Abstract
Excessive or inappropriate inflammatory responses can cause serious and even fatal diseases. The CCAAT/enhancer-binding protein alpha (CEBPA) gene encodes C/EBPα, a transcription factor that plays a fundamental role in controlling maturation of the myeloid lineage and is also expressed during the late phase of inflammatory responses when signs of inflammation are decreasing. MTL-CEBPA, a small activating RNA targeting for upregulation of C/EBPα, is currently being evaluated in a phase 1b trial for treatment of hepatocellular carcinoma. After dosing, subjects had reduced levels of pro-inflammatory cytokines, and we therefore hypothesized that MTL-CEBPA has anti-inflammatory potential. The current study was conducted to determine the effects of C/EBPα saRNA - CEBPA-51 - on inflammation in vitro and in vivo after endotoxin challenge. CEBPA-51 led to increased expression of the C/EBPα gene and inhibition of pro-inflammatory cytokines in THP-1 monocytes previously stimulated by E. coli-derived lipopolysaccharide (LPS). Treatment with MTL-CEBPA in an LPS-challenged humanized mouse model upregulated C/EBPα mRNA, increased neutrophils, and attenuated production of several key pro-inflammatory cytokines, including TNF-α, IL-6, IL-1β, and IFN-γ. In addition, a Luminex analysis of mouse serum revealed that MTL-CEBPA reduced pro-inflammatory cytokines and increased the anti-inflammatory cytokine IL-10. Collectively, the data support further investigation of MTL-CEBPA in acute and chronic inflammatory diseases where this mechanism has pathogenic importance.
Keywords: small activating RNA, saRNA, MTL-CEBPA, anti-inflammation, monocytes, humanized mice
MTL-CEBPA is a small activating RNA that targets the upregulation of transcription factor C/EBPα, currently being evaluated in a phase 1b trial for treatment of hepatocellular carcinoma. Zhou et al. demonstrate that MTL-CEBPA increases the expression of C/EBPα and displays anti-inflammatory effects in a tissue culture system and a humanized mouse model.
Introduction
Inflammation is an evolutionarily conserved host immune response to injury, infection, and stress.1 As a natural, self-protective, well-controlled reaction that removes harmful stimuli, inflammation can be beneficial; however, persistent chronic inflammation or an excessive inflammatory response is associated with diseases, such as certain cancers, rheumatoid arthritis, heart disease, type II diabetes, non-alcoholic steatohepatitis (NASH), and some neurodegenerative diseases (e.g., Alzheimer’s disease).2, 3, 4, 5 In addition, epidemiological studies and preclinical observations suggest that sustained inflammation is linked to a higher risk of cancer recurrence or metastasis after a period of clinical dormancy.6, 7 Therefore, in many diseases, it is crucial to control inflammation to reduce the risk of long-term tissue damage and disease relapse.8
CCAAT/enhancer-binding protein alpha (C/EBPα) is a basic leucine zipper-class transcription factor that is expressed in numerous tissues and cell types.9 It is required for differentiation of granulocytes and monocytes, independent of granulocyte colony-stimulating factor (G-CSF).10, 11 C/EBPα has been documented as a tumor suppressor.12, 13 Mutation or downregulation of C/EBPα expression is associated with various types of cancers, such as acute myeloid leukemia (AML), pancreatic cancer, lung cancer, breast cancer, and others, and correlates highly with tumor size and progression.14, 15 Moreover, ectopic expression of C/EBPα in various cancer cell lines results in tumor cell growth arrest.16, 17, 18, 19 Restoring endogenous C/EBPα expression has been shown to lead to a better therapeutic outcome in cancer.17, 20, 21 Upregulation of C/EBPα activity has the potential to improve liver function and limit hepatocellular carcinoma (HCC) growth.22, 23, 24 These findings demonstrated a critical role of C/EBPα expression in the regulation of cell motility, maturation, and tumorigenesis.13 The role of C/EBPα expression in inflammation is not well characterized25 although other CCAAT/EBPs such as C/EBPα have a clear role and are associated with activation of the NF-κB transcription factor.26 C/EBPβ and nuclear factor-κB (NF-κB; p65) form a ternary complex to regulate several promoters within the inflammation pathway, such as amyloid A, IL-6, and IL-8, and others.27, 28, 29
MTL-CEBPA is a small activating RNA (saRNA) that specifically targets C/EBPα and is formulated into a SMARTICLES liposomal nanoparticle (Sarker et al., 2017, J. Clin. Oncol.,abstract). It is currently being evaluated in patients with advanced liver cancer in a phase 1/2a trial (ClinicalTrials.gov: NCT02716012). Previous mechanistic studies have suggested that such short double-stranded RNAs that specifically target the promoter region of a gene can activate its transcription, a phenomenon termed transcription gene activation (TGA) which involves Argonaute-2 protein (Ago-2) activity.21, 30 The clinical observations revealed that intravenous (i.v.) administration of MTL-CEBPA induced an increase in CEBPA mRNA in white blood cells (WBCs) and a doubling of neutrophils within 24 h of dosing. Of note, MTL-CEBPA downregulated the “inflammatory signature” in circulating WBCs and neutrophils of patients, such as interferon-γ (IFN-γ), IL-6, and NK-κB, suggesting potential anti-inflammatory benefit of MTL-CEBPA (unpublished data).
Lipopolysaccharide (LPS)-induced inflammation, in in vitro cell lines and in animals, represents a standard paradigm for studying inflammation.31, 32 Also known as endotoxin, LPS is a major outer membrane component of Gram-negative bacteria. It is able to induce an inflammatory response through NLRP3 inflammasome activation that results in IL-1β and IL-18 production after activation of caspases,33 and subsequent production of other cytokines and mediators of inflammation by activated human immune cells, such as macrophages, monocytes, dendritic cells, T cells, and B cells.34, 35 LPS-stimulated macrophages and monocytes release multiple pro-inflammatory cytokines, such as tumor necrosis factor (TNF-α), interleukin-6 (IL-6), IL-1β, and IL-12, which have been known to play crucial roles in the inflammatory response.36
The aim of the present study was to evaluate the anti-inflammatory potential of MTL-CEBPA in LPS-stimulated THP-1 monocytes in vitro and LPS-challenged humanized NOD/SCID/IL2rγnull (hu-NSG) mice in vivo. Pro-inflammatory cytokine responses were determined by qRT-PCR, ELISA, and high-throughput Luminex assay. In the LPS-induced hu-NSG mouse model, we also analyzed the effect of MTL-CEBPA on different immune cell subsets in peripheral blood (PB) and bone marrow by flow cytometry, including macrophages, neutrophils, and T lymphocyte CD4+ and CD8+ T cells.
Results
CEBPA-51 Attenuates LPS-Induced Downregulation of C/EPBα and Inhibits Pro-inflammatory Cytokines In Vitro
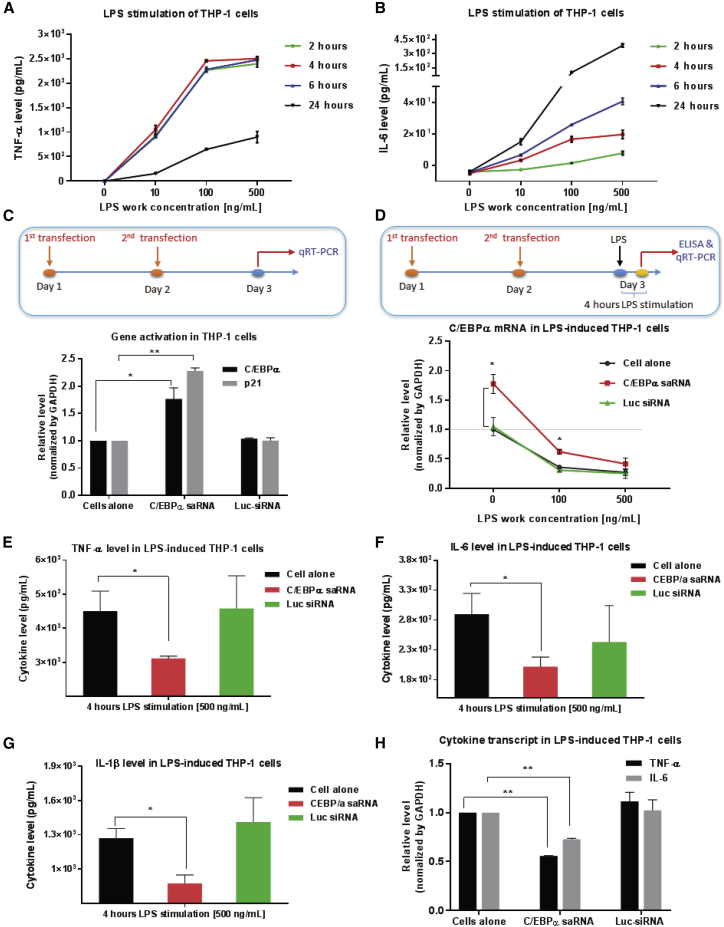
As an immortalized monocyte-like cell line with homogeneous genetic background, the THP-1 cell line represents a useful model system for investigating the structure and function of monocytes.37 It is well established that upon stimulation with bacterial LPS, THP-1 cells rapidly secrete high levels of several pro-inflammatory cytokines, such as TNF-α and IL-6, which are key inflammatory mediators.38, 39 Consistent with those studies,38, 39 time- and LPS dose-dependent TNF-α and IL-6 production was observed in THP-1 cells upon LPS stimulation (Figures 1A and 1B). Secretion of TNF-α after 4 h of LPS stimulation was defined as the optimal stimulation condition for in vitro evaluation of C/EBPα saRNA.
Figure 1.
CEBPA-51 Induces Specific Gene Activation and Suppresses Pro-inflammatory Cytokine Production in LPS-Stimulated THP-1 Monocytes
(A and B) LPS-mediated cytokines production was time and LPS dose dependent. THP-1 cells were treated with different concentrations of LPS. Cell-free supernatant was collected at various time points for quantitative analysis of the pro-inflammatory cytokines (A) TNF-α and (B) IL-6 by ELISA. (C) CEBPA-51 mediated specific gene activity in THP-1 cells. THP-1 cells were transfected with 10 nM CEBPA-51 or control Luc-siRNA twice with Lipofectamine 3000. At 24 h after the last transfection, total RNA was collected for quantitative analysis of target gene C/EBPα and its downstream gene p21 by qRT-PCR assay. (D) CEBPA-51 attenuated LPS-induced downregulation of C/EBPα. The THP-1 cells transfected with 10 nM of experimental RNAs twice were stimulated with different concentrations of LPS for 4 h. Total RNA was collected for qRT-PCR and cell-free supernatant was collected for ELISA. (E–G) CEBPA-51 inhibited the secretion of the soluble pro-inflammatory cytokines (E) TNF-α, (F) IL-6, and (G) IL-1β. (H) CEBPA-51 repressed the transcript RNA expression of cytokines TNF-α and IL-6. Each in vitro experiment was performed at least in triplicate. Data are presented as the mean ± SD. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. ns, no significant difference. Analysis with two-tailed Student’s t test.
