoside hydrolase (GH) families of enzymes that may cleave chitin (GH18, GH19, and GH20), β-glucan (GH55, GH81, and GH128), and α-mannan (GH38, GH76, GH92, GH99, and GH125) (8, 24).
In nature, bacteria are the major chitin degraders and its hydrolysis has been correlated with bacterial abundances in, e.g., soil communities (32). In fungus-growing termites, Bacteroidetes and Firmicutes bacteria appear to be the main producers of CAZymes putatively producing mycolytic enzymes, i.e., enzymes that lyse the fungal cell wall (8, 24). These studies remained preliminary, however, because they were based on either an unassembled low-coverage metagenome (8) or had limited functional predictions (24). Here, we sequenced the gut metagenome of the fungus-growing termite Odontotermes sp. and performed in silico analyses to elucidate its fungal and plant cell wall-degrading capacities at deeper functional levels (i.e., to EC numbers when possible), assigned putative enzyme functions to gut community members using peptide-based functional annotation where prediction of function was confirmed by more than one method. To investigate the link between termite diet and gut community composition, we compared our findings to metagenomes from the fungus-growing termite Macrotermes natalensis (24) and seven non-fungus-growing termite species feeding on plant material at different degrees of decomposition: the dung feeder Amitermes wheeleri (33), the two wood feeders Nasutitermes corniger and Microcerotermes parvus, a litter feeder Cornitermes sp., the two humus feeders Termes hospes and Neocapritermes taracua, and the soil feeder Cubitermes ugandensis (34). We reveal that the difference in gut community composition is associated with the presence of a mycolytic microbiota, providing insights into digestion and the role of gut communities in the fungus-growing termite symbiosis.
RESULTS
Taxonomic composition of fungus-growing termite gut microbiotas.
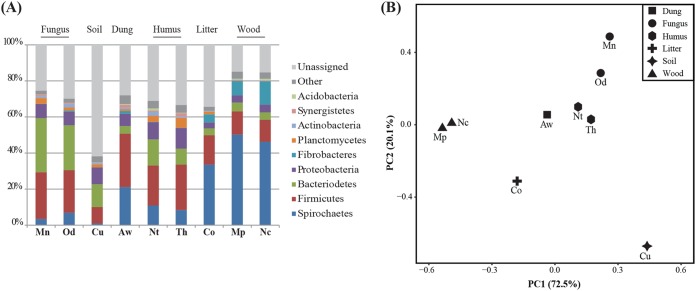
We assigned bacterial taxonomies to metagenome contigs by searching for the closest matches of protein-coding genes on each contig against the NR database in NCBI and compared the relative abundance of the contigs in each group to assess the composition of termite gut microbiotas. M. natalensis and Odontotermes sp. were distinct from other higher termites primarily being relatively richer in Bacteroidetes (Fig. 1A), corroborating previous work (20, 35), and termites in the same feeding group tend to be similar in gut microbiota composition (Fig. 1B; see Table S1 in the supplemental material). Taxonomic compositions at the phylum level were consistent with previous 16S rRNA amplicon surveys of fungus-growing termites (8, 20, 21). Firmicutes and Bacteroidetes dominated in Odontotermes sp. (24 and 25%, respectively) and M. natalensis (24 and 30%, respectively) (Fig. 1A; see also Table S1 in the supplemental material), comparable to an average 35 and 32% abundance in 16S-rRNA studies of Macrotermes subhyalinus and Odontotermes sp. in the Ivory Coast (21) and Odontotermes yunnanensis from Southwest China (8). Spirochaetes were relatively abundant in wood-feeding termites (46 to 50%) and Cornitermes sp. (32%), and they were low in relative abundance in fungus-growing termites (3 to 6%). At the genus and family levels, we identified the presence of genera that are part of the core gut microbial community in fungus-growing termites (e.g., Alistipes, Treponema, Dysgonomonas, Desulfovibrio, Ruminococcaceae, and Lachnospiraceae) (21). Alistipes and Bacteroides were most abundant in fungus-growing termites, representing 5 to 8% and 2 to 4% total abundances, respectively, sharply contrasting with other higher termites (on average, 0.1 and 0.5%, respectively) (Table S1). Treponema (Spirochaetes) was low in relative abundance in fungus growers and termites feeding on decaying plant material (3% and 6%, respectively), except for the litter feeder Cornitermes sp. (32%), while it was the most abundant taxon in wood feeders (42 to 45%) (Table S1). Comparisons to composition estimates from 16S rRNA studies (21, 22, 34) revealed that some genera were underrepresented in the metagenomes. Alistipes, for example, were found in much higher relative abundances (12% in average) from classifications using 16S rRNA than protein-coding genes (5 to 8%) (Table S3). Some taxonomic groups classified in 16S rRNA surveys, such as the TG3 phylum (20, 34) were not detected in the metagenomes (Table S1).
FIG 1.
(A) Relative abundances of bacterial phyla each comprising >1% of the microbiota in the guts of termite species with different diets. Termite species are arranged by the degree of plant degradation in the diet. (B) PCA of community similarities of termites with different diets. Mn, Macrotermes natalensis; Od, Odontotermes sp.; Nc, Nasutitermes corniger; Aw, Amitermes wheeleri; Mp, Microcerotermes parvus; Co, Cornitermes sp.; Th, Termes hospes; Nt, Neocapritermes taracua; Cu, Cubitermes ugandensis.
Bacterial community composition of metagenome contigs of gut metagenomes of nine termites with different diets at the phylum, class, order, family, and genus levels. Taxonomy classifications with more than 0.1% of the total abundance are shown. Mn, Macrotermes natalensis; Od, Odontotermes sp.; Nc, Nasutitermes corniger; Aw, Amitermes wheeleri; Mp, Microcerotermes parvus; Co, Cornitermes sp.; Th, Termes hospes; Nt, Neocapritermes taracua; Cu, Cubitermes ugandensis. Download Table S1, DOCX file, 0.03 MB (35.4KB, docx) .
Copyright © 2019 Hu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Fungus and plant cell wall-targeting enzymes.
To gain insights into the functional capacity for carbohydrates degradation, we first identified carbohydrate-active enzyme (CAZyme) families (30, 31) and classified the genes by their substrate target and thus putative enzyme function by assigning EC numbers using peptide-based functional annotation (36–38) (Table 1; see also Table S2 in the supplemental material). We focus our presentation and comparisons to enzymes, for which the prediction of function was confirmed by more than one method (for details, see Materials and Methods). Principle component analysis (PCA) of glycoside hydrolase (GH) family compositions (Fig. 2B) support that gut microbial enzyme capacities are similar for termites with similar diets. Fungus-growing termite guts were thus systematically different in GH family gene composition to the guts of termites feeding on plant biomass (Fig. 2B; see also Table S2 in the supplemental material), reflecting the differences in cell wall composition of plant and fungus material. Genes encoding GH enzymes that putatively target fungal cell wall components (detailed based on EC numbers in the paragraph below) were more abundant in fungus-growing termites, with the most marked differences being for GH125 (34-fold) and GH92 (14-fold) that likely target α-mannose (Table S2). Genes encoding GH families GH17, GH128, GH55, and GH87 that contain (among others) β-1,3-glucan-targeting enzymes and GH18 and GH19 that include several chitinases showed 2- to 12-fold higher relative abundance in fungus-growing termites (Table S2). In contrast and as expected, many GH family encoding genes primarily targeting plant cell wall components were low in relative abundance in fungus-growing termites but higher in wood-feeding termites (Table S2). Examples include GH94, GH10, and GH5 containing, e.g., cellulases, and GH74, GH120, GH39, GH26, GH10, and GH11 that contain hemi-cellulases, the genes of which were 3- to 5-fold more abundant in wood feeders than in other termites (Table S2).
TABLE 1.
