Vivax malaria is the second leading cause of malaria worldwide and the major cause of non-African malaria. Unfortunately, efforts to develop antimalarial vaccines specifically targeting Plasmodium vivax have been largely neglected, and few candidates have progressed into clinical trials. The Duffy binding protein is considered a leading blood-stage vaccine candidate because this ligand’s recognition of the Duffy blood group reticulocyte surface receptor is considered essential for infection. This study identifies a new target epitope on the ligand’s surface that may serve as the target of vaccine-induced binding-inhibitory antibody (BIAb). Understanding the potential targets of vaccine protection will be important for development of an effective vaccine.
KEYWORDS: DBPII, Plasmodium vivax, epitope mapping, malaria, vaccine
ABSTRACT
The Plasmodium vivax Duffy binding protein region II (DBPII) is a vital ligand for the parasite’s invasion of reticulocytes, thereby making this molecule an attractive vaccine candidate against vivax malaria. However, strain-specific immunity due to DBPII allelic variation in Bc epitopes may complicate vaccine efficacy, suggesting that an effective DBPII vaccine needs to target conserved epitopes that are potential targets of strain-transcending neutralizing immunity. The minimal epitopes reactive with functionally inhibitory anti-DBPII monoclonal antibody (MAb) 3C9 and noninhibitory anti-DBPII MAb 3D10 were mapped using phage display expression libraries, since previous attempts to deduce the 3C9 epitope by cocrystallographic methods failed. Inhibitory MAb 3C9 binds to a conserved conformation-dependent epitope in subdomain 3, while noninhibitory MAb 3D10 binds to a linear epitope in subdomain 1 of DBPII, consistent with previous studies. Immunogenicity studies using synthetic linear peptides of the minimal epitopes determined that the 3C9 epitope, but not the 3D10 epitope, could induce functionally inhibitory anti-DBPII antibodies. Therefore, the highly conserved binding-inhibitory 3C9 epitope offers the potential as a component in a broadly inhibitory, strain-transcending DBP subunit vaccine.
IMPORTANCE Vivax malaria is the second leading cause of malaria worldwide and the major cause of non-African malaria. Unfortunately, efforts to develop antimalarial vaccines specifically targeting Plasmodium vivax have been largely neglected, and few candidates have progressed into clinical trials. The Duffy binding protein is considered a leading blood-stage vaccine candidate because this ligand’s recognition of the Duffy blood group reticulocyte surface receptor is considered essential for infection. This study identifies a new target epitope on the ligand’s surface that may serve as the target of vaccine-induced binding-inhibitory antibody (BIAb). Understanding the potential targets of vaccine protection will be important for development of an effective vaccine.
INTRODUCTION
Plasmodium vivax is the second most prevalent cause of human malaria and is the most widely distributed, placing about 40% of the world’s population at risk (1). Although vivax malaria has historically been called “benign tertian malaria” (2), there have been increasing reports of clinical severity with emerging virulent forms of the parasite (3, 4), recurrent clinical episodes due to reactivation of the dormant forms in the liver (5), and widespread drug resistance (6–8), which potentially includes strains with low sensitivity to primaquine, the only drug against relapse, or in people with poor ability to metabolize the drug to its active form (9, 10). Therefore, there is an urgent need to develop new therapies, especially a vaccine to control and prevent vivax malaria.
Immunity to asexual blood-stage antigens plays an important role in controlling P. vivax infections, especially targets important for blocking blood-stage replication or reticulocyte reinfection. Merozoite antigens are believed to be an important component of naturally acquired immunity to blood-stage vivax malaria (11, 12), and they represent ideal candidates for vaccine-mediated immunity against blood-stage infection (13). The P. vivax Duffy binding protein (DBP) binds its cognate receptor, the Duffy antigen receptor for chemokines (DARC), on reticulocytes to form an irreversible junction critical for merozoite invasion of reticulocytes (14–19). Even though there have been a few P. vivax cases reported recently among Duffy-negative individuals (20–22), this phenomenon remains relatively restricted in prevalence. A gene duplication event, creating a paralog of the DBP ligand lost in some primate-adapted parasite strains (21, 23), seems to be an alternate invasion pathway that is secondary to or less efficient than DBP (24). Hence, DBPII is a prime candidate for vaccine-induced immunity against blood-stage P. vivax infection.
Plasmodium vivax DBP is a type I membrane protein localized to the merozoite’s micronemes and is a member of the Duffy binding-like erythrocyte binding protein (DBL-EBP) family (18). The DBL-EBPs are micronemal proteins that have similar sequences in their functional domains, including an N-terminal cysteine-rich ligand domain referred to as the Duffy binding-like domain (DBL) (17, 18, 25–28). For DBP, this principal determinant for receptor recognition is a 330-amino-acid also known as DBP region II (DBPII) (18, 29). Interestingly, crystal structural studies of DBPII, as well as the ligand domains of its Plasmodium falciparum homolog PfEBA175 (P. falciparum EBA175), revealed that the DBL domains form heterodimers as part of the receptor recognition process (26–28). It is the central region of DBPII important for dimerization that is also the most polymorphic region of DBP (30–32), occurring in a pattern consistent with high immune selection (32–35). Also, similar to other microbial ligands, the functionally important residues in the central region critical for erythrocyte binding are flanked by polymorphic residues not important for binding to the receptor (27, 35, 36). Therefore, the polymorphic nature of the central region of DBPII represents a potential challenge to developing a vaccine, since this variation contributes to strain specificity in naturally acquired immunity (35, 37–39).
In regions where malaria is endemic, individuals develop anti-DBPII antibodies, which increase with age, suggesting a boosting effect from repeated exposure to infection (40, 41). However, anti-DBPII antibodies tend to be weak, short-lived, and strain specific (41–43) with allelic differences appearing to be driven by immune selection (35, 38). Nonetheless, a few individuals do develop high titers of broadly inhibitory, strain-transcending anti-DBPII antibodies (30, 39, 41). These data are all consistent with the hypothesis that variation is an immune evasion mechanism responsible for strain-specific immunity and that stable, broadly inhibitory immunity is achieved when functionally inhibitory antibodies target conserved DBPII epitopes.
To map the immunoreactive surface of DBPII, we generated and characterized a panel of anti-DBPII murine monoclonal antibodies (MAbs) with different anti-DBPII titers and levels of inhibition (44). Using the most and least inhibitory anti-DBPII MAbs of this panel, we sought to identify a potential target of broadly binding-inhibitory antibody (BIAb). An important advance was achieved when MAb epitope cocrystal structures were solved for several of the highly inhibitory MAbs (45). Unfortunately, a high resolution of the epitope structure could not be elucidated for MAb 3C9, which is one of the most inhibitory MAbs.
In this study, we mapped the minimal reactive epitopes of anti-DBPII MAb 3C9 by screening DBPII gene fragment libraries expressed on M13 phage surface for minimal reactive peptide fragments. Phage display has been a useful tool for epitope mapping with the advantages of rapidly producing refolded protein, fused to phage coat, forming a traceable link back to the genotype (46, 47), and has been used to map epitopes on P. falciparum apical membrane antigen 1 (AMA1) (48–51), merozoite surface protein (MSP) (52), and circumsporozoite protein (CSP) (53–55). Additional immunogenicity studies demonstrated the ability of the 3C9 epitope, but not the 3D10 epitope, to elicit functionally inhibitory anti-DBPII serum antibodies that blocked DBPII-erythrocyte binding. The information derived from this study contributes to our understanding of a potential target for broadly inhibitory vaccine-elicited protective immunity that should be retained in the next generation of potent vaccines against asexual-stage vivax malaria. In particular, this study defines a potential immunogenic epitope of a DBPII vaccine that may be successful in boosting antibody responses targeted against conserved protective epitopes, with functional inhibition against broader allelic variants and diverse P. vivax strains.
RESULTS
Creation of DBPII gene random fragment phage displayed library.