We determined the effects of the C/EBPα saRNA CEBPA-51 on specific gene activation of C/EBPα and on pro-inflammatory cytokine expression in LPS-stimulated THP-1 cells. First, the experimental CEBPA-51 or unrelated control RNA (Luc-small interfering RNA [siRNA]) were twice transfected into THP-1 cells with the commercial transfection agent Lipofectamine 3000 (Figure 1C). Twenty-four hours after the second transfection, cells were pelleted for qRT-PCR assay. In the absence of LPS, the treatment of CEBPA-51 demonstrated an ability to significantly increase the expression of target C/EBPα gene by 1.8-fold and its downstream p21 gene by 2.2-fold relative to control. This confirmed an saRNA-mediated gene activity in non-LPS-stimulated THP-1 cells (Figure 1C). Increased expression of C/EBPα was also measured at the protein level by western blotting (Figure S1). Next, as shown in Figure 1D, the THP-1 cells transfected twice with experimental RNAs were stimulated with LPS for 4 h. As described above, cells were pelleted for qRT-PCR assay, and cell-free supernatants were collected for human cytokine ELISA. Of note, LPS stimulation (at 100 or 500 ng/mL) dramatically suppressed C/EBPα mRNA expression;40 however, the transient transfection of CEBPA-51 attenuated LPS-induced downregulation of C/EBPα and partially restored C/EBPα levels. Moreover, the ELISA results indicated that CEBPA-51 treatment in LPS-stimulated THP-1 cells significantly inhibited the levels of the pro-inflammatory cytokines TNF-α, IL-6, and IL-1β (Figures 1E–1G). Consistently, the transcript RNA of TNF-α and IL-6 was repressed by CEBPA-51 (Figure 1H).
LPS Inhibits C/EBPα Expression and Changes Immune Cell Subsets in hu-NSG Mice
Although LPS-induced inflammation studies have been investigated in many mouse models,41, 42, 43, 44 one limitation in those wild-type murine systems is the reliance on an entirely murine-based immune response to inflammation, thus resulting in different pathological conditions and some contradictory results in therapeutic efficacy studies when compared with those obtained in human patients. An LPS-induced inflammation animal model that can harbor human cells and mimic the human immune system may be useful to reduce and minimize the discrepancies between the murine and human immune systems, thus providing a better understanding of the human immune response and the effect of biologic therapy during LPS-induced inflammation. The NSG mouse45 is a well-established immunodeficient recipient murine strain that harbors various mutations,46 and it has been used successfully to achieve efficient, durable, systemic reconstitution of human cells in vivo—a process called “humanization.” Through the injection of human hematopoietic stem cells (HSCs) derived from fetal liver or umbilical cord blood or from mobilized adult CD34+ cells from PB, NSG newborn mice can be efficiently reconstituted with human lymphoid and myeloerythroid components. The “humanization” procedure results in a wide variety of human cells, such as CD4+ and CD8+ T cells, natural killer (NK) cells, monocytes and/or macrophages, neutrophils, and dendritic cells, in multiple organs of the resulting hu-NSG mice.47 hu-NSG mice have been widely used to investigate immune response and pathogenesis of various human diseases.48, 49, 50, 51 We used the human HSC-engrafted NSG mouse model to evaluate changes in several main human immune cell subsets of PB or bone marrow, including CD4+ and CD8+ T lymphocytes, monocytes and/or macrophages, and neutrophils, which regulate critical cytokine signaling,50 thus allowing study of key adaptive immune responses to LPS and the effects of MTL-CEBPA treatment. We established a hu-NSG mouse model as described previously.50 Flow cytometry was conducted to assess the human CD45+ cell population in the PB of NSG mice at 10–12 weeks after transplantation with fetal-liver-derived CD34+ HSCs (Figure S2). Experimental NSG mice that typically present 30%–75% of human CD45+ cell engraftment were chosen for our study.
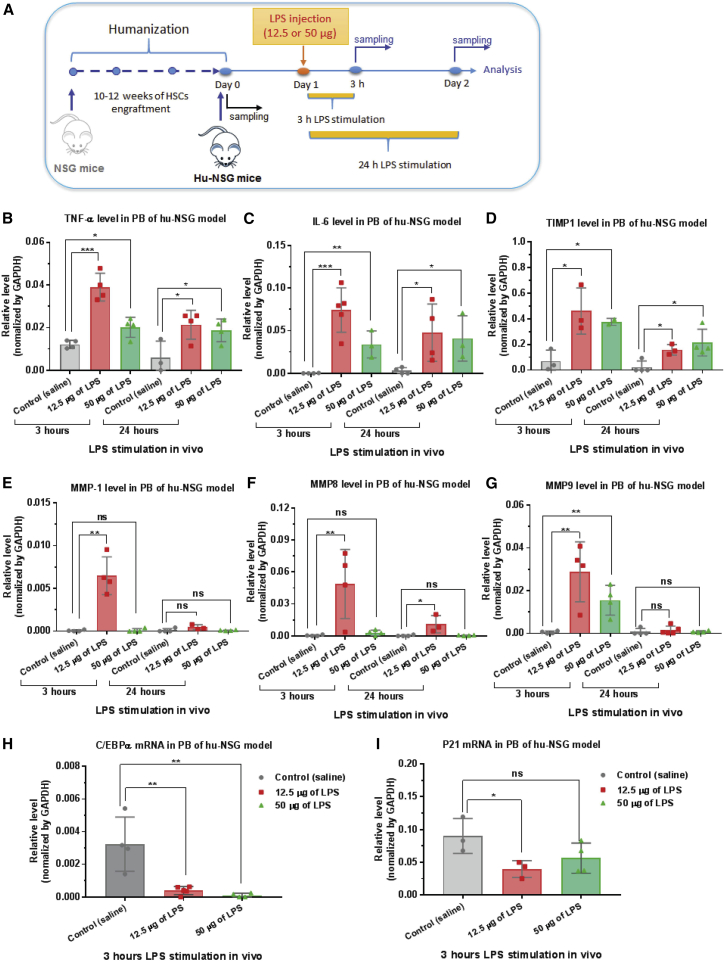
First, to identify the optimal dose and time of in vivo LPS stimulation, different doses of LPS (12.5 or 50 μg) were injected intraperitoneally into hu-NSG mice. At 3 or 24 h after LPS injection, PB was collected for qRT-PCR analysis (Figure 2A). Both doses of LPS stimulated production of the pro-inflammatory cytokines TNF-α and IL-6 (Figures 2B and 2C) in hu-NSG mice. The lower dose of LPS (12.5 μg) and shorter stimulation time (3 h) led to more pronounced cytokine expression in mouse PB. Moreover, the mRNA levels of several inflammatory mediators, such as TIMP1 (tissue inhibitor of metalloproteinase 1) and MMP1 (matrix metalloproteinase 1), -8, and -9, which have been reported to be upregulated after exposure of macrophages or monocytes to LPS stimulation in vitro, were also determined by qRT-PCR. The mRNA expression of these four tested genes were increased in PB after LPS injection (Figures 2D–2G). Similarly, a more marked upregulation was observed in the animals stimulated with a lower dose of LPS (12.5 μg) after 3 h of stimulation. Therefore, this dose was used to define the optimal stimulation condition for in vivo evaluation of MTL-CEBPA. In addition, we found that LPS stimulation inhibited the expression of C/EBPα mRNA and its downstream gene p21 in hu-NSG mice (Figures 2H and 2I), which was consistent with our observation in LPS-induced THP-1 cells and other previous studies.40, 52
Figure 2.
In Vivo LPS Stimulation Promotes Pro-inflammatory Cytokine Production and Inhibits C/EBPα Expression
(A) A schematic depicting the treatment of LPS-stimulated humanized NOD/SCID/IL2rγnull (hu-NSG) mice. After 10–12 weeks of CD34+ HSC engraftment, humanization of NSG mice was confirmed by flow cytometry. Different doses of LPS were injected intraperitoneally into hu-NSG mice. Peripheral blood (PB) was collected for analysis at various time points. (B and C) LPS stimulated the expression of pro-inflammatory cytokines in humanized NSG mice. Total RNA was isolated from PB for qRT-PCR analysis of cytokine (B) TNF-α and (C) IL-6 mRNAs. The lower dose of LPS (12.5 μg) and shorter stimulation time (3 h) led to more pronounced cytokine expression in mouse PB. (D–G) LPS upregulated the mRNA level of several inflammatory mediators: (D) TIMP1, (E) MMP1, (F) MMP8, and (G) MMP9. (H and I) In vivo LPS stimulation inhibited the expression of (H) C/EBPα mRNA and (I) its downstream gene p21 in humanized NSG mice. As described above, total RNA was isolated from PB for qRT-PCR analysis of the desired genes. Each determination analysis was performed in triplicate. Data are presented as the mean ± SD. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. ns, no significant difference. Analysis by two-tailed Student’s t test.
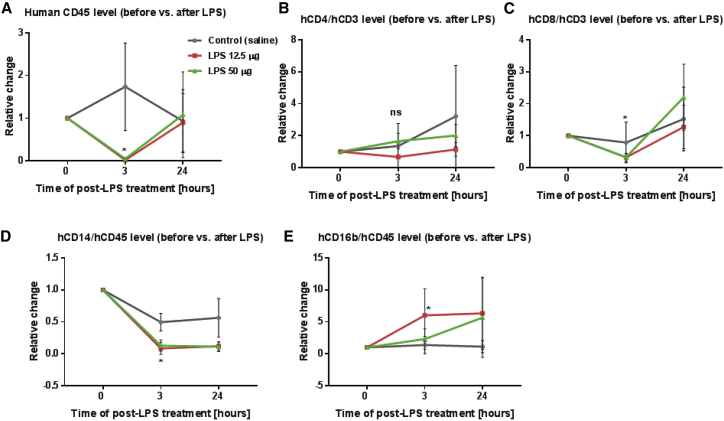
Next, to access the effect of LPS stimulation on immune cells in hu-NSG mice, leukocytes from PB were collected and stained with various antibodies for flow cytometric analysis (Figure S3). Because of mouse-to-mouse variation, we used the frequencies of each cell subset of individual mice before LPS injection (day 0) as a baseline and then calculated the relative change in frequency of each of the cell subsets. Therefore, the relative change of each cell subset before LPS-induced stimulation was defined as 1. The effect of LPS on cell subsets of PB is indicated in Figures 3A–3E. Compared to the baseline before LPS challenge, LPS administration induced a rapid, transient decline of human leukocytes (total hCD45 cells) at 3 h after LPS injection, and then returned to baseline levels at 24 h after LPS administration (Figure 3A). For lymphocytes, LPS also reduced the hCD8+ T cell subset, whereas it did not significantly affect the hCD4+ T cell subset at 3 h after LPS stimulation (Figures 3B and 3C). Furthermore, we observed different responses of monocytes and granulocytes to LPS exposure. Similar to previous studies, LPS challenge resulted in a decrease in the hCD14+ monocyte subset, while promoting the hCD16+ granulocyte subset (Figures 3D and 3E). Taken together, our results demonstrate that the in vivo LPS-stimulated hu-NSG mouse recapitulate several important aspects of acute inflammation and immune responses, thus supporting evaluation of the anti-inflammatory effects of MTL-CEBPA.
Figure 3.
In Vivo LPS Stimulation Affects Immune Cell Subsets in Humanized NSG Mice
As described in Figure 2A, leukocytes from PB were isolated at various time points and subsequently stained with various antibodies for flow cytometric analysis. Because of mouse-to-mouse variation, we used frequencies of each cell subset of individual mice before LPS injection (day 0) as a baseline and then calculated the relative frequency change of each cell subset at 3 and 24 h. The relative change of each cell subset before LPS stimulation was defined as 1. (A) Effect of LPS on human leukocytes. (B and C) Effect of LPS on T lymphocytes. (B) LPS did not significantly affect the human CD4+ T cell subset, but (C) it reduced the human CD8+ T cell subset at 3 h after LPS stimulation. (D and E) Effect of LPS on monocytes and granulocytes. (D) In vivo LPS stimulation resulted in a decrease in the human CD14+ monocyte subset, (E) but an increase in the human CD16+ granulocyte subset at 3 h after LPS stimulation. Flow analysis was performed in duplicate. Data are presented as the mean ± SD. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. ns, no significant difference. Analysis by two-tailed Student’s t test.
