Influenza A viruses are significant human pathogens that caused 83,000 deaths in the United States during 2017 to 2018, and there is need to understand the molecular correlates of illness and to identify prognostic markers of viral infection, symptom severity, and disease course. Preexisting antibodies against viral neuraminidase (NA) and hemagglutinin (HA) proteins play a critical role in lessening disease severity. We performed global gene expression profiling of peripheral blood leukocytes collected during acute and convalescent phases from a large cohort of people infected with A/H1N1pdm virus. Using statistical and machine-learning approaches, populations of genes were identified early in infection that correlated with active viral shedding, predicted length of shedding, or disease severity. Finally, these gene expression responses were differentially affected by increased levels of preexisting influenza antibodies, which could mask detection of these markers of contagiousness and disease severity in people with active clinical disease.
KEYWORDS: HA head antibodies, HA stalk antibodies, NA antibodies, antibody function, genomics, host response, human challenge, immune markers, immune response, influenza, microarrays, regulation of gene expression
ABSTRACT
In this study, we examined the relationships between anti-influenza virus serum antibody titers, clinical disease, and peripheral blood leukocyte (PBL) global gene expression during presymptomatic, acute, and convalescent illness in 83 participants infected with 2009 pandemic H1N1 virus in a human influenza challenge model. Using traditional statistical and logistic regression modeling approaches, profiles of differentially expressed genes that correlated with active viral shedding, predicted length of viral shedding, and predicted illness severity were identified. These analyses further demonstrated that challenge participants fell into three peripheral blood leukocyte gene expression phenotypes that significantly correlated with different clinical outcomes and prechallenge serum titers of antibodies specific for the viral neuraminidase, hemagglutinin head, and hemagglutinin stalk. Higher prechallenge serum antibody titers were inversely correlated with leukocyte responsiveness in participants with active disease and could mask expression of peripheral blood markers of clinical disease in some participants, including viral shedding and symptom severity. Consequently, preexisting anti-influenza antibodies may modulate PBL gene expression, and this must be taken into consideration in the development and interpretation of peripheral blood diagnostic and prognostic assays of influenza infection.
INTRODUCTION
Influenza A viruses (IAV) are a significant cause of morbidity and mortality. Annual estimates of death from seasonal IAV range up to 79,400 in the United States and 250,000 to 500,000 in industrial nations (1, 2). The 2009 H1N1 pandemic resulted in at least 200,000 deaths globally, while the 1918 H1N1 pandemic resulted in 50 to 100 million deaths (3–5). IAV infection in humans is typically an acute, self-limited disease associated with virus replication in the upper respiratory tract, which may progress to lower respiratory tract involvement in more severe cases, including secondary bacterial infections and pneumonia (6). Influenza illness typically presents with mild to moderate symptoms, including rhinorrhea, dry cough, malaise, headache, and fever (6). The onset of symptoms typically occurs after viral shedding begins; therefore, an infected person can be contagious for 1 to 2 days prior to symptom onset. This creates a significant challenge in limiting community spread using standard post-symptom onset diagnostic testing. In addition, complicated disease typically occurs after the initial onset of mild to moderate symptoms, making the task of early prediction and triage of patients who will progress to severe disease difficult. Identifying pre- or early symptomatic markers of infection, contagiousness, and disease severity is key to reducing morbidity and mortality by offering new tools to reduce the spread of disease through early detection and isolation of contagious individuals and early recognition of patients needing treatment or monitoring for severe disease and complication.
A critical host determinant affecting infection, shedding, and disease severity is preexisting influenza immunity, including both cellular immunity and anti-IAV antibodies arising from previous viral infection and/or vaccination (7–14). While cellular immunity and memory lymphocyte responses play essential roles in immunity to IAV, most studies of protection have focused on serum antibodies against IAV proteins. The most dominant IAV antigens are the surface glycoproteins hemagglutinin (HA) and neuraminidase (NA) (1). Many neutralizing antibodies bind to the head of the HA protein, which are typically measured using an HA inhibition (HAI) assay. For IAV vaccine development, a serum HAI antibody titer of 1:40 is the internationally accepted standard of protection and was demonstrated to protect 50% of individuals from IAV infection (15, 16). Recently, the role of non-HA head immunity has gained much interest, including antibodies against the IAV NA protein and a more conserved stalk (stem) region of the HA protein (17, 18). Previous studies using a human influenza A viral challenge model have shown the relative importance of these three types of antibody responses, and while antibodies against NA, HA head, and HA stalk are all correlated with a reduction in length of shedding, only prechallenge antibodies against NA were independently correlated with both a decreased length of shedding and symptom severity (11, 12).
This study was undertaken to examine the relationship between peripheral immune cell gene expression, clinical disease, and preexisting NA and HA serum antibody titer. Global gene expression profiles of peripheral blood leukocytes (PBL) from 83 human challenge study participants collected at 18 h, 42 h, 90 h, and 138 h after intranasal inoculation with A/H1N1pdm IAV were compared with viral shedding, symptom duration, symptom severity, and preexisting antibody levels. Using this approach, we were able to identify three distinct expression phenotypes as early as 42 h postinoculation and identify differentially expressed gene signatures at the pre- or early symptomatic phase that were predictive of shedding duration and clinical disease severity. This study shows the importance of preexisting antibodies against viral NA (NAI), HA head (HAI), and HA stalk (HA-S) in modulating PBL gene expression responses during human influenza viral challenge and identifies populations of genes that are predictive for disease severity and length of upper respiratory tract viral shedding.
RESULTS
Human influenza viral challenge model and participant cohort.
PBLs were collected for gene expression microarray analysis from 83 healthy volunteers inoculated with 107 TCID50 of a GMP-manufactured wild-type 2009 pandemic H1N1 IAV (A/H1N1pdm) in an established influenza viral challenge model (19). Previously published study participant demographics, clinical data, and baseline NAI, HAI, and HA stalk antibody titers are summarized in Table 1. A clinical study outcome denoted “mild-to-moderate influenza disease” (MMID) (19), was used and defined as viral shedding detected by clinical molecular testing, in addition to the onset of at least one acute influenza-like illness symptom following intranasal challenge. The No MMID participants either had symptoms in the absence of viral shedding, asymptomatic shedding, or neither symptoms nor shedding (Table 1).
TABLE 1.
Challenge participant demographics, clinical results, and preexisting antibody titers
| Characteristica | Value for characteristic | ||
|---|---|---|---|
| Mean age, yr (range) | 27 (18–47) | ||
| Sex | Male | Female | |
| No. of participants (%) | 44 (53.0) | 39 (47.0) | |
| Race | Asian | Black | White |
| No. of participants (%) | 5 (6.0) | 35 (41.7) | 43 (51.1) |
| Group | No MMID | MMID | |
| No. of participants | 39 | 44 | |
| Mean max symptom score (range) | 1.8 (0–7) | 5.6 (1–14) | |
| Mean no. of days of symptoms (range) | 3.6 (0–34) | 6.8 (2–11) | |
| Mean no. of days of shedding (range) | 0.15 (0–2) | 3.7 (1–9) | |
| Pre NAI titers (GMT) | 258.5 | 47.7 | |
| Pre HAI titers (GMT) | 22.4 | 4.2 | |
| Pre HA stalk titers (mean) | 66,227 | 30,430 | |
| Quartile | Lower quartile (Q1) | Median quartile (Q2) | Upper quartile (Q3) |
| Max symptom score | 1 | 3 | 6 |
| No. of days of symptoms | 3 | 5 | 7.75 |
| No. of days of shedding | 0 | 1 | 4 |
| Pre NAI titers (GMT) | 80 | 160 | 320 |
| Pre HAI titers (GMT) | 0 | 10 | 80 |
| Pre HA stalk titers (mean) | 23,605.5 | 52,014.5 | 82,366.5 |
max, maximum; Pre, preexisting (baseline).
Baseline PBL gene expression did not correlate with differential PBL gene expression responses or clinical outcome following challenge.
Expression microarrays were used to measure PBL global gene expression responses prior to viral challenge (0 h) and at multiple time points 18 h, 42 h, 90 h, and 138 h during the acute phase of IAV infection. Analysis of global preinfection gene expression profiles did not show significant differentiation of participants based on study outcome (MMID or No MMID), severity of illness (measured as maximum clinical symptom score), or length of viral shedding (see Fig. S1A to C in the supplemental material). Baseline expression levels of canonical antiviral response genes (20) did not show statistically significant differences between MMID and No MMID participants (n = 42) (Fig. S1D). Indeed, comparison of global profiles of preexposure samples failed to identify statistically significant gene expression differences between MMID and No MMID participants.
Baseline PBL gene expression does not predict participant outcome following IAV challenge. (A to C) Principal-component analysis of global day 0 gene expression profiles showing general outcome (A), illness severity (B), and days of shedding (C). (A) For general outcome, No MMID is shown in blue and MMID is shown in red. (B) Illness severity as measured by maximum symptom score (indicated using increasing green color gradient). (C) Days of shedding indicated using pink gradient. (D) Baseline expression levels of core antiviral response genes in MMID and No MMID participants. Results are shown as medians and standard deviations of intensity expression levels. Download FIG S1, TIF file, 5.4 MB (5.5MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Early gene expression changes correlated with active viral shedding.
