Abstract
Chimeric antigen receptor (CAR) engineering of T cells allows one to specifically target tumor cells via cell surface antigens. A candidate target in Ewing sarcoma is the ganglioside GD2, but heterogeneic expression limits its value. Here we report that pharmacological inhibition of Enhancer of Zeste Homolog 2 (EZH2) at doses reducing H3K27 trimethylation, but not cell viability, selectively and reversibly induces GD2 surface expression in Ewing sarcoma cells. EZH2 in Ewing sarcoma cells directly binds to the promoter regions of genes encoding for two key enzymes of GD2 biosynthesis, and EZH2 inhibition enhances expression of these genes. GD2 surface expression in Ewing sarcoma cells is not associated with distinct in vitro proliferation, colony formation, chemosensitivity, or in vivo tumorigenicity. Moreover, disruption of GD2 synthesis by gene editing does not affect its in vitro behavior. EZH2 inhibitor treatment sensitizes Ewing sarcoma cells to effective cytolysis by GD2-specific CAR gene-modified T cells. In conclusion, we report a clinically applicable pharmacological approach for enhancing efficacy of adoptively transferred GD2-redirected T cells against Ewing sarcoma, by enabling recognition of tumor cells with low or negative target expression.
Keywords: cellular immunotherapy, chimeric antigen receptors, combination therapy, epigenetic agents, Ewing sarcoma, gangliosides
Kailayangiri et al. report that small-molecule inhibitors of EZH2 methyltransferase activity reliably upregulate the ganglioside GD2 on the surface of Ewing sarcoma cells and render GD2-negative tumor cells susceptible to targeting with GD2-specific CAR T cells.
Introduction
Chimeric antigen receptors (CARs) are recombinant proteins that link antibody-derived antigen-binding domains to stimulatory T cell-signaling pathways. Expressed in immune effector cells, they induce activation responses to surface-expressed tumor target antigens, resulting in antigen-specific tumor cytolysis.1 T cells engineered to express CARs against the B lineage antigen CD19 are active to induce and maintain remissions in refractory B cell precursor leukemias2, 3, 4 and lymphomas.5, 6 The development of CAR T cells for solid tumors is limited by the paucity of target antigens reliably expressed at high densities on cancer cells, but not normal cells.
We seek to develop cellular therapies to treat Ewing sarcoma (EwS), an aggressive solid mesenchymal malignancy arising in bone and soft tissues.7 EwS is characterized by a specific chromosomal translocation, most commonly of chromosomes 22 and 11 (t(11,22)(q24;12)), resulting in the aberrant chimeric transcription factor EWSR1-FLI1.8 Our group and others have shown that EwS can express the disialoganglioside GD2 on the cell surface.9, 10 GD2 expression characterizes immature neuroectodermal cells, with restricted and low-level tissue expression after birth on neuronal cells in the CNS, peripheral nerves, and mesenchymal stroma cells (MSCs)11, 12 (reviewed in Rossig et al.13). GD2 was first evaluated as a therapeutic target in neuroblastoma, a cancer with abundant GD2 surface expression due to its tissue origin from neuroectoderm.14, 15
In early clinical studies, adoptive transfer of GD2-specific CAR T cells to neuroblastoma patients was safe with the first evidence of clinical activity (K. Straathof et al. 2018, AACR Cancer Res. abstract).16, 17, 18 Preclinical data support the use of GD2-specific CARs also for immunotherapy of EwS;10, 19, 20 however, only a proportion of EwS express GD2 at high levels,9, 19 which reduces the number of patients amenable to GD2-targeted therapies. Another limitation is the heterogeneity of GD2 antigen expression among tumor cells in individual EwS.19 Since effective CAR T cell-mediated cytolysis relies on high target densities,21, 22 GD2-low (GD2low) or GD2-negative (GD2neg) subpopulations escape CAR T cell targeting.
To enhance the impact of GD2 as an immune target in this cancer, we investigated a novel strategy to upregulate expression on the cell surface of EwS cells by an epigenetic agent, based on the following rationale. Biosynthesis of GD2 and other gangliosides during organ development is driven by stage-specific transcriptional activation of glycosyl transferases and underlies epigenetic regulation.23 Epigenetic reprogramming is highly relevant also in the pathology of EwS.24, 25, 26, 27, 28, 29 An important epigenetic regulator in EwS is Enhancer of Zeste Homolog 2 (EZH2), the catalytic component of the Polycomb Repressor Complex 2 (PRC2).24, 30 EZH2 acts as a histone methyltransferase, and it silences genes involved in cell differentiation in a highly context-dependent manner, by depositing repressive histone marks at histone 3 lysine 27 (H3K27me3).31 High-level EZH2 expression is induced in EwS as a direct consequence of EWSR1-FLI1,24 and it has a central role in maintaining self-renewal and tumorigenicity.24, 30 Epigenetic plasticity by EZH2-mediated gene regulation contributes to phenotypic heterogeneity among EwS cells.32 We hypothesized that EZH2 is involved in the regulation of synthesis of the non-protein neuroectodermal marker GD2 in this cancer, allowing us to sensitize EwS cells to GD2-targeted cell therapy by EZH2 inhibitors.
Results
Pharmacological Inhibition of EZH2 Selectively Upregulates Surface GD2 Expression in EwS Cells
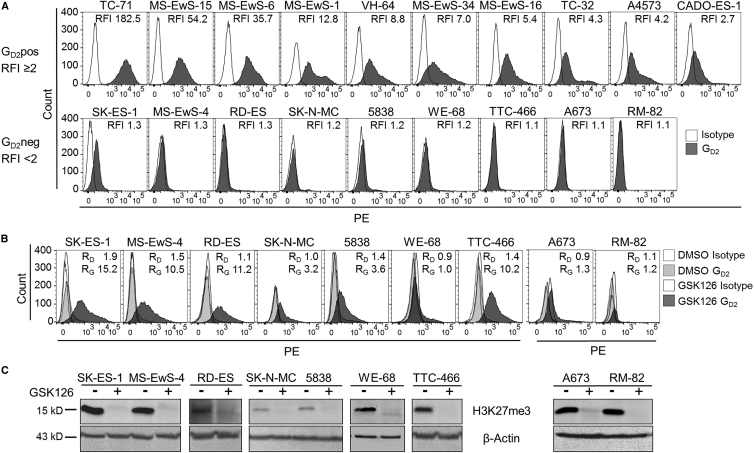
We classified 19 EwS cell lines by surface expression of GD2 using flow cytometry. Ten of 19 EwS cell lines expressed GD2 at a relative fluorescence intensity (RFI) ≥ 2 (median RFI 12.6, range 2.5–128.8), which we defined as GD2 positive (GD2pos), and 9 were GD2neg (median RFI 1.2, range 1.0–1.8) (Figure 1A). To investigate whether EZH2 inhibition can induce GD2 expression in GD2neg EwS, we cultured all GD2neg EwS cell lines in the presence of the small-molecule EZH2 inhibitor GSK126 at 10–12 μM. GSK126 upregulated GD2 expression to RFIs ≥ 2 in 6 of 9 cell lines within 7 or 14 days (Figure 1B). All cell lines tolerated GSK126 at these concentrations, with consistent viabilities (median 88.2%, range 71.3%–92.2%) during the 7- to 14-day cultures. Concomitant to GD2 upregulation, inhibition of EZH2 by GSK126 resulted in a decrease of H3K27me3 levels in all cell lines (Figure 1C). This confirms that GSK126 at these concentrations inhibits EZH2 methyltransferase activity in EwS cells.