MTL-CEBPA Activates C/EBPα and Induces Differential Effects on Immune Cells in Naive hu-NSG Mice
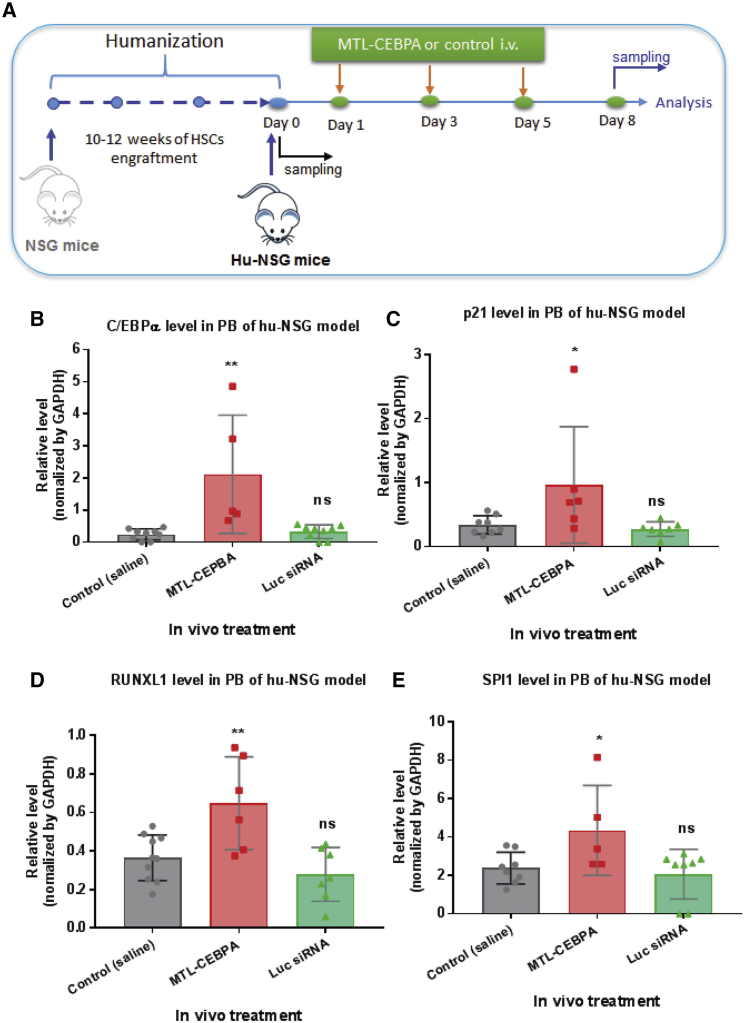
To investigate to what extent multiple doses of MTL-CEBPA may affect the level of target gene activation and cell differentiation, naive hu-NSG mice were treated three times on days 1, 3, and 5, with the respective MTL-CEBPA (3 mg/kg per injection) or unrelated control Luc-siRNA formulated with the same liposome nanoparticles (Figure 4A). We calculated human-equivalent doses of 3 mg/kg MTL-CEBPA on the basis of individual animal weights.22 In these conditions, the MTL-CEBPA administration via i.v. injection demonstrated the specific activation of its target C/EBPα mRNA and its downstream gene p21 (Figures 4B and 4C). In addition, we found that other downstream genes involved in myeloid differentiation, such as RUNX1 (Runt-related transcription factor 1) and SPI1 (transcription factor, PU.1), were also upregulated upon MTL-CEBPA treatment (Figures 4D and 4E).
Figure 4.
MTL-CEBPA Induces Specific Gene Activation in Naive Humanized NSG Mice
(A) A schematic depicting treatment with MTL-CEBPA in a naive humanized NSG mouse model. MTL-CEBPA or control Luc-siRNA (3 mg/kg per injection) was injected i.v. into humanized NSG mice three times on days 1, 3 and 5. Peripheral blood (PB) was collected for analysis at various time points (before and after treatment). (B and C) MTL-CEBPA administration mediated the specific activation of (B) its target C/EBPα mRNA and (C) its downstream gene p21. Total RNA was isolated from PB for qRT-PCR analysis of the desired genes as indicated. (D and E) MTL-CEBPA administration upregulated the expression of the downstream genes involved in myeloid differentiation: (D) RUNX1 and (E) SPI1. Each determination analysis was performed in triplicate. Data are presented as the mean ± SD. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. ns, no significant difference. Analysis by two-tailed Student’s t test.
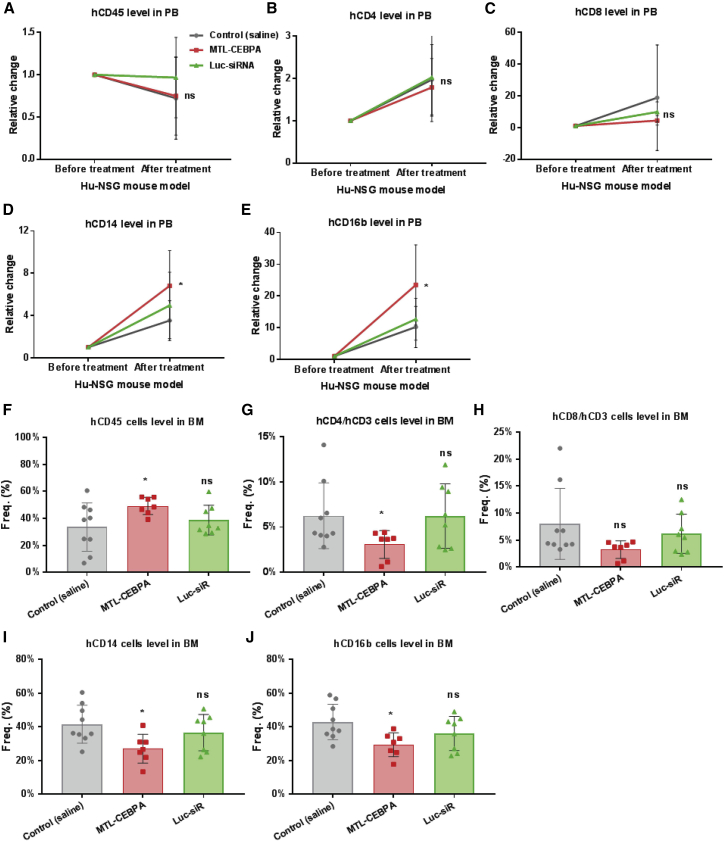
We also analyzed the dynamic response of immune cells in hu-NSG mice to MTL-CEBPA treatment. At 3 days after the last MTL-CEBPA treatment (day 8), leukocytes from PB and bone marrow were isolated for flow cytometric analysis, as described above (Figure 5). When cell subsets were normalized with the baseline prior to treatment (at day 0), we found that multiple doses of MTL-CEBPA did not significantly change total human CD45+ cells, and T lymphocytes (hCD4+ and hCD8+ T cell subsets) in PB (Figures 5A–5C). However, MTL-CEBPA treatment increased CD16+ granulocytes and CD14+ monocytes in PB (Figures 5D and 5E). Interestingly, the effect of MTL-CEBPA treatment on the cells from bone marrow was different. Compared to the control saline group, MTL-CEBPA treatment promoted total human CD45+ cells in bone marrow (Figure 5F), but reduced the number of hCD4+ T lymphocytes, CD14+ monocytes, and CD16+ granulocytes in bone marrow (Figures 5G–5J). These results suggest that MTL-CEBPA activates its target mRNA and related downstream genes and consequently promotes differentiation of myeloid cells, subsequently driving leukocyte trafficking from the bone marrow to the PB.
Figure 5.
MTL-CEBPA Leads to Differential Effects on Immune Cells in Naive Humanized NSG Mice
As described in Figure 4A, leukocytes from PB and cells from bone marrow (BM) were collected for flow cytometric analysis. Because of mouse-to-mouse variation, we used frequencies of each PB cell subset from individual mice before MTL-CEBPA injection (day 0) as a baseline and then calculated the relative frequency change of each cell subset at day 8. The relative change of each PB cell subset before MTL-CEBPA treatment was defined as 1. (A) Effect of MTL-CEBPA on human CD45+ leukocytes in PB. Multiple doses of MTL-CEBPA did not affect the (B) human CD4+ T cell subset and the (C) human CD8+ T cell subset in PB. MTL-CEBPA treatment increased the (D) human CD14+ monocyte subset and (E) human CD16+ granulocyte subset in PB. (F) Effect of MTL-CEBPA on human leukocytes in BM. MTL-CEBPA reduced (G) the human CD4+ T cell subset, but (H) did not significantly change the human CD8+ T cell subset in BM. MTL-CEBPA treatment reduced (I) the human CD14+ monocyte subset, and (J) the human CD16+ granulocyte subset in BM. Each flow analysis was performed in duplicate. Data are presented as the mean ± SD. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. ns, no significant difference. Analysis by two-tailed Student’s t test.
MTL-CEBPA Restores LPS-Induced Reduction of C/EBPα and Inhibits Pro-inflammatory Cytokines in LPS-Stimulated hu-NSG Mice
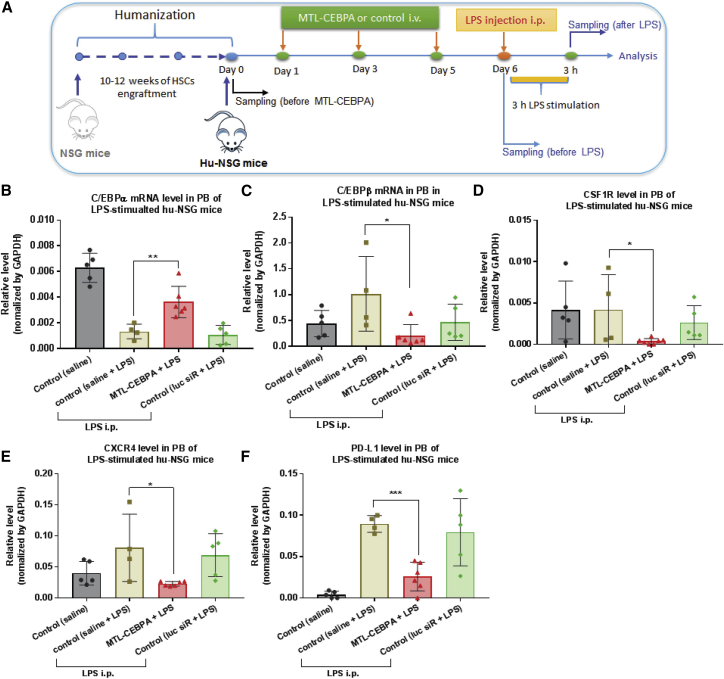
To investigate the in vivo anti-inflammatory potency and immune response of C/EBPα saRNA in the human immune system, we used the LPS-stimulated hu-NSG mouse as an acute inflammation model. After LPS challenge, pro-inflammatory cytokine releases and leukocyte trafficking from the bone marrow to the PB were traceable in the human NSG mice. As shown in Figure 6A, the hu-NSG mice were administered experimental RNAs—MTL-CEBPA or Luc-siRNA—with three doses of 3 mg/kg per i.v. injection. At 24 h after the last injection, one dose of LPS (12.5 μg) was injected intraperitoneally into hu-NSG mice, and the animals were sacrificed after 3 h of LPS stimulation (Figure 6A). Whole-blood samples were collected, and cellular and plasma fractions were separated for analysis. Consistent with our previous observation, although in vivo LPS stimulation suppressed C/EBPα mRNA level in PB, MTL-CEBPA restored C/EBPα expression (Figure 6B). Moreover, activating C/EBPα gene led to a decline of several genes associated with immune and inflammatory responses, such as C/EBPβ, CSF1R (Colony-Stimulating Factor 1 Receptor), CXCR4 (C-X-C Motif Chemokine Receptor 4) and PD-L1 (Programmed Cell Death 1 Ligand 1) (Figures 6C–6F).