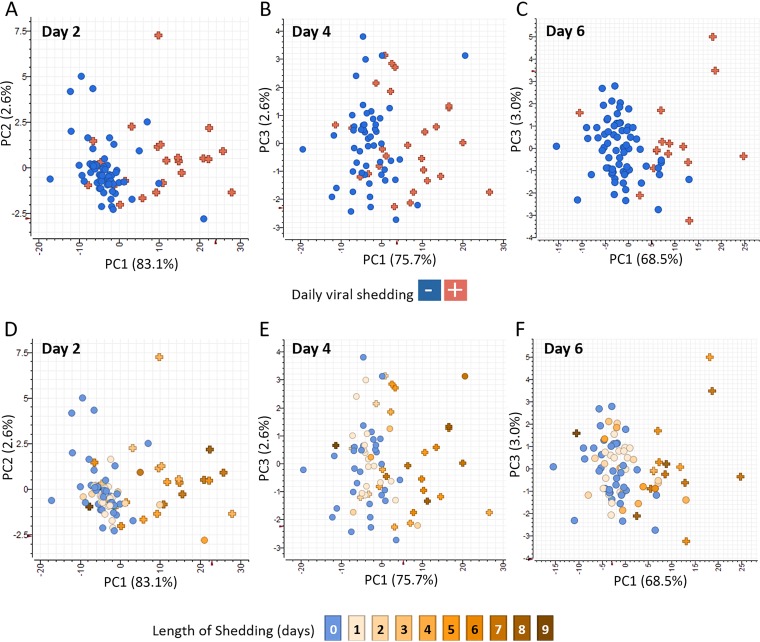
Expression values from each participant’s PBL sample collected postinfection (18 h, 42 h, 90 h, and 138 h) were normalized to their own preexposure time point (0 h). To identify genes associated with active viral replication and shedding but independent of clinical parameters, samples from participants that were positive for viral shedding in nasal washes were compared by two-tailed t test to samples from participants without evidence of shedding (see Tables S1 to S3 at https://systemsbiology.org/wp-content/uploads/Walters-et-al-Supplemental-Tables-S1-S9.xlsx). No genes were identified as significantly differentially expressed between shedders and nonshedders at 18 h postinfection. However, 151 genes were identified as significantly differentially expressed (two-fold difference in median expression value, P value ≤ 0.05) between shedders and nonshedders in at least one time point during the acute phase (42 h, 90 h, and 138 h postinfection). These genes include molecular pattern and type I IFN response-related genes (IFI27, TLR7, IFIT1-3, OASL, ISG15, IFI44, and OAS1-3), in addition to early TNFR responses, such as TRAIL, observed at 42 h. The expression profiles of genes correlated with viral shedding at 18 h, 42 h, 90 h, and 138 h are shown in Fig. S2A. Most viral shedding-associated genes were increased relative to preexposure levels (Fig. S2A). Consistent with a lack of significant differential expression at 18 h, a viral shedding gene expression signature was not observed at this time point in shedding participants. However, by 42 h postinfection, most shedders showed various levels of increased expression of viral shedding-associated genes relative to the nonshedders, and principal-component analysis using the 151 genes clearly showed participants grouping based on viral shedding status (Fig. 1A). A subset of shedders (n = 7) showed either very attenuated or no induction of viral shedding-associated gene expression signature despite shedding virus for between 4 and 9 days, while other participants (n = 4) not shedding virus at day 2 showed increased expression of viral shedding-associated genes at 42 h. Most of the nonshedding participants showing increased expression of viral shedding-associated genes shed virus either on day 1 or later time points, indicating that in some participants, the viral shedding-associated gene expression signature is detected prior to or after viral shedding even in the absence of symptoms (Fig. 1D).
FIG 1.
Viral shedding-associated PBL gene expression signature can be detected before and after detection of virus in nasal secretions. Principal-component analysis (PCA) of active viral shedding-associated gene expression identified by two-tailed t test comparisons of transcripts whose expression levels differed significantly (≥2-fold difference in median expression level, P value < 0.05) in PBLs from participants with influenza virus-positive versus -negative nasal washes at 42 h, 90 h, and 138 h after IAV challenge. Statistical tests were performed separately for each time point and identified 151 genes that were significant in at least one time point. (A to C) PCA of viral shedding-associated gene expression at 42 h, 90 h, and 138 h postinfection showing viral shedding status at each time point, respectively. Each participant is shown as a separate symbol with blue circles indicating participants not actively shedding virus and red plus symbols indicate individuals with virus-positive nasal washes on that day. (D to F) Longitudinal viral shedding. PCA of viral shedding-associated gene expression at 42 h, 90 h, and 138 h postinfection, respectively, showing duration of shedding during the course of the study with the total number of days of viral shedding for each participant indicated using a brown gradient with never shedding participants shown in blue.
Kinetics of viral shedding response during acute IAV infection in shedding outlier participants. (A) Expression profiles of viral shedding-associated genes (n = 151) in all participants (n = 83) during the acute phase of infection. Participant samples at each time point for active shedding IAV in nasal washes are indicated by red bars and samples negative for IAV are indicated by blue bars. (B) Expression of viral shedding-associated genes in subset of participants that show delayed or absent viral shedding-associated genes. Genes shown in red exhibited increased expression and genes shown in green exhibited decreased expression relative to each participant’s day 0 expression. Download FIG S2, TIF file, 5.4 MB (5.5MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Analysis of viral shedding-associated gene expression at 90 h and 138 h showed similar trends at each of these time points. Most shedders showed increased expression, and principal-component analysis demonstrated grouping of participants based on shedding status (Fig. 1B and C). However, participant subsets with discordant gene expression were again observed at each time point in both the shedding and nonshedding populations. In general, the participants without detectable viral shedding on day 4 that exhibited increased expression of viral shedding-associated genes shed virus either before or after the time point when increased expression of viral shedding-associated genes was observed. This was particularly evident at 138 h where many participants who were no longer shedding on day 6 still exhibited increased expression of viral shedding-associated genes at 138 h, likely due to residual effects from active viral shedding detected on prior days (Fig. S2A). Participants with active viral shedding who did not show induction of viral shedding-associated genes typically lacked expression of viral shedding-associated genes only on a single day when viral shedding was detected (Fig. S2B) but showed increased expression at later time points, suggesting a delayed host response to infection. However, several participants showed no induction of viral shedding associated at any time point during the acute phase of infection despite shedding for multiple days (Fig. S2B).
Biological pathway classification of viral shedding-associated genes at 42 h (n = 151), 90 h (n = 56), and 138 h (n = 87) was performed using the Panther gene ontology overrepresentation test (21, 22). Pathway activation was dominated by antiviral responses and stress responses, including interferon alpha/beta signaling, and cytokine signaling (see Tables S1 to S3 at https://systemsbiology.org/wp-content/uploads/Walters-et-al-Supplemental-Tables-S1-S9.xlsx). Responses limited to only 42 h postinfection included TRAF3-dependent IRF activation, NF-κB activation through the FADD/RIP-1 pathway, TRAF6-mediated IRF7 and NF-κB activation, as well as RIG-I/MDA5 activation (see Table S4 at the above URL). Collectively, these data demonstrate that a PBL viral shedding-associated gene signature was present in most participants at time points where IAV was present in nasal washes and in some participants prior to or following viral shedding (Fig. 1D to F). However, there was a small subset of participants who failed to initiate a host response despite multiple days of active viral replication.
PBL viral shedding-associated gene expression phenotype correlated with clinical symptoms, length of viral shedding, and preexisting antibody titers.
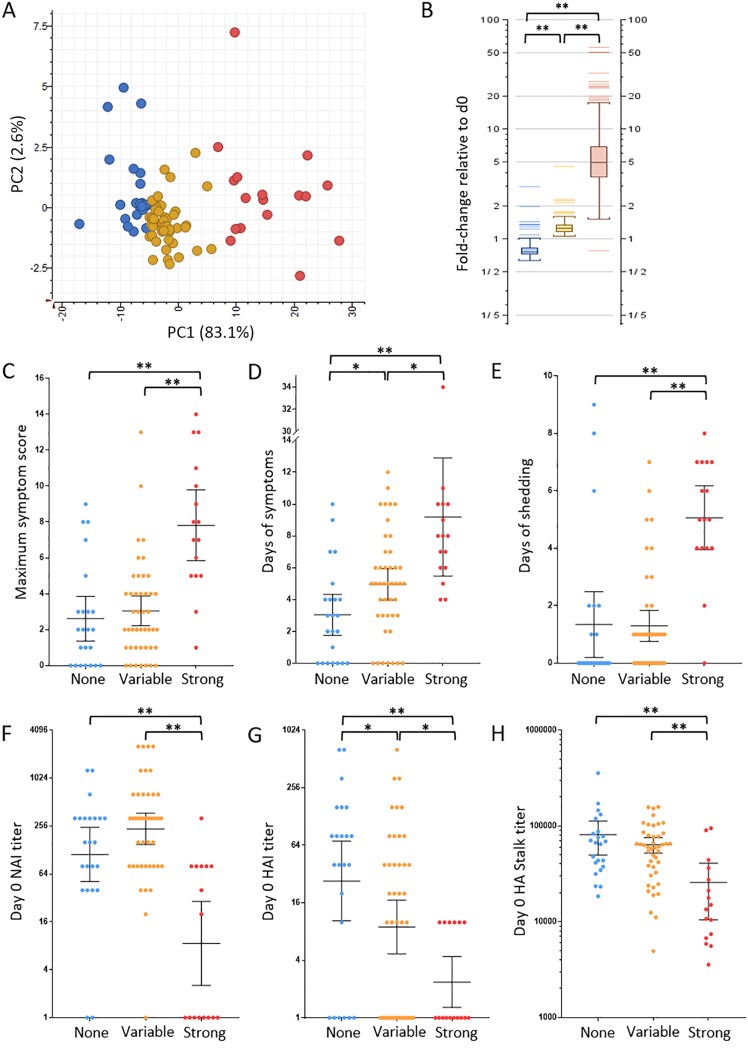
To identify the factors driving phenotypic gene expression variation, unsupervised clustering was used to group participants based on their PBL viral shedding associated gene expression response independent of clinical parameters. At each time point during acute infection, k-means clustering analysis identified three main subgroups of participants based on the magnitude of their PBL viral shedding-associated gene expression responses: (i) strong; (ii) variable, and (iii) no response (Fig. S3). As shown in Fig. 2A, principal-component analysis was used to visualize participant grouping based on viral shedding-associated gene expression at 42 h, and average expression levels of viral shedding-associated genes were significantly different between the strong, variable, and no response k-means phenotypes (Fig. 2B). Statistical analysis by rank sum t test of clinical illness metrics, including maximum symptom score, number of days of symptoms, and duration of viral shedding was performed to determine whether the three participant subgroups correlated with clinical outcome. The strong response population at 42 h had significantly more severe illness and longer duration of illness (median maximum symptom score = 7.5; median = 8 days symptoms) compared to those with either variable (median maximum symptom score = 2; median = 5 days symptoms) or no response (median maximum symptom score = 2; median = 3 days of symptoms) (Fig. 2C and D). Participants with a strong PBL viral shedding-associated gene expression response also shed virus for longer (median = 5 days) than those with either variable (median shedding = 1 day) or no response (median shedding = 0 days) (Fig. 2E). Analysis of participant groups at 90 h and 138 h revealed similar statistically significant differences in clinical disease (Fig. S4). Some participants with variable response at 42 h also had significantly longer illness (median = 5 days) compared to those with no response (median = 3 days) but were not significantly different in terms of severity of illness or duration of shedding. No significant differences in clinical metrics were observed between the variable and no response groups at 90 h and 138 h postinfection (Fig. S4).