Figure 1.
EZH2 Inhibition Induces GD2 Expression in GD2neg EwS Cell Lines In Vitro
(A) GD2 surface expression by flow cytometry in 19 EwS cell lines. According to RFI, cell lines were categorized as GD2pos (RFI ≥ 2) or GD2neg (RFI < 2). Representative experiments of two or three are shown. (B) GD2 surface expression in 9 GD2neg EwS cell lines cultured with 10 μM GSK126 or equivalent volumes of DMSO for 7 days (RD-ES, WE-68, and TTC-466) or 14 days (A673 and RM-82) or with 12 μM GSK126 or DMSO for 7 days (SK-ES-1, MS-EwS-4, SK-N-MC, and 5838). RD, RFI after incubation with DMSO; RG, RFI after incubation with GSK126. (C) Histone H3K27me3 levels in lysates from the EwS cell cultures in (B) at the end of incubation with GSK126 (+) or under control conditions with DMSO alone (−) by western blot analysis.
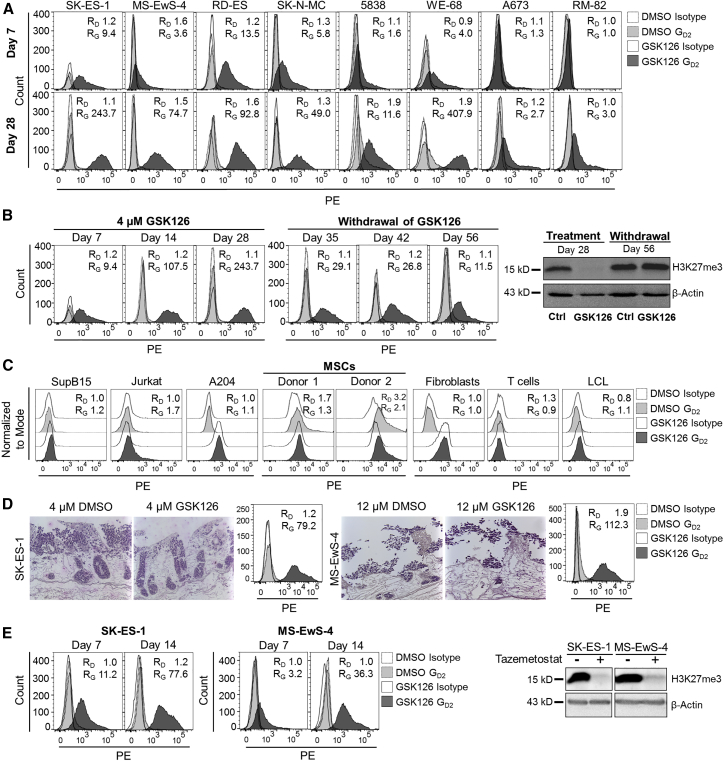
At a lower concentration of 4 μM, GSK126 was effective to induce GD2 expression in 5 cell lines on day 7 (Figure 2A). In continued cultures in the presence of the inhibitor for up to 28 or 29 days, GD2 expression exceeded RFIs ≥ 2 in 8 of 8 cell lines (Figure 2A). To understand the kinetics of GD2 upregulation and histone methylase activity during prolonged exposure of EwS cells to the EZH2 inhibitor, we determined GD2 surface expression and histone trimethylation at weekly intervals during in vitro culture of the GD2neg cell line SK-ES-1 with 4 μM GSK126. GD2 expression gradually increased until day 28, and withdrawal of the agent reduced GD2 surface expression (Figure 2B, left panel). GD2 up- and downregulation in the presence and absence of GSK126 corresponded to loss and recovery of H3K27me3 by western blot analysis, respectively (Figure 2B, right panel). Culturing the EwS cell lines in the presence of 4 μM GSK126 for 14 days did not significantly reduce their in vitro expansion (Figure S1A) or colony formation (Figure S1B). Thus, pharmacological inhibition of EZH2 at non-toxic doses effective to reduce H3K27me3 selectively upregulates GD2 surface expression in a majority of GD2neg EwS cell lines.
Figure 2.
Upregulation of GD2 Expression by EZH2 Inhibition Is Reversible and Limited to EwS Cell Lines
(A) GD2 surface expression by flow cytometry in 8 GD2neg EwS cell lines cultured with 4 μM GSK126 or equivalent volumes of DMSO (control) for 7 days (upper panel) and for 28 days (lower panel). RD, RFI after incubation with DMSO; RG, RFI after incubation with GSK126. (B) GD2 surface expression by weekly flow cytometry and H3K27me3 methylation by western blot analysis (days 28 and 56) in SK-ES-1 cells cultured with 4 μM GSK126 or DMSO for 28 days, followed by withdrawal of GSK126 from the culture medium. Ctrl, control. (C) GD2 surface expression on leukemia cell lines (SupB15 and Jurkat) and rhabdoid tumor cell line A204 and on mesenchymal stroma cells (MSCs), fibroblasts, T cells, and LCL from healthy human donors after culture with 4 μM GSK126 or DMSO for 7 days (MSCs) or 14 days (all others). (D) Immunohistochemical H&E staining (left) and GD2 surface expression by flow cytometry (right) of SK-ES-1 and MS-EwS-4 cells cultured on a biologic tissue matrix in a dynamic 3D culture model in the presence or absence of 4 or 12 μM GSK126, as indicated, or respective volumes of DMSO for 14 days. (E) GD2 surface expression by flow cytometry (days 7 and 14) and H3K27me3 methylation by western blot analysis (day 14) in SK-ES-1 and MS-EwS-4 cells cultured in the presence of 1 μM tazemetostat or equivalent volumes of DMSO for 14 days.