Relative abundances of enzymes putatively targeting fungal and plant cell wall polysaccharides in the gut metagenomes of nine termites with different diets
| EC no. | Enzyme | Substrate | Relative abundancea
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Fungus |
Soil | Dung | Humus |
Litter | Wood |
||||||
| Mn | Od | Cu | Aw | Nt | Th | Co | Mp | Nc | |||
| 3.2.1.14 | Chitinase | Chitin | 209.59 | 33.14 | 3.38 | 16.89 | 5.97 | 8.74 | 14.70 | 39.06 | 43.58 |
| 3.2.1.52 | β-N-Acetylhexosaminidase | Chitin | 317.18 | 208.60 | 23.51 | 172.84 | 84.44 | 67.93 | 127.47 | 362.88 | 357.39 |
| 3.5.1.41 | Chitin deacetylase | Chitin | 1.79 | 5.46 | 2.09 | 16.89 | 8.71 | 3.38 | 11.30 | 24.00 | 4.98 |
| 3.2.1.132 | Chitosanase | Chitin | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| 3.2.1.165 | Exo-β-glucosaminidase | Chitin | 34.37 | 18.35 | 0.43 | 0.00 | 4.56 | 1.68 | 0.00 | 1.35 | 0.28 |
| 3.2.1.39 | Endo-1,3-β-glucanase | β-Glucan | 45.73 | 36.28 | 1.75 | 0.00 | 0.68 | 2.35 | 4.05 | 26.47 | 13.15 |
| 3.2.1.58 | Exo-1,3-β-glucanase | β-Glucan | 21.12 | 18.12 | 3.36 | 0.00 | 3.92 | 2.68 | 2.67 | 20.37 | 2.86 |
| 3.2.1.75 | Endo-1,6-β-glucosidase | β-Glucan | 48.53 | 16.34 | 0.61 | 6.14 | 3.96 | 3.17 | 0.00 | 1.34 | 0.00 |
| 3.2.1.21 | Exo-β-glucosidase | β-Glucan | 415.89 | 251.02 | 4.78 | 75.52 | 38.20 | 47.17 | 95.79 | 249.86 | 207.43 |
| 3.2.1.59 | Endo-1,3-α-glucosidase | α-Glucan | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| 3.2.1.84 | Exo-1,3-α-glucanase | α-Glucan | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| 3.2.1.20 | Endo-1,4-α-glucanase | α-Glucan | 6.59 | 21.58 | 2.27 | 0.00 | 2.40 | 1.67 | 0.35 | 0.57 | 27.73 |
| 3.2.1.3 | Exo-1,4-α-glucanase | α-Glucan | 19.64 | 16.52 | 1.25 | 0.00 | 0.30 | 0.00 | 0.00 | 1.02 | 0.31 |
| 3.2.1.4 | Endo-1,4-β-glucanase | Cellulose | 135.62 | 218.16 | 16.71 | 27.63 | 40.83 | 41.57 | 243.08 | 685.00 | 793.69 |
| 3.2.1.176 | Cellobiohydrolase (reducing end) | Cellulose | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| 3.2.1.91 | Cellobiohydrolase (nonreducing end) | Cellulose | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 1.42 | 0.00 | 0.00 | 0.00 |
| 1.1.99.18 | Cellobiose dehydrogenase | Cellulose | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| 1.1.99.29 | Pyranose dehydrogenase | Cellulose | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| LPMO | Lytic polysaccharide monooxygenase | Cellulose | 0.00 | 0.00 | 0.18 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 1.11 |
| 3.2.1.8 | Endo-1,4-β-xylanase | Hemicellulose | 127.75 | 204.86 | 11.58 | 247.14 | 57.88 | 44.17 | 426.90 | 591.74 | 780.19 |
| 3.2.1.37 | Exo-β-1,4-xylosidase | Hemicellulose | 314.18 | 308.09 | 5.81 | 142.76 | 23.47 | 58.63 | 146.37 | 290.59 | 199.88 |
| 3.2.1.131 | Xylan α-1,2-glucuronosidase | Hemicellulose | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| 3.2.1.177 | α-Xylosidase | Hemicellulose | 167.41 | 158.69 | 4.18 | 19.96 | 8.43 | 40.55 | 30.39 | 75.39 | 59.00 |
| 3.2.1.151 | Xyloglucan endo-β-1,4-glucanase | Hemicellulose | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| 3.1.1.72 | Acetyl xylan esterase | Hemicellulose | 69.49 | 114.59 | 4.62 | 0.00 | 9.04 | 3.26 | 10.34 | 35.56 | 88.05 |
| 3.1.1.6 | Acetylesterase | Hemicellulose | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| 3.1.1.73 | Feruloyl esterase | Hemicellulose | 0.00 | 0.00 | 0.00 | 0.00 | 0.29 | 0.00 | 0.00 | 0.00 | 0.62 |
| 3.2.1.55 | α-Arabinofuranosidase | Hemicellulose | 184.50 | 211.25 | 9.03 | 27.02 | 28.15 | 48.96 | 57.67 | 175.31 | 138.77 |
| 3.2.1.99 | Arabinan endo-α-1,5-arabinanase | Hemicellulose | 31.75 | 20.05 | 0.77 | 0.00 | 6.81 | 2.73 | 1.78 | 0.60 | 0.15 |
| 1.10.3.2 | Laccase | Lignin | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.25 | 5.63 |
| 3.1.1.- | 4-O-Methyl-glucuronoyl methylesterase | Lignin | 58.92 | 104.66 | 11.20 | 27.63 | 39.18 | 35.32 | 24.60 | 34.79 | 131.45 |
| 1.11.1.16 | Versatile peroxidase | Lignin | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| 4.2.2.2 | Pectate lyase | Pectin | 36.00 | 53.84 | 0.00 | 0.00 | 1.97 | 3.70 | 11.00 | 24.54 | 7.81 |
| 4.2.2.10 | Pectin lyase | Pectin | 0.72 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| 3.1.1.- | Pectin acetyl esterase/rhamnogalacturonan acetylesterase |
Pectin | 106.96 | 80.44 | 6.96 | 39.91 | 12.42 | 9.49 | 2.25 | 11.76 | 25.42 |
| 3.1.1.11 | Pectin methyl esterase | Pectin | 72.57 | 56.74 | 0.00 | 0.00 | 0.28 | 1.12 | 5.31 | 0.94 | 0.00 |
| 3.2.1.15 | Endopolygalacturonases | Pectin | 92.05 | 61.05 | 0.30 | 38.68 | 8.83 | 6.42 | 11.63 | 20.46 | 20.44 |
| 3.2.1.67 | Exopolygalacturonases | Pectin | 0.00 | 0.92 | 0.00 | 0.00 | 1.22 | 0.64 | 0.00 | 0.71 | 0.00 |
| 3.2.1.174 | Rhamnogalacturonan rhamnohydrolase | Pectin | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| 3.2.1.40 | α-Rhamnosidase | Pectin | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| 3.2.1.- | Endo-1,2-α-mannanase | Mannan | 91.35 | 92.10 | 8.77 | 0.00 | 29.61 | 24.99 | 15.19 | 34.57 | 34.26 |
| 3.2.1.24 | Exo-α-mannosidase | Mannan | 20.32 | 24.65 | 1.32 | 0.00 | 11.59 | 13.28 | 14.53 | 26.42 | 10.72 |
| 3.2.1.101 | Endo-1,6-α-mannanase | Mannan | 9.73 | 7.04 | 0.46 | 0.00 | 1.58 | 0.59 | 0.00 | 0.00 | 0.00 |
| 3.2.1.- | Exo-1,6-α-mannosidase | Mannan | 39.84 | 34.46 | 1.13 | 0.00 | 1.07 | 2.10 | 0.14 | 1.14 | 1.95 |
| 3.2.1.78 | Endo-1,4-β-mannanase | Mannan | 48.81 | 48.74 | 11.00 | 43.29 | 17.61 | 15.77 | 44.69 | 117.11 | 109.08 |
| 3.2.1.25 | β-Mannosidase | Mannan | 31.94 | 21.11 | 1.16 | 0.00 | 12.34 | 24.07 | 12.09 | 45.02 | 36.08 |
| 3.2.1.22 | Exo-α-galactosidase | Galactan | 116.54 | 112.55 | 14.60 | 59.87 | 57.90 | 76.76 | 77.72 | 162.74 | 152.89 |
| 3.2.1.23 | β-Galactosidase | Galactan | 533.54 | 390.10 | 13.60 | 18.73 | 64.91 | 71.15 | 92.29 | 175.80 | 181.17 |
| 3.2.1.49 | α-N-Acetylgalactosaminidase | Galactan | 138.91 | 85.11 | 22.08 | 7.37 | 106.47 | 67.98 | 10.30 | 10.49 | 2.58 |
Relative abundances are scaled by multiplication with 106 to improve visualization. Mn, Macrotermes natalensis; Od, Odontotermes sp.; Nc, Nasutitermes corniger; Aw, Amitermes wheeleri; Mp, Microcerotermes parvus; Co, Cornitermes sp.; Th, Termes hospes; Nt, Neocapritermes taracua; Cu, Cubitermes ugandensis.