Figure 6.
MTL-CEBPA Restores LPS-Induced Reduction of C/EBPα and Downregulates Several Genes Associated with Immune and Inflammatory Response in an LPS-Stimulated Humanized NSG Model
(A) A schematic depicting the treatment scheme of MTL-CEBPA for LPS-stimulated humanized NSG mouse model. MTL-CEBPA or control Luc-siRNA (3 mg/kg per injection) were injected i.v. into humanized NSG mice three times on days 1, 3 and 5. At 24 h after the last injection (day 6), a single dose of LPS (12.5 μg) was injected intraperitoneally into humanized NSG mice. After 3 h of in vivo LPS-stimulation, the animals were sacrificed for sampling. PB was collected for analysis at various time points. At the end of the experiment, BM cells were collected. (B) MTL-CEBPA treatment restored LPS-induced reduction of C/EBPα mRNA expression. (C–F) MTL-CEBPA treatment downregulated the expression of several genes associated with immune and inflammatory responses: (C) C/EBPβ, (D) CSF1R, (E) CXCR4, and (F) PD-L1. Total RNA was isolated from PB cells for qRT-PCR analysis of the desired genes, as indicated. Each determination analysis was performed in triplicate. Data are presented as the mean ± SD. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. ns, no significant difference. Analysis by two-tailed Student’s t test.
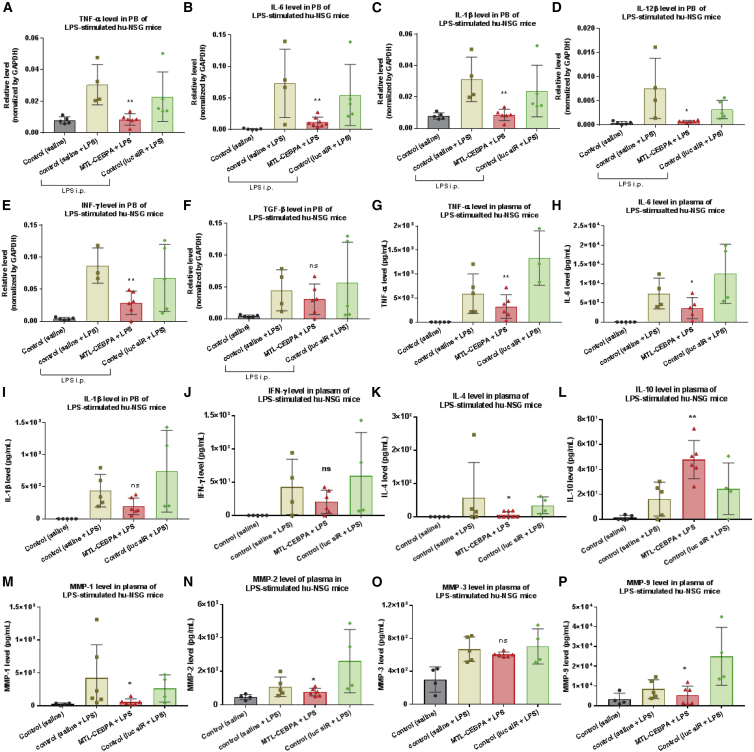
To determine to what extent MTL-CEBPA actively modulated the inflammatory response in the LPS-stimulated hu-NSG mice, total RNAs and mouse plasma from PB were accessed for cytokine profiling analysis by qRT-PCR and Luminex assay, respectively. As expected, mRNA expression of cellular pro-inflammatory cytokines was markedly upregulated after LPS injection (Figures 7A–7F). However, MTL-CEBPA treatment suppressed their expression, including TNF-α, IL-6, IL-1β, IL-12β, and IFN-γ, although no significant change was observed in TGF-β (Transforming Growth Factor-beta) mRNA (Figures 7A–7F). Furthermore, to validate the anti-inflammatory effect of MTL-CEBPA, secretion of inflammatory mediators in mouse plasma, such as soluble cytokines and MMPs, was measured by Luminex assay (Figures 7G–7P). Consistently, our results revealed that MTL-CEBPA treatment resulted in a decline of pro-inflammatory cytokines (TNF-α, IL-6, and IL-4) and MMPs (MMP1, -2, and -9). There was no significant change in the production of IL-1β, IFN-γ, and MMP3. Interestingly, compared with the LPS-stimulated control groups (saline or Luc-siRNA), we found that the production of IL-10, an anti-inflammatory cytokine, was increased in the animals treated with MTL-CEBPA (Figure 7L). Our data suggested that MTL-CEBPA effectively repressed LPS-triggered inflammatory response in vivo.
Figure 7.
MTL-CEBPA Inhibits Pro-inflammatory Cytokine Production in an LPS-Stimulated Humanized NSG Model
As described in Figure 6A, at the end of LPS stimulation, animals were sacrificed, whole-blood samples were collected, and cellular and plasma fractions were separated for cytokine profiling analysis by qRT-PCR and Luminex assay, respectively. (A–F) MTL-CEBPA treatment downregulated mRNA expression of the cellular pro-inflammatory cytokines (A) TNF-α, (B) IL-6, (C) IL-1β, (D) IL-12β, and (E) IFN-γ, but did not significantly change (F) TGF-β. (G–P) The effect of MTL-CEBPA treatment on secretion of soluble inflammatory mediators in mouse plasma was measured by Luminex assay. MTL-CEBPA treatment resulted in a decline of the pro-inflammatory cytokines (G) TNF-α, (H) IL-6, and (K) IL-4 and the MMPs (M) MMP1, (N) MMP2, and (P) MMP9. There was no significant change in the production of (I) IL-1β, (J) IFN-γ, and (O) MMP3. (L) MTL-CEBPA promoted the production of IL-10, an anti-inflammatory cytokine. Each determination analysis was performed in duplicate. Data are presented as the mean ± SD. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. ns, no significant difference. Analysis by two-tailed Student’s t test.
MTL-CEBPA Promotes CD8+ T-Lymphocytes and CD16+ Granulocytes in LPS-Stimulated hu-NSG Mice
We further evaluated the impact of systematic administration of MTL-CEBPA on various immune cells in LPS-stimulated hu-NSG mice, we conducted flow cytometry to measure the change of cell subsets in the PB and BM (Table 1). Because of mouse-to-mouse variation, we used the frequency of each cell subset of individual mice before LPS injection as a baseline and then calculated the relative frequency change of each cell subset. Therefore, the relative change of each cell subset before LPS stimulation was defined as 1. The effects of LPS and MTL-CEBPA on cell subsets of PB are indicated in Figures 8A–8L. Compared with untreated control mice (saline+LPS), we observed an increase in total human CD45 cells (Figure 8A) and in the human CD8+ T lymphocyte subset in PB after MTL-CEBPA treatment (Figure 8C), although the human CD4+ T lymphocyte subset was not significantly changed (Figure 8B). In the absence of MTL-CEBPA, LPS stimulation promoted the PD-1+ CD4+ and PD-1+ CD8+ T lymphocyte subsets (Figures 8D and 8E). However, MTL-CEBPA treatment reversed the LPS-induced increase and reduced of PD-1+ T lymphocyte subsets in PB. In addition, MTL-CEBPA treatment activated T lymphocytes, indicating an increase in the CD25+, CD69+ and CD45RO+ subsets (Figures 8F–8H). Although MTL-CEBPA treatment had no significant impact on the CD14+ monocyte or CD11b+CD14+ macrophage subsets (Figures 8I and 8J), it actually expanded CD16+ granulocytes, including the CD10+ neutrophils subset (Figures 8K and 8L). Similar to the observations in MTL-CEBPA-treated naive hu-NSG mice, MTL-CEBPA treatment resulted in differential immune response on PB cells versus BM cells (Table 1). The effect on BM cells at the endpoint is shown in Figures 8M–8X. In the presence of MTL-CEBPA, total human CD45 cells in BM was reduced (Figure 8M), which was the opposite of the observation in human CD45 cells in PB. There was no significant effect on the human CD4+ T lymphocyte subset (Figure 8N). However, increases in the human CD8+ T lymphocyte subset (Figure 8O) and activated T lymphocyte subsets (Figures 8R–8T) were observed in the group treated with MTL-CEBPA. In contrast to the effect on PD-1+ subsets in PB, MTL-CEBPA treatment increased PD-1+ T lymphocyte subsets in BM. Moreover, we found that MTL-CEBPA treatment also expanded CD14+ monocytes and/or macrophages (Figures 8U and 8V) and CD16+ granulocytes and/or neutrophils, including the CD10+ neutrophil subset (Figures 8W and 8X). Collectively, our data demonstrate that MTL-CEBPA led to differential effects on immune cells from PB and BM in LPS-stimulated hu-NSG mice. It promoted differentiation of myeloid cells to monocytes and granulocytes, and it activated CD8+ T lymphocytes.
Table 1.
The Effect of MTL-CEBPA on Various Immune Cell Subsets of LPS-Stimulated hu-NSG Mice
| Subsets | Cells from LPS-Stimulated Hu-NSG Mice | PB Cells | BM Cells |
|---|---|---|---|
| T lymphocytes | Hu-CD45+ | ↑ | ↓ |
| Hu-CD4+/hu-CD3+ | ns | ns | |
| Hu-CD8+/hu-CD3+ | ↑ | ↑ | |
| Hu-PD-1+/hu-CD4+ | ↓ | ↑ | |
| Hu-PD-1+/hu-CD8+ | ↓ | ↑ | |
| Hu-CD25+/hu-CD3+ | ↑ | ↑ | |
| Hu-CD69+/hu-CD3+ | ↑ | ns | |
| Hu-CD45RO+/hu-CD3+ | ↑ | ns | |
| Macrophages and granulocytes | Hu-CD14+/CD45+ (monocytes) | ns | ↑ |
| Hu-CD11b+/CD14+ (macrophage) | ns | ↑ | |
| Hu-CD16b+/CD45+ (granulocytes) | ↑ | ↑ | |
| Hu-CD10+/CD16b+/CD14− (mature neutrophils) | ↑ | ↑ |
ns, no significant change; ↑, increase; ↓, decrease; PB, peripheral blood; BM, bone marrow.
Figure 8.