To investigate whether GD2 upregulation by EZH2 inhibition is restricted to EwS compared to other types of cancer and to normal cells, we cultured the B cell precursor leukemia cell line SupB15, the T cell leukemia cell line Jurkat, and the rhabdoid tumor cell line A204 in the presence of GSK126 (4 μM), and we determined GD2 expression levels on day 14. None of the 3 cell lines expressed GD2 at any time point before or after culture with GSK126 (Figure 2C). We further investigated GD2 expression in MSCs, the proposed cell of origin for EwS, fibroblasts, T cells, and B-lymphoblastic cell cultures, all derived from healthy human donors. GD2 was not upregulated by GSK126 treatment in vitro in any of these normal human cell populations (Figure 2C).
We further assessed the capacity of EZH2 inhibition to upregulate GD2 expression in EwS in a 3D tumor model mimicking in vivo conditions for T cell migration into solid tumor tissues.33 EwS cells were seeded onto a biological tissue matrix consisting of decellularized small-intestine submucosa and mucosa (SISmuc), and they were cultured in a dynamic bioreactor system in the presence or absence of 4 μM (SK-ES-1) or 12 μM (MS-EwS-4) GSK126 for 14 days. Histochemistry analysis confirmed the formation of multilayered tumor tissue on the matrix (Figure 2D). GSK126 effectively upregulated cell surface expression of GD2 on EwS cells also in the 3D model (Figure 2D).
To obtain further evidence that GD2 upregulation by GSK126 is mediated by inhibition of the epigenetic target EZH2, we reproduced our findings with an alternative EZH2 inhibitor, tazemetostat. This agent is undergoing clinical investigation as an anticancer agent, including for pediatric sarcoma patients. Tazemetostat at the pharmacologically relevant concentration of 1 μM34 effectively upregulated GD2 surface expression in the two GD2neg EwS cell lines SK-ES-1 and MS-EwS-4 while reducing H3K27 methylation (Figure 2E).
We conclude that EZH2 inhibition selectively and reliably upregulates ganglioside GD2 on the cell surface of EwS cells, also in a complex 3D tumor model and using different pharmacological inhibitors.
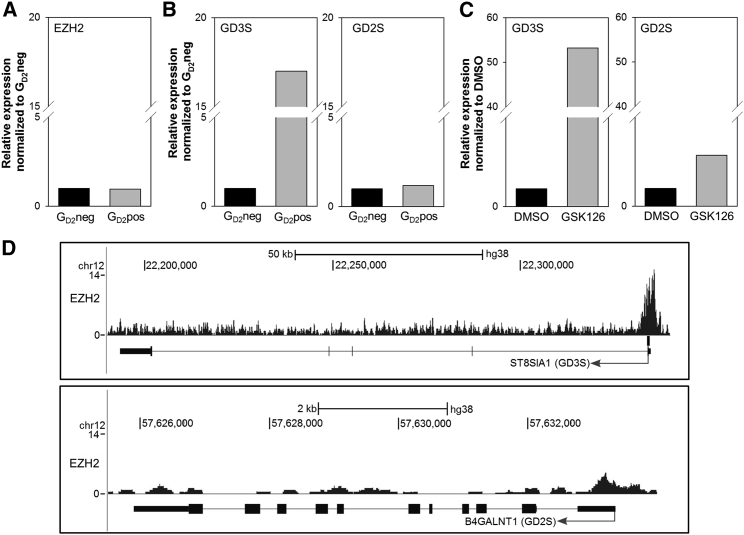
EZH2 Modulates GD2 Expression in EwS Cells by Regulating the Expression of Genes Involved in GD2 Biosynthesis
Expression of GD2 during development is regulated through stage- and tissue-specific expression of glycosyltransferases, GD3 synthase (GD3S), and GD2 synthase (GD2S), which synthesize GD3 from its precursor GM3 and convert GD3 to GD2, respectively.35 To understand the mechanism by which epigenetic modification affects the expression of GD2 in EwS, we quantified transcripts of EZH2 and of GD3S and GD2S in 9 GD2neg and 10 GD2pos EwS cell lines. EZH2 expression was not different in GD2neg versus GD2pos cell lines (Figure 3A). Of the two enzymes upstream of GD2, GD3S, but not GD2S, was expressed at higher levels in GD2pos compared to GD2neg EwS cell lines (Figure 3B). Treatment with 4 μM GSK126 enhanced the gene expression of GD3S by a mean of 53-fold (±44.6) and of GD2S by a mean of 2.9-fold (±2.0) in GD2neg EwS cell lines (Figure 3C). Analysis of a publicly available chromatin immunoprecipitation sequencing (ChIP-seq) dataset showed strong binding of EZH2 to the promoter of GD3 synthase gene ST8SIA1 in the EwS cell line SK-N-MC (Figure 3D). The promoter of GD2 synthase gene B4GALNT1 displayed a visible, but not significant, EZH2 peak (Figure 3D). This indicates that EZH2 directly represses the genes involved in GD2 biosynthesis. Together these findings support a mechanism by which EZH2 inhibition in EwS removes repressive histone marks on the genes encoding for GD3S and GD2S, resulting in enhanced biosynthesis of GD2.
Figure 3.
Expression of EZH2 and Ganglioside Synthases GD3S and GD2S in EwS Cells
(A) EZH2 gene expression in 9 GD2neg and 10 GD2pos EwS cell lines by qRT-PCR. (B) GD3S and GD2S gene expression in 9 GD2neg and 10 GD2pos EwS cell lines by qRT-PCR. (C) GD3S and GD2S gene expression in 9 GD2neg EwS cell lines following 14 days of incubation in the presence of 4 μM GSK126 or DMSO (control) by qRT-PCR. (D) Screenshots of ChIP-seq data analysis of SK-N-MC EwS cells using the UCSC Genome Browser.
GD2pos and GD2neg EwS Cell Lines and Subpopulations of EwS Cells Have Comparable In Vitro Proliferation Rates, Clonogenicity, Tumorigenicity, and Chemosensitivity
Safe therapeutic upregulation of GD2 requires knowledge of the functional significance of GD2 expression in EwS. In breast cancer, GD2 was found to define a malignant population with a higher capacity to self-renew and reinitiate tumor growth than GD2-negative cells.36, 37 Inducing GD2 expression by epigenetic regulation could thus promote a cell population with high aggressiveness and metastatic properties.