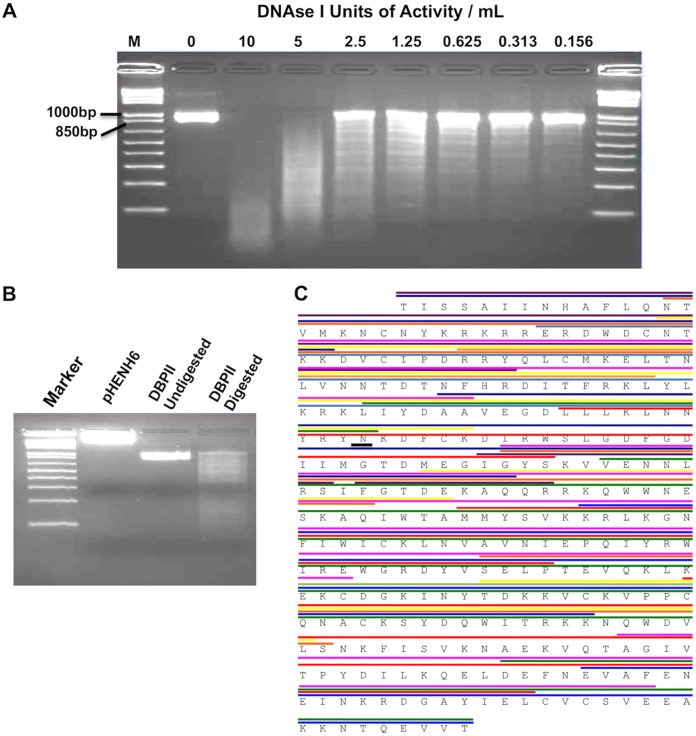
Variable-length DBPII gene fragments were created by digestion of DNA encoding the complete open reading frame of DBPII with DNase I (Fig. 1). First, a pilot experiment was used to determine the best possible concentration of the enzyme to obtain the broadest spread of the DBPII fragments (Fig. 1A), and 5 U/ml was determined to be the optimal concentration (Fig. 1B). The digested fragments were cloned into the pHENH6 vector and successfully used to express a large diversity of random-length fragments of DBPII. The DBPII fragments were expressed on the surface of the engineered phagemid as chimeric proteins fused with a C-terminal c-myc epitope tag acting as an expression marker and the pIII minor coat protein. The recombinant phagemid library for DBPII was estimated to have 7 × 105 PFU/μg of plasmid DNA gene fragments. Sequencing of 40 random clones from the library revealed that the fragments in the library spanned the entire coding sequence, and no bias toward any particular region was observed (Fig. 1C), although most of the fragments were ligated out of frame or in the incorrect orientation in accordance with random chance. Phage stocks generated from the gene fragment libraries were biopanned on the highly inhibitory MAb 3C9 and the poorly inhibitory MAb 3D10 to enrich for high-affinity binders to each of these antibodies.
FIG 1.
Generation of DBPII gene fragment library. DNA gels of DBPII Sal1 digested to make gene fragments. (A) The optimal DNase I concentration was evaluated to obtain the broadest spread of DBPII fragments. Lane M contains markers. (B) DBPII Sal1 fragments obtained by digestion with 5 U/ml of DNase I and blunt ended ligated into the pHENH6 phagemid vector and transformed into E. coli TG1. (C) Sequencing of PCR products generated by screening 40 individual clones revealed that the gene fragments in the library spanned the entire coding sequence, with no bias toward any particular region. The different colors represent the lengths of the various gene fragments identified.
Biopanning of DBPII phage displayed fragment library for minimal epitopes reactive with anti-DBPII MAbs.
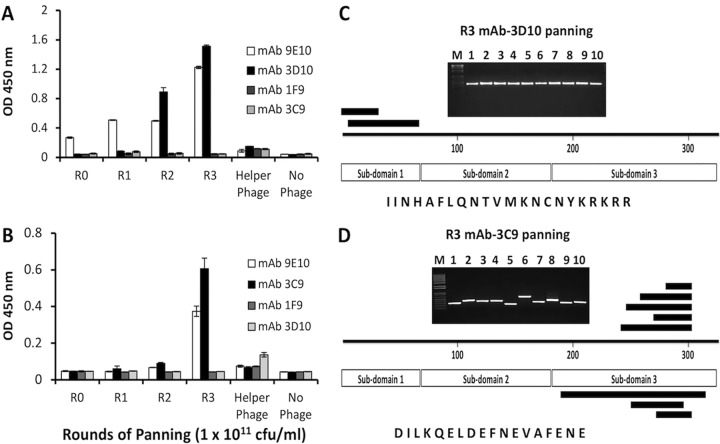
Affinity selection of reactive phage to each target antibody was conducted through sequential rounds of panning by standard procedures. An antibody to the c-myc epitope tag, MAb 9E10, was used in ELISAs as a control to standardize library titer in each round of panning, and an anti-PfAMA1 antibody, MAb 1F9, served as a negative control (Fig. 2), while MAb 3D10 was used as a control to validate the phage display as a platform for epitope identification. Panning on MAb 3D10 selected for phage clones that bound well to MAb 3D10, but poorly to MAbs 3C9 and 1F9 (Fig. 2A). Inversely, panning on MAb 3C9 selected for phage clones that bound well to MAb 3C9 and poorly to MAbs 3D10 and 1F9 (Fig. 2B). Phage clones isolated from panning on all the antibodies bound well to MAb 9E10, indicating that the procedure had enriched for clones possessing DBPII coding sequence in frame and in the correct orientation.
FIG 2.
DBPII gene fragments identified through biopanning. (A and B) ELISA reactivity of phage clones enriched by successive panning on MAb 3D10 (A) and MAb 3C9 (B). A pool of phage from each round of panning was tested for binding to anti-DBPII MAbs 3D10 and 3C9 and the anti-c-myc epitope tag antibody MAb 9E10. The PfAMA-1-specific MAb 1F9 served as a negative-control antibody. The bars represent mean ODs of triplicate wells, and error bars indicate the standard deviations (SD). Individual clones (n = 10) from round 3 (R3) of panning on each of the MAbs were PCR amplified and sequenced. The positions of the various peptides identified are indicated in panels C and D for MAbs 3D10 and 3C9, respectively. The common consensus sequence identified from the phage clones by panning on each of the antibodies is shown.
Sequence analysis of 10 clones from each round of panning was used to identify the length and specificity of DBPII CDS inserts. Clones from the last rounds of panning tended to have inserts of similar sizes, while those of the early rounds tended to be more variable. Sequencing results further showed that all clones in the last round of panning (R3) were in frame and in the right orientation, corroborating ELISA results. Ten clones from the last rounds of panning on MAb 3D10 contained only two different sequences comprising fragments of subdomain 1. These overlapping fragments, which identified the minimal 3D10-reactive epitope as IINHAFLQNTVMKNCNYKRKRR represented surface residues on subdomain 1 of DBPII (Fig. 2C) and overlapped the previously identified minimal epitope determined by X-ray crystallography (45). From the panning on MAb 3C9, eight unique sequence fragments were obtained comprising overlapping fragments of subdomain 3, with DILKQELDEFNEVAFENE as the minimal reactive epitope (Fig. 2D). This epitope is highly conserved in P. vivax field isolates.
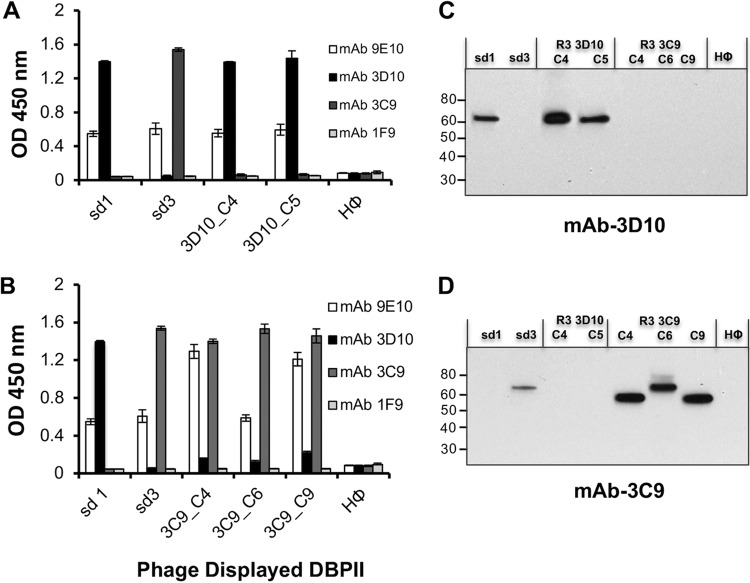
Two and three phage clones from the DBPII gene fragment library enriched during round 3 panning on MAbs 3D10 and 3C9, respectively, were cultured, and each clone was tested for reactivity with the selecting MAb by ELISA and Western blotting (Fig. 3). Results from both assays indicated that phage clones isolated from panning on MAb 3D10 reacted specifically with the 3D10 MAb (Fig. 3A and C). Similarly, MAb 3C9 phage clones were found to react specifically with MAb 3C9 (Fig. 3B and D). The observed specificity suggested that the isolated phage clones displayed the minimal fragments of DBPII needed for the MAbs to bind their respective epitopes.
FIG 3.
Cross reactivity of isolated phage clones with MAbs 3D10 and 3C9. (A to D) Two phage clones (clone 4 [C4] and C5) and three phage clones (C4, C6, and C9) from round 3 panning of the DBPII gene fragment library on MAbs 3D10 and 3C9, respectively, and phage clones expressing sd1 and sd3 fragments of DBPII were tested for cross-reactivity with the homologous and heterologous antibodies by ELISA (A and B) and immunoblot analysis (C and D). MAb 3D10 binds specifically to MAb 3D10-isolated phage clones and the sd1-expressing clones, while MAb 3C9 binds only to MAb 3C9-isolated clones and sd3-expressing clones. MAb 1F9 is a nonspecific anti-DBPII antibody used as a negative control, and MAb 9E10 is specific to the c-myc epitope of the phagemid. Each bar represents the mean OD450 for triplicate wells, and error bars represent SD.