FIG 2.
Differential PBL expression of viral shedding-associated gene expression at 42 h correlates with clinical symptoms, length of viral shedding, and preexisting immunity. (A) Principal-component analysis of the three PBL viral shedding-associated gene expression response groups at 42 h postinfection identified using k-means analysis. The three PBL viral shedding-associated gene expression response groups are no response (blue), variable response (yellow), and strong response (red). (B) Graph of expression response group average expression of genes associated with viral shedding at 42 h relative to day 0 (d0). (C to E) Statistical analysis of clinical illness, including maximum symptom score (C), days of symptoms (D), and days of viral shedding (E) between the three PBL response groups. (F to H) Statistical analysis of preexisting antibodies between PBL response groups as measured by day 0 NAI titer (F), HAI titer (G), and HA stalk ELISA titer (H). For NAI and HAI titers, the geometric mean titer (GMT) is shown with 95% confidence intervals, while HA stalk titer is shown as the median titer with 95% confidence intervals. Values that were statistically significant different for the different groups were assessed using rank sum t test and indicated by a bar and asterisk as follows: *, P ≤ 0.05; **, P < 0.01.
k-means analysis of viral shedding-associated genes identifies three main participant response groups. Expression profiles of viral shedding (VS)-associated genes (n = 151) at 42 h, 90 h, and 138 h demonstrating three general response groups: strong, variable, and none. Genes shown in red exhibited increased expression and genes shown in green exhibited decreased expression relative to each participant’s day 0 expression. Download FIG S3, TIF file, 5.4 MB (5.5MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Statistical analysis of clinical parameters between three viral shedding-associated gene expression response groups at 90 h and 138 h postinfection. Statistical significance between groups was assessed using rank sum t test and indicated by asterisks as follows: *, P ≤ 0.05; **, P < 0.01. Download FIG S4, TIF file, 5.4 MB (5.5MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
To examine the relationships between preexisting serum NAI, HAI, and HA stalk titer and PBL gene expression responses, statistical analysis by rank sum t test of each participant’s day 0 levels of serum antibodies directed against the viral NA by inhibition titer (NAI), the HA head by inhibition titer (HAI), and the HA stalk domain by ELISA was performed. Analysis of preexisting NAI titers showed that the participants in the 42 h viral shedding-associated gene expression strong response group had a statistically significant lower geometric mean titer (GMT) of 8.57 compared to the variable and no response groups, with NAI GMTs of 236 and 112.6, respectively (Fig. 2F). No statistical difference in NAI titer was observed between the variable and no response groups. Analysis of day 0 HAI titers showed the 42 h strong response participants had statistically significant lower HAI titers with a group GMT of 2.37 compared to the variable and no response groups that had HAI GMTs of 8.94 and 27.1, respectively (Fig. 2G). Measurement of group I HA stalk antibody ELISA titer on day 0 similarly showed that participants with a strong viral shedding-associated gene expression response at 42 h had statistically significant lower HA stalk titers with a mean HA stalk antibody titer of 25,700 compared to the variable and no response participant groups, which had mean titers of 63,867 and 81,176, respectively (Fig. 2H). Unlike NAI and HA stalk day 0 titer, day 0 HAI titers were also significantly different between the variable and no response groups, with a higher GMT seen in the no response compared with the variable response group. Analysis of preexisting immunity on the 90 h and 138 h viral shedding-associated gene expression phenotypes populations showed similar statistical differences in NAI, HAI, and HA stalk antibody titers between the viral shedding-associated strong gene expression response group and the variable and no response participants (Fig. S5). However, in contrast to 42 h, no statistical differences in HAI titer were observed between the variable and no response groups at 90 h and 138 h (Fig. S5).
Statistical analysis of preexisting immunity between three viral shedding-associated gene expression response groups at 90 h and 138 h postinfection. Statistical significance between groups was assessed using rank sum t test and indicated by asterisks as follows: *, P ≤ 0.05; **, P < 0.01. Download FIG S5, TIF file, 5.4 MB (5.5MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
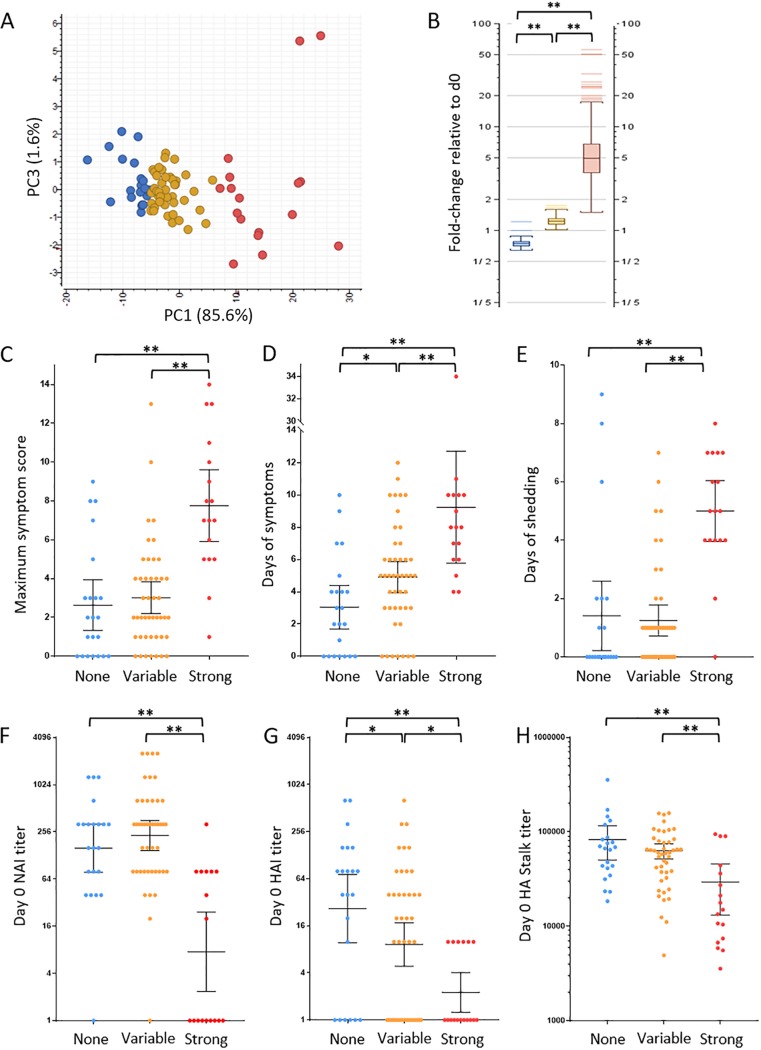
A longitudinal logistic regression model was also developed to determine which gene expression changes were correlated with active viral shedding. This model identified differential expression genes by fitting a mixed-effect model that included gene expression, number of days, and age as covariates, and interaction terms between gene expressions and days were considered (see Materials and Methods). Daily viral shedding was used as the outcome variable, and its measurements over 4 days are modeled as longitudinal data. A list of 151 top-ranked genes was selected based on the rankings of the model fitting results (see Table S5 at https://systemsbiology.org/wp-content/uploads/Walters-et-al-Supplemental-Tables-S1-S9.xlsx). The decision to limit this analysis to the top 151 logistic model ranked genes was made to allow for comparison of clinical, immune, and response pathway classification with the 151 total viral shedding-associated gene set. As shown in Fig. 3A and B, k-means analysis grouped participants based on expression levels of logistic model identified genes into strong, variable, and no response groups that showed significant differences in expression (Fig. S6). Increasing expression of logistic regression viral shedding-associated genes also correlated with maximum symptom score, days of symptoms, and days of shedding (Fig. 3C to E). These results were in agreement with the viral shedding-associated genes shown in Fig. 2C to E.
FIG 3.
Logistic regression determined differential PBL gene expression correlated with viral shedding. (A) Principal-component analysis of participant PBL expression of viral shedding-associated genes identified by logistic regression at 42 h postchallenge. The response group was determined by k-means analysis: no response (blue), strong response (red), and variable response (yellow). (B) Graph of response group average gene expression at 42 h genes relative to day 0. (C to E) Statistical analysis of clinical illness, including maximum symptom score (C), days of symptoms (D), and days of viral shedding (E) between the 42 h PBL expression response groups. (F to H) Statistical analysis of preexisting antibodies between the three PBL response groups at 42 h postinfection as measured by day 0 NAI titer (F), HAI titer (G), and HA stalk ELISA titer (H). For NAI and HAI titers, the geometric mean titer (GMT) is shown with 95% confidence intervals, while HA stalk titer is shown as median with 95% confidence intervals. Statistical significance between groups was assessed using rank sum t test and indicated as follows: *, P ≤ 0.05; **, P < 0.01.
k-means analysis of logistic regression-derived viral shedding-associated genes grouped participants into three main participant response groups. Expression profiles of logistic regression derived (LR-VS) gene expression (n = 151) at 42 h, 90 h, and 138 h demonstrating three general response groups: strong, variable, and none. Genes shown in red to yellow exhibited increased expression, and genes shown in blue to black exhibited decreased expression relative to each participant’s day 0 expression. Download FIG S6, TIF file, 5.4 MB (5.5MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Examination of preinfection (day 0) NAI, HAI, and HA stalk antibody titers in the strong, variable, and no response participants at 42 h who showed strong expression of the logistic regression viral shedding-associated genes had a significantly lower NAI GMT of 7.56 compared to the variable and no response groups, with NAI GMTs of 230.4 and 158.4, respectively (Fig. 3F). No statistical difference in NAI titer between the variable and no response groups was observed. As shown in Fig. 3G, at 42 h, HAI GMT titers in the strong logistic regression viral shedding-associated gene response group (2.25) were significantly lower than both the variable and no response groups (GMT of 9.24 and 26.61, respectively). The HAI titer in the logistic regression viral shedding-associated variable gene expression response group was also significantly lower than the no response group. Comparison of baseline group I HA stalk antibody titer also demonstrated that participants in the 42 h strong response group expression group had significantly lower HA stalk titers, with a median HA stalk antibody titer of 17,774, compared to the variable and no response participant groups, with median titers of 50,912 and 63,949, respectively (Fig. 3H). Analysis of preexisting NAI and HA stalk based immunity at 90 h and 138 h in the logistic regression viral shedding-associated strong gene expression groups showed statistical differences with variable and no response participants like those observed for viral shedding-associated gene expression (Fig. S7). Analysis of HAI titers in the response group at 90 h showed significantly lower HAI titer in the strong response group compared to the no response group, while HAI titers of the strong logistic regression viral shedding response group at 138 h was significantly lower than the both the variable and no response groups. These results both confirmed the importance of preexisting NA and HA stalk antibody correlation results show in Fig. 2F and G. Taken together, the clinical disease and immune correlates of logistic regression viral shedding-associated gene expression were nearly identical to the results described for expression of daily t test-derived viral shedding-associated genes shown in Fig. 2. Comparison of viral shedding-associated genes derived from t test and logistic regression showed an 80.1% overlap of genes with nearly identical pathway classification results (see Table S6 at https://systemsbiology.org/wp-content/uploads/Walters-et-al-Supplemental-Tables-S1-S9.xlsx).