MTL-CEBPA Promotes CD8+ T Lymphocytes and CD16+ Granulocytes in an LPS-Stimulated Humanized NSG Model
As described in Figure 6A, cells from PB and BM were collected for flow cytometric analysis. (A) MTL-CEBPA increased human CD45+ cells in PB. It did not affect (B) human CD4+ T cell subset, but (C) increased human CD8+ T cell subset in PB. MTL-CEBPA reduced the LPS-induced increase of (D) PD-1+ CD4+ and (E) PD-1+CD8+ T lymphocyte subsets in PB. MTL-CEBPA activated the (F) human CD25+, (G) CD69+, and (H) CD45RO+ T lymphocyte subsets. MTL-CEBPA treatment had no significant impact on (I) CD14+ monocyte or (J) CD11b+ CD14+ macrophage subset in PB. It promoted the (K) CD16+ granulocyte and (L) CD10+CD16+CD14− neutrophil subsets in PB. (M) MTL-CEBPA reduced human CD45+ cells in BM. (N) MTL-CEBPA did not significantly change the human CD4+ T lymphocyte subset, but (O) it increased the human CD8+ T lymphocyte subset in BM. It increased the (P) PD-1+ CD4+ T lymphocyte and (Q) PD-1+CD8+ T lymphocyte subsets in BM. It also activated T lymphocytes in BM: (R) human CD25+, (S) CD69+, and (T) CD45RO+ subsets. MTL-CEBPA treatment expanded the (U) human CD14+ monocyte and (V) CD11b+CD14+ macrophage subsets and the (W) CD16+ granulocyte and (X) CD10+CD16+CD14− neutrophil subsets in bone marrow. Each determination analysis was performed in duplicate. Data are presented as the mean ± SD. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. ns, no significant difference. Analysis by two-tailed Student’s t test.
Our results demonstrate that C/EBPα saRNA can downregulate pro-inflammatory cytokine production in human immune cells previously stimulated by LPS. In LPS-challenged humanized mice, we observed that MTL-CEBPA administered by i.v. injection upregulated C/EBPα mRNA, expanded CD14–CD16+ neutrophils and attenuated production of several key pro-inflammatory cytokines, including TNF-α, IL-6, IL-1β, IL-12β, and IFN-γ. Moreover, Luminex analysis of mouse serum revealed that MTL-CEBPA significantly inhibited pro-inflammatory cytokines and MMP expression (e.g., TNF-α; IL-6; IL-4; and MMP-1, -2, and -9), and increased anti-inflammatory cytokine production (IL-10).
Taken together, these data show that MTL-CEBPA treatment mediated the suppression of LPS-induced pro-inflammatory cytokines and the associated changes in inflammatory cell subsets through the regulation of the expression of multiple genes involved in the inflammatory response. Additional molecular and mechanistic studies are necessary to address specific pathways involved in the MTL-CEBPA mediated anti-inflammatory response.
Discussion
The data presented here demonstrate that increased expression of C/EBPα at the time of endotoxin (LPS) stimulation of THP-1 cells and endotoxin challenge in humanized mice resulted in reduced expression of inflammatory cytokines and increased levels of the anti-inflammatory cytokine IL-10. Endotoxin (LPS) challenge in mice activates the NLRP3 inflammasome pathway, leading to transient cytokine release and a systemic acute-phase response, with subsequent downregulation of cytokines and normalization of acute-phase response biomarkers.33 During the acute phase of the response to endotoxin, without interventions that artificially increase C/EBPα expression, our data and the literature demonstrate increased C/EBPβ expression and decreased or unchanged C/EBPα expression.26, 53 However, with prolonged LPS stimulation, the late phase of this response is associated with reduced inflammasome activity that in part is mediated by production of IL-10.54 Our results therefore suggest that interventions that increase C/EBPα expression during an acute inflammatory response downregulate that response by mimicking transcriptional changes that naturally occur in the late phases of an NLRP3 inflammasome-driven response when inflammation is decreasing. In our study, we clearly showed that treatment with MTL-CEBPA prior to LPS stimulation blunted the expression of C/EBPβ, increased C/EBPα expression, and decreased the pro-inflammatory cytokine response, while increasing levels of the anti-inflammatory cytokine, IL-10.55 To our knowledge, this is the first direct evidence that helps explain the biological reason for decreased C/EBPα expression during the early phases of an acute inflammatory response. If instead C/EBPα expression had been increased, the inflammatory response may have been aborted at an early stage and may have blunted the host’s protective response to infection.
In addition, C/EBPα saRNA-mediated downregulation of cytokines has also been observed in humans and in other pre-clinical studies. In a case study of a patient who was treated with MTL-CEBPA (ClinicalTrials.gov: NCT02716012), the patient’s tumor decreased by 75% on computed tomographic (CT) scan and magnetic resonance imaging (MRI). It is of interested to note that white blood cell (WBC) cytokines showed a massive decrease in expression of IFN-γ, TNF-α, and IL-6r (V.R, R.H., and N.H., unpublished data). In a liver inflammation and/or failure rat model, the MTL-CEBPA was demonstrated to improve survival and prevent liver failure with normalization of liver enzyme.22 Moreover, in a diethylnitrosamine (DEN)-induced cirrhotic HCC rat model, C/EBPα saRNA delivered by a transferrin receptor (TfR) aptamer improved liver function and downregulated cytokines, including IFN-α, IL-6, IL-1β, and TGF-β1 (K.W.H., R.H., and N.H., unpublished data). Consistently, these data demonstrate the beneficial effect of the use of MTL-CEBPA in an actual disease scenario.
The transcription factor βα is known to play a critical role during embryogenesis and subsequently in adipogenesis, in homeostatic glucose metabolism, and, importantly, in myeloid development.12 We know that the binding sites for the C/EBP family of transcription factors are present in the promoter regions of numerous genes and downstream pathways, with significant influence on maintenance of cellular function and response to injury in specific cellular populations.9, 56 C/EBPα is also known to be a master regulator of hematopoiesis by inducing myeloid differentiation. It has been shown that myeloid lineage-specific deletion of C/EBPα results in significantly enhanced myeloid-derived suppressor cell (MDSC) proliferation and expansion and in an increase in myeloid progenitors and a decrease in mature cells13. Specifically, C/EBPα is a myeloid-specific transcription factor that couples lineage commitment to terminal differentiation and cell cycle arrest.10 Mutations in the gene predispose mice to a granulocytic myeloproliferative disorder characterized by a block in granulocyte differentiation, accumulation of myeloblasts and promyelocytes, and expansion of myeloid progenitor populations.11 The data presented in this study also demonstrate that the anti-inflammatory effects of MTL-CEBPA are associated with the differentiation of myeloid cells to monocytes and granulocytes and activation of CD4 and CD8 T lymphocytes.
In addition to the reduced inflammatory response to endotoxin, treatment of humanized mice with MTL-CEBPA challenged with LPS resulted in an increased number of T cells expressing CD25, CD45RO, and CD69, suggesting the presence in blood of higher levels of activated T cells. T cells also had reduced expression of PD-1 that would reduce the immunosuppressive effects of PD-L1. This result is compatible with the finding of higher levels of activated T cells in blood. The exact mechanism behind this increase in the context of an inflammatory response is not clear; however, previously published data demonstrating that C/EBPα restricts IFN-γ expression in T cells to allow proper class switching by B cells are also interesting to consider in this context.57
These data suggest that the anti-inflammatory effects of MTL-CEBPA are also differentiated from those of other anti-inflammatory agents that target pro-inflammatory cytokines, such as TNF-α and IL-1β. For example, anti-TNF-α biologics reduce T cell and macrophage immune function and increase the risk of tuberculosis reactivation.58 Although the risk of tuberculosis seems to be lower with newer biologics, serious infections caused by viral and fungal pathogens and by other intracellular pathogens occur more often in patients with rheumatoid arthritis treated with these agents.6, 58, 59 MTL-CEBPA may therefore be a safer anti-inflammatory agent compared to those targeting specific cytokines important in host defense. In the inflammatory bowel disorders (Crohn’s disease and ulcerative colitis), there is evidence suggesting that the cytokine profiles that trigger an acute episode are disease specific and relate to either Th1- or Th2-like differentiation processes, resulting in differing cytokine responses.60 The limitation of current biologics may therefore be that they target the inflammatory process too far down the pathway and as a result can either lack efficacy or result in undesirable side effects. Similarly, in other important inflammatory conditions such as gout, biological therapeutics have focused on targeting IL-1β or IL-6.61 The advantage of targeting a broader range of cytokines farther up in the pathway appears desirable in this context and represents exciting further areas of research for MTL-CEBPA.
The role of C/EBPα in IL-6 and G-CSF production to induce granulopoiesis has been previously described.62 The data presented in the current study similarly show that C/EBPα saRNA attenuates LPS-induced downregulation of C/EBPα and inhibits IL-6 (and other pro-inflammatory cytokine) production in LPS-stimulated THP-1 monocytes.
Yan et al.25 have shown in a similar LPS-induced septic shock model that C/EBPα-deficient macrophages produce less IL-10 in response to LPS and that this process is regulated by the downstream pleiotropic protein progranulin. They also identified that mice deficient in C/EBPα in hematopoietic cells are highly vulnerable to LPS-induced septic shock.25 In our study, production of IL-10 increased in LPS-challenged mice treated with MTL-CEBPA compared with an LPS-challenged control group, further supporting the previously published data, and showing that IL-10 is an important downregulating mechanism in NLRP3 inflammasome-driven inflammation.55
Based on clinical data from the phase 1 trial evaluating patients with HCC, we observed a consistent increase in expression of C/EBPα mRNA in the total WBC population and a reversible increase in WBCs and neutrophil counts following administration of MTL-CEBPA (Sarker et al., 2017, J. Clin. Oncol., abstract; Sarker et al., 2018, J. Clin. Oncol., abstract). Ma et al.63 showed that higher levels of C/EBPα are required for granulocyte and lower levels for monocyte lineage specification and that this myeloid bifurcation may be facilitated by increased C/EBPα gene expression in granulocyte compared with monocyte progenitors. Friedman et al.64 proposed that, although C/EBPα is required for development of immature granulocyte-monocyte progenitors, it subsequently inhibits monopoiesis and induces granulopoiesis. The relevance of these findings in relation to the efficacy of anti-cancer treatment are unclear, but the effects are likely to originate in the myeloid cells of the tumor immune microenvironment and may be linked to the anti-inflammatory myeloid cell population phenotype.
Although therapy with MTL-CEBPA is currently being evaluated in cancer, our data support further investigation of MTL-CEBPA in diseases characterized by poorly controlled inflammation associated with excessive inflammasome activation.65 In particular, acute or chronic inflammatory diseases associated with NLRP3 inflammasome activation, such as sepsis with organ dysfunction,66 NASH,67 and rheumatoid arthritis68 should be considered.
Materials and Methods
Chemicals and Reagents
Unless otherwise noted, all chemicals, including LPS and E. coli O111:B4 (1 mg/mL solution), were purchased from Sigma-Aldrich; all restriction enzymes were obtained from New England BioLabs (NEB); and all cell culture products were purchased from Gibco (Gibco-BRL, a division of Life Technologies). Human TNF-α, human IL-6 ELISA kits, and anticoagulant tubes were purchased from BD Biosciences. Sources for the other reagents were as follows: Lipofectamine 3000 (Thermo Fisher Scientific); QuantiNova Reverse Transcription kit (QIAGEN); human CD34+ enrichment by magnetic-activated cell sorting (MACS) system (Miltenyi Biotec); DNeasy blood and tissue kit (QIAGEN); SsoAdvanced Universal SYBR Green Supermix (Bio-Rad); and THP-1 monocytes (NIH AIDS Research and Reference Reagent Program).
DNA oligos, primers, siRNAs, and saRNAs were purchased from Integrated DNA Technologies (IDT). QuantiTect Primers used for qRT-PCR were purchased from QIAGEN. MTL-CEBPA (liposome-formulated C/EBPα saRNA nanoparticles), and the control liposome-formulated Luc-siRNA nanoparticles were provided by MiNA Therapeutics, London, UK.22
RNA Sequences
C/EBPα saRNA (CEBPA-51):30 sense, 5′-GCmG GmUC mAUmU GmUC mACmU GmGU CmUmU-3′ (special: 2′-OMe modified), and antisense, 5′-GAC CAG UGA CAA UGA CCG CmUmU-3′ (special: 2′-OMe modified 3′-UU overhang). Unrelated control siRNA: sense, 5′-GCG GAG ACA GCG ACG AAG AGC UCA UCA-3′, and antisense, 5′-UGA GCU CUU CGU CGC UGU CUC CGC UU-3′.