FIG 2.
(A) Simplified schematic of fungal (top) and plant (bottom) cell wall structures and targets of enzymes identified in the metagenomes. (B) PCA of relative abundances of carbohydrate-active enzymes in worker gut metagenomes. Shapes represent termite feeding groups. Mn, Macrotermes natalensis; Od, Odontotermes sp.; Nc, Nasutitermes corniger; Aw, Amitermes wheeleri; Mp, Microcerotermes parvus; Co, Cornitermes sp.; Th, Termes hospes; Nt, Neocapritermes taracua; Cu, Cubitermes ugandensis. (C) Higher (red) and lower (blue) relative abundances of enzymes putatively targeting chitin, β-glucan, and α-glucan from the fungal cell wall and cellulose and hemicellulose in nine metagenomes shown as log2-transformed fold changes to the average relative abundance across nine species. Mannanases and galactosidases that putatively target plant and fungal cell wall components, respectively, are grouped separately.
Relative abundance of glycoside hydrolase families in gut metagenomes of nine species of termites with different diets. Mn, Macrotermes natalensis; Od, Odontotermes sp.; Nc, Nasutitermes corniger; Aw, Amitermes wheeleri; Mp, Microcerotermes parvus; Co, Cornitermes sp.; Th, Termes hospes; Nt, Neocapritermes taracua; Cu, Cubitermes ugandensis. Download Table S2, DOCX file, 0.04 MB (44.2KB, docx) .
Copyright © 2019 Hu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Assigning EC numbers to the CAZy genes where possible allowed us to get one step closer to identify putative fungus and plant cell wall substrate targets (Table 1 and Fig. 2C). In fungus-growing termites, the genes of enzymes targeting fungus cell wall polysaccharides were higher in relative abundance, while genes of enzymes targeting plant cell wall components were relatively low (Table 1 and Fig. 2C). For example, the genes encoding chitinase with EC number 3.2.1.14 that endo-hydrolyzes chitin was 5-fold higher in relative abundance in M. natalensis (Table 1 and Fig. 2C). Similarly, genes of glucanases targeting 1,3- and 1,6-β-glucan (endo-1,3-β-glucanase, exo-1,3-β-glucanase, and endo-1,6-β-glucosidase) of the fungus cell wall were 2- to 5-fold more abundant in fungus farmers (Table 1 and Fig. 2C). Genes putatively encoding endo-1,2-α-mannanase, exo-α-mannosidase, endo-1,6-α-mannanase, and exo-1,6-α-mannosidase that target α-mannan of the fungus cell wall were more abundant in fungus growers than genes encoding endo-1,4-β-mannanase and β-mannosidase that target the 1,4-β-mannan of hemicellulose in plant cell walls (Fig. 2C). In wood-feeding termites, genes encoding endo-1,4-β-glucanase and endo-1,4-β-xylanase that endo-hydrolyze cellulose and xylan were the most abundant plant cell wall-targeting enzyme genes (Table 1), and they were 2- to 3-fold higher in relative abundance than in other termite guts (Fig. 2C). Genes encoding laccase (EC 1.10.3.2), which plays a role in the cleavage of lignin, were found only in wood-feeding termite guts but in low relative abundance. Genes of several enzymes with exohydrolysis activity, for example, exo-β-glucosidase (EC 3.2.1.21) that hydrolyzes the nonreducing end of glucoses, exo-β-1,4-xylosidase (EC 3.2.1.37) that targets the nonreducing terminal of xylan, and β-N-acetylhexosaminidase (EC 3.2.1.52) that hydrolyzes free-end N-acetyl-d-hexosamine residues after chitin breakdown were relatively abundant in both fungus-growing and wood-feeding termites (Table 1). Genes encoding galactosidase (3.2.1.22, 3.2.1.23, and 3.2.1.49) that target galactose from the mannan of the fungus cell wall and pectin or hemicellulose components from the plant cell wall were relatively high in abundance in both fungus-growing termites and wood feeding termite guts (Table 1 and Fig. 2C).
The bacteria encoding the most abundant cell wall-targeting enzymes.
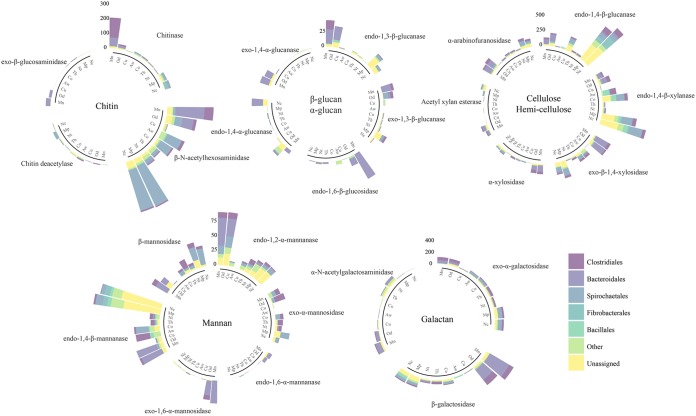
To gain further insight into the functional contribution of gut microbiota members to carbohydrate degradation, we grouped enzymes targeting polysaccharides from fungus and plant cell wall components by their taxonomy (Fig. 3). Clostridiales and Bacteroidales contributed most of the cell wall-degrading enzyme genes in fungus-growing termites (83% in M. natalensis and 68% in Odontotermes sp.). The two orders may, however, differ in what enzymes they contribute. The majority of the endo-1,2-α-mannanases, endo-1,6-α-mannanases, and exo-1,6-α-mannosidases (60%) that target α-mannan of the fungus cell wall were coded for by members of Bacteroidales, whereas the exo-α-mannosidases were mainly contributed by members of Clostridiales (Fig. 3). Members from both orders contribute genes for endo-1,3-β-glucanase and exo-1,3-β-glucanase that target β-glucan in the fungus cell wall, but only Bacteroidales contribute endo-1,6-β-glucosidase.
FIG 3.
Relative abundances of enzymes putatively targeting chitin, β-glucan, and α-glucan of fungal cell wall and cellulose and hemicellulose encoded by the five most abundant bacterial orders in the nine termite species. Mannanases and galactosidases that target plant and fungal cell wall components, respectively, are shown separately. The relative abundances are scaled by multiplication with 106 to improve visualization. Note that only bar length reflects relative abundance, not bar width or area. Mn, Macrotermes natalensis; Od, Odontotermes sp.; Nc, Nasutitermes corniger; Aw, Amitermes wheeleri; Mp, Microcerotermes parvus; Co, Cornitermes sp.; Th, Termes hospes; Nt, Neocapritermes taracua; Cu, Cubitermes ugandensis.
In wood feeders, genes encoding enzymes for cellulose and hemicellulose cleavage were coded for by all abundant members of the gut microbiotas (Fig. 3). Spirochaetales contributed 32% of the cell wall-degrading enzyme genes (Fig. 3), but notably, a large proportion of these genes (36%) could not be assigned to bacterial orders. Genes putatively encoding chitinases and β-N-acetylhexosaminidases for chitin degradation were contributed by both Clostridiales and Bacteroidales and, to a lesser extent, Spirochaetales in wood feeders. In termite species that were neither fungus nor wood feeding, most of the fungus cell wall-degrading enzyme genes were also contributed by Clostridiales, Bacteroidales, and Spirochaetales (17 to 92%), but the relative abundances of these enzyme genes were far lower than those observed in fungus and wood feeders (Table 1 and Fig. 3).
DISCUSSION
Diet is a major driver of taxonomic and functional composition of gut microbial communities (39–41). Our characterization of cell wall degrading enzyme from higher termites confirms that the functional profiles of gut microbial communities are tightly linked with termite diet, which is consistent with previous work that has focused on community compositions (21, 22, 35), and preliminary functional associations with microbiota structure in the Macrotermitinae (8, 24). Fungus-growing termite diets consist of fungal hyphae and plant material that is partly degraded by the symbiotic fungus (22). The gut microbiota thus not only functionally complements final plant biomass decomposition by providing oligosaccharide-targeting enzymes (8, 24) but also provides key enzymes for the digestion of fungal biomass. The mycolytic potential of the gut microbiota of the South African Odontotermes sp. and M. natalensis (this study) was comparable to O. yunnanensis from Southwest China (8), suggesting conserved functions across space and time and highlighting the robust link between taxonomy and function of the intimate interactions between gut community members and termite hosts.