Statistical analysis of preexisting immunity between three logistic regression-derived viral shedding-associated (LR-VS) gene expression response groups at 90 h and 138 h postinfection. Statistical significance between groups was assessed using rank sum t test and indicated by asterisks as follows: *, P ≤ 0.05; **, P < 0.01. Download FIG S7, TIF file, 5.4 MB (5.5MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
PBL expression signatures predictive of length of viral shedding and differential role of HA stalk antibodies.
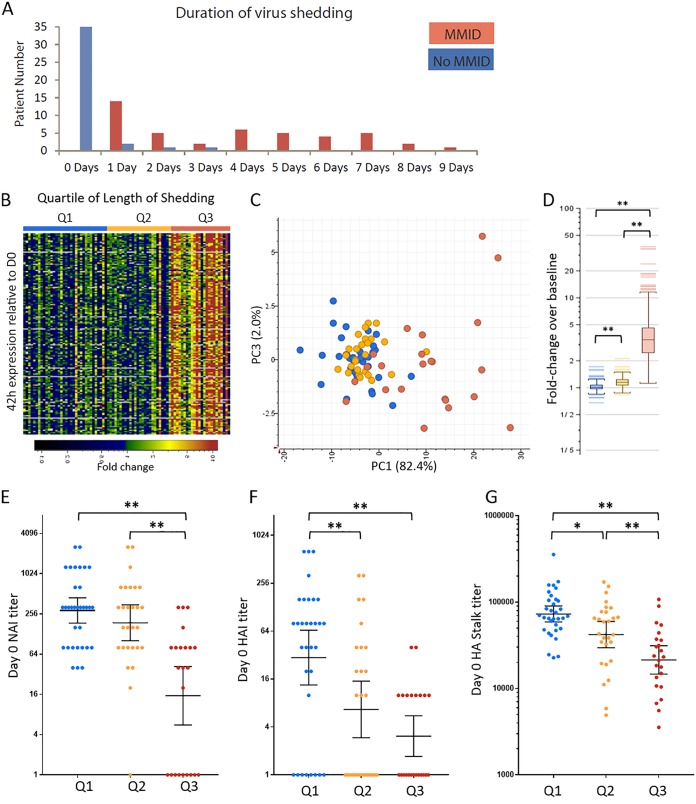
In most participants, onset of detectable viral shedding in nasal wash samples was 18 h postchallenge (31/44 MMID participants and 4/4 of the No MMID participants that asymptomatically shed virus), although in some participants, it was delayed for up to 4 to 5 days (Fig. S8A). As shown in Fig. 4A, duration of viral shedding was highly variable, ranging from 1 to 9 days in the MMID population and 1 to 3 days in the No MMID population. The total number of days of viral shedding in nasal passages of each participant was used as a reference to identify genes whose expression level correlated with how long an individual was potentially contagious. The goal was to identify genes detectable between 42 h and 90 h that could predict duration of viral shedding to help limit spread of infection. Pearson correlation analysis (r ≥ 0.5) identified 229 and 177 genes with expression levels that positively correlated with length of shedding at 42 h and 90 h, respectively, with 125 genes common between the two time points. No genes that negatively correlated with length of shedding were identified. Higher expression of these length of shedding-correlated genes (n = 125) correlated with quartile of shedding (Table 1), as shown in Fig. 4B and C. Statistical analysis demonstrated significant differences in expression levels of length of shedding-correlated genes at 42 h postinfection, with participants in the upper quartile of shedding showing the highest expression (Fig. 4D). In a hospital/clinical setting, individual preexposure samples would not be available for use as a baseline. To determine whether the length of shedding signature could also be used to predict duration of shedding in the absence of individual baselines, a pool of the entire cohort’s (n = 83) baseline prechallenge expression was instead used as the baseline to determine length of shedding-correlated gene expression levels at 42 h and showed similar results (Fig. S8B and C).
FIG 4.
PBL expression signatures present at 42 h correlate with length of viral shedding. (A) Graph showing length of shedding in days in mild-to-moderate influenza disease (MMID) and No MMID participants. (B) Heatmap showing postinfection expression profiles of transcripts (n = 125) at 42 h postinfection that correlate (r ≥ 0.5) positively with length of shedding in participants grouped by quartile of length of shedding. Genes shown in green to red were increased and genes shown in blue to black were decreased relative to each participant’s day 0 expression. (C) Principal-component analysis of length of shedding correlated genes at 42 h postinfection colored by quartile of length of shedding (Table 1). (D) Graph of the average 42 h expression of length of shedding correlated genes relative to day 0 grouped by quartile of length of shedding. (E to G) Statistical analysis of preexisting antibodies between participants grouped by quartile of length of shedding as measured by day 0 NAI titer (E), HAI titer (F), and HA stalk ELISA titer (G). Statistical significance between groups was assessed using rank sum t test with t test and indicated as follows: *, P ≤ 0.05; **, P < 0.01.
Shedding-correlated gene expression. (A) Graph showing distribution of shedding onset day in MMID and no MMID participants. (B) Expression of length of shedding-correlated genes using pooled day 0 baseline. Heatmap showing expression profiles of length of shedding (n = 125 genes) at 42 h in participants (n = 83) during the acute phase of infection grouped by quartile of length of shedding. Genes shown in red exhibited increased expression and genes shown in green exhibited decreased expression relative to each participant’s day 0 expression. Download FIG S8, TIF file, 5.4 MB (5.5MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Analysis of NA, HA head, and HA-S antibodies in participants grouped by quartile of length of nasal wash viral shedding demonstrated that NAI titers were significantly lower in participants with the highest quartile of viral shedding with a GMT of 15.3 compared to both median and lowest quartile shedders with GMTs of 188.8 and 288.1, respectively (Fig. 4E). Analysis of HAI titer showed that the highest quartile shedders had significantly lower titers (GMT = 3.1) compared to the lowest quartile shedders with (GMT = 29.8), while median quartile shedders had significantly lower HAI titer compared to the lowest quartile shedders (GMT = 6.65) (Fig. 4F). Comparison of preexisting HA stalk antibodies revealed a statistical difference between all three quartiles of length of shedding, as shown in Fig. 4G. Participants in the highest quartile for shedding had a HA stalk mean titer of 30,245 compared to those in the middle quartile with a mean titer of 57,929 and the lowest quartile shedders with a mean titer of 87,747. A subset of participants in the highest quartile of shedding group showed no or little change in expression level of the length of shedding signature (Fig. 5C). Unlike most participants in the highest quartile of shedding, these participants generally had higher levels of at least one antibody directed against NA, HA, or HA stalk (Fig. S9).
FIG 5.
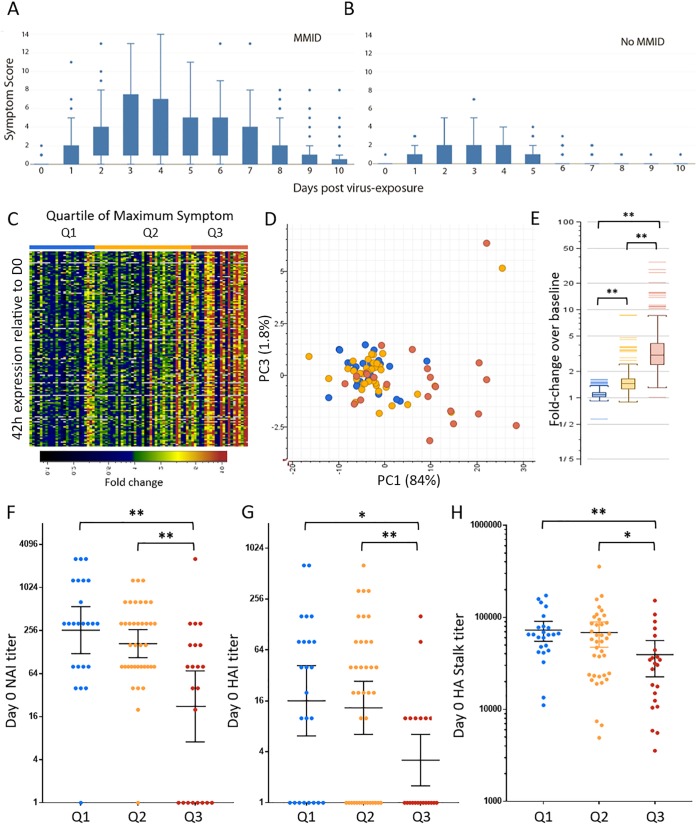
PBL expression signatures present at 42 h correlated with later disease severity. (A and B) Box plot of range of daily symptom scores in participants with mild-to-moderate influenza disease (MMID) (A) and participants with No MMID (B). (C) Heatmap showing expression profiles of transcripts (n = 126) at 42 h postinfection that correlate (r ≥ 0.5) positively with maximum symptom score. Genes shown in green to red were increased and genes shown in blue to black were decreased relative to each participant’s day 0 expression. (D) Principal-component analysis of maximum symptom-correlated genes at 42 h postinfection colored by quartile of maximum symptom score. (E) Graph of average 42 h expression of maximum symptom-correlated genes relative to day 0 grouped by quartile of maximum symptom score. (F to H) Statistical analysis of preexisting antibodies between participants grouped by quartile of maximum symptom score as measured by day 0 NAI titer (F), HAI titer (G), and HA stalk ELISA titer (H). Statistical significance between groups was assessed using rank sum t test and indicated as follows: *, P ≤ 0.05; **, P < 0.01.