To further characterize the linear epitope for MAb 3D10, a random 20-mer peptide library termed Adlib1 (Adalta Pty Ltd.) was also used to pan on the antibody. The level of reactivity was assessed by ELISA after three rounds of panning. Rounds 1 to 3 reacted well with MAb 3D10, while its reactivity remained relatively poor to the negative isotype control, anti-AMA1 MAb 5G8 (see Fig. S1A in the supplemental material). Ten clones from round 3 of panning were sequenced, and three different sequences (mimotopes) were identified, each containing what may be a degenerate sequence motif (Fig. S1B). A recurrent motif within the 3D10 mimotopes was a hydrophilic three-residue motif YK(R/Y/E). Although the Adlib1 peptide library is unrelated to DBPII, the YKR motif matches a similar sequence motif within the 22-amino-acid sequence in subdomain 1 selected by MAb 3D10 on the DBPII gene fragment library (Fig. 2C).
Panning a random peptide library on MAb 3D10. (A) ELISA showing reactivity of phage clones from rounds of panning on MAb 3D10. MAb 5G8 served as a negative-control antibody, while a MAb 5G8-positive binding phage clone served as a positive control (Pos C). The bars represent means for triplicate wells, while the error bars represent standard deviations (SD). (B) Alignment of a sequence of the MAb 3D10 binding epitope on DBPII sd1 from gene fragment library (top) and sequences of three mimotopes (M1, M2, and M3) from random peptide library with affinity for MAb 3D10. The residues in boldface type (underlined) show 3-amino-acid motifs common to the DBPII epitope and the mimotope sequences. Download FIG S1, TIF file, 0.3 MB (296KB, tif) .
Copyright © 2019 George et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Mutational analysis of the 3C9 epitope.
To confirm the epitope identified through panning on MAb 3C9, DBPII-Sal1 with deleted residues at the beginning, middle, and end of the epitope were evaluated for binding to MAbs 3C9 and 3D10. Deletion of the DILKQ residues at the beginning (mutant 1) and EFNE residues in the middle (mutant 2) of the epitope, respectively, showed about 25% and 50% reduction in binding of MAb 3C9, while deletion of the FENE residues (mutant 3) at the end of the epitope completely inhibits MAb 3C9 binding (Fig. 4). On the other hand, MAbs 3D10 bound all three mutants and the wild type. Similarly, two other DBP inhibitory MAbs (2D10 and 2H2), located upstream of the 3C9 epitope also bound to all mutants and the wild type.
FIG 4.
Binding of DBP monoclonal antibodies to DBPII-Sal1 mutants. (Left) Western blot analysis showing binding of MAbs 3D10, 3C9, 2D10, and 2H2 to Escherichia coli (BL21 star) expressing DBPII with mutations within the 3C9 epitope (left). Lanes: WT, wild type; M1, mutant 1; M2, mutant 2; M3, mutant 3; rSal1, purified rDBPII-Sal1. (Right) Sequences of a portion of subdomain 3 of the three mutants and the wild type are shown. The 3C9 epitope is underlined (solid line), 2D10 and 2H2 share an epitope (dotted line), and deleted residues within the 3C9 epitope are indicated by dashes.
Evaluation of immunogenicity of DBPII epitope peptides.
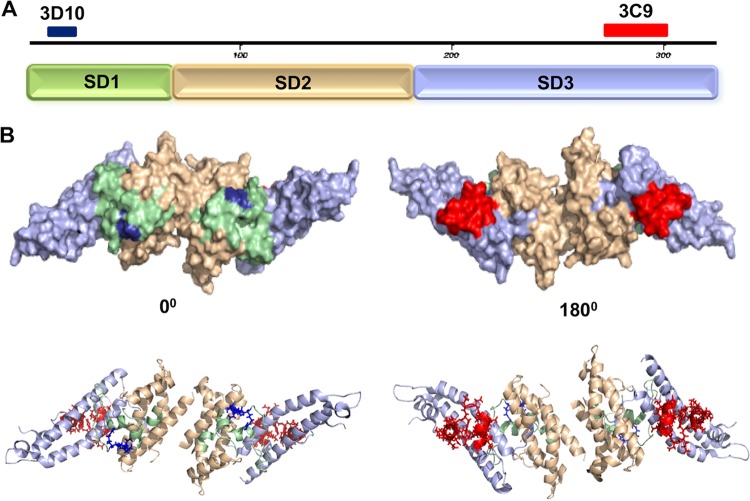
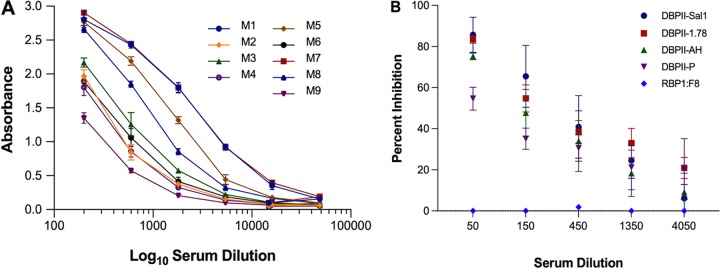
Both the 3D10 and 3C9 epitopes mapped to residues determined to be on the surface of the DBPII 3D crystal structure (Fig. 5). Since it appeared that the 3C9 epitope may retain its alpha helical secondary structure present in the native DBPII structure and 3D10 is at least partly a linear epitope, we sought to determine whether peptide immunogens of these minimal reactive epitopes could induce antibodies reactive with native DBPII. Particularly for the 3C9 epitope, we sought to determine whether the synthetic peptide of the epitope could retain sufficient native structure to elicit functional antibodies reactive to the native epitope and inhibitory against DBPII’s erythrocyte binding activity. The minimal peptides identified from panning with DBPII gene fragment library and the random peptide library were synthesized and conjugated to KLH carrier protein to immunize mice. ELISAs were used to determine reactivity of the antipeptide sera with the minimal peptide epitopes of MAbs 3D10 and 3C9. Serum from each mouse was tested for affinity to its homologous peptide. All peptides were immunogenic in mice and produced antibodies to the homologous peptides. Further, each antiserum was tested for affinity to rDBPII, and interestingly, mice immunized with 3C9 epitope peptide produced a measurable antibody response reactive with rDBPII (Table 1 and Fig. 6A).
FIG 5.
Putative epitopes of MAbs 3C9 and 3D10 mapped on 3D structure of DBPII. (A) The three subdomains (subdomain 1 [SD1] to SD3) are highlighted by color along with the putative epitopes of the inhibitory MAb 3C9 (red) on SD3 and the noninhibitory MAb 3D10 (blue) on SD1. (B) The 3D structure of DBPII is shown as the dimer required for DARC receptor recognition. The two views represent the front and back of the ligand for the surface model (top) and for secondary structure cartoon (bottom).
TABLE 1.
Epitopes identified through biopanning phage libraries on MAbs 3C9 and 3D10
| Epitope | Peptide sequencea | ELISA data |
|
|---|---|---|---|
| Peptide | rDBPII Sal1 | ||
| 3C9 | DILKQELDEFNEVAFENE | + | + |
| 3D10 | IINHAFLQNTVMKNCNYKRKRR | + | − |
| 3D10-M1b | VGNLDFSRFHKSSLDYKRGQ | + | − |
| 3D10-M2b | VKFTDRYKYSSMKGYARQGR | + | − |
| 3D10-M3b | KINMYKEVRTRQLSVRPSPE | + | − |
| Control | TPDERYRELDSHAQNESC | + | − |
Degenerate sequence is highlighted in boldface type.
Mimotopes identified by panning on random peptide library.
FIG 6.
Immunogenicity of 3C9 epitope peptide. (A) Mice immunized with the peptide corresponding to the 3C9 epitope-generated antibodies reactive with rDBPII by ELISA. Each curve represents antibody response from individual mice. Each point on the curve represents the mean OD for triplicate wells, and error bars indicate standard deviations (SD). (B) Pooled mouse sera were tested by endpoint dilution for inhibition of DBPII-erythrocyte binding against allelic DBPII variants by standard in vitro COS7 assay. Each curve on the chart represents the means from two independent experiments with each dilution tested in triplicate. Error bars represent SD.
Characterization of specificity and functional activity of anti-DBPII peptide serum antibodies.
To assess the anti-3C9 epitope peptide antibody activity, the peptide serum was evaluated for its potential to inhibit COS7 cell surface-expressed variant DBPII alleles (Sal1, P, 7.18, and AH) (see Table S1 in the supplemental material), from binding to DARC-positive erythrocytes. A concentration-dependent inhibition of DBPII-erythrocyte binding was observed (Fig. 6B).