Fungus-growing termite gut mycolytic enzymes were primarily coded for by Clostridiales and Bacteroidales, which also dominate the core microbiota of the termite subfamily (20, 21, 35, 43, 44). The ancestor of the Macrotermitinae likely had a bacterial gut microbiota similar to those of lower termites (but without protists) (6, 9). Fungiculture exposed the gut microbiota to larger amounts of fungal biomass than the ancestral lignocellulolytic diet, likely resulting in gut microbiotas that became more similar to those observed in extant cockroaches (20, 21). This is likely a product of several factors. First, bacterial strains present in the Macrotermitinae ancestor that could utilize a fungal diet were likely selected for and consequently increased in relative abundance (e.g., Alistipes, Dysgonomonas, and members of the Ruminococcaceae) (20, 21). Many mycolytic microbes were conceivably already present in termite guts prior to the origin of fungiculture, since the ancestral termites fed on partially degraded plant substrates containing fungal biomass. Second, bacteria that do not contribute to the breakdown of fungal material in the Macrotermitinae ancestor were likely outcompeted/selected against in the fungal-material-rich environment. Third, novel lineages adopted from other termites or the environment were likely coopted when fungiculture evolved. This is consistent with recent work demonstrating rampant horizontal transmission of gut bacterial lineages associated with termites (45). Collectively, this led to a gut microbiota with relatively more mycolytic enzymes (i.e., α-mannanases, β-1,3/1,6-glucanases, and chitinases) and relatively fewer lignocellulolytic enzymes (i.e., cellulase and hemicellulose).
In sharp contrast to the fungus growers, lignocellulose-degrading enzymes dominated the gut microbiota of wood-feeding termites (N. corniger and M. parvus), as expected from the requirement for the breakdown of recalcitrant plant components, which in fungus farmers is handled by Termitomyces (24; although lignin cleavage may be initiated during the first gut passage [27]). Interestingly, however, the relatively high abundance of chitinolytic enzymes of Spirochaetales origin in wood-feeding termites suggests that the decaying wood these species feed on harbors fungal biomass that gut bacteria, or the termite host, likely utilize. However, fungal cell wall-targeting enzymes such as β-1,3-glucanase may also serve to protect against fungal infections (cf. reference 46). Experimental work targeting the expression of these enzymes in the presence of fungal pathogens, ideally combined with a varying fungal biomass diet content, have the potential to shed light on their relative defensive and dietary roles.
An important caveat of our study is that we were limited by comparisons of bacterial abundances and encoded enzymes between whole-gut DNA extractions and metagenomes in the two fungus-growing termites with P3 compartment of wood- and litter-feeding termites and the lumen metagenome of the dung feeder. This may have biased our comparative analyses somewhat, but we believe that at least the P3 comparison to whole guts is reliable for the following reasons. First, the P3 compartment is expected to contain the vast majority of microbial cells of termite guts (e.g., >97% in the wood feeder N. corniger [47]). Consequently, even if relative abundances may differ across gut sections (cf. reference 34), the overall community structure and functions should be primarily driven by P3-residing bacteria. Nevertheless, the contribution of enzyme genes from some member such as Clostridiales, which is consistently more abundant in the P1 compartment (48), may have been underestimated when only the P3 region was sequenced. Second, comparisons of community structure between whole gut 16S rRNA from fungus-growing termite (21) with P3 community analyses (20) found nearly identical community structure (see Fig. 4 in reference 26). More importantly, although community composition of the gut fluid of N. corniger (33) largely resemble hindgut and P3 compartment analyses (20, 34, 47, 48), we advocate that the comparisons to the lumen metagenome of A. wheeleri is taken with a grain of salt for two main reasons. First, lumen and gut wall-associated microbiomes may differ substantially (49, 50). Second, although we normalize our comparisons by metagenome size, the low coverage of A. wheeleri most likely undersamples the true composition of the metagenome, thus probably precluding the identification of low-abundance bacteria and enzyme genes (Table 1). Third, although the impact is expected to be minor (cf. reference 42), discrepancies could be impacted by sequencing protocols (454 versus Illumina) due to potential sequencing biases associated with GC content, sequencing errors, and differences in read length.
The shotgun metagenomics data showed distinct compositional profiles of cell wall-degrading enzyme genes in termite guts consistent with expected gut community functions. However, without experimental data of the bacteria densities and complete bacteria genomes, our comparisons cannot reflect enzyme quantities and in situ activities (cf. reference 51). A large fraction of the contigs also do not have a taxonomic assignment (Table S1), and although identification of all abundant genera was consistent with previous 16S rRNA studies (21, 22, 34), some bacteria were absent or underrepresented in the metagenomes compared to 16S rRNA surveys and vice versa (Table S3). The lack of identification of the presence of members of the TG3 phylum in the metagenome is likely an artifact, as the phylum lies within the Fibrobacteres in the NCBI sequence database we used for taxonomic identification. The genera Chitinispirillum and Ruminiclostridium we identified in the metagenomes were absent in 16S rRNA analyses (21, 22, 34) because there were no rRNA gene references for these two genera in the database at the time they were analyzed and published (52). Also, the lack of appropriate reference genomes for termite-associated strains limits the classification of the metagenome to genus level, which is likely the cause for the underrepresentation of the genus Alistipes. Some Alistipes sequences may not have been identified because they do not match to the reference genome which are mostly isolated from the human microbiome (53–56). Thus, limited by reference-dependent methods that require closely related sequences in the public databases, the contribution of cell wall-degrading enzymes of some taxa is likely to be underestimated. Deeper metagenome sequencing that can enable binning of sequences to improve taxonomic classification and functional predictions, coupled with functional studies of the gut-compartment-specific expression of bacterial enzymes, are thus warranted.
Comparison of the taxonomic assignments of 16S rRNA studies (21, 22, 34) and metagenomic sequences (this study) at phylum (top) and genus (bottom) levels. Taxonomy classifications with more than 0.1% of the total abundance are shown. Taxonomy with no 16S rRNA gene references are marked as “NA.” Download Table S3, XLSX file, 0.02 MB (18.7KB, xlsx) .
Copyright © 2019 Hu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Similarities in host diet have been shown to drive convergence in the functional potential of gut microbes in other organisms (57, 58). Selection for particular physiological traits may, however, not necessarily be directly linked to specific phylogenetic groups of microbes. Exploring communities associated with diverse fungus-growing hosts (and their associated fungi) would allow us to explicitly test for convergent evolution of mycolytic microbial communities, even if these were likely comprised of different microbial consortia. A number of other insects utilize fungus material as a nutrient source, including fungus-growing ants (59, 60), some Drosophila species (61), the Malaysian mushroom-harvesting ant Euprenolepis (62), Sirex wood wasps (63, 64), and ambrosia beetles (65–67). Different bacteria are likely to be involved, but predictions would be that predominantly fungal diets should select for microbial communities with comparable mycolytic capacities. Recent work on mycophagous Drosophila supports that this may be so. Bost et al. (68) demonstrated that gut bacteria in mycophagous Drosophila are implicated in fungal cell wall metabolism, with cysteine and methionine metabolism enzymes originating from Bacteroidetes, Firmicutes, and Proteobacteria gut microbiota members, phyla that are also abundant and mycolytic in fungus-growing termites.
The prevalence of microbial communities with ample mycolytic capacities in the guts of fungus-growing termite species suggests that the adoption of a fungal diet has been associated with a functional and compositional change in gut microbial communities at the onset of fungiculture in termites. The high relative abundance of fungal cell wall-degrading enzymes indicates adaptations to the decomposition of fungal diet, which is consistent with this capacity being absent or less in termites with predominantly plant-based diets. An exception is wood-feeding termites, for which wood-degrading fungi may also comprise an appreciable component of the termite diet. To improve our understanding of the link between the digestive function and the gut microbes in fungus-growing termites, further work will be needed to elucidate whether the functional capacities of the gut microbiota reflect the amount of fungal biomass in the diet and differences in properties of the fungal species fed on. Estimates of bacterial densities and enzyme activities are also needed to fully understand the capacity of biomass degradation in termite gut. Metagenomic analysis, including more fungus-growing termite species, improved coverage, and longer reads, paired with reference-free taxonomy binning methods and functional annotation, will also largely enhance our understanding of the property of gut microbes in a functional angle.
MATERIALS AND METHODS
Odontotermes sp. collection.