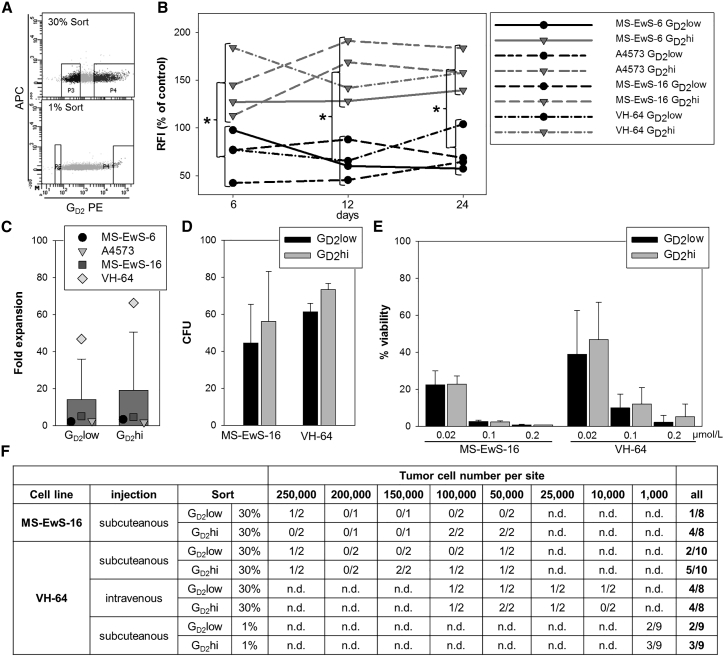
First we compared the in vitro expansion and colony-forming capacities of EwS cell lines expressing high or low densities of GD2 (Figure 1A). In vitro proliferation was not significantly different between GD2pos and GD2neg cell lines (Figure S2A). EwS cells from GD2pos and GD2neg cell lines had comparable capacities to form colonies (Figure S2B). Moreover, GD2pos and GD2neg cell lines did not differ in their sensitivity to the cytotoxic agent doxorubicin (Figure S2C). To compare the functional properties of GD2pos and GD2neg tumor cells within individual EwS cell lines, we separated the 30% highest and lowest GD2-expressing subpopulations (GD2hi versus GD2low) from the GD2pos bulk cell lines MS-EwS-6, A4573, MS-EwS-16, and VH-64 by fluorescence-activated cell sorting (Figure 4A). During subsequent in vitro culture for 24 days, the subpopulations from all 4 cell lines maintained significantly different GD2 expression levels (Figure 4B). In vitro expansion rates under standard growth conditions were comparable between the two subpopulations (Figure 4C). In two cell lines, MS-EwS-16 and VH-64, we compared the capacities of GD2hi and GD2low subpopulations to form colonies in vitro and initiate tumors in vivo and their chemosensitivities. GD2hi and GD2low EwS cells formed comparable numbers of colonies in vitro (Figure 4D). The two subpopulations had similar sensitivities to increasing concentrations of doxorubicin (Figure 4E).
Figure 4.
High GD2 Surface Expression in EwS Cells Does Not Affect In Vitro Proliferation, Colony Formation, Chemosensitivity, and In Vivo Tumorigenicity
(A) FACS selection strategy of tumor cell subpopulations with the highest (GD2hi, 30% or 1%) and lowest (GD2low, 30% or 1%) GD2 surface expression, exemplified by cell line VH-64. (B) GD2 surface expression on sorted GD2hi and GD2low subpopulations (30%) determined by flow cytometry every 6 days during subsequent cell culture over a period of 3 weeks. The median RFIs of GD2 expression in the two subpopulations are shown in relation to bulk cells from the same cell lines. *p < 0.02 (t test). (C) In vitro expansion of tumor cells from the sorted subpopulations (30%) for 6 days of standard monolayer cultures, quantified by trypan blue exclusion and cell counting. (D) Colony formation of tumor cells from sorted subpopulations (30%) in semisolid media after 7 or 8 days of culture. (E) Viabilities of tumor cells from the sorted subpopulations (30%) following incubation with the indicated concentrations of doxorubicin by luminometry. (F) Tumor formation of GD2hi and GD2low subpopulations selected from VH-64 and MS-EwS-16 cells after subcutaneous or intravenous transplantation into NSG mice at the indicated cell numbers. All error bars indicate SDs.
In xenografting experiments in NOD Scid gamma (NSG) mice, MS-EwS-16 cells, irrespective of GD2hi and GD2low expression, only rarely initiated tumor xenografts (Figure 4F). The capacity of low numbers of VH-64 cells to initiate tumors after subcutaneous or intravenous administration was not superior in the GD2hi population (Figure 4F). To further reproduce this finding, we sorted the 1% lowest and highest GD2-expressing tumor cells from bulk VH-64 cells (Figure 4A), and we injected 1,000 cells/flank into 9 NSG mice. Again, GD2hi and GD2low subsets of VH-64 initiated tumors at comparable rates of 2 versus 3 of 9 mice (Figure 4F). Thus, GD2hi expression within individual EwS cell lines does not confer enhanced tumor-initiating capacities.
Loss of GD2 by Genetic Disruption of GD3 Synthase in EwS Cells Does Not Affect In Vitro Growth, Clonogenicity, and Chemosensitivity
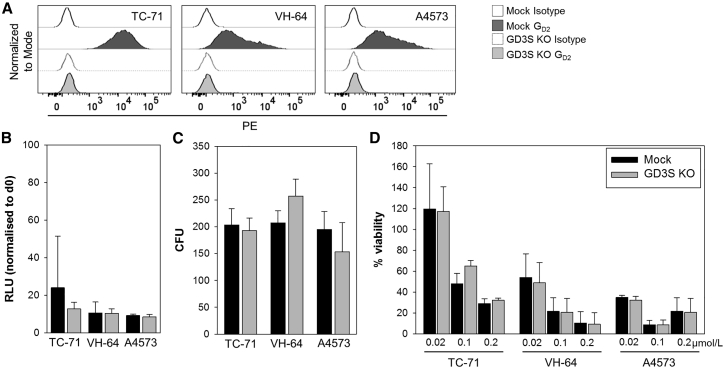
To provide direct evidence that GD2 in EwS cells does not affect their functional characteristics, we eliminated GD2 expression by gene editing. Since GD2 as a carbohydrate is not directly targetable, we disrupted the GD3S gene by CRISPR/Cas9 genome editing. GD3S-targeted mutagenesis effectively eliminated GD2 surface expression in the GD2pos EwS cell lines TC-71, VH-64, and A4573 (Figure 5A). Loss of GD2 expression by gene editing did not affect the capacity of EwS cells to proliferate (Figure 5B) or form colonies in vitro (Figure 5C) or their chemosensitivity (Figure 5D) compared to mock control cells. We conclude that GD2 expression in EwS cells does not confer distinct functional properties of in vitro growth and sensitivity to cytotoxic drugs.
Figure 5.
Disruption of GD2 Surface Expression in EwS Cells by GD3S Gene Editing Does Not Affect Colony Formation, Tumorigenicity, and Chemosensitivity
(A) GD2 expression in 3 EwS cell lines following disruption of the GD3S gene by CRISPR/Cas9 genome editing (GD3SKO) or in wild-type cells (mock control) by flow cytometry. (B) Proliferation of GD3SKO or wild-type EwS cells after 3-day in vitro culture by luminometry. (C) Colony formation of GD3SKO or wild-type EwS cells in semisolid media. (D) Viabilities of GD3SKO and wild-type cells following incubation with the indicated concentrations of doxorubicin by luminometry. All error bars indicate SDs.