Expression of length of shedding-correlated genes and relationship with preexisting anti-NA, anti-HA, and anti-HA stalk antibody levels. Heatmap showing expression profiles of length of shedding (n = 125 genes) at 42 h in participants (n = 83) during the acute phase of infection grouped by the quartile of length of shedding. Participant baseline (day 0) HAI and NAI titers are shown above as a gradient with blue color intensity increasing with increasing HAI and NAI titers and undetectable titers shown in gray. Genes shown in red exhibited increased expression and genes shown in green exhibited decreased expression relative to each participant’s day 0 expression. Participants in the lower quartile of shedding (0 days) have little induction of biomarker genes, and all have preexisting anti-NA and/or anti-HA. Some participants in the median quartile (virus shedding detected 1 to 3 days) have relatively high expression of length of shedding signature. Participants that do not express high levels of length of shedding-correlated genes have high preexisting immunity to either HA or NA. Participants in the upper quartile (virus shedding detected 5 to 9 days) generally show high expression of length of shedding signature. However, some participants (indicated using black lines) show little induction of length of shedding-correlated genes and have high preexisting HA or NA immunity. Download FIG S9, TIF file, 5.4 MB (5.5MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
PBL expression signatures predictive of illness severity.
Although the onset of symptoms was day 1 for most participants, peak illness, defined by the maximum symptom score during each participant’s course of illness, occurred between days 3 and 4 postinfection in both MMID and No MMID populations (Fig. S10A and B), with the MMID participants reporting markedly higher daily symptom scores (Fig. 5A and B). Day 3 postinfection was the most common time point when MMID participants experienced their peak illness (n = 14). Among the No MMID population, day 3 was also the most common day for peak illness (n = 11). However, in contrast to the MMID population, a greater proportion of the No MMID participants exhibited their highest symptoms scores on days 1 and 2 (n = 4 and n = 7, respectively), suggesting that peak illness occurs earlier in the No MMID population. While the kinetics of symptom onset and peak illness was consistent within the MMID population, the severity of illness varied considerably, as shown in both the range in daily reported symptoms and the maximum symptom score for participants (Fig. 5A), while both the range and maximum symptom scores were markedly smaller in the No MMID population (Fig. 5B).
Maximum symptom score-correlated gene expression. (A and B) Graph depicting distribution of symptom onset day and peak illness in MMID and No MMID participants, respectively. (C) Expression of maximum symptom-correlated genes using pooled day 0 baseline. Heatmap showing expression profiles of maximum symptom (n = 151 genes) at 42 h following challenge in participants (n = 83) during the acute phase of infection grouped by the quartile of length of shedding. Genes shown in red exhibited increased expression and genes shown in green exhibited decreased expression relative to each participant’s day 0 expression. Download FIG S10, TIF file, 5.4 MB (5.5MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Given the range in disease severity following IAV exposure, the ability to triage patients during acute infection and identify those at risk of progressing to severe illness could decrease morbidity/mortality associated with influenza complications. To accomplish this, the maximum symptom score was used as a reference to identify genes whose expression level correlated with a participant’s disease severity. Correlation analysis (r ≥ 0.5) identified 145 and 196 genes with expression levels that positively correlated with maximum symptom scores using expression data from 42 h and 90 h, respectively, with 126 genes common between the two time points. No genes that negatively correlated with maximum symptom score were identified. As shown in Fig. 5C and D, expression levels of these 126 genes at 42 h correlated with the quartile of overall maximum symptom score, with statistically higher average expression of these genes in participants in the top quartile of maximum symptom score (Fig. 5E), even though at 42 h, most had not yet reached peak illness. Again, the maximum symptom signature was assessed using a pool of the entire cohort’s day 0 (n = 83) as the reference baseline to determine expression levels of the maximum symptom-correlated genes (Fig. S10C and D).
Analysis of preexisting NA antibodies in participants grouped by quartile of maximum symptom score (Fig. 5F) showed that expression of these genes also correlated with significantly lower NAI titer in participants in the highest quartile of maximum symptom score (GMT = 22.3) compared to both median and lowest quartile participants with NAI GMTs of 168 and 259, respectively. No statistical difference in NAI was observed between median and lowest quartile maximum symptom score participants. PBL expression of maximum symptom-correlated genes also correlated with preexisting HA head antibodies (Fig. 5G), with participants in the top quartile of maximum symptom score having the statistically significant lowest titers (HAI GMT = 3.2) compared to participants with median and lowest quartile maximum symptom scores with HAI GMT of 13.3 and 169.1, respectively. No statistical difference in HAI titer was observed between the median and lowest quartile maximum symptom score participants. Analysis of preexisting HA stalk antibody titers showed statistically lower titers in participants in the top quartile of maximum symptom scores (mean titer = 39,394) compared to the median and lowest quartile participants (mean titers of 68,406 and 72,801, respectively) (Fig. 5H). No difference in serum HA stalk antibody titer was observed between lowest and median quartiles. Pathway classification analysis of the length of shedding and maximum symptom-correlated gene sets showed nearly identical results (see Tables S7 and S8 at https://systemsbiology.org/wp-content/uploads/Walters-et-al-Supplemental-Tables-S1-S9.xlsx), with TRAF3-dependent IRF activation mapped only to the maximum symptom-correlated genes (see Table S9 at the above URL).
DISCUSSION
The present study was designed to correlate clinical outcome and preexisting NA and HA antibody titers with PBL transcriptional responses in influenza A virus-infected participants with a wide range of preexisting serum antibody titers to NA, HA head, and HA stalk. Expression analysis showed unique and common populations of genes that were correlated with a positive clinical microbiology test for viral shedding in nasal washes on days 2, 4, and 6 postinfection. k-means clustering analysis of the active viral shedding genes showed three main PBL gene expression phenotypes, with some participants showing a “strong” expression phenotype, while others had a “variable response” or “no response.” The PBL expression phenotype of viral shedding-associated genes after viral infection was not predicated by day 0 PBL expression values but instead was correlated with preexisting serum antibody titers against NA, HA head, and HA stalk. While preexisting NA, HA head, and HA stalk antibody titers all correlated significantly with PBL viral shedding-associated gene expression phenotype, maximum clinical symptom score, and days of symptoms and days of shedding, only HA head antibody titer (as measured by HAI) was shown to be statistically significant between variable and no response participant groups at 42 h postinfection. We were further able to identify populations of genes that were correlated with and were predictive of length of viral shedding and illness severity (as measured by maximum symptom score).
Previous studies have examined PBL transcriptional profiling following IAV challenge to identify host-based prognostic and diagnostic markers (23, 24). One study examining whole-blood mRNA signatures in a large cohort of adults hospitalized with confirmed influenza infection found that interferon-related signatures were generally associated with less severe infection, while bacterial response-related signatures were associated with more severe infection (requiring mechanical ventilation) (25). Woods et al., performed transcriptional profiling on peripheral blood samples from 41 healthy volunteers experimentally challenged with H1N1 or H3N2 IAV, 18 of whom developed symptomatic infection (defined as a modified Jackson score of ≥6 for 5 consecutive days plus viral shedding for at least 2 consecutive days), and a gene expression signature was identified in these challenge participants that could detect the majority of infected cases (23). Using the same cohort of H3N2 IAV-challenged participants, biomarker signatures distinguishing early from later phases of infection were identified (26). However, these studies included only participants without detectable baseline HA head antibody titers and antibodies to NA and HA stalk were not measured, so it is unclear how preexisting immunity would affect detection of these biomarker signatures. An important finding of the current study is that the analysis of peripheral blood, which is distal to the site of infection, for influenza diagnostic/prognostic testing is affected by preexisting immunity with antibody titers adequate to suppress PBL gene expression responses but insufficient to prevent viral shedding and symptoms (see Fig. S9 in the supplemental material). This masking of gene expression biomarkers will reduce the predictive accuracy of point-of-care devices. While clinical symptoms, such as disease severity and presence of pneumonia, and demographic data, such as age and presence of comorbidities, are correlates of length of shedding (27–31), many of these clinical disease correlates of medically attended influenza do not manifest until many days after infection and have limited prognostic value. In contrast, significant differential PBL gene expression can be observed before 48 h postexposure just as symptoms are beginning to appear. Thus, the development of PBL gene expression-based disease prediction currently has its greatest clinical potential during outbreaks or pandemics where little protective immunity is present in the population.
The dominant influenza virus surface antigens are NA and HA proteins (1). In addition to targeting immune cell responses, such as opsonization and antibody-dependent cellular phagocytosis (ADCP), complement-mediated cytotoxicity (CMC), and antibody-dependent cell-mediated cytotoxicity (ADCC), antibodies can also directly inhibit viral protein function (32, 33). Antibodies against the head region of HA, which contains the receptor binding domain (RBD), are neutralizing both in vitro and in vivo, and antibodies directed against the HA stalk region have been shown to be more broadly protective and can be neutralizing both in vitro and in vivo (17, 34, 35). HA head immunity has also been shown to be an independent predictor of MMID and viral shedding, but not symptom severity, and HA stalk immunity was an independent predictor only of MMID (11). Clinical studies have shown that NA immunity is an independent predictor of MMID (i.e., shedding with symptoms), viral shedding, and symptom severity. Further, vaccination with recombinant NA proteins has shown potent clinical efficacy, and increasing NA antibody responses remain an attractive target for vaccine development (17, 34, 35, 36, 51). Of the three antibody titers measured in this study, HA head immunity was shown to be the most effective antibody correlate of differential PBL gene expression as a predictor of both length of shedding and number of symptoms, which is likely attributable to the potent neutralizing ability of HA head antibodies.