Termites from an Odontotermes sp. colony (code Od127) were collected at the Experimental Farm of the University of Pretoria, Pretoria, South Africa (−25.742700, 28.256517). The species identity of this colony had been previously established as by mitochondrial gene COII barcoding (22). Fifty old major workers were sampled, and entire guts were dissected and pooled in a 1.5-ml Eppendorf tube and stored at –80°C until DNA extraction.
Gut microbiota DNA extraction.
Guts were ground in liquid nitrogen, and DNA was extracted using DNeasy blood and tissue kits (Qiagen, Hilden, Germany) according to the manufacturer’s description, except that a chloroform extraction step was included after incubation with protease K. After proteinase K digestion, 1 volume chloroform-isoamyl alcohol (24/1) was added. The tubes were incubated for 15 min on a slowly rotating wheel and then centrifuged at 3,000 × g for 10 min. The supernatant was transferred to spin columns, and the remainder of the manufacturer’s protocol was followed. The quality and purity of samples were determined using NanoDrop (Thermo Scientific, Wilmington, DE).
Metagenome sequencing and assembly.
DNA was sheared to ∼350-bp fragments, end repaired, A tailed, and ligated with Illumina paired-end adaptors (Illumina). The ligated fragments were selected from the desired size on agarose gels and amplified by ligation-mediated PCR, and libraries were sequenced with 150-bp read lengths on an Illumina HiSeq2500. The quality of raw sequencing reads was assessed before assembly. Reads containing the adaptor, >10% N, or >50% low-quality bases (Q-score ≥ 5) were removed. To exclude sequences from Termitomyces and the termite hosts, quality-controlled reads were mapped to the M. natalensis and Termitomyces genomes (24) using the Burrows-Wheeler Aligner v0.7.15 (69) BWA-MEM algorithm; any aligned reads were filtered.
Clean reads were assembled by IDBA-UD v1.1.2 (70, 71) with an iterative set up from k-mer size of 19 to 99 at step of 10 (–pre_correction –mink 19 –maxk 99 –step 10). Unassembled reads were picked out by mapping reads back to the initial assembly and assembled separately with the same setup. Redundancies of sequences from the same organism within the metagenome were removed by clustering all contigs at 95% identity with CD-hit v4.6.6 (72), and only the longest contig per cluster was kept. For comparison, high-quality reads of M. natalensis old major worker gut microbiota from Poulsen et al. (24) were also reassembled using the same procedure. Genes in both assemblies were predicted by Prodigal v2.6.3 (73) with metagenomics parameters (–c –p meta).
Non-fungus-growing termite gut metagenomes.
We obtained seven published non-fungus-growing termite gut metagenomes. These included a dung feeder, Amitermes wheeleri (33); two wood feeders, Nasutitermes corniger and Microcerotermes parvus; a litter feeder, Cornitermes sp.; two humus feeders, Termes hospes and Neocapritermes taracua; and a soil feeder, Cubitermes ugandensis (34). Contigs and protein coding genes were downloaded from JGI IMG/M (https://img.jgi.doe.gov/) (Table 2).
TABLE 2.
Summary of metagenome data
| Termite species | Diet | Reference | Accession no. | Sample data size (Gbp) |
Assembled bases (%) |
Assembly size (Mbp) |
Assembly N50 (bp) |
No. of genes |
|---|---|---|---|---|---|---|---|---|
| Odontotermes sp. | Fungus | NAa | PRJNA476694 | 8.6 | 57.9 | 797.15 | 1,133 | 753,265 |
| Macrotermes natalensis | Fungus | 24 | PRJNA193472 | 8.5 | 50.9 | 498.71 | 1,569 | 486,207 |
| Amitermes wheeleri | Dung | 33 | PRJNA173365 | 0.3 | 57.7 | 139.30 | 512 | 321,461 |
| Nasutitermes corniger | Wood | 34 | PRJNA366361 | 46.8 | 20.5 | 634.52 | 980 | 1,338,688 |
| Microcerotermes parvus | Wood | 34 | PRJNA271983 | 43.2 | 25.0 | 712.35 | 945 | 1,419,719 |
| Cornitermes sp. | Litter | 34 | PRJNA405701 | 45.9 | 25.3 | 131.61 | 519 | 2,945,692 |
| Termes hospes | Humus | 34 | PRJNA405704 | 34.2 | 16.7 | 1,212.37 | 424 | 2,975,319 |
| Neocapritermes taracua | Humus | 34 | PRJNA366256 | 28.3 | 8.8 | 885.58 | 454 | 2,115,406 |
| Cubitermes ugandensis | Soil | 34 | PRJNA366375 | 32.0 | 10.8 | 1,121.61 | 475 | 3,061,751 |
NA, not applicable.
Relative abundances of contigs within metagenomes.
Clean reads of fungus-growing termites gut microbiota were mapped to the contigs by Bowtie v2.2.9 (74) with subsequent masked duplication, taking the best match for each read. Contig coverage was first estimated by the contig length and the number of mapped reads per contigs. Mapped read numbers were scaled to an equivalent of 10 Gb of sequence per metagenome, after which the relative abundance of each contig was calculated as the coverage of the contig divided by the sum of coverages of all contigs (cf. reference 75). For non-fungus-growing termites, coverage information of contigs was obtained from JGI (76, 77), and the relative abundances were estimated as described above.
Metagenome taxonomic assignment.
Taxonomic assignment of protein-coding genes was carried out using Diamond (18) alignment against the NR database in NCBI. Alignments with E values >1e–5 and sequences with identities <30% were removed. Taxonomic information of the top hit was assigned to each gene. Contigs referring to taxonomical levels were determined by a modified lowest common ancestor (LCA)-based algorithm implemented in MEGAN (78). Taxonomic classification supported by <10% of the genes on each contig were first filtered, and the LCAs for the taxonomic classification of the rest of the genes were assigned to the contig. The relative abundance of contigs belonging to the same taxonomic group was summed up to represent the taxonomic composition of that taxonomic group in the microbiota. Taxonomic composition at the phylum level for metagenomes was centered log ratio (CLR) transformed and then compared and visualized by principle component analysis (PCA) (79) using R v3.3.2 (80).
Carbohydrate-active enzyme analysis.
We classified genes in CAZyme families by searching against CAZyme hidden Markov profiles in the dbCAN database (v7; CAZyDB accessed 31 July 2018 [81]) using HMMer v3.1b (E ≤ 1e–5) (82). The longest matched profile was selected if CAZyme domain overlapped with peptide pattern recognition using HotPep (36–38). CAZyme family classifications that were supported by both approaches were kept. The composition of GH families in termite species was CLR transformed, and the differences between termite species were visualized using PCA (79) in R v3.3.2 (80). Enzyme functions of genes in each CAZyme family were determined by BLASTp searches (E ≤ 1e–5) against the ExPASy enzyme records, CCD searches against the COG database (83), and GhostKOALA searches using the KEGG database online tool (84). Genes coding for enzymes related to fungal and plant cell wall degradation were selected, and their substrate targets and bacterial taxonomy were assigned and manually checked. To compare the fungal and plant cell wall-targeting enzymes across termite species, the relative abundance profiles were hierarchically clustered and are shown as a dendrogram in Fig. 2C. For each enzyme, the relative abundances were compared to the average across the nine termite species, and the fold change was log2 transformed and is presented in a heatmap in Fig. 2C.
Data availability.
Clean reads and metagenome assembly have been submitted to the SRA and GenBank under BioProject accession numbers PRJNA476694 and PRJNA193472.
ACKNOWLEDGMENTS
We thank Z. Wilhelm de Beer, Michael J. Wingfield, and the staff and students at the Forestry and Agricultural Biotechnology Institute, University of Pretoria, for hosting field work and contributions to the research environment, and we thank Christine Beemelmanns, René Benndorf, Saria Otani, Margo Wisselink, Sabine M. E. Vreeburg, Duur K. Aanen, Lennart van de Peppel, Nina Kreuzenbeck, and Victoria L. Challinor for help with excavations.
This study was funded by the CAPES Foundation, Ministry of Education of Brazil (BEX: 13240/13-7; to R.R.D.C.), and by a Villum Kann Rasmussen Young Investigator Fellowship (VKR10101; to M.P.).
We declare no conflict of interest.
H.H. and R.R.D.C. designed the experiments and analyses, collected material in the field, extracted DNA, sent samples for sequencing, and drafted the first version of the figures, tables, and manuscript. H.H. carried out the assembly of the metagenome and the functional analyses. B.P. and L.L. carried out HotPep analyses and contributed to data interpretation. M.S. contributed to data analyses and interpretations. M.P. supervised H.H. and R.R.D.C., helped design the study, and contributed with comments on analyses and the first versions of figures, tables, and text. All authors contributed to writing the manuscript.