Upregulation of Surface GD2 in EwS Cells by Pharmacological Inhibition of EZH2 Enables Effective Targeting by GD2-Specific CAR T Cells
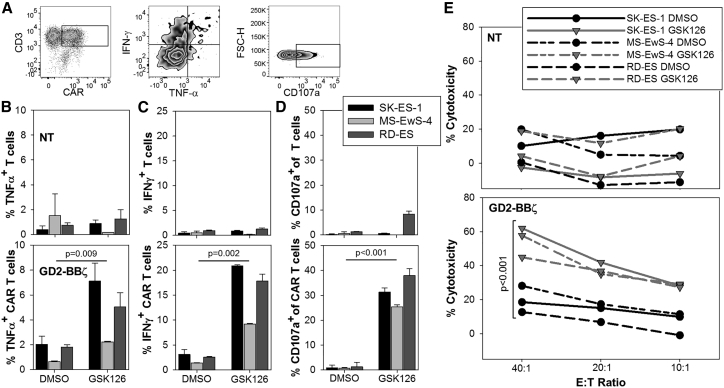
To investigate the capacity of EZH2 inhibition to sensitize EwS cells to treatment with GD2-specific CAR T cells, human T cells from 2 donors were gene modified to express the GD2-specific CAR GD2-BBζ,10 resulting in CAR surface expression in 59.7% and 63.1% of T cells by anti-idiotype staining (Figure 6A). Three GD2neg EwS cell lines (SK-ES-1, MS-EwS-4, and RD-ES) were cultured in the presence of 4 μM GSK126 dissolved in DMSO or DMSO alone for 14 days, followed by coincubation with CAR T cells. The capacity of the tumor cells to induce antigen-specific, CAR-mediated T cell activation was assessed by quantification of the intracellular cytokines interferon-γ (IFN-γ) and tumor necrosis factor alpha (TNF-α) and of CD107a, a marker of degranulation and a prerequisite for perforin-mediated toxicity. Whereas GD2neg EwS cells pretreated with DMSO alone failed to induce activation responses by GD2-BBζ-transduced T cells above the low background of non-transduced T cells, GSK126-pretreated EwS cells mediated significant secretion of TNF-α (Figures 6A and 6B) and IFN-γ (Figures 6A and 6C) and CD107 upregulation (Figures 6A and 6D) by the GD2-BBζ CAR-expressing T cells. Moreover, GSK126 pretreatment significantly enhanced cytolysis of initially GD2neg EwS cells by GD2-BBζ-transduced T cells (Figure 6E). Thus, EZH2 inhibition sensitizes GD2neg EwS cells to antigen-specific functional interactions with GD2-redirected CAR T cells and to CAR T cell-mediated in vitro cytolysis.
Figure 6.
EZH2 Inhibitor Pretreatment Sensitizes GD2-Negative EwS Cell Lines to In Vitro Cytolysis by GD2-Specific CAR T Cells
(A) Exemplary gating strategies for the quantification of intracellular cytokine expression and CD107a expression in CAR (GD2-BBζ)-transduced T cells. (B and C) Quantification of intracellular expression of IFN-γ (B) and TNF-α (C) in non-transduced (NT) or GD2-BBζ-transduced T cells coincubated for 6 h with EwS cell lines pretreated with 4 μM GSK126 or DMSO alone for 14 days. Shown is one representative experiment of two. (D) Degranulation responses by CD107a expression in NT or GD2-BBζ-transduced T cells after 3-h coincubation with EwS cell lines pretreated as above. Shown is one representative experiment of two. Statistical analysis using t test. (E) Cytolysis of EwS cells after 4-h coincubation with NT or GD2-BBζ-transduced T cells. EwS cells had been pretreated as in (B)–(D). Shown is one representative experiment of two. Statistical analysis by paired t test for all E:T ratios. All error bars indicate SDs.
Discussion
GD2 was one of the first antigens used to retarget T cells to tumor cells by CARs,38, 39 and it remains an attractive CAR target. As an oncofetal antigen, GD2 has highly restricted normal tissue expression.11, 12 A concern has been low-level antigen expression on neuronal cells, but GD2-directed CAR T cell therapy has been safe in early clinical trials in patients with neuroblastoma, now including studies with signal-enhanced CARs, use of lymphodepleting chemotherapy, and clear evidence of activity by cytokine release and/or tumor responses (K. Straathof et al. 2018, AACR Cancer Res. abstract).16, 17, 18 Extending the therapeutic potential of GD2-specific CAR T cells beyond neuroblastoma is challenged by low levels and high heterogeneity of GD2 antigen expression in other cancers expressing GD2.9, 10, 40 Here we report a strategy by which EwS cells with low or absent GD2 expression can be sensitized to GD2-directed CAR T cell therapy. At pharmacologically relevant concentrations that effectively reduce H3K27me3, the EZH2 inhibitors GSK126 and tazemetostat significantly upregulated GD2 surface expression in a majority of GD2neg EwS cell lines, including a multilayered 3D tumor model. Following epigenetic upregulation of GD2 in this manner, EwS cells induced antigen-specific activation responses by GD2-retargeted CAR T cells, and they were effectively lysed by the CAR T cells in vitro.
We did not attempt to demonstrate activity of the combination therapy in an in vivo model for the following reason: in mouse xenograft models of EwS, GD2-specific CAR T cells or CAR natural killer (NK) cells had only modest antitumor activity, despite the consistent expression of GD2.9, 10, 20 Further therapeutic elements will have to be included to break barriers in the tumor stroma against effective in vivo tumor control. CAR T cells fail to access tumors at sufficient numbers, and upon activation they upregulate immune-inhibitory molecules, such as PD-L141 and HLA-G,42 and they induce large populations of myeloid suppressor cells.9 But although reliable expression of the CAR target antigen is insufficient to establish efficacy in mouse models and in clinical settings, it remains an indispensable prerequisite for the action of CAR T cells.