Cell Lines and Cell Culture
THP-1 cells, a human monocytic cell line, obtained through the NIH AIDS Research and Reference Reagent Program, is a suspension cell line and was grown in RPMI-1640 medium supplemented with 10% FBS, 1.0 mM sodium pyruvate, and 0.05 mM 2-mercaptoethanol. Cells were cultured in a humidified 5% CO2 incubator at 37°C and split 1:10 once per week upon reaching confluence.
In Vitro Evaluation in LPS-Stimulated THP-1 Cell Model
In Vitro Small RNA Transfection and LPS-Stimulation of THP-1 Cells
On the experimental day, the THP-1 cells were grown in 24-well plates with seeding at 2 × 105 in 400 μL complete culture medium. Subsequently, the cells were transfected with 10 nM experimental saRNA or control siRNA using Lipofectamine 3000 (Thermo Fisher Scientific) in accordance with the manufacturer’s instructions. At 24 h after the first transfection, the cells were transfected with the same experimental RNAs again, as described above. At 48 h after first transfection, the cells were stimulated with 0, 100, or 500 ng/mL LPS from E. coli (O111:B4; Sigma). Stimulations were carried out for 2, 4, 6, and 24 h. Afterward, the cell suspensions were pelleted by centrifugation. The cell-free supernatants (conditional media) were transferred into a new tube and stored at −80°C until quantification of cytokines by ELISA or Luminex assay. The cell pellets were used for total RNA extraction with Trizol (Thermo Fisher Scientific), according to the manufacturer’s instructions.
Measurement of Human Cytokine Production by ELISA and qRT-PCR Assay
Concentrations of the pro-inflammatory cytokines TNF-α and IL-6 in the cell-free supernatant were determined with a BD Biosciences ELISA kit. The gene expression of cytokines in the total RNA was quantified with a qRT-PCR assay. cDNA synthesis was performed with the QuantiNova Revese Transcription kit (QIAGEN), according to the manufacturer’s recommended protocol. Expression of the TNF-α and IL-6 coding RNAs was analyzed by qRT-PCR using SsoAdvanced Universal SYBR Green Supermix (Bio-Rad), and specific QuantiTect Primers sets (QIAGEN) at a final concentration of 400 nM. GAPDH expression was used for normalization of the qRT-PCR data. QuantiTect Primers sets were listed as TNF-α (cat no. QT00029162), IL-6 (cat no. QT00083720), and GAPDH (cat no. QT00079247).
Quantitation of Target mRNA C/EBPα and Its Downstream Gene p21 by qRT-PCR Assay
Total RNA was extracted from THP-1 cells transfected with experimental small RNAs 48 h after the first transfection. The cDNA was produced using 2 μg of the total RNA. Reverse transcription was carried out using the QuantiNova Reverse Transcription kit (QIAGEN), according to the manufacturer’s recommended protocol. Expression of the target mRNA C/EBPα and its downstream gene p21 was analyzed by qRT-PCR using SsoAdvanced Universal SYBR Green Supermix (Bio-Rad), and specific QuantiTect Primers sets (QIAGEN) at a final concentration of 400 nM, as described above. GAPDH expression was used for normalization of the qRT-PCR data. QuantiTect Primers sets are listed as CEBPA (cat no. QT00203357), P21 (CDKN1A; cat no. QT00062090), and GAPDH (cat no. QT00079247).
Generation and Characterization of hu-NSG
Establishment and characterization of CD34+ HSC-engrafted NSG mice have been described previously.50
Ethics Statement
All animal care and procedures were performed according to protocols reviewed and approved by the City of Hope Institutional Animal Care and Use Committee (IACUC) held by the senior author of this paper ( John Rossi, IACUC 12034). Human fetal liver tissue was obtained from Advance Bioscience Resources (Alameda, CA), a nonprofit organization, in accordance with federal and state regulations. The vendor has its own Institutional Review Board (IRB) and is compliant with human subject protection requirements.
The research involved blood specimens from anonymous human subjects with no identifiers to age, race, ethnicity, or gender. All human tissue specimens were obtained from healthy, anonymous donors by third-party sources. We used discarded PB from anonymous, healthy, adult donors from the City of Hope Blood Donor Center (Duarte, CA), for isolation of PB mononuclear cells (PBMCs) and subsequent primary CD4+ T cell cultures. The information provided for the above was evaluated and determined not to involve research in human subjects (45 Code of Federal Regulations [CFR] § 46.102 (d)(f)). Therefore, it does not have to be approved, nor does it have to undergo continuing review by the Institutional Review Board (IRB) of the City of Hope (IRB/REF: 97071/075546).
In Vivo Evaluation in hu-NSG Mouse Model
In Vivo MTL-CEBPA Treatment and LPS Stimulation
To evaluate the effect of MTL-CEBPA on naive hu-NSG mouse model, we administered the experimental saRNA (MTL-CEBPA) or control Luc-siRNA to hu-NSG mice in the absence of LPS stimulation by i.v. injection at a 3 mg/kg dose in a 200 μL injection volume. The injections were administered every 2 days three times (on days 1, 3, and 5). At 1 d before MTL-CEBPA treatment (day 0), mice were bled by the retro-orbital route, and PB was collected for total RNA extraction and WBC isolation. At 3 days after the last MTL-CEBPA treatment (day 8), the mice were euthanized, and PB and bone marrow were collected for total RNA extraction and WBC isolation. The expression of desired genes, such as cytokines, target C/EBPα, and related mediators, and the inflammatory cell subpopulations were subsequently assayed. Three groups of animals were randomly assigned for the experiments. n represents the number of tested mice per group. Group 1: saline (n = 7); group 2: MTL-CEBPA (n = 6); and group 3: Luc-siRNA control (n = 7).
In addition, the effect of MTL-CEBPA on the LPS-induced hu-NSG mouse model was determined. Mice were randomly allocated to the following groups: saline, saline plus LPS, MTL-CEBPA plus LPS, and Luc-siRNA plus LPS. Mice from the control and LPS groups received an equal volume of PBS. As described above, the treatment of experimental RNAs was conducted three times (on days 1, 3, and 5). At 21 h after the last MTL-CEBPA treatment (day 6), LPS (12.5 μg per mice) was administered via intraperitoneal (i.p.) injection. At 3 h after LPS induction, mice were euthanized, and blood and bone marrow samples were collected as described above. Mouse serum was collected and stored at −80°C until use for the Luminex assay. Four groups of animals were randomly assigned for the test. n represents the number of tested mice per group. Group 1: saline (n = 5); group 2: saline+LPS (n = 5); group 3: MTL-CEBPA+LPS (n = 6); and group 4: Luc-siRNA control+LPS (n = 5).
Quantitation of Desired Gene Expression by qRT-PCR Assay
Total RNA was extracted from 50–140 μL of EDTA-treated plasma with the QIAamp Viral RNA kit (QIAGEN). The cDNA synthesis was performed with a QuantiNova Reverse Transcription kit (QIAGEN), according to the manufacturer’s recommended protocol. Expression of the desired RNAs was analyzed by qRT-PCR using SsoAdvanced Universal SYBR Green Supermix (Bio-Rad) and specific QuantiTect Primers sets (QIAGEN) at a final concentration of 400 nM. GAPDH expression was used for normalization of the qRT-PCR data. QuantiTect Primers sets are listed as CEBPA (cat. no. QT00203357); P21 (CDKN1A; cat. no. QT00062090); TNF-α (cat. no. QT00029162); IL-6 (cat. no. QT00083720); IL-4 (cat. no. QT00012565); IL-1β (cat. no. QT00021385); IL-12β (cat. no. QT00000364); TGF-β (cat. no. QT00000728); IFN-γ (cat. no. QT00000525); NOS1 (cat. no. QT00043372); CEBPB (cat. no. QT00237580); CXCR4 (cat. no. QT00223188); PD-L1 (CD274; cat. no. QT00082775); CSF1R (cat. no. QT00073276); RUNX1 (cat. no. QT00026712); SPl1 (cat. no. QT00082222); GAPDH (cat. no. QT00079247); TIMP1 (cat. no. QT00084168); MMP1 (cat. no. QT00014581); MMP8 (cat. no. QT00029820); and MMP9 (cat. no. QT00040040).
Measurement of Human Cytokine Production by Luminex Assay
As described above, hu-NSG mice were treated with experimental RNAs at days 1, 3, and 5. At day 6, the animals were stimulated with LPS via i.p. injection for 3 h to induce an acute inflammatory response. Subsequently, the animals were euthanized and blood and bone marrow samples obtained. A Luminex X-MAP bead array-based assay was performed at the Analytical Pharmacology Core Facility (APCF), City of Hope, to simultaneously quantify multiple cytokines and inflammatory mediators in mouse serum. The Luminex kit covered TNF-α, IL-6, IL-4, IL-10, IFN-γ, MMP-1, MMP-2, MMP-3, MMP-9, and IL-1β.
Analysis of Immune Cell Subsets by Flow Cytometry
As described above, whole blood was collected from experimental animals before treatment (at day 0) and after treatment (at day 6), including pre-LPS stimulation (without LPS) and post-LPS stimulation (with 3 h LPS induction). Red blood cells were lysed as reported. Flow cytometric analysis of human cells was performed in the Analytical Cytometry Core, City of Hope, on a FACSCalibur instrument (BD Biosciences) with either BD Fortessa (BD Biosciences) or FlowJo software, version 8.8.6. The gating strategy performed was an initial forward-scatter versus side-scatter (FSC/SSC) gating to exclude debris, followed by a human CD45 cell gate. For analysis of lymphocyte populations in PB, a further lymphoid gate (low side scatter) was also applied to exclude cells of monocytic origin. All antibodies used were fluorochrome conjugated and human specific and obtained from BD Biosciences: BB515-conjugated CD45 (clone HI30); phycoerythrin (PE)-Cy7-conjugated CD3 (clone SK7); Pacific Blue-conjugated CD4 (clone RPA-T4); BUV395-conjugated CD8 (clone RPA-T8); PE-conjugated PD-1 (clone MIH 4); BB700-conjugated CD45RO (clone UCHL1); BUV737-conjuagted CD25 (clone 2A3); APC-conjugated CD69 (clone FN50); BUV395-conjugated CD10 (clone HI10a); PE-Cy7-conjugated CD11b (clone ICRF44); Alexa 700-conjugated CD14 (clone M5E2); and BV421-conjugated CD16b (clone CLB-gran11.5). Gates were set using fluorescence − 1 controls, where cells were stained with all antibodies except the one of interest. Specificity was also confirmed by using isotype-matched nonspecific antibodies (BD Biosciences) and tissues from animals that had not been engrafted with human cells.
Statistical Analysis
Unless otherwise noted, error bars in all figures represent SD or SEM. GraphPad Prism software was used for statistical analyses by unpaired Student’s t test (two-tailed) or one-way ANOVA, and differences were considered statistically significant when p < 0.05.