REFERENCES
- 1.Buczkowski G, Bertelsmeier C. 2017. Invasive termites in a changing climate: a global perspective. Ecol Evol 7:974–985. doi: 10.1002/ece3.2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Eggleton P. 2000. Global patterns of termite diversity, p 25–51. In Abe T, Bignell DE, Higashi M (ed), Termites: evolution, sociality, symbioses, ecology. Kluwer Academic Publishers, Dordrecht, The Netherlands. [Google Scholar]
- 3.Eggleton P. 2001. Termites and trees: a review of recent advances in termite phylogenetics. Insectes Sociaux 48:187–193. doi: 10.1007/PL00001766. [DOI] [Google Scholar]
- 4.Kambhampati S, Eggleton P. 2000. Taxonomy and phylogeny of termites, p 1–23. In Abe T, Bignell DE, Higashi M (ed), Termites: evolution, sociality, symbioses, ecology. Kluwer Academic Publishers, Dordrecht, Netherlands. [Google Scholar]
- 5.Inward DJ, Vogler AP, Eggleton P. 2007. A comprehensive phylogenetic analysis of termites (Isoptera) illuminates key aspects of their evolutionary biology. Mol Physiol Evol 44:953–967. doi: 10.1016/j.ympev.2007.05.014. [DOI] [PubMed] [Google Scholar]
- 6.Brune A. 2014. Symbiotic digestion of lignocellulose in termite guts. Nat Rev Microbiol 12:168–180. doi: 10.1038/nrmicro3182. [DOI] [PubMed] [Google Scholar]
- 7.Thongaram T, Hongoh Y, Kosono S, Ohkuma M, Trakulnaleamsai S, Noparatnaraporn N, Kudo T. 2005. Comparison of bacterial communities in the alkaline gut segment among various species of higher termites. Extremophiles 9:229–238. doi: 10.1007/s00792-005-0440-9. [DOI] [PubMed] [Google Scholar]
- 8.Liu N, Zhang L, Zhou H, Zhang M, Yan X, Wang Q, Long Y, Xie L, Wang S, Huang Y, Zhou Z. 2013. Metagenomic insights into metabolic capacities of the gut microbiota in a fungus-cultivating termite Odontotermes yunnanensis. PLoS One 8:e69184. doi: 10.1371/journal.pone.0069184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brune A, Dietrich C. 2015. The gut microbiota of termites: digesting the diversity in the light of ecology and evolution. Annu Rev Microbiol 69:145–166. doi: 10.1146/annurev-micro-092412-155715. [DOI] [PubMed] [Google Scholar]
- 10.Ni J, Tokuda G. 2013. Lignocellulose-degrading enzymes from termites and their symbiotic microbiota. Biotech Adv 31:838–850. doi: 10.1016/j.biotechadv.2013.04.005. [DOI] [PubMed] [Google Scholar]
- 11.Watanabe H, Tokuda G. 2010. Cellulolytic systems in insects. Annu Rev Entomol 55:609–613. doi: 10.1146/annurev-ento-112408-085319. [DOI] [PubMed] [Google Scholar]
- 12.Breznak JA. 1982. Intestinal microbiota of termites and other xylophagous insects. Annu Rev Microbiol 36:323–343. doi: 10.1146/annurev.mi.36.100182.001543. [DOI] [PubMed] [Google Scholar]
- 13.Higashi M, Abe T, Burns TP. 1992. Carbon-nitrogen balance and termite ecology. Proc R Soc B 249:303–308. [Google Scholar]
- 14.Sapountzis P, de Verges J, Rousk K, Cilliers M, Vorster BJ, Poulsen M. 2016. Potential for nitrogen fixation in the fungus-growing termite symbiosis. Front Microbiol 7:1993. doi: 10.3389/fmicb.2016.01993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Um S, Fraimout A, Sapountzis P, Oh DC, Poulsen M. 2013. The fungus-growing termite Macrotermes natalensis harbors bacillaene-producing Bacillus sp. that inhibit potentially antagonistic fungi. Sci Rep 3:3250. doi: 10.1038/srep03250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Round JL, Mazmanian SK. 2009. The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol 9:313–323. doi: 10.1038/nri2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Viaud S, Daillere R, Boneca IG, Lepage P, Langella P, Chamaillard M, Pittet MJ, Ghiringhelli F, Trinchieri G, Goldszmid R, Zitvogel L. 2015. Gut microbiome and anticancer immune response: really hot sh*t! Cell Death Differ 22:199–214. doi: 10.1038/cdd.2014.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Buchfink B, Xie C, Huson D. 2015. Fast and sensitive protein alignment using DIAMOND. Nat Methods 12:59–60. doi: 10.1038/nmeth.3176. [DOI] [PubMed] [Google Scholar]
- 19.Bourguignon T, Lo N, Cameron SL, Šobotník J, Hayashi Y, Shigenobu S, Watanabe D, Roisin Y, Miura T, Evans TA. 2015. The evolutionary history of termites as inferred from 66 mitochondrial genomes. Mol Biol Evol 32:406–421. doi: 10.1093/molbev/msu308. [DOI] [PubMed] [Google Scholar]
- 20.Dietrich C, Köhler T, Brune A. 2014. The cockroach origin of the termite gut microbiota: patterns in bacterial community structure reflect major evolutionary events. Appl Environ Microbiol 80:2261–2269. doi: 10.1128/AEM.04206-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Otani S, Mikaelyan A, Nobre T, Hansen LH, Koné NA, Sørensen SJ, Aanen DK, Boomsma JJ, Poulsen M. 2014. Identifying the core microbial community in the gut of fungus-growing termites. Mol Ecol 18:4631–4644. doi: 10.1111/mec.12874. [DOI] [PubMed] [Google Scholar]
- 22.Otani S, Zhukova M, Kone NA, da Costa RR, Mikaelyan A, Sapountzis P, Poulsen M. 2019. Gut microbial compositions mirror caste-specific diets in a major lineage of social insects. Environ Microbiol Rep 11:196–205. doi: 10.1111/1758-2229.12728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nobre T, Rouland-Lefèvre C, Aanen DK. 2011. Comparative biology of fungus cultivation in termites and ants, p 193–210. In Bignell D, Roisin Y, Lo N (ed), Biology of termites: a modern synthesis, 2nd ed Springer, Dordrecht, The Netherlands. [Google Scholar]
- 24.Poulsen M, Hu H, Li C, Chen Z, Xu L, Otani S, Nygaard S, Korb J, Aanen DK, Wang J, Boomsma JJ, Zhang G. 2014. Complementary symbiont contributions to plant decomposition in a fungus-farming termite. Proc Natl Acad Sci U S A 40:14500–14505. doi: 10.1073/pnas.1319718111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leuthold RH, Badertscher S, Imboden H. 1989. The inoculation of newly formed fungus comb with Termitomyces in Macrotermes colonies (Isoptera, Macrotermitinae). Ins Soc 36:328–338. doi: 10.1007/BF02224884. [DOI] [Google Scholar]
- 26.Poulsen M. 2015. Towards an integrated understanding of the consequences of fungus domestication on the fungus-growing termite gut microbiota. Environ Microbiol 17:2562–2572. doi: 10.1111/1462-2920.12765. [DOI] [PubMed] [Google Scholar]
- 27.Li H, Yelled DJ, Lie C, Yang M, Ke J, Zhang R, Liu Y, Zhu N, Liang S, Mo X, Ralph J, Currie CR, Mo J. 2017. Lignocellulose pretreatment in a fungus-cultivating termite. Proc Natl Acad Sci U S A 144:4709–4714. doi: 10.1073/pnas.1618360114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hyodo F, Tayasu I, Inoue T, Azuma TI, Kudo T, Abe T. 2003. Differential role of symbiotic fungi in lignin degradation and food provision for fungus-growing termites (Macrotermitinae: Isoptera). Funct Ecol 17:186–193. doi: 10.1046/j.1365-2435.2003.00718.x. [DOI] [Google Scholar]
- 29.Reid ID, Bartnicki-Garcia S. 1976. Cell-wall composition and structure of yeast cells and conjugation tubes of Tremella mesenterica. J Gen Microbiol 96:35–50. doi: 10.1099/00221287-96-1-35. [DOI] [PubMed] [Google Scholar]