Expression of GD2 and its regulation by an epigenetic mechanism in EwS is well in line with the pathogenesis of this cancer. According to current models, EwS arises in MSCs as a consequence of the disease-defining chromosomal translocation.30 Since surface GD2 characterizes both neuroectodermal and MSCs,11, 43 expression in EwS could be a reminiscent feature of its histogenic origin. In normal MSCs, GD2 expression is associated with an immature phenotype and is lost during non-neuronal lineage differentiation,44, 45 likely by epigenetic gene repression, which has a central role in modulating ganglioside expression during normal neural development.23 Epigenetic gene regulation is highly relevant also in the pathogenesis of EwS. In the permissive cellular environment of the mesenchymal cell, the aberrant transcription factor EWSR1-FLI1 reprograms the epigenome to induce the oncogenic phenotype.25, 27
Among the various epigenetic mechanisms involved in EwS pathogenesis, we focused on the PRC enzyme EZH2, since it is a direct target of EWSR1-FLI1 and a critical mediator of malignant cell growth.24, 30 Our observation that the inhibition of EZH2 in GD2neg EwS cells induces GD2 expression leads us to propose that EZH2-mediated repression of genes involved in GD2 biosynthesis modulate GD2 expression in this cancer, resulting in the observed intratumor and interpatient heterogeneity of expression. This hypothesis is supported by our following findings: first, GD2 upregulation by EZH2 inhibition is associated with the removal of methylation marks at histone H3K27, and it is reversible by withdrawal of the agent; second, EZH2 directly binds to the promoters of genes encoding for the two critical enzymes in GD2 biosynthesis, GD3S and GD2S; and, finally, induction of GD2 by EZH2 inhibition is associated with upregulated gene expression of GD3S and, to a lesser extent, GD2S.
Epigenetic regulation of GD2 expression in tumor cells likely involves more than a single genetic target. Synthesis of GD2 and other complex gangliosides and their precursors relies on various enzymatic steps, each of which alone or, more likely, in combination could be a target for modulation by EZH2 and other epigenetic regulators. Our finding of a key significance of GD3S for GD2 expression is well in line with data obtained in breast cancer.36 In addition, PRC2-independent effects of EZH2 and even activation of genes involved in the degradation of GD2 could contribute to the molecular mechanism.46
While we are the first to show upregulation of GD2 by EZH2 inhibition in EwS, another example supports epigenetic regulation of GD2 expression in cancer: in GD2-positive neuroblastoma cells, the histone deacetylase (HDAC) inhibitor vorinostat was reported to further enhance expression of the antigen.47 GD2 upregulation by vorinostat was associated with increased levels of GD2S protein, but not transcripts, and GD3S was not investigated.
Since gangliosides have important biological functions in both normal and malignant cells, we were concerned that the manipulation of GD2 expression in EwS cells could affect their malignant phenotype. Intriguingly, GD2-expressing cells in breast cancer define a malignant population with a higher capacity to self-renew and reinitiate tumor growth36, 48 and a molecular profile associated with stem cell function and epithelial-mesenchymal transition (EMT). Inhibition of GD2 biosynthesis in breast cancer hampers self-renewal, mammosphere formation, tumor initiation, and cell motility, suggesting a functional contribution of GD2 to stem cell features.36, 48 In comparative experiments with EwS subpopulations selected for high and low or absent GD2 expression and by disruption of GD2 expression using targeted gene editing, we provide clear evidence that GD2 expression in EwS is functionally irrelevant for proliferation, stem cell-associated functions, and chemosensitivity. These findings support the safety of upregulating this antigen on EwS cells.
By enhancing GD2 surface expression, EZH2 inhibitors emerge as promising new candidates for effective combination regimens with GD2-targeted therapies in EwS. Since H3K27 methylation by EZH2 regulates various cellular functions, the use of EZH2 inhibitors as sensitizers for CAR T cell targeting requires consideration of additional effects both on tumor cells and on T cells. Due to its role in cell differentiation and tumorigenicity in EwS, EZH2 was suggested as a therapeutic target.24, 30 As a single agent in a preclinical in vivo model, the inhibitor tazemetostat had only low activity against EwS xenografts.49 Still, the antitumor effects of epigenetic therapy and GD2-specific immune targeting could add up to a potent combination strategy. In T cells, EZH2 inhibition was found to increase the cytotoxic activity of effector T cells and reprogram regulatory functional profiles of suppressive T cell populations,50 further encouraging the combination of EZH2 inhibitors with adoptive T cell therapies. Importantly, pharmacological EZH2 inhibition may reverse the limited T cell trafficking into the tumor microenvironment of solid cancers caused by epigenetic silencing of chemokines CXCL9 and CXCL10,51 and, thereby, it may overcome one of the most significant barriers to CAR T cell targeting of solid tumors.
We conclude that a combination of GD2-specific CAR T cell therapy with pharmacological EZH2 inhibition deserves investigation as a new therapeutic strategy in EwS and potentially other cancers with heterogeneous GD2 expression, including osteosarcoma, various soft tissue sarcomas, melanoma, lung cancer, and breast cancer. Re-biopsies or repeated GD2 antibody scans of EwS patients treated with EZH2 inhibitors would be highly informative to confirm the effects of EZH2 inhibitors on GD2 expression levels and their tumor selectivity in patients. Studying epigenetic regulation of carbohydrate antigen expression in cancer could lead to additional combination strategies also for other non-protein CAR target antigens.
Materials and Methods
Cell Lines
The EwS cell lines VH-64, RM-82, and WE-68 were gifts from Frans van Valen’s laboratory at the Institute of Experimental Orthopedics of the University of Muenster, Germany. MS-EwS-1 (previously described as MS-PES-119), MS-EwS-4 (originally described as MS-PES-410), MS-EwS-6 (originally described as MS-PES-642), MS-EwS-16 (originally described as DC-ES-652), MS-EwS-15 (originally described as DC-ES-1553), and MS-EwS-34 were established from biopsy material of individual patients obtained at metastatic relapse, as reported previously.19, 42, 52 A4573, 5838, TTC-466, and TC-32 were a gift from the Children’s Hospital Los Angeles. A673, TC-71, SK-ES-1, RD-ES, and CADO-ES-1 were purchased from DSMZ (Braunschweig, Germany). SK-N-MC was purchased from ATCC. A204 is a rhabdoid tumor cell line purchased from DSMZ (Braunschweig, Germany). Normal human fibroblasts generated from skin biopsies were obtained from Cliona Rooney (Houston, TX, USA). MSCs of two donors were cultured as described.54 The B cell precursor leukemia cell line SupB15 and the T cell leukemia cell line Jurkat were purchased from ATCC. T cell cultures and B-lymphoblastoid cell lines (LCLs) were generated from healthy donors as described.55 The identity of the cell lines was confirmed by short tandem repeat (STR) profiling (Table S1). For standard adherent growth, tumor cells were cultured in collagen-coated 25- or 75-cm2 tissue culture flasks (MS-EwS-1, MS-EwS-4, MS-EwS-6, MS-EwS-15, MS-EwS-16, MS-EwS-34, TC-71, TC-32, VH-64, CADO-ES-1, SK-ES-1, SK-N-MC, RD-ES, and WE-68) or in uncoated flasks (all others) in RPMI 1640 medium (Invitrogen, Germany), supplemented with 10% heat-inactivated fetal calf serum (FCS; Thermo Fisher Scientific) and 2 mM L-glutamine (Sigma-Aldrich, Germany), and they were maintained at 37°C and 5% CO2. For in vivo experiments, VH-64 cells were lentivirally transduced to express an enhanced GFP-luciferase fusion protein.10
General Lab Operation
The assays were performed by experienced individuals throughout the course of the study. The study was performed using established laboratory protocols covering the processing, freezing, storage, and thawing of cells as well as the staining procedure, data acquisition, and gating strategy. Raw data can be provided upon request.