Many of the gene expression signatures for viral shedding, length of shedding, and maximum symptom consisted of genes associated with elevated antiviral responses, notably type I IFN responses, consistent with a recent report associating strong induction of antiviral response-related gene signatures in patients with less severe influenza infection (25). In the current study, several antiviral pathways, including pathogen-associated molecular pattern (PAMP) receptor responses, including both TLR and RIG-I (DDX58) signaling responses, as well as TNF receptor superfamily responses, were observed only early in infection (by 42 h postinoculation), namely, the strong induction of genes in the TNFRSF signaling pathway early in infection, including increased expression of IRF7 and NF-κB activation downstream of TRAF3/6 signaling from TNFRSF members, as well as significantly increased expression of TNFSF10 (TRAIL) in participants with active viral shedding. TRAIL can interact with several TNFRSF members, which results in the activation of MAPK8/JNK, and CASP3/8. Death receptor interaction with TRAIL has been reported to lead to death domain (DD)-mediated interaction with FADD and activation of the death effector domain (DED) or FADD (37–39). Activated natural killer (NK) cells and CD8+ cells have been reported to express both TRAIL and cognate receptors, although they themselves have been shown to be resistant to the cytotoxic effect of TRAIL (40). The role of TRAIL in early PBL responses to influenza infection is unclear, nor is it clear whether these signals originate in the PBL NK and CD8+ T cells or from the respiratory epithelium where infection is occurring, and additional studies will be needed to address the roles of these observations in clinical disease.
The ability to predict IAV contagiousness and illness severity through examination of PBL gene expression responses and subsequent development of point-of-care diagnostic/prognostic assays is an important goal that could have significant impact on public health, particularly regarding patient triage, isolation, drug treatment, and medical intervention (including mechanical ventilation and antivirals or initiation of antibiotics for secondary bacterial pneumonias) during an epidemic or pandemic (41), but the question of how PBL and serum antibody responses relate to influenza virus infection of upper respiratory tract epithelial cells remains unanswered. The strongest gene expression responses observed in this study were present in PBLs from challenge participants with the longest length of shedding and most severe symptoms; however, several participants (7 of 83) were symptomatic shedders but did not show an elevated PBL gene expression response. An explanation for this observation is that increased disease is associated with a larger antiviral response in the respiratory epithelium with higher concentrations of antiviral cytokines (e.g., type I IFNs) that leads to a “spillover” effect into the peripheral blood. It would be expected that participants with disease that lacked a PBL signal were mounting antiviral and immune responses in the nasopharynx arising from infection of nasal epithelial cells. Examination of mucosal immune responses, including characterization of epithelial (42, 43) and resident immune cell responses, measurement of antibodies (including secreted IgA) and antiviral and immune mediators in nasal epithelial and airway samples will be particularly important to understand the relationships between respiratory and peripheral cellular responses. Moreover, characterization of responses in different immune cell populations (44), the relationships between antibody responses and cellular immunity (45), and how influenza exposure history (from vaccination and/or natural infection) affects antibody and T cell receptor repertoires (46) in challenge studies and community-acquired infection cohorts will be critical for the understanding of clinical disease and for vaccine development and testing.
MATERIALS AND METHODS
Clinical study.
Two healthy volunteer wild-type (H1N1)pdm09 influenza challenge studies were performed at the National Institutes of Health (NIH) Clinical Center, as previously described (12, 19). Briefly, healthy volunteers (ages ranged from 18 to 50 years) were intranasally inoculated with 107 50% tissue culture infective doses (TCID50) of a GMP-manufactured wild-type 2009 influenza A (H1N1)pdm virus. Daily viral shedding in nasal wash samples was determined using the BioFire FilmArray multiplex respiratory pathogen assay (19, 47). Daily clinical symptoms were documented by both physician- and patient self-assessment and included nasal/sinus congestion, fatigue, headache, rhinorrhea, sore throat, myalgia, dry cough, productive cough, arthralgia, chills, diarrhea, nausea, fever (>38°C), and sweats as described in reference 19. PBLs were collected for RNA extraction and expression microarray analysis immediately before inoculation and then at 18 h, 42 h, 90 h, and 138 h after infection. Multiple clinical endpoints were reported including the presence or absence of mild to moderate influenza disease (MMID), defined as a positive molecular clinical test for influenza plus symptoms. In addition, symptom severity (defined as the number of influenza-related symptoms), presence or absence of symptoms or shedding alone, duration of symptoms, and duration of shedding were recorded. The studies (clinicaltrials.gov identifier NCT01971255 and NCT01646138) were approved by the NIAID Institutional Review Board and conducted in accordance with the provisions of the Declaration of Helsinki and good clinical practice guidelines.
Measurement of serum anti-HA, anti-HA head, and anti-HA stalk antibodies.
Measurements of neuraminidase inhibition (NAI), hemagglutination inhibition (HAI), and viral shedding used for this analysis were detailed previously and used standard methods (11). Measurement of antihemagglutinin (anti-HA) stalk antibody titers were using an enzyme-linked immunosorbent assay (ELISA) method as previously described (11).
RNA isolation and expression microarray analysis.
Total RNA was isolated from whole blood using PAXgene Blood RNA kit IVD (Qiagen, Gaithersburg, MD). RNA quality was assessed using a BioAnalyzer (Agilent Technologies, CA). Gene expression profiling experiments were performed using Agilent Human Whole Genome 44K microarrays. Fluorescent probes were prepared using Agilent QuickAmp labeling kit according to the manufacturer’s instructions. Each RNA sample was labeled and hybridized to individual arrays. Spot quantitation was performed using Agilent’s Feature Extractor software, and all data were then entered into a custom-designed database, SLIMarray (http://slimarray.systemsbiology.net), and then uploaded into Genedata Analyst 9.0 (Genedata, Basel, Switzerland).
Statistics.
Data normalization was performed in Genedata Analyst using central tendency, followed by relative normalization using each individual participant’s baseline (day 0) as a reference or using pooled day 0 data (n = 83). Pearson correlation, Student’s t test, k-means and principal-component analyses were performed using Genedata Analyst 9.0 and GraphPad Prism. All statistical analysis was performed using individual replicates, and the Benjamini-Hochberg procedure was used to correct for the false-positive rate in multiple comparisons. For immunity correlation analysis, a rank sum t test was performed using R. Panther (21, 48) was used for gene ontology and Reactome pathway analysis using Bonferroni correction. A longitudinal logistic regression was also used to model and rank genes that best explain the variations in the daily viral shedding (as determined by BioFire assay positivity). Briefly, we model the daily binary viral shedding indicator using a logistic model with age, time, and gene as covariates and a subject-specific random intercept term ui to addresses the correlation among multiple measurements. Let the response yij denote the daily BioFire assay positivity at the jth day for subject i, the longitudinal logistic regression model is described as follows:
Marginal (regression) screening was used to determine which dependent variables (differentially expressed genes) were correlated with the PBL gene expression response. A random intercept term was used to address the correlation among different measures of the same subjects. This model was applied to each gene separately and recorded the −2 log likelihood as the model fitting result. After ranking the genes, we selected the top 151 genes. This number of genes was chosen to match the total number of genes identified by t test correlation of daily PBL gene expression with daily viral shedding.
Data availability.
The complete MIAME-compliant (49) microarray data set has been deposited in NCBI’s Gene Expression Omnibus (50) and is accessible through GEO Series accession number GSE118223.
ACKNOWLEDGMENTS
This work was supported in part by the intramural research program of the National Institute of Allergy and Infectious Diseases (AI000986-12 and AI001157-07) to J.K.T. and (AI001157-07) to M.J.M., Defense Advanced Research Projects Agency (DARPA) Prometheus Program (HR0011831160) to J.C.K., and the National Center for Supercomputing Applications University of Illinois Urbana-Champaign. We acknowledge the LID Clinical Studies Unit, H. Clifford Lane, Richard T. Davey, the NIH Clinical Center Special Clinical Studies Unit staff, and the Department of Laboratory Medicine for their support of the clinical protocols.
K.-A.W., M.J.M., J.K.T., and J.C.K. conceived and conducted experiments and performed data analysis. R.Z., M.W., Z.R., H.W., L.A., C.B., and M.L. performed data analysis. K.S., J.K.P., D.B., T.B., L.A.R., and A.C.M. conducted experiments. L.C., A.H., and M.J.M. performed clinical studies. K.-A.W., M.J.M., J.K.T., and J.C.K. wrote the paper, and all authors had final approval of the submitted and published versions.
We declare that we have no conflict of interest.
Footnotes
Citation Walters K-A, Zhu R, Welge M, Scherler K, Park J-K, Rahil Z, Wang H, Auvil L, Bushell C, Lee MY, Baxter D, Bristol T, Rosas LA, Cervantes-Medina A, Czajkowski L, Han A, Memoli MJ, Taubenberger JK, Kash JC. 2019. Differential effects of influenza virus NA, HA head, and HA stalk antibodies on peripheral blood leukocyte gene expression during human infection. mBio 10:e00760-19. https://doi.org/10.1128/mBio.00760-19.
Contributor Information
Keith P. Klugman, Emory University.
Harry Greenberg, Stanford University.
Stacey Schultz-Cherry, St. Jude Children's Research Hospital.
REFERENCES
- 1.Krammer F, Smith GJD, Fouchier RAM, Peiris M, Kedzierska K, Doherty PC, Palese P, Shaw ML, Treanor J, Webster RG, García-Sastre A. 2018. Influenza. Nat Rev Dis Primers 4:3. doi: 10.1038/s41572-018-0002-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Centers for Disease Control and Prevention. 2018. Estimated influenza illnesses, medical visits, hospitalizations, and deaths in the United States – 2017-2018 influenza season. Centers for Disease Control and Prevention, Atlanta, GA. https://www.cdc.gov/flu/about/burden/2017-2018.htm.