- 30.Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, Henrissat B. 2009. The carbohydrate-active enzymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res 37:D233–D238. doi: 10.1093/nar/gkn663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lombard V, Golaconda Ramulu H, Drula E, Coutinho PM, Henrissat B. 2014. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res 42:D490–495. doi: 10.1093/nar/gkt1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kielak AM, Cretoiu MS, Semenov AV, Sørensen SJ, van Elsas JD. 2013. Bacterial chitinolytic communities respond to chitin and pH alteration in soil. Appl Environ Microbiol 79:263–272. doi: 10.1128/AEM.02546-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.He S, Ivanova N, Kirton E, Allgaier M, Bergin C, Scheffrahn RH, Kyrpides NC, Warnecke F, Tringe SG, Hugenholtz P. 2013. Comparative metagenomic and metatranscriptomic analysis of hindgut paunch microbiota in wood- and dung-feeding higher termites. PLoS One 8:e61126. doi: 10.1371/journal.pone.0061126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rossmassler K, Dietrich C, Thompson C, Mikaelyan A, Nonoh JO, Scheffrahn RH, Sillam-Dussès D, Brune A. 2015. Metagenomic analysis of the microbiota in the highly compartmented hindguts of six wood- or soil-feeding higher termites. Microbiome 3:56. doi: 10.1186/s40168-015-0118-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mikaelyan A, Dietrich C, Köhler T, Poulsen M, Sillam-Dussès D, Brune A. 2015. Diet is the primary determinant of bacterial community structure in the guts of higher termites. Mol Ecol 24:5284–5295. doi: 10.1111/mec.13376. [DOI] [PubMed] [Google Scholar]
- 36.Busk PK, Lange L. 2013. Function-based classification of carbohydrate-active enzymes by recognition of short, conserved peptide motifs. Appl Environ Microbiol 79:3380–3391. doi: 10.1128/AEM.03803-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Busk PK, Lange M, Pilgaard B, Lange L. 2014. Several genes encoding enzymes with the same activity are necessary for aerobic fungal degradation of cellulose in nature. PLoS One 9:e114138. doi: 10.1371/journal.pone.0114138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Busk PK, Pilgaard B, Lezyk MJ, Meyer AS, Lange L. 2017. Homology to peptide pattern for annotation of carbohydrate-active enzymes and prediction of function. BMC Bioinformatics 18:214. doi: 10.1186/s12859-017-1625-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, Ling AV, Devlin AS, Varma Y, Fischbach MA, Biddinger SB, Dutton RJ, Turnbaugh PJ. 2014. Diet rapidly and reproducibly alters the human gut microbiome. Nature 505:559–563. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Perez-Cobas AE, Maiques E, Angelova A, Carrasco P, Moya A, Latorre A. 2015. Diet shapes the gut microbiota of the omnivorous cockroach Blattella germanica. FEMS Microbiol Ecol 91:fiv022. doi: 10.1093/femsec/fiv022. [DOI] [PubMed] [Google Scholar]
- 41.Bodawatta KH, Sam K, Jønsson KA, Poulsen M. 2018. Comparative analyses of the digestive tract microbiota of New Guinean passerine birds. Front Microbiol 9:1830. doi: 10.3389/fmicb.2018.01830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Luo C, Tsementzi D, Kyrpides N, Read T, Konstantinidis KT. 2012. Direct comparisons of Illumina vs. Roche 454 sequencing technologies on the same microbial community DNA sample. PLoS ONE 7:e30087. doi: 10.1371/journal.pone.0030087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hongoh Y, Deevong P, Inoue T, Moriya S, Trakulnaleamsai S, Ohkuma M, Vongkaluang C, Noparatnaraporn N, Kudo T. 2005. Intra- and interspecific comparisons of bacterial diversity and community structure support coevolution of gut microbiota and termite host. Appl Environ Microbiol 71:6590–6599. doi: 10.1128/AEM.71.11.6590-6599.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hongoh Y. 2010. Diversity and genomes of uncultured microbial symbionts in the termite gut. Biosci Biotechnol Biochem 74:1145–1151. doi: 10.1271/bbb.100094. [DOI] [PubMed] [Google Scholar]
- 45.Bourguignon T, Lo N, Dietrich C, Šobotník J, Sidek S, Roisin Y, Brune A, Evans TA. 2018. Rampant host switching shaped the termite gut microbiome. Curr Biol 28:649–654. doi: 10.1016/j.cub.2018.01.035. [DOI] [PubMed] [Google Scholar]
- 46.Rosengaus RB, Schultheis KF, Yalonetskaya A, Bulmer MS, DuComb WS, Benson RW, Godoy-Carter V. 2014. Symbiont-derived β-1,3-glucanases in a social insect: mutualism beyond nutrition. Front Microbiol 5:607. doi: 10.3389/fmicb.2014.00607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kohler T, Dietrich C, Scheffrahn RH, Brune A. 2012. High-resolution analysis of gut environment and bacterial microbiota reveals functional compartmentation of the gut in wood-feeding higher termites (Nasutitermes spp.). Appl Environ Microbiol 78:4691–4701. doi: 10.1128/AEM.00683-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mikaelyan A, Meuser K, Brune A. 2017. Microenvironmental heterogeneity of gut compartments drives bacterial community structure in wood- and humus-feeding higher termites. FEMS Microbiol Ecol 93:fiw210. doi: 10.1093/femsec/fiw210. [DOI] [PubMed] [Google Scholar]
- 49.Berchtold M, Chatzinotas A, Schonhuber W, Brune A, Amann R, Hahn D, Konig H. 1999. Differential enumeration and in situ localization of microorganisms in the hindgut of the lower termite Mastotermes darwiniensis by hybridization with rRNA-targeted probes. Arch Microbiol 172:407–416. doi: 10.1007/s002030050778. [DOI] [PubMed] [Google Scholar]
- 50.Li H, Dietrich C, Zhu N, Mikaelyan A, Ma B, Pi R, Liu Y, Yang M, Brune A, Mo J. 2016. Age polyethism drives community structure of the bacterial gut microbiota in the fungus-cultivating termite Odontotermes formosanus. Environ Microbiol 18:1440–1451. doi: 10.1111/1462-2920.13046. [DOI] [PubMed] [Google Scholar]
- 51.Nayfach S, Pollard KS. 2016. Toward accurate and quantitative comparative metagenomics. Cell 166:1103–1116. doi: 10.1016/j.cell.2016.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mikaelyan A, Kohler T, Lampert N, Rohland J, Boga H, Meuser K, Brune A. 2015. Classifying the bacterial gut microbiota of termites and cockroaches: a curated phylogenetic reference database (DictDb). Syst Appl Microbiol 38:472–482. doi: 10.1016/j.syapm.2015.07.004. [DOI] [PubMed] [Google Scholar]
- 53.Browne HP, Forster SC, Anonye BO, Kumar N, Neville BA, Stares MD, Goulding D, Lawley TD. 2016. Culturing of ‘unculturable’ human microbiota reveals novel taxa and extensive sporulation. Nature 533:543. doi: 10.1038/nature17645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pfleiderer A, Mishra AK, Lagier J-C, Robert C, Caputo A, Raoult D, Fournier P-E. 2014. Non-contiguous finished genome sequence and description of Alistipes ihumii sp. nov. Stand Genomic Sci 9:1221–1235. doi: 10.4056/sigs.4698398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shkoporov AN, Chaplin AV, Khokhlova EV, Shcherbakova VA, Motuzova OV, Bozhenko VK, Kafarskaia LI, Efimov BA. 2015. Alistipes inops sp. nov. and Coprobacter secundus sp. nov., isolated from human faeces. Int J Syst Evol Microbiol 65:4580–4588. doi: 10.1099/ijsem.0.000617. [DOI] [PubMed] [Google Scholar]
- 56.Song Y, Könönen E, Rautio M, Liu C, Bryk A, Eerola E, Finegold SM. 2006. Alistipes onderdonkii sp. nov. and Alistipes shahii sp. nov., of human origin. Int J Syst Evol Microbiol 56:1985–1990. doi: 10.1099/ijs.0.64318-0. [DOI] [PubMed] [Google Scholar]
- 57.Muegge BD, Kuczynski J, Knights D, Clemente JC, González A, Fontana L, Henrissat B, Knight R, Gordon JI. 2011. Diet drives convergence in gut microbiome functions across mammalian phylogeny and within humans. Science 332:970–974. doi: 10.1126/science.1198719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Delsuc F, Metcalf JL, Wegener Parfrey L, Song SJ, González A, Knight R. 2014. Convergence of gut microbiomes in myrmecophagous mammals. Mol Ecol 6:1301–1317. doi: 10.1111/mec.12501. [DOI] [PubMed] [Google Scholar]