Flow Cytometry and Cell Sorting
For the analysis of GD2 expression, 50,000 tumor cells were stained with Phycoerythrin (PE)-conjugated monoclonal antibody (mAb) against GD2 (14.G2a), and dead cells were excluded from analysis by staining with Zombie Violet (both BioLegend, Heidelberg, Germany). Surface expression of the CAR was determined by the staining of 250,000 T cells with CF488-conjugated anti-idiotype antibody ganglidiomab56 (100 ng). CF488 conjugation of ganglidiomab was done with the Mix-n-Stain kit, according to the manufacturer’s recommendation (Sigma-Aldrich, Germany). All antibodies were titrated and tested on known positive and negative cells prior to use, and all samples were acquired either directly or not later than 24 h after staining (fixed samples). For each sample, at least 5,000 cells within the respective gates were analyzed with FACS Diva 8.0 using FACS Canto or FACS Celesta flow cytometers (all BD Biosciences, Germany). Subsequent analysis and figure creation were done with FlowJo version (v.)10 (FlowJo, USA). RFIs were calculated by dividing median fluorescence intensities of mAb-stained cells by those obtained with isotype antibodies. For the fluorescence-activated cell sorting (FACS) experiments, 2–4 × 107 tumor cells were harvested, washed with magnetic activated cell sorting (MACS) buffer, and stained with PE-conjugated anti-GD2 antibody 14.G2a for 15 min at room temperature (RT) in the dark. After two additional washing steps, cells were resuspended in 2.5 mL MACS buffer; filtered using FACS tubes with filter and lid; sorted by gating on the 30% highest and 30% lowest or 1% highest and 1% lowest GD2-expressing cells, respectively, using FACS Aria II (BD Biosciences, Germany); and subsequently cultured as described in the respective assays.
Dynamics of GD2 Expression
EwS cell lines were sorted into GD2low (30%) and GD2hi (30%) subpopulations as described above, and they were seeded with 1 × 106 cells in 25-mm2 culture flasks. Every 6 days, cells were harvested and stained with PE-conjugated GD2 antibody 14.G2a over a period of 3 weeks. The RFI of GD2 expression in each subpopulation was calculated in relation to control cells having undergone the sorting procedure without selection of subpopulations.
Treatment with EZH2 Inhibitor
EwS tumor cells were harvested, counted, and seeded in collagen-coated 6-well plates (Sarstedt, Germany) at 0.1–0.3 × 106 cells/well in a total volume of 2 mL. After 2–4 h, the EZH2 inhibitor GSK126 (Active Biochemicals, USA) or tazemetostat (Cayman Chemicals, USA) dissolved in DMSO or DMSO alone as a control was added at a concentration of 10–12, 4, or 1 μM, as indicated in the figures. After 4 days of incubation at 37°C and 5% CO2, the medium was changed, and the EZH2 inhibitor was added again at the same concentrations. Every 7 days, cells were pooled, harvested, counted, and analyzed for the GD2 expression described as above, or lysates for western blot analysis were generated as detailed below.
Western Blot Analysis
EZH2 inhibitor- or DMSO-treated tumor cells were homogenized in 30–100 μL ice-cold radioimmunoprecipitation assay (RIPA) buffer (0.1% DTT, Sigma-Aldrich) with fresh protease inhibitor cocktail (Roche, Germany), shortly fractured in liquid nitrogen, thawed on ice, and then clarified by spinning for 15 min at 4°C and 20,000 × g. After measuring the protein concentration with Bradford reagent, 50 μg sample was separated by electrophoresis on an SDS 15% polyacrylamide gel and then electroblotted onto a nitrocellulose membrane (Bio-Rad). Blocking was done in Tris-buffered saline with Tween20 (TBST) buffer containing 5% BSA for 1 h, followed by incubation with anti-H3K27me3 antibody (Abcam, Germany) diluted 1:1,000 in TBST containing 5% BSA for 12 h at 4°C. After washing, the membrane was incubated with horseradish peroxidase (HRP)-linked anti-mouse immunoglobulin G (IgG) whole Ab (GE Healthcare, Germany) at 1:2,000 in TBST 5% BSA for 1 h at RT, followed by treatment with enhanced chemiluminescence reagent (ECL, Plus Western Blotting Detection System, GE Healthcare), and either exposed to Hyperfilm ECL film (GE Healthcare) for 1 min or directly analyzed by the Imager (ECL Chemostar, INTAS Science Imaging, Germany). Equal protein loading was determined by Ponceau staining. Equal protein loading was also determined by shortly washing the membrane in TBST buffer, followed by incubation with 7 mL Restor Plus stripping buffer (Thermo Scientific) for 15 min at RT. The membrane was then washed twice in 20 mL TBST buffer for 10 min at RT, followed by blocking with TBST buffer with 5% nonfat dry milk for 1 h and detection with a β-actin-specific antibody (Cell Signaling Technology, Germany) diluted 1:5,000 in TBST with 5% BSA for 12 h at 4°C. After washing, the membrane was incubated with HRP-linked anti-rabbit antibody (GE Healthcare) 1:2,000 in TBST 5% milk for 1 h, followed by detection as described above.
In Vitro Expansion Assay
For short-term proliferation, 5,000 cells each were seeded into 3–6 wells of a collagen-coated white-bottom 96-well plate in a total volume of 100 μL. To determine the initial ATP levels for each cell line (day 0), the plate was incubated at RT for 30 min. Then 100 μL Cell-Titer Glo was added, and the plate was placed on an orbital shaker for 2 min and incubated for another 10 min at RT in the dark. The amount of metabolized ATP was quantified by determining the RLUs with GloMax Discover (Promega, Mannheim, Germany). A second plate was then incubated for 96 h at 37°C and 5% CO2, and the same procedure was performed. The relative luminescence units (RLUs) measured after 96 h of incubation were divided by the means of the RLUs on day 0 for each cell line. The results represent the rises in ATP levels indicating proliferation. To assess long-term proliferation, tumor cells were sorted into GD2low (30%) and GD2hi (30%) subpopulations, as described above, and seeded at 1 × 106 cells in 25-mm2 culture flasks. On day 6, cells were harvested, resuspended in RPMI media, and counted using trypan blue staining and a microscope. Fold expansion was calculated by dividing the absolute cell numbers on day 6 by the seeded cell numbers.