Author Contributions
J.Z. and J.J.R. conceived the study, designed the experiments, and wrote the main manuscript; J.Z. prepared the animal IACUC protocol and data analysis and prepared the table and figures; H.L. conducted the in vitro test, including RNA transfection, qRT-PCR, ELISA, and western blot analysis; H.L., and X.X. performed the human cell isolation, engraftment, and establishment and the characterization of the hu-NSG mice and the LPS-stimulated hu-NSG mice; H.L., and X.X. performed the in vivo experiments, including injection, tissue/cell isolation, total RNA isolation, the qRT-PCR assay, and the flow cytometry analysis; A.H. and N.P. helped with the isolation of human cells and the flow cytometry analysis; V.R. provided the MTL-CEBPA materials and designed the flow cytometry protocol; B.H.L. helped with the interpretation of the results, provided relevant literature, and contributed to the writing of the Discussion in the manuscript, in addition to making general comments on the manuscript; V.R., M.H.S., K-W.H., D.B., N.A.H., and D.D. provided helpful advice and comments on the manuscript; and J.J.R., N.A.H., and R.H. provided funding. All authors reviewed the manuscript.
Conflicts of Interest
J.J.R, N.A.H, V.R., R.H., K-W.H., D.B., and D.D own stock in MiNA (Holdings) Ltd. All remaining authors declare no competing interests.
Acknowledgments
We would like to thank the City of Hope Animal Resources Center (ARC) for the support of animal use and care, the Analytical Cytometry Core for cell sorting and technical assistance in the flow cytometry analyses, the Pathology Core for the postmortem analysis of blood and tissue specimens, and the Analytical Pharmacology Core for the Luminex analyses of multiple cytokines in mouse serum. Finally, we would like to thank Drs. Marina Botto and Shiranee Sriskandan, Imperial College London, UK, and Dr. Steve Felstead, Deljay Consulting, Ltd., Earlston, UK, for reviewing the draft manuscript and providing helpful advice and comments. This project was supported by the NIH (grant numbers R01AI29329, R01AI42552, and R01HL07470 to J.J.R.), the National Cancer Institute of the NIH (grant number P30CA033572 to support City of Hope Integrative Genomics, Analytical Pharmacology, and Analytical Cytometry Cores), and MiNA Therapeutics Ltd. Funding for the open access charge was provided by the NIH. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. We obtained THP-1 cells from the NIH AIDS Research and Reference Reagent Program.
Footnotes
Supplemental Information can be found with this article online at https://doi.org/10.1016/j.ymthe.2019.02.018.
Contributor Information
Nagy A. Habib, Email: nagy.habib@imperial.ac.uk.
John J. Rossi, Email: jrossi@coh.org.
Supplemental Information
References
- 1.Eming S.A., Krieg T., Davidson J.M. Inflammation in wound repair: molecular and cellular mechanisms. J. Invest. Dermatol. 2007;127:514–525. doi: 10.1038/sj.jid.5700701. [DOI] [PubMed] [Google Scholar]
- 2.Kumar R., Clermont G., Vodovotz Y., Chow C.C. The dynamics of acute inflammation. J. Theor. Biol. 2004;230:145–155. doi: 10.1016/j.jtbi.2004.04.044. [DOI] [PubMed] [Google Scholar]
- 3.Caughey G.H. Mast cell tryptases and chymases in inflammation and host defense. Immunol. Rev. 2007;217:141–154. doi: 10.1111/j.1600-065X.2007.00509.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coussens L.M., Werb Z. Inflammation and cancer. Nature. 2002;420:860–867. doi: 10.1038/nature01322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Parimisetty A., Dorsemans A.C., Awada R., Ravanan P., Diotel N., Lefebvre d’Hellencourt C. Secret talk between adipose tissue and central nervous system via secreted factors-an emerging frontier in the neurodegenerative research. J. Neuroinflammation. 2016;13:67. doi: 10.1186/s12974-016-0530-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Singh J.A., Cameron C., Noorbaloochi S., Cullis T., Tucker M., Christensen R., Ghogomu E.T., Coyle D., Clifford T., Tugwell P., Wells G.A. Risk of serious infection in biological treatment of patients with rheumatoid arthritis: a systematic review and meta-analysis. Lancet. 2015;386:258–265. doi: 10.1016/S0140-6736(14)61704-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.De Cock J.M., Shibue T., Dongre A., Keckesova Z., Reinhardt F., Weinberg R.A. Inflammation Triggers Zeb1-Dependent Escape from Tumor Latency. Cancer Res. 2016;76:6778–6784. doi: 10.1158/0008-5472.CAN-16-0608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mantovani A., Allavena P., Sica A., Balkwill F. Cancer-related inflammation. Nature. 2008;454:436–444. doi: 10.1038/nature07205. [DOI] [PubMed] [Google Scholar]
- 9.Darlington G.J., Wang N., Hanson R.W. C/EBP alpha: a critical regulator of genes governing integrative metabolic processes. Curr. Opin. Genet. Dev. 1995;5:565–570. doi: 10.1016/0959-437x(95)80024-7. [DOI] [PubMed] [Google Scholar]
- 10.Radomska H.S., Huettner C.S., Zhang P., Cheng T., Scadden D.T., Tenen D.G. CCAAT/enhancer binding protein alpha is a regulatory switch sufficient for induction of granulocytic development from bipotential myeloid progenitors. Mol. Cell. Biol. 1998;18:4301–4314. doi: 10.1128/mcb.18.7.4301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Porse B.T., Bryder D., Theilgaard-Mönch K., Hasemann M.S., Anderson K., Damgaard I., Jacobsen S.E., Nerlov C. Loss of C/EBP alpha cell cycle control increases myeloid progenitor proliferation and transforms the neutrophil granulocyte lineage. J. Exp. Med. 2005;202:85–96. doi: 10.1084/jem.20050067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lourenço A.R., Coffer P.J. A tumor suppressor role for C/EBPα in solid tumors: more than fat and blood. Oncogene. 2017;36:5221–5230. doi: 10.1038/onc.2017.151. [DOI] [PubMed] [Google Scholar]
- 13.Mackert J.R., Qu P., Min Y., Johnson P.F., Yang L., Lin P.C. Dual negative roles of C/EBPα in the expansion and pro-tumor functions of MDSCs. Sci. Rep. 2017;7:14048. doi: 10.1038/s41598-017-12968-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tseng H.H., Hwang Y.H., Yeh K.T., Chang J.G., Chen Y.L., Yu H.S. Reduced expression of C/EBP alpha protein in hepatocellular carcinoma is associated with advanced tumor stage and shortened patient survival. J. Cancer Res. Clin. Oncol. 2009;135:241–247. doi: 10.1007/s00432-008-0448-5. [DOI] [PubMed] [Google Scholar]
- 15.Tomizawa M., Watanabe K., Saisho H., Nakagawara A., Tagawa M. Down-regulated expression of the CCAAT/enhancer binding protein alpha and beta genes in human hepatocellular carcinoma: a possible prognostic marker. Anticancer Res. 2003;23(1A):351–354. [PubMed] [Google Scholar]
- 16.Bennett K.L., Hackanson B., Smith L.T., Morrison C.D., Lang J.C., Schuller D.E., Weber F., Eng C., Plass C. Tumor suppressor activity of CCAAT/enhancer binding protein alpha is epigenetically down-regulated in head and neck squamous cell carcinoma. Cancer Res. 2007;67:4657–4664. doi: 10.1158/0008-5472.CAN-06-4793. [DOI] [PubMed] [Google Scholar]
- 17.Gery S., Tanosaki S., Bose S., Bose N., Vadgama J., Koeffler H.P. Down-regulation and growth inhibitory role of C/EBPalpha in breast cancer. Clin. Cancer Res. 2005;11:3184–3190. doi: 10.1158/1078-0432.CCR-04-2625. [DOI] [PubMed] [Google Scholar]
- 18.Wu Y.V., Okada T., DeCarolis P., Socci N., O’Connor R., Geha R.C., Joy Somberg C., Antonescu C., Singer S. Restoration of C/EBPα in dedifferentiated liposarcoma induces G2/M cell cycle arrest and apoptosis. Genes Chromosomes Cancer. 2012;51:313–327. doi: 10.1002/gcc.21917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Halmos B., Huettner C.S., Kocher O., Ferenczi K., Karp D.D., Tenen D.G. Down-regulation and antiproliferative role of C/EBPalpha in lung cancer. Cancer Res. 2002;62:528–534. [PubMed] [Google Scholar]
- 20.Yin H., Lowery M., Glass J. In prostate cancer C/EBPalpha promotes cell growth by the loss of interactions with CDK2, CDK4, and E2F and by activation of AKT. Prostate. 2009;69:1001–1016. doi: 10.1002/pros.20947. [DOI] [PubMed] [Google Scholar]
- 21.Setten R.L., Lightfoot H.L., Habib N.A., Rossi J.J. Development of MTL-CEBPA: Small Activating RNA Drug for Hepatocellular Carcinoma. Curr. Pharm. Biotechnol. 2018;19:611–621. doi: 10.2174/1389201019666180611093428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reebye V., Huang K.W., Lin V., Jarvis S., Cutilas P., Dorman S., Ciriello S., Andrikakou P., Voutila J., Saetrom P. Gene activation of CEBPA using saRNA: preclinical studies of the first in human saRNA drug candidate for liver cancer. Oncogene. 2018;37:3216–3228. doi: 10.1038/s41388-018-0126-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhao X., Voutila J., Ghobrial S., Habib N.A., Reebye V. Treatment of Liver Cancer by C/EBPA saRNA. Adv. Exp. Med. Biol. 2017;983:189–194. doi: 10.1007/978-981-10-4310-9_13. [DOI] [PubMed] [Google Scholar]
- 24.Reebye V., Sætrom P., Mintz P.J., Huang K.W., Swiderski P., Peng L., Liu C., Liu X., Lindkaer-Jensen S., Zacharoulis D. Novel RNA oligonucleotide improves liver function and inhibits liver carcinogenesis in vivo. Hepatology. 2014;59:216–227. doi: 10.1002/hep.26669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yan W., Ding A., Kim H.J., Zheng H., Wei F., Ma X. Progranulin Controls Sepsis via C/EBPα-Regulated Il10 Transcription and Ubiquitin Ligase/Proteasome-Mediated Protein Degradation. J. Immunol. 2016;197:3393–3405. doi: 10.4049/jimmunol.1600862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Alam T., An M.R., Papaconstantinou J. Differential expression of three C/EBP isoforms in multiple tissues during the acute phase response. J. Biol. Chem. 1992;267:5021–5024. [PubMed] [Google Scholar]
- 27.Shimizu H., Yamamoto K. NF-kappa B and C/EBP transcription factor families synergistically function in mouse serum amyloid A gene expression induced by inflammatory cytokines. Gene. 1994;149:305–310. doi: 10.1016/0378-1119(94)90166-x. [DOI] [PubMed] [Google Scholar]
- 28.Ray A., Ray B.K. Lipopolysaccharide-mediated induction of the bovine interleukin-6 gene in monocytes requires both NF-kappa B and C/EBP binding sites. DNA Cell Biol. 1995;14:795–802. doi: 10.1089/dna.1995.14.795. [DOI] [PubMed] [Google Scholar]