- 59.Schultz TR, Brady SG. 2008. Major evolutionary transitions in ant agriculture. Proc Natl Acad Sci U S A 105:5435–5440. doi: 10.1073/pnas.0711024105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schiøtt M, Rogowska-Wrzesinska A, Roepstorff P, Boomsma JJ. 2010. Leaf-cutting ant fungi produce cell wall degrading pectinase complexes reminiscent of phytopathogenic fungi. BMC Biol 8:156. doi: 10.1186/1741-7007-8-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jaenike J, James AC. 1991. Aggregation and the coexistence of mycophagous Drosophila. J Anim Ecol 60:913–928. doi: 10.2307/5421. [DOI] [Google Scholar]
- 62.Witte V, Maschwitz U. 2008. Mushroom harvesting ants in the tropical rain forest. Naturwissenschaften 95:1049–1054. doi: 10.1007/s00114-008-0421-9. [DOI] [PubMed] [Google Scholar]
- 63.Madden JL, Coutts MP. 1979. The role of fungi in the biology and ecology of wood wasps (Hymenoptera: Siricidae), p 165–174. In Batra LF. (ed), Insect-fungus symbiosis: nutrition, mutualism, and commensalism. Allanheld Osmun, London, United Kingdom. [Google Scholar]
- 64.Hajek AE, Nielsen C, Kepler RM, Long SJ, Castrillo L. 2013. Fidelity among Sirex woodwasps and their fungal symbionts. Microb Ecol 65:753–762. doi: 10.1007/s00248-013-0218-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Batra LR. 1966. Ambrosia fungi: extent of specificity to ambrosia beetles. Science 153:193–195. doi: 10.1126/science.153.3732.193. [DOI] [PubMed] [Google Scholar]
- 66.Hulcr J, Cognato AI. 2010. Repeated evolution of crop theft in fungus-farming ambrosia beetles. Evolution 64:3205–3212. doi: 10.1111/j.1558-5646.2010.01055.x. [DOI] [PubMed] [Google Scholar]
- 67.De Fine Licht HH, Biedermann PH. 2012. Patterns of functional enzyme activity in fungus farming ambrosia beetles. Front Zool 9:13. doi: 10.1186/1742-9994-9-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bost A, Martinson VG, Franzenburg S, Adair KL, Albasi A, Wells MT, Douglas AE. 2018. Functional variation in the gut microbiome of wild Drosophila populations. Mol Ecol 27:2834–2845. doi: 10.1111/mec.14728. [DOI] [PubMed] [Google Scholar]
- 69.Li H, Durbin R. 2010. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 15:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Peng Y, Leung HCM, Yiu SM, Chin FYL. 2010. IDBA: a practical iterative de Bruijn graph de novo assembler, p 426–440. In Berger B. (ed), Research in computational molecular biology: RECOMB lecture notes in computer science, vol 6044 Springer, Berlin, Germany. [Google Scholar]
- 71.Peng Y, Leung HC, Yiu SM, Chin F. 2011. Meta-IDBA: a de Novo assembler for metagenomic data. Bioinformatics 27:i94–i101. doi: 10.1093/bioinformatics/btr216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li W, Godzik A. 2006. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22:1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- 73.Hyatt D, Chen GL, Locascio PF, Land ML, Larimer FW, Hauser LJ. 2010. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, Nielsen T, Pons N, Levenez F, Yamada T, Mende DR, Li J, Xu J, Li S, Li D, Cao J, Wang B, Liang H, Zheng H, Xie Y, Tap J, Lepage P, Bertalan M, Batto J-M, Hansen T, Le Paslier D, Linneberg A, Nielsen HB, Pelletier E, Renault P, Sicheritz-Ponten T, Turner K, Zhu H, Yu C, Li S, Jian M, Zhou Y, Li Y, Zhang X, Li S, Qin N, Yang H, Wang J, Brunak S, Doré J, Guarner F, Kristiansen K, Pedersen O, Parkhill J, Weissenbach J, Bork P, Ehrlich SD, Wang J. 2010. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Nordberg H, Cantor M, Dusheyko S, Hua S, Poliakov A, Shabalov I, Smirnova T, Grigoriev IV, Dubchak I. 2014. The genome portal of the Department of Energy Joint Genome Institute: 2014 updates. Nucleic Acids Res 42:D26–D31. doi: 10.1093/nar/gkt1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Grigoriev IV, Nordberg H, Shabalov I, Aerts A, Cantor M, Goodstein D, Kuo A, Minovitsky S, Nikitin R, Ohm RA, Otillar R, Poliakov A, Ratnere I, Riley R, Smirnova T, Rokhsar D, Dubchak I. 2012. The genome portal of the Department of Energy Joint Genome Institute. Nucleic Acids Res 40:D26–D31. doi: 10.1093/nar/gkr947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Huson DH, Mitra S, Ruscheweyh H-J, Weber N, Schuster SC. 2011. MEGAN analysis of metagenomic data. Genomic Res 21:1552–1560. doi: 10.1101/gr.120618.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gloor GB, Macklaim JM, Pawlowsky-Glahn V, Egozcue JJ. 2017. Microbiome datasets are compositional: and this is not optional. Front Microbiol 8:2224. doi: 10.3389/fmicb.2017.02224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.R Core Team. 2013. R: a language and environment for statistical computing. http://www.R-project.org/.
- 81.Yin Y, Mao X, Yang J, Chen X, Mao F, Xu Y. 2012. dbCAN: a web resource for automated carbohydrate-active enzyme annotation. Nucleic Acids Res 40:W445–W451. doi: 10.1093/nar/gks479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Eddy SR, Mitchison G, Durbin R. 1995. Maximum discrimination hidden Markov modules of sequence consensus. J Comp Biol 2:9–23. doi: 10.1089/cmb.1995.2.9. [DOI] [PubMed] [Google Scholar]
- 83.Marchler-Bauer A, Bo Y, Han L, He J, Lanczycki CJ, Lu S, Chitsaz F, Derbyshire MK, Geer RC, Gonzales NR, Gwadz M, Hurwitz DI, Lu F, Marchler GH, Song JS, Thanki N, Wang Z, Yamashita RA, Zhang D, Zheng C, Geer LY, Bryant SH. 2017. CDD/SPARCLE: functional classification of proteins via subfamily domain architectures. Nucleic Acids Res 45:D200–D203. doi: 10.1093/nar/gkw1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kanehisa M, Sato Y, Morishima K. 2016. BlastKOALA and GhostKOALA: KEGG tools for functional characterization of genome and metagenome sequences. J Mol Biol 428:726–731. doi: 10.1016/j.jmb.2015.11.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Bacterial community composition of metagenome contigs of gut metagenomes of nine termites with different diets at the phylum, class, order, family, and genus levels. Taxonomy classifications with more than 0.1% of the total abundance are shown. Mn, Macrotermes natalensis; Od, Odontotermes sp.; Nc, Nasutitermes corniger; Aw, Amitermes wheeleri; Mp, Microcerotermes parvus; Co, Cornitermes sp.; Th, Termes hospes; Nt, Neocapritermes taracua; Cu, Cubitermes ugandensis. Download Table S1, DOCX file, 0.03 MB (35.4KB, docx) .
Copyright © 2019 Hu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Relative abundance of glycoside hydrolase families in gut metagenomes of nine species of termites with different diets. Mn, Macrotermes natalensis; Od, Odontotermes sp.; Nc, Nasutitermes corniger; Aw, Amitermes wheeleri; Mp, Microcerotermes parvus; Co, Cornitermes sp.; Th, Termes hospes; Nt, Neocapritermes taracua; Cu, Cubitermes ugandensis. Download Table S2, DOCX file, 0.04 MB (44.2KB, docx) .
Copyright © 2019 Hu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Comparison of the taxonomic assignments of 16S rRNA studies (21, 22, 34) and metagenomic sequences (this study) at phylum (top) and genus (bottom) levels. Taxonomy classifications with more than 0.1% of the total abundance are shown. Taxonomy with no 16S rRNA gene references are marked as “NA.” Download Table S3, XLSX file, 0.02 MB (18.7KB, xlsx) .
Copyright © 2019 Hu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Data Availability Statement
Clean reads and metagenome assembly have been submitted to the SRA and GenBank under BioProject accession numbers PRJNA476694 and PRJNA193472.