DBPII variant alleles used in COS7 cell binding assay. Differences in amino acid residues between alleles compared to DBPII-Sal1 are indicated by letters, while similar residues are represented by periods. Download Table S1, PDF file, 0.1 MB (127.3KB, pdf) .
Copyright © 2019 George et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Statistical analysis using Dunn’s multiple-comparison analysis showed no significant differences in the overall inhibitory responses between the different alleles (P > 0.05). These results suggest that the 3C9 peptide produced a functional antibody response to this conserved region of DBPII within these alleles, although significant differences in inhibition were observed between the Sal1 and P alleles at high antibody concentrations (P = 0.03).
DISCUSSION
DBP is vital for P. vivax merozoite invasion of Duffy-positive reticulocytes, making it a leading vaccine candidate against blood-stage vivax malaria (14). As the prime representative member of the DBL-EBP (18, 25), DBP has helped define the functional and immunological properties of this important family of malarial ligands (17, 27, 29, 32). One of the biologically important conundrums of DBP is that the conserved structure of the N-terminal cysteine-rich DBL ligand domain essential for receptor recognition is under immune selective pressure driving allelic variation that alters the antigenic character of epitope targets of protective antibodies (26, 56). This variation is mainly constrained to polymorphisms in the DBL domain that exhibits some variation by geographic region (57–59). Some polymorphic residues on DBPII are unique to a certain geographical region, while others are common among global vivax alleles, like residues K371E, D384G, E385K, K386N, N417K, L424I, W437R, and I503K (57, 58, 60, 61). Generally, variation occurs in nonessential residues flanking residues critical for receptor recognition, and variation in some residues is more important than in others (26, 35). Overall evidence indicates that variation plays an important role in strain-specific immunity to P. vivax DBP.
In previous studies, we determined that one of the primary dominant Bc epitopes reactive with functionally inhibitory human immune sera contains some of the most polymorphic residues (27, 30), and this epitope was subsequently determined to be a major determinant of strain-specific immunity in DBPII (37, 62, 63). In a recent study, we established that a synthetic DBPII-based vaccine termed DEKnull-2, which lacked the polymorphic residues in the ligand domain of DBPII was able to induce broadly binding-inhibitory anti-DBPII antibodies, capable of blocking merozoite invasion of reticulocytes (64). This suggests that targeting immune response to conserved functional epitopes within DBPII, which are targets of broadly BIAb is key to overcoming the challenge of strain-specific immunity associated with a DBP vaccine. Using X-ray crystallography, we identified the epitopes of three inhibitory anti-DBPII MAbs (2D10, 2H2, and 2C6) and a noninhibitory MAb (3D10) (44, 45). In this study, we used a different platform (phage display) to identify the epitope of a broadly inhibitory anti-DBP MAb 3C9 (44) for which a high-resolution epitope structure could not be elucidated. This platform is a great tool for elucidating the molecular nature of protein-protein interactions (46–48) and was previously used to successively map functional epitopes on PfAMA1, a leading candidate for asexual-stage P. falciparum malaria vaccine (48), and we have established the methodology for DBPII. Recombinant filamentous phage was engineered to display DBPII-Sal1 on its surface as part of the pIII capsid protein. Selective panning of recombinant phage libraries on antibodies was used to isolate target epitopes or peptide mimics. Positive epitope-containing clones reactive with the anti-DBPII antibodies were enriched by successive panning assays. Importantly, panning on MAb 3D10 identified an epitope previously identified and confirmed by X-ray crystallography (45), located in subdomain 1, thus validating this platform.
The specificity of the epitope identified for MAb 3C9, which is located in subdomain 3, was confirmed by mutational analysis (Fig. 4). Essential residues for MAb 3C9 binding were located toward the end of the epitope. Binding of MAbs 3D10, 2D10, and 2H2 to all three mutants validates the mutants and suggest that the mutations within the 3C9 epitope did not compromise the structure and conformation of the rest of the protein domains. It is important to note that MAbs 2D10 and 2H2, also located in subdomain 3 share an epitope, located about 35 residues upstream of the 3C9 epitope (45).
A surprising outcome from this study and our previous study mapping the epitopes of inhibitory anti-DBPII MAb (45) is the discovery that Bc epitope targets of highly inhibitory anti-DBP are located in subdomain 3 away from the dimer interface and residues determined to be critical for erythrocyte binding. The subdomain 3 epitope identified by the 3C9 MAb is independent of the epitopes identified by X-ray crystallography, further validating that this subdomain of the DBP ligand is sensitive to antibody functional inhibition and may serve as a key target for vaccine-elicited antibody.
Our previous analyses indicated that most inhibitory antibodies to DBPII are dependent on 3D conformation and reactivity as well as functional immunogenicity are lost when DBPII is denatured (40, 44, 64). However, the alpha helical nature of the 3C9 epitope suggested that it may retain some of its native conformation in a synthetic peptide. Therefore, to further evaluate its potential as a subunit synthetic vaccine, peptides representing the 3C9 and 3D10 epitopes were characterized by vaccination studies. The immunogenicity of the “protective” 3C9 epitope was compared to the minimal peptide epitope of 3D10. Although 3D10 represented the highest titer MAb within the anti-DBP panel created, and reacted equally to all DBP alleles tested by ELISA, it has virtually no ability to block DBPII-erythrocyte binding (44).
Various studies have examined the functional efficacy of vaccine-induced anti-DBP antibodies to block DBP-erythrocyte binding or inhibition of merozoite invasion of erythrocytes (62, 65–67). As a general rule, properly refolded rDBPII capable of erythrocyte binding activity is required for induction of inhibitory anti-DBPII antibodies. Indeed, the epitope targets of the protective MAb 3C9 (conformational) and nonprotective MAb 3D10 (partially linear) are consistent with this observation. In our approach, the DBP peptide immunogens, consisting of the synthetic linear peptides of the 3C9 or 3D10 epitopes conjugated to carrier KLH, were used to immunize mice. The 3D10 immunogens included mimotope peptides of epitope targets isolated from a random peptide library. Surprisingly, we found that a 3C9 epitope peptide successfully induced an antibody response reactive to the immunizing peptide, the refolded protein, as well as inhibitory to DBPII-erythrocyte binding. This result suggests that the 3C9 linear peptide has some inherent structural tendency to assume its 3D conformation displayed on the surface of native DBP and is the target of inhibitory antibody. Although this epitope is conserved among currently available sequences from clinical isolates, in our original characterization of MAb 3C9 (44), its reactivity to different DBP allelic variants tested showed some degree of variability, indicating that the antibody’s specificity may be altered by conformational changes due to variation outside the targeted epitope.
Structure-based immunogen design for malaria vaccine development (structural vaccinology) using specific functional epitopes of antigens has been attracting much attention recently (68). The down side of this approach is the low immunogenicity of the peptide epitopes used. Generally, both B-cell and T-cell responses play a role in naturally acquired immunity to malaria. Thus, the immunogenicity and functional activity of epitopes such as the 3C9 epitope can be enhanced (i) by incorporating a universal T-helper epitope (such as the P2 tetanus toxoid-derived T-cell epitope) (69), (ii) by using adjuvants such as TLR agonists (TLR4 and TLR5) (70) for immunization, which have been successfully used to enhance immunogenicity of a B-cell epitope of PvMSP9 and the prophylactic L2-based HPV vaccine, respectively, or (iii) by packaging the epitope on viruses and virus-like particles (VLPs) by presenting the peptide as an immunogen at high densities on the surface of the viral particle which elicit stronger, and longer-lasting immune responses. This platform has been used as the basis for developing the human papillomavirus (HPV) vaccine (71).
Thus, conserved epitopes like the 3C9 epitope and other previously identified epitopes capable of eliciting protective antibodies (45), provide critical motifs that should be retained in a DBP-based subunit vaccine design against asexual-stage vivax malaria. This can be achieved by creating an epitope scaffold immunogen with flexible backbones (multiepitope peptide vaccine) through conjugation of the peptide epitopes in a single construct (72–74). Such a vaccine will elicit inhibitory antibodies specific for each conserved epitope and reduce immune evasion mechanisms associated with genetic variants in the DBPII gene.
MATERIALS AND METHODS
Generation of phage-displayed DBPII gene random fragment library.