- 29.Stein B., Baldwin A.S., Jr. Distinct mechanisms for regulation of the interleukin-8 gene involve synergism and cooperativity between C/EBP and NF-kappa B. Mol. Cell. Biol. 1993;13:7191–7198. doi: 10.1128/mcb.13.11.7191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Voutila J., Reebye V., Roberts T.C., Protopapa P., Andrikakou P., Blakey D.C., Habib R., Huber H., Saetrom P., Rossi J.J., Habib N. Development and Mechanism of Small Activating RNA Targeting CEBPA, a Novel Therapeutic in Clinical Trials for Liver Cancer. Mol. Ther. 2017;25:2705–2714. doi: 10.1016/j.ymthe.2017.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Raetz C.R., Whitfield C. Lipopolysaccharide endotoxins. Annu. Rev. Biochem. 2002;71:635–700. doi: 10.1146/annurev.biochem.71.110601.135414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nazem A., Sankowski R., Bacher M., Al-Abed Y. Rodent models of neuroinflammation for Alzheimer’s disease. J. Neuroinflammation. 2015;12:74. doi: 10.1186/s12974-015-0291-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.He Y., Hara H., Núñez G. Mechanism and Regulation of NLRP3 Inflammasome Activation. Trends Biochem. Sci. 2016;41:1012–1021. doi: 10.1016/j.tibs.2016.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rankin J.A. Biological mediators of acute inflammation. AACN Clin. Issues. 2004;15:3–17. doi: 10.1097/00044067-200401000-00002. [DOI] [PubMed] [Google Scholar]
- 35.Morrison D.C., Ryan J.L. Endotoxins and disease mechanisms. Annu. Rev. Med. 1987;38:417–432. doi: 10.1146/annurev.me.38.020187.002221. [DOI] [PubMed] [Google Scholar]
- 36.Beutler B., Rietschel E.T. Innate immune sensing and its roots: the story of endotoxin. Nat. Rev. Immunol. 2003;3:169–176. doi: 10.1038/nri1004. [DOI] [PubMed] [Google Scholar]
- 37.Bosshart H., Heinzelmann M. THP-1 cells as a model for human monocytes. Ann. Transl. Med. 2016;4:438. doi: 10.21037/atm.2016.08.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chanput W., Mes J., Vreeburg R.A., Savelkoul H.F., Wichers H.J. Transcription profiles of LPS-stimulated THP-1 monocytes and macrophages: a tool to study inflammation modulating effects of food-derived compounds. Food Funct. 2010;1:254–261. doi: 10.1039/c0fo00113a. [DOI] [PubMed] [Google Scholar]
- 39.Dreskin S.C., Thomas G.W., Dale S.N., Heasley L.E. Isoforms of Jun kinase are differentially expressed and activated in human monocyte/macrophage (THP-1) cells. J. Immunol. 2001;166:5646–5653. doi: 10.4049/jimmunol.166.9.5646. [DOI] [PubMed] [Google Scholar]
- 40.Xiang X., An W., Jiang C., Zhao J., Wang X., Sun G., Li Y., Zhang W. Lipopolysaccharide inhibits the expression of resistin in adipocytes. J. Mol. Endocrinol. 2013;51:287–299. doi: 10.1530/JME-13-0117. [DOI] [PubMed] [Google Scholar]
- 41.Khajuria V., Gupta S., Sharma N., Kumar A., Lone N.A., Khullar M., Dutt P., Sharma P.R., Bhagat A., Ahmed Z. Anti-inflammatory potential of hentriacontane in LPS stimulated RAW 264.7 cells and mice model. Biomed. Pharmacother. 2017;92:175–186. doi: 10.1016/j.biopha.2017.05.063. [DOI] [PubMed] [Google Scholar]
- 42.Huo M., Gao R., Jiang L., Cui X., Duan L., Deng X., Guan S., Wei J., Soromou L.W., Feng H., Chi G. Suppression of LPS-induced inflammatory responses by gossypol in RAW 264.7 cells and mouse models. Int. Immunopharmacol. 2013;15:442–449. doi: 10.1016/j.intimp.2013.01.008. [DOI] [PubMed] [Google Scholar]
- 43.Ernst W., Zimara N., Hanses F., Männel D.N., Seelbach-Göbel B., Wege A.K. Humanized mice, a new model to study the influence of drug treatment on neonatal sepsis. Infect. Immun. 2013;81:1520–1531. doi: 10.1128/IAI.01235-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Srinivasan A., Salazar-Gonzalez R.M., Jarcho M., Sandau M.M., Lefrancois L., McSorley S.J. Innate immune activation of CD4 T cells in salmonella-infected mice is dependent on IL-18. J. Immunol. 2007;178:6342–6349. doi: 10.4049/jimmunol.178.10.6342. [DOI] [PubMed] [Google Scholar]
- 45.Nehls M., Pfeifer D., Schorpp M., Hedrich H., Boehm T. New member of the winged-helix protein family disrupted in mouse and rat nude mutations. Nature. 1994;372:103–107. doi: 10.1038/372103a0. [DOI] [PubMed] [Google Scholar]
- 46.Bosma G.C., Custer R.P., Bosma M.J. A severe combined immunodeficiency mutation in the mouse. Nature. 1983;301:527–530. doi: 10.1038/301527a0. [DOI] [PubMed] [Google Scholar]
- 47.Ohbo K., Suda T., Hashiyama M., Mantani A., Ikebe M., Miyakawa K., Moriyama M., Nakamura M., Katsuki M., Takahashi K. Modulation of hematopoiesis in mice with a truncated mutant of the interleukin-2 receptor gamma chain. Blood. 1996;87:956–967. [PubMed] [Google Scholar]
- 48.Marsden M.D., Zack J.A. Humanized Mouse Models for Human Immunodeficiency Virus Infection. Annu. Rev. Virol. 2017;4:393–412. doi: 10.1146/annurev-virology-101416-041703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Holt N., Wang J., Kim K., Friedman G., Wang X., Taupin V., Crooks G.M., Kohn D.B., Gregory P.D., Holmes M.C., Cannon P.M. Human hematopoietic stem/progenitor cells modified by zinc-finger nucleases targeted to CCR5 control HIV-1 in vivo. Nat. Biotechnol. 2010;28:839–847. doi: 10.1038/nbt.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Satheesan S., Li H., Burnett J.C., Takahashi M., Li S., Wu S.X., Synold T.W., Rossi J.J., Zhou J. HIV Replication and Latency in a Humanized NSG Mouse Model during Suppressive Oral Combinational Antiretroviral Therapy. J. Virol. 2018;92 doi: 10.1128/JVI.02118-17. e02118-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Misharin A.V., Haines G.K., 3rd, Rose S., Gierut A.K., Hotchkiss R.S., Perlman H. Development of a new humanized mouse model to study acute inflammatory arthritis. J. Transl. Med. 2012;10:190. doi: 10.1186/1479-5876-10-190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ejarque-Ortiz A., Tusell J.M., Serratosa J., Saura J. CCAAT/enhancer binding protein-alpha is down-regulated by toll-like receptor agonists in microglial cells. J Neurosci Res. 2007;85:985–993. doi: 10.1002/jnr.21195. [DOI] [PubMed] [Google Scholar]
- 53.Poli V. The role of C/EBP isoforms in the control of inflammatory and native immunity functions. J. Biol. Chem. 1998;273:29279–29282. doi: 10.1074/jbc.273.45.29279. [DOI] [PubMed] [Google Scholar]
- 54.Habib N., Khan I.U., Salam N., Xiao M., Ahmed I., Zhi X.Y., Li W.J. Tepidimonas sediminis sp. nov. and Tepidimonas alkaliphilus sp. nov., two novel moderately thermophilic species isolated from a hot spring. Antonie van Leeuwenhoek. 2018;111:1023–1031. doi: 10.1007/s10482-017-1002-8. [DOI] [PubMed] [Google Scholar]
- 55.Gurung P., Li B., Subbarao Malireddi R.K., Lamkanfi M., Geiger T.L., Kanneganti T.D. Chronic TLR Stimulation Controls NLRP3 Inflammasome Activation through IL-10 Mediated Regulation of NLRP3 Expression and Caspase-8 Activation. Sci. Rep. 2015;5:14488. doi: 10.1038/srep14488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang N.D., Finegold M.J., Bradley A., Ou C.N., Abdelsayed S.V., Wilde M.D., Taylor L.R., Wilson D.R., Darlington G.J. Impaired energy homeostasis in C/EBP alpha knockout mice. Science. 1995;269:1108–1112. doi: 10.1126/science.7652557. [DOI] [PubMed] [Google Scholar]
- 57.Tanaka S., Tanaka K., Magnusson F., Chung Y., Martinez G.J., Wang Y.H., Nurieva R.I., Kurosaki T., Dong C. CCAAT/enhancer-binding protein α negatively regulates IFN-γ expression in T cells. J. Immunol. 2014;193:6152–6160. doi: 10.4049/jimmunol.1303422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dobler C.C. Biologic Agents and Tuberculosis. Microbiol. Spectr. 2016;4 doi: 10.1128/microbiolspec.TNMI7-0026-2016. TNMI7-0026-2016. [DOI] [PubMed] [Google Scholar]
- 59.Li S., Yu Y., Yue Y., Zhang Z., Su K. Microbial Infection and Rheumatoid Arthritis. J. Clin. Cell. Immunol. 2013;4:174. doi: 10.4172/2155-9899.1000174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Strober W., Fuss I.J. Proinflammatory cytokines in the pathogenesis of inflammatory bowel diseases. Gastroenterology. 2011;140:1756–1767. doi: 10.1053/j.gastro.2011.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cavagna L., Taylor W.J. The emerging role of biotechnological drugs in the treatment of gout. BioMed Res. Int. 2014;2014:264859. doi: 10.1155/2014/264859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang P., Iwama A., Datta M.W., Darlington G.J., Link D.C., Tenen D.G. Upregulation of interleukin 6 and granulocyte colony-stimulating factor receptors by transcription factor CCAAT enhancer binding protein alpha (C/EBP alpha) is critical for granulopoiesis. J. Exp. Med. 1998;188:1173–1184. doi: 10.1084/jem.188.6.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ma O., Hong S., Guo H., Ghiaur G., Friedman A.D. Granulopoiesis requires increased C/EBPα compared to monopoiesis, correlated with elevated Cebpa in immature G-CSF receptor versus M-CSF receptor expressing cells. PLoS One. 2014;9:e95784. doi: 10.1371/journal.pone.0095784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Friedman A.D., Keefer J.R., Kummalue T., Liu H., Wang Q.F., Cleaves R. Regulation of granulocyte and monocyte differentiation by CCAAT/enhancer binding protein alpha. Blood Cells Mol. Dis. 2003;31:338–341. doi: 10.1016/s1079-9796(03)00135-9. [DOI] [PubMed] [Google Scholar]
- 65.Wilson S.P., Cassel S.L. Inflammasome-mediated autoinflammatory disorders. Postgrad. Med. 2010;122:125–133. doi: 10.3810/pgm.2010.09.2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cinel I., Opal S.M. Molecular biology of inflammation and sepsis: a primer. Crit. Care Med. 2009;37:291–304. doi: 10.1097/CCM.0b013e31819267fb. [DOI] [PubMed] [Google Scholar]
- 67.Wu X., Dong L., Lin X., Li J. Relevance of the NLRP3 Inflammasome in the Pathogenesis of Chronic Liver Disease. Front. Immunol. 2017;8:1728. doi: 10.3389/fimmu.2017.01728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mathews R.J., Robinson J.I., Battellino M., Wong C., Taylor J.C., Eyre S., Churchman S.M., Wilson A.G., Isaacs J.D., Hyrich K., Biologics in Rheumatoid Arthritis Genetics and Genomics Study Syndicate (BRAGGSS) Evidence of NLRP3-inflammasome activation in rheumatoid arthritis (RA); genetic variants within the NLRP3-inflammasome complex in relation to susceptibility to RA and response to anti-TNF treatment. Ann. Rheum. Dis. 2014;73:1202–1210. doi: 10.1136/annrheumdis-2013-203276. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.