- 3.Dawood FS, Iuliano AD, Reed C, Meltzer MI, Shay DK, Cheng P-Y, Bandaranayake D, Breiman RF, Brooks WA, Buchy P, Feikin DR, Fowler KB, Gordon A, Hien NT, Horby P, Huang QS, Katz MA, Krishnan A, Lal R, Montgomery JM, Mølbak K, Pebody R, Presanis AM, Razuri H, Steens A, Tinoco YO, Wallinga J, Yu H, Vong S, Bresee J, Widdowson M-A. 2012. Estimated global mortality associated with the first 12 months of 2009 pandemic influenza A H1N1 virus circulation: a modelling study. Lancet Infect Dis 12:687–695. doi: 10.1016/S1473-3099(12)70121-4. [DOI] [PubMed] [Google Scholar]
- 4.Morens DM, Taubenberger JK, Harvey HA, Memoli MJ. 2010. The 1918 influenza pandemic: lessons for 2009 and the future. Crit Care Med 38:e10–e20. doi: 10.1097/CCM.0b013e3181ceb25b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Taubenberger JK, Morens DM. 2006. 1918 influenza: the mother of all pandemics. Emerg Infect Dis 12:15–22. doi: 10.3201/eid1201.050979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Uyeki TM. 2017. Influenza. Ann Intern Med 167:ITC33–ITC48. doi: 10.7326/AITC201709050. [DOI] [PubMed] [Google Scholar]
- 7.Monto AS, Petrie JG, Cross RT, Johnson E, Liu M, Zhong W, Levine M, Katz JM, Ohmit SE. 2015. Antibody to influenza virus neuraminidase: an independent correlate of protection. J Infect Dis 212:1191–1199. doi: 10.1093/infdis/jiv195. [DOI] [PubMed] [Google Scholar]
- 8.Nussing S, Sant S, Koutsakos M, Subbarao K, Nguyen THO, Kedzierska K. 2018. Innate and adaptive T cells in influenza disease. Front Med 12:34–47. doi: 10.1007/s11684-017-0606-8. [DOI] [PubMed] [Google Scholar]
- 9.Sant AJ, Richards KA, Nayak J. 2018. Distinct and complementary roles of CD4 T cells in protective immunity to influenza virus. Curr Opin Immunol 53:13–21. doi: 10.1016/j.coi.2018.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schmidt ME, Varga SM. 2018. The CD8 T cell response to respiratory virus infections. Front Immunol 9:678. doi: 10.3389/fimmu.2018.00678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Park JK, Han A, Czajkowski L, Reed S, Athota R, Bristol T, Rosas LA, Cervantes-Medina A, Taubenberger JK, Memoli MJ. 2018. Evaluation of preexisting anti-hemagglutinin stalk antibody as a correlate of protection in a healthy volunteer challenge with influenza A/H1N1pdm virus. mBio 9:e02284-17. doi: 10.1128/mBio.02284-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Memoli MJ, Shaw PA, Han A, Czajkowski L, Reed S, Athota R, Bristol T, Fargis S, Risos K, Powers JH, Davey RT Jr, Taubenberger JK. 2016. Evaluation of antihemagglutinin and antineuraminidase antibodies as correlates of protection in an influenza A/H1N1 virus healthy human challenge model. mBio 7:e00417-16. doi: 10.1128/mBio.00417-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eichelberger MC, Morens DM, Taubenberger JK. 2018. Neuraminidase as an influenza vaccine antigen: a low hanging fruit, ready for picking to improve vaccine effectiveness. Curr Opin Immunol 53:38–44. doi: 10.1016/j.coi.2018.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krammer F, Fouchier RAM, Eichelberger MC, Webby RJ, Shaw-Saliba K, Wan H, Wilson PC, Compans RW, Skountzou I, Monto AS. 2018. NAction! How can neuraminidase-based immunity contribute to better influenza virus vaccines? mBio 9:e02332-17. doi: 10.1128/mBio.02332-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hobson D, Curry RL, Beare AS, Ward-Gardner A. 1972. The role of serum haemagglutination-inhibiting antibody in protection against challenge infection with influenza A2 and B viruses. J Hyg 70:767–777. doi: 10.1017/S0022172400022610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cox RJ. 2013. Correlates of protection to influenza virus, where do we go from here? Hum Vaccin Immunother 9:405–408. doi: 10.4161/hv.22908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Steel J, Lowen AC, Wang TT, Yondola M, Gao Q, Haye K, García-Sastre A, Palese P. 2010. Influenza virus vaccine based on the conserved hemagglutinin stalk domain. mBio 1:e00018-10. doi: 10.1128/mBio.00018-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang TT, Tan GS, Hai R, Pica N, Ngai L, Ekiert DC, Wilson IA, Garcia-Sastre A, Moran TM, Palese P. 2010. Vaccination with a synthetic peptide from the influenza virus hemagglutinin provides protection against distinct viral subtypes. Proc Natl Acad Sci U S A 107:18979–18984. doi: 10.1073/pnas.1013387107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Memoli MJ, Czajkowski L, Reed S, Athota R, Bristol T, Proudfoot K, Fargis S, Stein M, Dunfee RL, Shaw PA, Davey RT, Taubenberger JK. 2014. Validation of the wild-type influenza A human challenge model H1N1pdMIST: an A(H1N1)pdm09 dose-finding investigational new drug study. Clin Infect Dis 60:693–702. doi: 10.1093/cid/ciu924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fabregat A, Jupe S, Matthews L, Sidiropoulos K, Gillespie M, Garapati P, Haw R, Jassal B, Korninger F, May B, Milacic M, Roca CD, Rothfels K, Sevilla C, Shamovsky V, Shorser S, Varusai T, Viteri G, Weiser J, Wu G, Stein L, Hermjakob H, D'Eustachio P. 2018. The Reactome Pathway Knowledgebase. Nucleic Acids Res 46:D649–D655. doi: 10.1093/nar/gkx1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mi H, Muruganujan A, Casagrande JT, Thomas PD. 2013. Large-scale gene function analysis with the PANTHER classification system. Nat Protoc 8:1551–1566. doi: 10.1038/nprot.2013.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thomas PD, Kejariwal A, Campbell MJ, Mi H, Diemer K, Guo N, Ladunga I, Ulitsky-Lazareva B, Muruganujan A, Rabkin S, Vandergriff JA, Doremieux O, 2003. PANTHER: a browsable database of gene products organized by biological function, using curated protein family and subfamily classification. Nucleic Acids Res 31:334–341. doi: 10.1093/nar/gkg115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Woods CW, McClain MT, Chen M, Zaas AK, Nicholson BP, Varkey J, Veldman T, Kingsmore SF, Huang Y, Lambkin-Williams R, Gilbert AG, Hero AO, Ramsburg E, Glickman S, Lucas JE, Carin L, Ginsburg GS. 2013. A host transcriptional signature for presymptomatic detection of infection in humans exposed to influenza H1N1 or H3N2. PLoS One 8:e52198. doi: 10.1371/journal.pone.0052198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zaas AK, Chen M, Varkey J, Veldman T, Hero AO, Lucas J, Huang Y, Turner R, Gilbert A, Lambkin-Williams R, Øien NC, Nicholson B, Kingsmore S, Carin L, Woods CW, Ginsburg GS. 2009. Gene expression signatures diagnose influenza and other symptomatic respiratory viral infections in humans. Cell Host Microbe 6:207–217. doi: 10.1016/j.chom.2009.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dunning J, Blankley S, Hoang LT, Cox M, Graham CM, James PL, Bloom CI, Chaussabel D, Banchereau J, Brett SJ, Moffatt MF, O’Garra A, Openshaw PJM. 2018. Progression of whole-blood transcriptional signatures from interferon-induced to neutrophil-associated patterns in severe influenza. Nat Immunol 19:625–635. doi: 10.1038/s41590-018-0111-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang Y, Zaas AK, Rao A, Dobigeon N, Woolf PJ, Veldman T, Øien NC, McClain MT, Varkey JB, Nicholson B, Carin L, Kingsmore S, Woods CW, Ginsburg GS, Hero AO. 2011. Temporal dynamics of host molecular responses differentiate symptomatic and asymptomatic influenza A infection. PLoS Genet 7:e1002234. doi: 10.1371/journal.pgen.1002234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meschi S, Selleri M, Lalle E, Bordi L, Valli MB, Ferraro F, Ippolito G, Petrosillo N, Lauria FN, Capobianchi MR. 2011. Duration of viral shedding in hospitalized patients infected with pandemic H1N1. BMC Infect Dis 11:140. doi: 10.1186/1471-2334-11-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Khoury J, Szwarcwort M, Kra-Oz Z, Saffuri M, Seh K, Yahalomi T, Braun E, Azzam ZS, Paul M, Neuberger A. 2018. Duration of viral shedding and factors associated with prolonged shedding among inpatients with influenza treated with oseltamivir: a prospective cohort study. Eur J Clin Microbiol Infect Dis 37:319–323. doi: 10.1007/s10096-017-3135-0. [DOI] [PubMed] [Google Scholar]
- 29.Wang B, Russell ML, Fonseca K, Earn DJD, Horsman G, Van Caeseele P, Chokani K, Vooght M, Babiuk L, Walter SD, Loeb M. 2017. Predictors of influenza A molecular viral shedding in Hutterite communities. Influenza Other Respir Viruses 11:254–262. doi: 10.1111/irv.12448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leekha S, Zitterkopf NL, Espy MJ, Smith TF, Thompson RL, Sampathkumar P. 2007. Duration of influenza A virus shedding in hospitalized patients and implications for infection control. Infect Control Hosp Epidemiol 28:1071–1076. doi: 10.1086/520101. [DOI] [PubMed] [Google Scholar]
- 31.Fielding JE, Kelly HA, Mercer GN, Glass K. 2014. Systematic review of influenza A(H1N1)pdm09 virus shedding: duration is affected by severity, but not age. Influenza Other Respir Viruses 8:142–150. doi: 10.1111/irv.12216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Biburger M, Lux A, Nimmerjahn F. 2014. How immunoglobulin G antibodies kill target cells: revisiting an old paradigm. Adv Immunol 124:67–94. doi: 10.1016/B978-0-12-800147-9.00003-0. [DOI] [PubMed] [Google Scholar]
- 33.Corti D, Cameroni E, Guarino B, Kallewaard NL, Zhu Q, Lanzavecchia A. 2017. Tackling influenza with broadly neutralizing antibodies. Curr Opin Virol 24:60–69. doi: 10.1016/j.coviro.2017.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.DiLillo DJ, Tan GS, Palese P, Ravetch JV. 2014. Broadly neutralizing hemagglutinin stalk-specific antibodies require FcgammaR interactions for protection against influenza virus in vivo. Nat Med 20:143–151. doi: 10.1038/nm.3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ekiert DC, Friesen RHE, Bhabha G, Kwaks T, Jongeneelen M, Yu W, Ophorst C, Cox F, Korse HJWM, Brandenburg B, Vogels R, Brakenhoff JPJ, Kompier R, Koldijk MH, Cornelissen LAHM, Poon LLM, Peiris M, Koudstaal W, Wilson IA, Goudsmit J. 2011. A highly conserved neutralizing epitope on group 2 influenza A viruses. Science 333:843–850. doi: 10.1126/science.1204839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Couch RB, Kasel JA, Gerin JL, Schulman JL, Kilbourne ED. 1974. Induction of partial immunity to influenza by a neuraminidase-specific vaccine. J Infect Dis 129:411–420. doi: 10.1093/infdis/129.4.411. [DOI] [PubMed] [Google Scholar]
- 37.Boatright KM, Salvesen GS. 2003. Mechanisms of caspase activation. Curr Opin Cell Biol 15:725–731. doi: 10.1016/j.ceb.2003.10.009. [DOI] [PubMed] [Google Scholar]
- 38.Fulda S. 2014. Tumor-necrosis-factor-related apoptosis-inducing ligand (TRAIL). Adv Exp Med Biol 818:167–180. doi: 10.1007/978-1-4471-6458-6_8. [DOI] [PubMed] [Google Scholar]