The full-length PCR product (20 μg) from DBPII Sal1 haplotype was digested with DNase I at 5 U/ml. The resulting fragments were purified by sodium acetate precipitation, and ragged ends were blunted with a Vent polymerase (NEB). The pHENH6 vector was digested with PstI (NEB) and also blunted with Vent polymerase. The resulting products from the randomly fragmented DBPII were cloned into the treated pHENH6 vector and then into Escherichiacoli TG1 by electroporation. The distribution of the fragments in the library was determined by PCR and sequencing of 40 random clones. The library was then grown in 20 ml of 2× Tryptone-yeast extract broth containing 50 μg/ml of ampicillin at 37°C with shaking until an optical density at 600 nm (OD600) of 0.6 was attained. M13K07 helper phage (1 × 1011 PFU) was added and cultured for an extra 1 h at 37°C without shaking to allow infection. The entire culture was then transferred into 400 ml of super broth containing 70 μg/ml kanamycin and 50 μg/ml ampicillin and incubated at 37°C with shaking overnight. The bacteria were pelleted by centrifugation at 8,000 × g for 15 min, and the phage was precipitated from the supernatant with PEG-NaCl as described previously (75, 76). The phage was suspended in 1 ml of PBS and used subsequently or stored at −80°C.
Panning of phage-displayed DBPII gene fragment library.
Affinity panning of DBPII Sal1 gene fragment to anti-DBPII MAbs 3D10 and 3C9 was conducted as described previously (48). Briefly, 10 wells of Maxisorp-Nunc ELISA plate were coated overnight at 4°C with 100 μl per well of test MAb at a final concentration of 2.5 μg/ml diluted in PBS. The plates were washed twice with PBS, and unbound surfaces were blocked with 200 μl of 5% skim milk in PBS for 2 h at room temperature. After another round of washing, approximately 1 × 1011 phage/well in 1% skim milk in PBS were added to wells and incubated for 1 h at room temperature. Nonadherent phage particles were washed off twice with PBS/0.05% Tween 20. The bound phage was eluted with 100 μl of 0.1 M glycine (pH 2.2) and immediately added to 10 ml of log-phase E. coli TG1 cells and cultured for 30 min at 37°C for infection to occur. Ampicillin was added to 50 μg/ml, and the culture was incubated for another 30 min at 37°C before addition of the M13 helper phage and incubated for another 1 h at 37°C. The culture was then transferred to 200 ml super broth containing 50 μg/ml of ampicillin and 70 μg/ml of kanamycin and incubated at 37°C overnight with shaking at 200 rpm. The phage was harvested by centrifugation at 8,000 × g for 15 min, and another round of panning was conducted for a total of up to four panning cycles per antibody. Ten clones from each round of panning were selected, and their inserts were amplified by PCR and sequenced to determine the identity of the fragments of DBPII that bind to each MAb.
Panning a random peptide library on MAb 3D10.
Panning on MAb 3D10 was performed using a random 20-mer peptide library (courtesy of M. Foley and R. F. Anders, La Trobe University, Australia; Adalta Pty Ltd.) as described previously (48, 75). Briefly, phage peptide library at 1011 phage/well was incubated with MAb 3D10-coated wells for 1 h at room temperature. Eluted phage was allowed to reinfect E. coli K91 grown to log phase in 10 ml Tryptone-yeast extract medium at 37°C for 1 h and then grown overnight in 200 ml of super broth with 40 μg/ml of tetracycline. Phage was harvested, and further rounds of panning were conducted as described above. Clones isolated from round 3 were sequenced to determine the peptide sequences.
ELISA of phage displayed rounds of panning.
Enrichment ELISA was performed as described previously (48). Briefly, 100 μl of anti-DBP MAbs at 2.5 μg/ml in PBS was coated onto the wells of 96-well microtiter plates (Maxisorp-Nunc) overnight at 4°C. Anti-PfAMA1 specific MAb 1F9 or 5G8 (48) was coated on each plate as a negative-control antibody. The plates were washed twice with PBS 0.05% Tween 20 and blocked for 2 h at room temperature with 200 μl/well of 10% skim milk diluted in wash buffer. After three washes, phage diluted in PBS/1% milk to 1 × 1011 PFU/ml was added in triplicates and incubated for 2 h at room temperature. The plates were washed four times, and the wells were incubated with HRP-conjugated anti-M13 MAb (GE Life Sciences) for 1 h at room temperature. The plates were then washed five times, and bound phage was detected by incubating wells with 100 μl of H2O2-activated TMB substrate (Sigma). The reaction was stopped with 100 μl of 2 N H2SO4, and the plates were read at 450-nm absorbance.
Immunoblots.
Approximately 1 × 1012 phage particles were boiled in SDS-PAGE sample buffer for 3 min, separated by SDS-PAGE, and electrophoretically transferred onto a nitrocellulose membrane. The membrane was blocked overnight in 10% skim milk diluted in PBS/Tween 20 and probed with anti-DBPII MAbs. The membrane was then washed with PBS/Tween 20 before incubating with an HRP-conjugated anti-mouse secondary antibody. The bound antibody was detected by enhanced chemiluminescence (ECL) substrate (Amersham).
Mutational analysis.
With the DBPII-Sal1 strain as the template for mutagenesis, three different mutants were created within the 3C9 epitope by deleting the DILKQ, EFNE, and FENE residues at the beginning, middle, and end of the epitope, respectively, using the Q5 site-directed mutagenesis kit (NEB) according to the manufacturer’s protocol. Mutations were confirmed by sequencing. The mutants expressed in Escherichia coli BL21 Star (NEB) were screened by Western blotting for reactivity with MAbs 3C9, 3D10, and two other DBP inhibitory antibodies (MAbs 2D10 and 2H2), previously shown by cocrystallization studies to share an epitope (45).
Immunizations.
Peptides corresponding to sequences of the epitopes identified from panning on the gene fragment library (3C9, from DBPII subdomain 3; 3D10, from DBPII subdomain 1) and mimotopes from panning on the random peptide (3D10-M1, 3D10-M2, and 3D10-M3) (Table 1) were commercially synthesized (Pacific Immunology) and conjugated to keyhole limpet hemocyanin (KLH) using maleimide-activated mcKLH (ThermoScientific), following the manufacturer’s specifications. The conjugated peptides were used to immunize 6- to 8-week-old BALB/c mice (Harlan) to raise immune sera. All animals were handled in compliance with good animal practice, and all experimental procedures were performed in accordance with IACUC protocols approved by the Division of Research Integrity and Compliance, University of South Florida. Briefly, mice in all groups (n = 10) were bled for preimmune sera, and each mouse was immunized three times at 3-week intervals with 5 μg of KLH-conjugated peptide emulsified in Titermax Gold adjuvant. Each animal received a 50-μl antigen-adjuvant mix administered subcutaneously at the base of the tail. Mice immunized with KLH and adjuvant alone served as control. All mice were bled for final serum 4 weeks after the second boost.
Measurement of antibody titers.
Total anti-peptide IgG titers in the sera of each group was evaluated by endpoint titration ELISA against recombinant DBPII Sal1 and homologous peptide conjugated to bovine serum albumin (BSA). Imject Maleimide-Activated BSA Spin kit (ThermoScientific) was used as specified by the manufacturer for peptide conjugation. ELISAs were conducted as described previously (44), 0.1 μg of peptide conjugated to BSA or 0.25 μg of recombinant antigen was used to coat each well of a microtiter plate overnight and blocked with 5% skim milk PBS/0.05% Tween 20 detected by threefold dilution of mouse sera was used starting at 1:200. Bound antibodies were detected using alkaline phosphatase-conjugated anti-mouse antibody (Kirkegaard & Perry Laboratories). Preimmune serum, at the lowest dilution, served as background and was subtracted.
Measurement of functional inhibition of DBP-erythrocyte binding.
Pooled immune serum from mice that recognized rDBPII Sal1 by ELISA was tested further for inhibition of DBPII-erythrocyte binding by a modified version of the standard in vitro COS7 cell assay (29, 40). An expression plasmid, pEGFP-N1 (where EGFP is enhanced green fluorescent protein) (Clontech), was used to target variant DBPII alleles (Sal1, P, 7.18, and AH) (44, 62) (see Table S1 in the supplemental material) onto the surfaces of transfected COS7 cells as fusion proteins to the N terminus of EGFP. The expressed protein mimics the native protein on the surface of the parasite. Details of the assay and the different DBPII alleles were previously reported (62). COS7 cells transfected with plasmid expressing reticulocyte binding protein 1 (RBP1) and an empty plasmid were used as controls. Rosettes (COS7 cells with adherent erythrocytes) were determined by microscopy as positive when adherent erythrocytes covered at least 50% of the cell surface. The ability of the serum to inhibit binding of erythrocytes to DBPII expressed on the surfaces of transfected COS7 cells was determined by incubating transfected COS7 cells with triple fold dilutions of the serum prior to incubation with DARC-positive human erythrocytes. Binding inhibition was scored as the percentage of rosettes in wells of transfected COS7 cells in the presence of immune serum relative to wells incubated with preimmune serum (62). Differences in the inhibitory responses of the immune serum to the different alleles was compared by one-way analysis of variance (ANOVA) and multiple-comparison analysis by Tukey’s test using SAS software.
ACKNOWLEDGMENTS
This study was supported by National Institutes of Health grant R01AI064478 (J.H.A.).
We declare that we have no commercial or other association that poses a conflict of interest.
REFERENCES
- 1.Gething PW, Elyazar IR, Moyes CL, Smith DL, Battle KE, Guerra CA, Patil AP, Tatem AJ, Howes RE, Myers MF, George DB, Horby P, Wertheim HF, Price RN, Mueller I, Baird JK, Hay SI. 2012. A long neglected world malaria map: Plasmodium vivax endemicity in 2010. PLoS Negl Trop Dis 6:e1814. doi: 10.1371/journal.pntd.0001814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Garnham PC. 1951. Some effects on the community of malaria eradication with special reference to the relapse phenomenon. East Afr Med J 28:6–10. [PubMed] [Google Scholar]
- 3.Kochar DK, Das A, Kochar SK, Saxena V, Sirohi P, Garg S, Kochar A, Khatri MP, Gupta V. 2009. Severe Plasmodium vivax malaria: a report on serial cases from Bikaner in northwestern India. Am J Trop Med Hyg 80:194–198. doi: 10.4269/ajtmh.2009.80.194. [DOI] [PubMed] [Google Scholar]
- 4.Alexandre MA, Ferreira CO, Siqueira AM, Magalhaes BL, Mourao MP, Lacerda MV, Alecrim MG. 2010. Severe Plasmodium vivax malaria, Brazilian Amazon. Emerg Infect Dis 16:1611–1614. doi: 10.3201/eid1610.100685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Krotoski WA, Collins WE, Bray RS, Garnham PC, Cogswell FB, Gwadz RW, Killick-Kendrick R, Wolf R, Sinden R, Koontz LC, Stanfill PS. 1982. Demonstration of hypnozoites in sporozoite-transmitted Plasmodium vivax infection. Am J Trop Med Hyg 31:1291–1293. doi: 10.4269/ajtmh.1982.31.1291. [DOI] [PubMed] [Google Scholar]
- 6.Rieckmann KH, Davis DR, Hutton DC. 1989. Plasmodium vivax resistance to chloroquine? Lancet 2:1183–1184. [DOI] [PubMed] [Google Scholar]
- 7.Baird JK, Basri H, Purnomo Bangs MJ, Subianto B, Patchen LC, Hoffman SL. 1991. Resistance to chloroquine by Plasmodium vivax in Irian Jaya, Indonesia. Am J Trop Med Hyg 44:547–552. doi: 10.4269/ajtmh.1991.44.547. [DOI] [PubMed] [Google Scholar]
- 8.Price RN, von Seidlein L, Valecha N, Nosten F, Baird JK, White NJ. 2014. Global extent of chloroquine-resistant Plasmodium vivax: a systematic review and meta-analysis. Lancet Infect Dis 14:982–991. doi: 10.1016/S1473-3099(14)70855-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baird JK, Hoffman SL. 2004. Primaquine therapy for malaria. Clin Infect Dis 39:1336–1345. doi: 10.1086/424663. [DOI] [PubMed] [Google Scholar]
- 10.Bennett JW, Pybus BS, Yadava A, Tosh D, Sousa JC, McCarthy WF, Deye G, Melendez V, Ockenhouse CF. 2013. Primaquine failure and cytochrome P-450 2D6 in Plasmodium vivax malaria. N Engl J Med 369:1381–1382. doi: 10.1056/NEJMc1301936. [DOI] [PubMed] [Google Scholar]
- 11.Yazdani SS, Mukherjee P, Chauhan VS, Chitnis CE. 2006. Immune responses to asexual blood-stages of malaria parasites. Curr Mol Med 6:187–203. doi: 10.2174/156652406776055212. [DOI] [PubMed] [Google Scholar]
- 12.Marsh K, Kinyanjui S. 2006. Immune effector mechanisms in malaria. Parasite Immunol 28:51–60. doi: 10.1111/j.1365-3024.2006.00808.x. [DOI] [PubMed] [Google Scholar]
- 13.Mueller I, Galinski MR, Tsuboi T, Arevalo-Herrera M, Collins WE, King CL. 2013. Natural acquisition of immunity to Plasmodium vivax: epidemiological observations and potential targets. Adv Parasitol 81:77–131. doi: 10.1016/B978-0-12-407826-0.00003-5. [DOI] [PubMed] [Google Scholar]
- 14.Miller LH, Mason SJ, Clyde DF, McGinniss MH. 1976. The resistance factor to Plasmodium vivax in blacks. The Duffy-blood-group genotype, FyFy. N Engl J Med 295:302–304. doi: 10.1056/NEJM197608052950602. [DOI] [PubMed] [Google Scholar]
- 15.Wertheimer SP, Barnwell JW. 1989. Plasmodium vivax interaction with the human Duffy blood group glycoprotein: identification of a parasite receptor-like protein. Exp Parasitol 69:340–350. doi: 10.1016/0014-4894(89)90083-0. [DOI] [PubMed] [Google Scholar]
- 16.Miller LH, Mason SJ, Dvorak JA, McGinniss MH, Rothman IK. 1975. Erythrocyte receptors for (Plasmodium knowlesi) malaria: Duffy blood group determinants. Science 189:561–563. doi: 10.1126/science.1145213. [DOI] [PubMed] [Google Scholar]
- 17.Adams JH, Hudson DE, Torii M, Ward GE, Wellems TE, Aikawa M, Miller LH. 1990. The Duffy receptor family of Plasmodium knowlesi is located within the micronemes of invasive malaria merozoites. Cell 63:141–153. doi: 10.1016/0092-8674(90)90295-P. [DOI] [PubMed] [Google Scholar]
- 18.Adams JH, Sim BK, Dolan SA, Fang X, Kaslow DC, Miller LH. 1992. A family of erythrocyte binding proteins of malaria parasites. Proc Natl Acad Sci U S A 89:7085–7089. doi: 10.1073/pnas.89.15.7085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chitnis CE, Chaudhuri A, Horuk R, Pogo AO, Miller LH. 1996. The domain on the Duffy blood group antigen for binding Plasmodium vivax and P. knowlesi malarial parasites to erythrocytes. J Exp Med 184:1531–1536. doi: 10.1084/jem.184.4.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cavasini CE, Mattos LC, Couto AA, Bonini-Domingos CR, Valencia SH, Neiras WC, Alves RT, Rossit AR, Castilho L, Machado RL. 2007. Plasmodium vivax infection among Duffy antigen-negative individuals from the Brazilian Amazon region: an exception? Trans R Soc Trop Med Hyg 101:1042–1044. doi: 10.1016/j.trstmh.2007.04.011. [DOI] [PubMed] [Google Scholar]
- 21.Menard D, Chan ER, Benedet C, Ratsimbasoa A, Kim S, Chim P, Do C, Witkowski B, Durand R, Thellier M, Severini C, Legrand E, Musset L, Nour BY, Mercereau-Puijalon O, Serre D, Zimmerman PA. 2013. Whole genome sequencing of field isolates reveals a common duplication of the Duffy binding protein gene in Malagasy Plasmodium vivax strains. PLoS Negl Trop Dis 7:e2489. doi: 10.1371/journal.pntd.0002489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ryan JR, Stoute JA, Amon J, Dunton RF, Mtalib R, Koros J, Owour B, Luckhart S, Wirtz RA, Barnwell JW, Rosenberg R. 2006. Evidence for transmission of Plasmodium vivax among a duffy antigen negative population in Western Kenya. Am J Trop Med Hyg 75:575–581. doi: 10.4269/ajtmh.2006.75.575. [DOI] [PubMed] [Google Scholar]
- 23.Chan ER, Barnwell JW, Zimmerman PA, Serre D. 2015. Comparative analysis of field-isolate and monkey-adapted Plasmodium vivax genomes. PLoS Negl Trop Dis 9:e0003566. doi: 10.1371/journal.pntd.0003566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ntumngia FB, Thomson-Luque R, Torres Lde M, Gunalan K, Carvalho LH, Adams JH. 2016. A novel erythrocyte binding protein of Plasmodium vivax suggests an alternate invasion pathway into Duffy-positive reticulocytes. mBio 7:e01261-16. doi: 10.1128/mBio.01261-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Adams JH, Blair PL, Kaneko O, Peterson DS. 2001. An expanding ebl family of Plasmodium falciparum. Trends Parasitol 17:297–299. doi: 10.1016/S1471-4922(01)01948-1. [DOI] [PubMed] [Google Scholar]
- 26.Batchelor JD, Malpede BM, Omattage NS, DeKoster GT, Henzler-Wildman KA, Tolia NH. 2014. Red blood cell invasion by Plasmodium vivax: structural basis for DBP engagement of DARC. PLoS Pathog 10:e1003869. doi: 10.1371/journal.ppat.1003869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Batchelor JD, Zahm JA, Tolia NH. 2011. Dimerization of Plasmodium vivax DBP is induced upon receptor binding and drives recognition of DARC. Nat Struct Mol Biol 18:908–914. doi: 10.1038/nsmb.2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tolia NH, Enemark EJ, Sim BK, Joshua-Tor L. 2005. Structural basis for the EBA-175 erythrocyte invasion pathway of the malaria parasite Plasmodium falciparum. Cell 122:183–193. doi: 10.1016/j.cell.2005.05.033. [DOI] [PubMed] [Google Scholar]