- 39.Martinez-Lostao L, de Miguel D, Al-Wasaby S, Gallego-Lleyda A, Anel A. 2015. Death ligands and granulysin: mechanisms of tumor cell death induction and therapeutic opportunities. Immunotherapy 7:883–898. doi: 10.2217/imt.15.56. [DOI] [PubMed] [Google Scholar]
- 40.Mirandola P, Ponti C, Gobbi G, Sponzilli I, Vaccarezza M, Cocco L, Zauli G, Secchiero P, Manzoli FA, Vitale M. 2004. Activated human NK and CD8+ T cells express both TNF-related apoptosis-inducing ligand (TRAIL) and TRAIL receptors but are resistant to TRAIL-mediated cytotoxicity. Blood 104:2418–2424. doi: 10.1182/blood-2004-04-1294. [DOI] [PubMed] [Google Scholar]
- 41.Morens DM, Taubenberger JK. 2018. Influenza cataclysm, 1918. N Engl J Med 379:2285–2287. doi: 10.1056/NEJMp1814447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Meliopoulos VA, Van de Velde L-A, Van de Velde NC, Karlsson EA, Neale G, Vogel P, Guy C, Sharma S, Duan S, Surman SL, Jones BG, Johnson MDL, Bosio C, Jolly L, Jenkins RG, Hurwitz JL, Rosch JW, Sheppard D, Thomas PG, Murray PJ, Schultz-Cherry S. 2016. An epithelial integrin regulates the amplitude of protective lung interferon responses against multiple respiratory pathogens. PLoS Pathog 12:e1005804. doi: 10.1371/journal.ppat.1005804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tan KS, Yan Y, Koh WLH, Li L, Choi H, Tran T, Sugrue R, Wang Y, Chow VT. 2018. Comparative transcriptomic and metagenomic analyses of influenza virus-infected nasal epithelial cells from multiple individuals reveal specific nasal-initiated signatures. Front Microbiol 9:2685. doi: 10.3389/fmicb.2018.02685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Seong Y, Lazarus NH, Sutherland L, Habtezion A, Abramson T, He XS, Greenberg HB, Butcher EC. 2017. Trafficking receptor signatures define blood plasmablasts responding to tissue-specific immune challenge. JCI Insight 2:e90233. doi: 10.1172/jci.insight.90233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.He X-S, Holmes TH, Sasaki S, Jaimes MC, Kemble GW, Dekker CL, Arvin AM, Greenberg HB. 2008. Baseline levels of influenza-specific CD4 memory T-cells affect T-cell responses to influenza vaccines. PLoS One 3:e2574. doi: 10.1371/journal.pone.0002574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Emerson RO, DeWitt WS, Vignali M, Gravley J, Hu JK, Osborne EJ, Desmarais C, Klinger M, Carlson CS, Hansen JA, Rieder M, Robins HS. 2017. Immunosequencing identifies signatures of cytomegalovirus exposure history and HLA-mediated effects on the T cell repertoire. Nat Genet 49:659–665. doi: 10.1038/ng.3822. [DOI] [PubMed] [Google Scholar]
- 47.Babady NE. 2013. The FilmArray(R) respiratory panel: an automated, broadly multiplexed molecular test for the rapid and accurate detection of respiratory pathogens. Expert Rev Mol Diagn 13:779–788. doi: 10.1586/14737159.2013.848794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mi H, Muruganujan A, Thomas PD. 2012. PANTHER in 2013: modeling the evolution of gene function, and other gene attributes, in the context of phylogenetic trees. Nucleic Acids Res 41:D377–D386. doi: 10.1093/nar/gks1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brazma A, Hingamp P, Quackenbush J, Sherlock G, Spellman P, Stoeckert C, Aach J, Ansorge W, Ball CA, Causton HC, Gaasterland T, Glenisson P, Holstege FC, Kim IF, Markowitz V, Matese JC, Parkinson H, Robinson A, Sarkans U, Schulze-Kremer S, Stewart J, Taylor R, Vilo J, Vingron M. 2001. Minimum information about a microarray experiment (MIAME)-toward standards for microarray data. Nat Genet 29:365–371. doi: 10.1038/ng1201-365. [DOI] [PubMed] [Google Scholar]
- 50.Edgar R, Domrachev M, Lash AE. 2002. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Murphy BR, Kasel JA, Chanock RM. 1972. Association of serum anti-neuraminidase antibody with resistance to influenza in man. N Engl J Med 286:1329–1332. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Baseline PBL gene expression does not predict participant outcome following IAV challenge. (A to C) Principal-component analysis of global day 0 gene expression profiles showing general outcome (A), illness severity (B), and days of shedding (C). (A) For general outcome, No MMID is shown in blue and MMID is shown in red. (B) Illness severity as measured by maximum symptom score (indicated using increasing green color gradient). (C) Days of shedding indicated using pink gradient. (D) Baseline expression levels of core antiviral response genes in MMID and No MMID participants. Results are shown as medians and standard deviations of intensity expression levels. Download FIG S1, TIF file, 5.4 MB (5.5MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Kinetics of viral shedding response during acute IAV infection in shedding outlier participants. (A) Expression profiles of viral shedding-associated genes (n = 151) in all participants (n = 83) during the acute phase of infection. Participant samples at each time point for active shedding IAV in nasal washes are indicated by red bars and samples negative for IAV are indicated by blue bars. (B) Expression of viral shedding-associated genes in subset of participants that show delayed or absent viral shedding-associated genes. Genes shown in red exhibited increased expression and genes shown in green exhibited decreased expression relative to each participant’s day 0 expression. Download FIG S2, TIF file, 5.4 MB (5.5MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
k-means analysis of viral shedding-associated genes identifies three main participant response groups. Expression profiles of viral shedding (VS)-associated genes (n = 151) at 42 h, 90 h, and 138 h demonstrating three general response groups: strong, variable, and none. Genes shown in red exhibited increased expression and genes shown in green exhibited decreased expression relative to each participant’s day 0 expression. Download FIG S3, TIF file, 5.4 MB (5.5MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Statistical analysis of clinical parameters between three viral shedding-associated gene expression response groups at 90 h and 138 h postinfection. Statistical significance between groups was assessed using rank sum t test and indicated by asterisks as follows: *, P ≤ 0.05; **, P < 0.01. Download FIG S4, TIF file, 5.4 MB (5.5MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Statistical analysis of preexisting immunity between three viral shedding-associated gene expression response groups at 90 h and 138 h postinfection. Statistical significance between groups was assessed using rank sum t test and indicated by asterisks as follows: *, P ≤ 0.05; **, P < 0.01. Download FIG S5, TIF file, 5.4 MB (5.5MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
k-means analysis of logistic regression-derived viral shedding-associated genes grouped participants into three main participant response groups. Expression profiles of logistic regression derived (LR-VS) gene expression (n = 151) at 42 h, 90 h, and 138 h demonstrating three general response groups: strong, variable, and none. Genes shown in red to yellow exhibited increased expression, and genes shown in blue to black exhibited decreased expression relative to each participant’s day 0 expression. Download FIG S6, TIF file, 5.4 MB (5.5MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Statistical analysis of preexisting immunity between three logistic regression-derived viral shedding-associated (LR-VS) gene expression response groups at 90 h and 138 h postinfection. Statistical significance between groups was assessed using rank sum t test and indicated by asterisks as follows: *, P ≤ 0.05; **, P < 0.01. Download FIG S7, TIF file, 5.4 MB (5.5MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Shedding-correlated gene expression. (A) Graph showing distribution of shedding onset day in MMID and no MMID participants. (B) Expression of length of shedding-correlated genes using pooled day 0 baseline. Heatmap showing expression profiles of length of shedding (n = 125 genes) at 42 h in participants (n = 83) during the acute phase of infection grouped by quartile of length of shedding. Genes shown in red exhibited increased expression and genes shown in green exhibited decreased expression relative to each participant’s day 0 expression. Download FIG S8, TIF file, 5.4 MB (5.5MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Expression of length of shedding-correlated genes and relationship with preexisting anti-NA, anti-HA, and anti-HA stalk antibody levels. Heatmap showing expression profiles of length of shedding (n = 125 genes) at 42 h in participants (n = 83) during the acute phase of infection grouped by the quartile of length of shedding. Participant baseline (day 0) HAI and NAI titers are shown above as a gradient with blue color intensity increasing with increasing HAI and NAI titers and undetectable titers shown in gray. Genes shown in red exhibited increased expression and genes shown in green exhibited decreased expression relative to each participant’s day 0 expression. Participants in the lower quartile of shedding (0 days) have little induction of biomarker genes, and all have preexisting anti-NA and/or anti-HA. Some participants in the median quartile (virus shedding detected 1 to 3 days) have relatively high expression of length of shedding signature. Participants that do not express high levels of length of shedding-correlated genes have high preexisting immunity to either HA or NA. Participants in the upper quartile (virus shedding detected 5 to 9 days) generally show high expression of length of shedding signature. However, some participants (indicated using black lines) show little induction of length of shedding-correlated genes and have high preexisting HA or NA immunity. Download FIG S9, TIF file, 5.4 MB (5.5MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Maximum symptom score-correlated gene expression. (A and B) Graph depicting distribution of symptom onset day and peak illness in MMID and No MMID participants, respectively. (C) Expression of maximum symptom-correlated genes using pooled day 0 baseline. Heatmap showing expression profiles of maximum symptom (n = 151 genes) at 42 h following challenge in participants (n = 83) during the acute phase of infection grouped by the quartile of length of shedding. Genes shown in red exhibited increased expression and genes shown in green exhibited decreased expression relative to each participant’s day 0 expression. Download FIG S10, TIF file, 5.4 MB (5.5MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Data Availability Statement
The complete MIAME-compliant (49) microarray data set has been deposited in NCBI’s Gene Expression Omnibus (50) and is accessible through GEO Series accession number GSE118223.