- 29.Chitnis CE, Miller LH. 1994. Identification of the erythrocyte binding domains of Plasmodium vivax and Plasmodium knowlesi proteins involved in erythrocyte invasion. J Exp Med 180:497–506. doi: 10.1084/jem.180.2.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chootong P, Ntumngia FB, VanBuskirk KM, Xainli J, Cole-Tobian JL, Campbell CO, Fraser TS, King CL, Adams JH. 2010. Mapping epitopes of the Plasmodium vivax Duffy binding protein with naturally acquired inhibitory antibodies. Infect Immun 78:1089–1095. doi: 10.1128/IAI.01036-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ampudia E, Patarroyo MA, Patarroyo ME, Murillo LA. 1996. Genetic polymorphism of the Duffy receptor binding domain of Plasmodium vivax in Colombian wild isolates. Mol Biochem Parasitol 78:269–272. doi: 10.1016/S0166-6851(96)02611-4. [DOI] [PubMed] [Google Scholar]
- 32.Tsuboi T, Kappe SH, Al-Yaman F, Prickett MD, Alpers M, Adams JH. 1994. Natural variation within the principal adhesion domain of the Plasmodium vivax Duffy binding protein. Infect Immun 62:5581–5586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Baum J, Thomas AW, Conway DJ. 2003. Evidence for diversifying selection on erythrocyte-binding antigens of Plasmodium falciparum and P. vivax. Genetics 163:1327–1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baum J, Ward RH, Conway DJ. 2002. Natural selection on the erythrocyte surface. Mol Biol Evol 19:223–229. doi: 10.1093/oxfordjournals.molbev.a004075. [DOI] [PubMed] [Google Scholar]
- 35.VanBuskirk KM, Cole-Tobian JL, Baisor M, Sevova ES, Bockarie M, King CL, Adams JH. 2004. Antigenic drift in the ligand domain of Plasmodium vivax Duffy binding protein confers resistance to inhibitory antibodies. J Infect Dis 190:1556–1562. doi: 10.1086/424852. [DOI] [PubMed] [Google Scholar]
- 36.Xainli J, Adams JH, King CL. 2000. The erythrocyte binding motif of Plasmodium vivax duffy binding protein is highly polymorphic and functionally conserved in isolates from Papua New Guinea. Mol Biochem Parasitol 111:253–260. doi: 10.1016/S0166-6851(00)00315-7. [DOI] [PubMed] [Google Scholar]
- 37.Ntumngia FB, Adams JH. 2012. Design and immunogenicity of a novel synthetic antigen based on the ligand domain of the Plasmodium vivax Duffy binding protein. Clin Vaccine Immunol 19:30–36. doi: 10.1128/CVI.05466-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McHenry AM, Barnes SJ, Ntumngia FB, King CL, Adams JH. 2011. Determination of the molecular basis for a limited dimorphism, N417K, in the Plasmodium vivax Duffy-binding protein. PLoS One 6:e20192. doi: 10.1371/journal.pone.0020192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cole-Tobian JL, Michon P, Biasor M, Richards JS, Beeson JG, Mueller I, King CL. 2009. Strain-specific Duffy binding protein antibodies correlate with protection against infection with homologous compared to heterologous Plasmodium vivax strains in Papua New Guinean children. Infect Immun 77:4009–4017. doi: 10.1128/IAI.00158-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Michon P, Fraser T, Adams JH. 2000. Naturally acquired and vaccine-elicited antibodies block erythrocyte cytoadherence of the Plasmodium vivax Duffy binding protein. Infect Immun 68:3164–3171. doi: 10.1128/IAI.68.6.3164-3171.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.King CL, Michon P, Shakri AR, Marcotty A, Stanisic D, Zimmerman PA, Cole-Tobian JL, Mueller I, Chitnis CE. 2008. Naturally acquired Duffy-binding protein-specific binding inhibitory antibodies confer protection from blood-stage Plasmodium vivax infection. Proc Natl Acad Sci U S A 105:8363–8368. doi: 10.1073/pnas.0800371105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ceravolo IP, Sanchez BA, Sousa TN, Guerra BM, Soares IS, Braga EM, McHenry AM, Adams JH, Brito CF, Carvalho LH. 2009. Naturally acquired inhibitory antibodies to Plasmodium vivax Duffy binding protein are short-lived and allele-specific following a single malaria infection. Clin Exp Immunol 156:502–510. doi: 10.1111/j.1365-2249.2009.03931.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xainli J, Cole-Tobian JL, Baisor M, Kastens W, Bockarie M, Yazdani SS, Chitnis CE, Adams JH, King CL. 2003. Epitope-specific humoral immunity to Plasmodium vivax Duffy binding protein. Infect Immun 71:2508–2515. doi: 10.1128/IAI.71.5.2508-2515.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ntumngia FB, Schloegel J, Barnes SJ, McHenry AM, Singh S, King CL, Adams JH. 2012. Conserved and variant epitopes of Plasmodium vivax Duffy binding protein as targets of inhibitory monoclonal antibodies. Infect Immun 80:1203–1208. doi: 10.1128/IAI.05924-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen E, Salinas ND, Huang Y, Ntumngia F, Plasencia MD, Gross ML, Adams JH, Tolia NH. 2016. Broadly neutralizing epitopes in the Plasmodium vivax vaccine candidate Duffy binding protein. Proc Natl Acad Sci U S A 113:6277–6282. doi: 10.1073/pnas.1600488113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Smith GP. 1985. Filamentous fusion phage: novel expression vectors that display cloned antigens on the virion surface. Science 228:1315–1317. doi: 10.1126/science.4001944. [DOI] [PubMed] [Google Scholar]
- 47.Wilson DR, Finlay BB. 1998. Phage display: applications, innovations, and issues in phage and host biology. Can J Microbiol 44:313–329. doi: 10.1139/w98-015. [DOI] [PubMed] [Google Scholar]
- 48.Coley AM, Campanale NV, Casey JL, Hodder AN, Crewther PE, Anders RF, Tilley LM, Foley M. 2001. Rapid and precise epitope mapping of monoclonal antibodies against Plasmodium falciparum AMA1 by combined phage display of fragments and random peptides. Protein Eng 14:691–698. doi: 10.1093/protein/14.9.691. [DOI] [PubMed] [Google Scholar]
- 49.Sabo JK, Keizer DW, Feng Z-P, Casey JL, Parisi K, Coley AM, Foley M, Norton RS. 2007. Mimotopes of apical membrane antigen 1: structures of phage-derived peptides recognized by the inhibitory monoclonal antibody 4G2dc1 and design of a more active analogue. Infect Immun 75:61–73. doi: 10.1128/IAI.01041-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Casey JL, Coley AM, Anders RF, Murphy VJ, Humberstone KS, Thomas AW, Foley M. 2004. Antibodies to malaria peptide mimics inhibit Plasmodium falciparum invasion of erythrocytes. Infect Immun 72:1126–1134. doi: 10.1128/IAI.72.2.1126-1134.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Narum DL, Ogun SA, Batchelor AH, Holder AA. 2006. Passive immunization with a multicomponent vaccine against conserved domains of apical membrane antigen 1 and 235-kilodalton rhoptry proteins protects mice against Plasmodium yoelii blood-stage challenge infection. Infect Immun 74:5529–5536. doi: 10.1128/IAI.00573-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Demangel C, Lafaye P, Mazie JC. 1996. Reproducing the immune response against the Plasmodium vivax merozoite surface protein 1 with mimotopes selected from a phage-displayed peptide library. Mol Immunol 33:909–916. doi: 10.1016/S0161-5890(96)00058-2. [DOI] [PubMed] [Google Scholar]
- 53.Greenwood J, Willis AE, Perham RN. 1991. Multiple display of foreign peptides on a filamentous bacteriophage. Peptides from Plasmodium falciparum circumsporozoite protein as antigens. J Mol Biol 220:821–827. doi: 10.1016/0022-2836(91)90354-9. [DOI] [PubMed] [Google Scholar]
- 54.Willis AE, Perham RN, Wraith D. 1993. Immunological properties of foreign peptides in multiple display on a filamentous bacteriophage. Gene 128:79–83. doi: 10.1016/0378-1119(93)90156-W. [DOI] [PubMed] [Google Scholar]
- 55.Monette M, Opella SJ, Greenwood J, Willis AE, Perham RN. 2001. Structure of a malaria parasite antigenic determinant displayed on filamentous bacteriophage determined by NMR spectroscopy: implications for the structure of continuous peptide epitopes of proteins. Protein Sci 10:1150–1159. doi: 10.1110/ps.35901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ntumngia FB, King CL, Adams JH. 2012. Finding the sweet spots of inhibition: understanding the targets of a functional antibody against Plasmodium vivax Duffy binding protein. Int J Parasitol 42:1055–1062. doi: 10.1016/j.ijpara.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ju HL, Kang JM, Moon SU, Kim JY, Lee HW, Lin K, Sohn WM, Lee JS, Kim TS, Na BK. 2012. Genetic polymorphism and natural selection of Duffy binding protein of Plasmodium vivax Myanmar isolates. Malar J 11:60. doi: 10.1186/1475-2875-11-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hwang SY, Kim SH, Kho WG. 2009. Genetic characteristics of polymorphic antigenic markers among Korean isolates of Plasmodium vivax. Korean J Parasitol 47Suppl:S51–S58. doi: 10.3347/kjp.2009.47.S.S51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Premaratne PH, Aravinda BR, Escalante AA, Udagama PV. 2011. Genetic diversity of Plasmodium vivax Duffy binding protein II (PvDBPII) under unstable transmission and low intensity malaria in Sri Lanka. Infect Genet Evol 11:1327–1339. doi: 10.1016/j.meegid.2011.04.023. [DOI] [PubMed] [Google Scholar]
- 60.Babaeekho L, Zakeri S, Djadid ND. 2009. Genetic mapping of the duffy binding protein (DBP) ligand domain of Plasmodium vivax from unstable malaria region in the Middle East. Am J Trop Med Hyg 80:112–118. doi: 10.4269/ajtmh.2009.80.112. [DOI] [PubMed] [Google Scholar]
- 61.Ju HL, Kang JM, Moon SU, Bahk YY, Cho PY, Sohn WM, Park YK, Park JW, Kim TS, Na BK. 2013. Genetic diversity and natural selection of Duffy binding protein of Plasmodium vivax Korean isolates. Acta Trop 125:67–74. doi: 10.1016/j.actatropica.2012.09.016. [DOI] [PubMed] [Google Scholar]
- 62.Ntumngia FB, Schloegel J, McHenry AM, Barnes SJ, George MT, Kennedy S, Adams JH. 2013. Immunogenicity of single versus mixed allele vaccines of Plasmodium vivax Duffy binding protein region II. Vaccine 31:4382–4388. doi: 10.1016/j.vaccine.2013.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chen E, Salinas ND, Ntumngia FB, Adams JH, Tolia NH. 2015. Structural analysis of the synthetic Duffy binding protein (DBP) antigen DEKnull relevant for Plasmodium vivax malaria vaccine design. PLoS Negl Trop Dis 9:e0003644. doi: 10.1371/journal.pntd.0003644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ntumngia FB, Pires CV, Barnes SJ, George MT, Thomson-Luque R, Kano FS, Alves JRS, Urusova D, Pereira DB, Tolia NH, King CL, Carvalho LH, Adams JH. 2017. An engineered vaccine of the Plasmodium vivax Duffy binding protein enhances induction of broadly neutralizing antibodies. Sci Rep 7:13779. doi: 10.1038/s41598-017-13891-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Devi YS, Mukherjee P, Yazdani SS, Shakri AR, Mazumdar S, Pandey S, Chitnis CE, Chauhan VS. 2007. Immunogenicity of Plasmodium vivax combination subunit vaccine formulated with human compatible adjuvants in mice. Vaccine 25:5166–5174. doi: 10.1016/j.vaccine.2007.04.080. [DOI] [PubMed] [Google Scholar]
- 66.Arevalo-Herrera M, Castellanos A, Yazdani SS, Shakri AR, Chitnis CE, Dominik R, Herrera S. 2005. Immunogenicity and protective efficacy of recombinant vaccine based on the receptor-binding domain of the Plasmodium vivax Duffy binding protein in Aotus monkeys. Am J Trop Med Hyg 73:25–31. doi: 10.4269/ajtmh.2005.73.5_suppl.0730025. [DOI] [PubMed] [Google Scholar]
- 67.Grimberg BT, Udomsangpetch R, Xainli J, McHenry A, Panichakul T, Sattabongkot J, Cui L, Bockarie M, Chitnis C, Adams J, Zimmerman PA, King CL. 2007. Plasmodium vivax invasion of human erythrocytes inhibited by antibodies directed against the Duffy binding protein. PLoS Med 4:e337. doi: 10.1371/journal.pmed.0040337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Negahdaripour M, Golkar N, Hajighahramani N, Kianpour S, Nezafat N, Ghasemi Y. 2017. Harnessing self-assembled peptide nanoparticles in epitope vaccine design. Biotechnol Adv 35:575–596. doi: 10.1016/j.biotechadv.2017.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rodrigues-da-Silva RN, Correa-Moreira D, Soares IF, de-Luca PM, Totino PRR, Morgado FN, Oliveira Henriques MDG, Peixoto Candea AL, Singh B, Galinski MR, Moreno A, Oliveira-Ferreira J, Lima-Junior JDC. 2019. Immunogenicity of synthetic peptide constructs based on PvMSP9E795-A808, a linear B-cell epitope of the P. vivax merozoite surface protein-9. Vaccine 37:306–313. doi: 10.1016/j.vaccine.2018.10.016. [DOI] [PubMed] [Google Scholar]
- 70.Negahdaripour M, Eslami M, Nezafat N, Hajighahramani N, Ghoshoon MB, Shoolian E, Dehshahri A, Erfani N, Morowvat MH, Ghasemi Y. 2017. A novel HPV prophylactic peptide vaccine, designed by immunoinformatics and structural vaccinology approaches. Infect Genet Evol 54:402–416. doi: 10.1016/j.meegid.2017.08.002. [DOI] [PubMed] [Google Scholar]
- 71.Tumban E, Peabody J, Peabody DS, Chackerian B. 2011. A pan-HPV vaccine based on bacteriophage PP7 VLPs displaying broadly cross-neutralizing epitopes from the HPV minor capsid protein, L2. PLoS One 6:e23310. doi: 10.1371/journal.pone.0023310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Correia BE, Bates JT, Loomis RJ, Baneyx G, Carrico C, Jardine JG, Rupert P, Correnti C, Kalyuzhniy O, Vittal V, Connell MJ, Stevens E, Schroeter A, Chen M, Macpherson S, Serra AM, Adachi Y, Holmes MA, Li Y, Klevit RE, Graham BS, Wyatt RT, Baker D, Strong RK, Crowe JE, Johnson PR, Schief WR. 2014. Proof of principle for epitope-focused vaccine design. Nature 507:201–206. doi: 10.1038/nature12966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Patarroyo ME, Patarroyo MA. 2008. Emerging rules for subunit-based, multiantigenic, multistage chemically synthesized vaccines. Acc Chem Res 41:377–386. doi: 10.1021/ar700120t. [DOI] [PubMed] [Google Scholar]
- 74.Azoitei ML, Correia BE, Ban YE, Carrico C, Kalyuzhniy O, Chen L, Schroeter A, Huang PS, McLellan JS, Kwong PD, Baker D, Strong RK, Schief WR. 2011. Computation-guided backbone grafting of a discontinuous motif onto a protein scaffold. Science 334:373–376. doi: 10.1126/science.1209368. [DOI] [PubMed] [Google Scholar]
- 75.Adda CG, Tilley L, Anders RF, Foley M. 1999. Isolation of peptides that mimic epitopes on a malarial antigen from random peptide libraries displayed on phage. Infect Immun 67:4679–4688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Coley AM, Parisi K, Masciantonio R, Hoeck J, Casey JL, Murphy VJ, Harris KS, Batchelor AH, Anders RF, Foley M. 2006. The most polymorphic residue on Plasmodium falciparum apical membrane antigen 1 determines binding of an invasion-inhibitory antibody. Infect Immun 74:2628–2636. doi: 10.1128/IAI.74.5.2628-2636.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Panning a random peptide library on MAb 3D10. (A) ELISA showing reactivity of phage clones from rounds of panning on MAb 3D10. MAb 5G8 served as a negative-control antibody, while a MAb 5G8-positive binding phage clone served as a positive control (Pos C). The bars represent means for triplicate wells, while the error bars represent standard deviations (SD). (B) Alignment of a sequence of the MAb 3D10 binding epitope on DBPII sd1 from gene fragment library (top) and sequences of three mimotopes (M1, M2, and M3) from random peptide library with affinity for MAb 3D10. The residues in boldface type (underlined) show 3-amino-acid motifs common to the DBPII epitope and the mimotope sequences. Download FIG S1, TIF file, 0.3 MB (296KB, tif) .
Copyright © 2019 George et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
DBPII variant alleles used in COS7 cell binding assay. Differences in amino acid residues between alleles compared to DBPII-Sal1 are indicated by letters, while similar residues are represented by periods. Download Table S1, PDF file, 0.1 MB (127.3KB, pdf) .
Copyright © 2019 George et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.