Colony Formation Assay
EwS cells were plated in triplicate in 35-mm tissue culture dishes (Thermo Scientific, USA) in methylcellulose-enriched media (1.9% methylcellulose, 15% fetal bovine serum, 0.23% BSA, 1% penicillin/streptomycin [pen/strep], and 82% Iscove’s modified Dulbecco’s media). Cells were plated at 450 cells/mL in 1-mL volumes and incubated for 7–8 days at 37°C and 5% CO2. Numbers of colonies from each culture dish were calculated using an inverted microscope and a scoring grid. The mean colony numbers (colony-forming units [CFUs]) of the triplicate dishes were used for the graphic analyses.
Chemosensitivity Assay
The chemosensitivity of EwS cells was determined by quantification of the numbers of viable cells using the CellTiter Glo Luminescent Cell Viability Assay (Promega, Mannheim, Germany). Tumor cells were seeded in opaque-walled 96-well plates at 5,000 cells/well in 50 μL growth medium, and they were incubated with doxorubicin at dilutions of 0.02, 0.1, 0.2, 0.5, and 1 μmol/L at 37°C and 5% CO2. After 72 h of incubation, 100 μL CellTiter Glo reagent was added to each well. The plates were placed on an orbital shaker for 2 min, then incubated for 10 min at RT in the dark. The amount of metabolized ATP was quantified by determining the RLUs with GloMax Discover (Promega, Mannheim, Germany). Percent viability was determined by dividing the RLUs of doxorubicin-treated cells with untreated cells × 100.
Dynamic 3D Tumor Cell Culture Model
The biological scaffold SISmuc was prepared from porcine gut as previously described,33 followed by removal of mesentery and vascular tree from completely decellularized explants. The matrix is registered under the trade mark BioVaSc-TERM and after seeding with tumor cells under OncVaSc-TERM. All explantations were in compliance with the German Animal Protection Laws (§4 Abs. 3), and the institute’s animal protection officer regularly informed the responsible authorities. The animals received proper attention and humane care in compliance with the Guide for Care and Use of Laboratory Animals (NIH 85e23), and the study was approved by the institutional animal protection board.
For the treatment with EZH2 inhibitor, 1 × 105 EwS cells were seeded on the luminal side of the SISmuc. First, static culture was performed for 3 days by administering 2.5 mL cell-specific media (RPMI 1640 + 10% FCS + Na-Pyruvat + pen/strep) to the cell crowns. After 3 days, the reseeded SISmuc scaffolds were placed into the chambers of customized flow bioreactors and attached to a tubing system containing 45 mL cell-specific media connected to a peristaltic pump, which provides a slow media flow of 3–4 mL/min. 3D tumors on the scaffolds were treated with the EZH2 inhibitor or control for 14 days, then extracted from the bioreactor and cut in half to fix one part in 4% formaldehyde for further paraffin embedding and to extract the cells from the other half for flow cytometry analysis using PBS-EDTA and Trypsin. GD2 staining was performed as described above.
H&E Staining
Samples from the 3D culture model were fixed in 4% paraformaldehyde, washed in PBS, and embedded in paraffin. Paraffin-embedded sections of 4–5 μM were stained with hematoxylin (Merck, Germany) for 3 min and eosin (Roth, Germany) for 2 min. The tissue sections were examined under a light microscope (Leica CTR5500, Germany) after mounting with Vitrocloud (Langenbrink, Germany).
Mouse Model
Mouse experiments were approved by the animal care committee of the local government (LANUV, Recklinghausen, Az. 87-51.04.2010.A117). NSG mice were used from own breeding in the central animal experimental facility Münster (ZTE), originally purchased from Charles River (Germany), and they were housed in pathogen-free rooms in type-2L (long) individually ventilated cages (Charles River) with a maximum of six animals per cage. They were allowed access to sterile food and water ad libitum. The RT was held constantly at 21°C. 8- to 12-week-old NSG mice of both genders were used for the experiments.
For subcutaneous (s.c.) tumor growth, VH-64 or MS-EwS-16 cells were sorted into GD2low (30%) and GD2hi (30%) subpopulations as described above, and they were injected at 1–25 × 104 into the right and left flanks of NSG mice. Tumor growth was monitored at regular intervals and measured using a caliper. Mice were anesthetized with isoflurane and sacrificed when the tumor volumes reached the experimental endpoint. If no tumor was detectable, mice were sacrificed after 12 weeks. Additionally, VH-64 cells sorted into the 1% lowest and highest GD2-expressing cells were injected at 1 × 103 cells each into the right and left flanks of NSG mice. Tumor volume measurement was done as above.
For intravenous (i.v.) tumor growth, VH-64 cells transduced with firefly luciferase were sorted into the 30% lowest and highest GD2-expressing cells, and they were injected at 1–25 × 104 cells into the tail veins of NSG mice. EwS engraftment and growth were monitored weekly starting 2 weeks after transplantation by bioluminescence imaging (BLI), using an IVIS Spectrum Imaging System (PerkinElmer). Mice were injected intraperitoneally (i.p.) with D-luciferin (150 mg/kg; Synchem OHG, s039). At 3 min after injection, mice were anesthetized with isoflurane (2% isoflurane and 0.5 L/min oxygen), and images were acquired after 6 min in dorsal and ventral positions (f/stop, 1; binning, 4; and exposure time, automatic). Mice were sacrificed after tumor volumes reached the experimental endpoints. If no tumor was detectable, mice were sacrificed after 12 weeks.
Real-Time qPCR
RNA was isolated using the RNeasy-Kit (QIAGEN, Germany), according to the manufacturer’s instructions. RNA concentration and purity were determined using Nanodrop analysis (Thermo Scientific, Germany). The purity of all samples was above 1.8 A260/A280. RNA was either stored at −80°C prior to cDNA synthesis or directly used. cDNA was synthesized using the Quick-Start Protocol of QuantiTect Reverse Transcription Kit (QIAGEN, Germany), according to the manufacturer’s recommendations. PCR reactions of 10 μL contained 1 μL cDNA, 5 μL 2× QuantiTect SYBR Green PCR Master Mix (QIAGEN, Germany), and 0.5 μM primers. Primers for EZH2 (QT00054614), ST8SIA1 (GD3 synthase, QT00054159), and B4GALNT1 (GD2 synthase, QT02564009) were purchased from QIAGEN. Primers for the reference gene HPRT1 (forward primer 5′-TGAGGATTTGGAAAGGGTGT-3′, reverse primer 5′-GAGCACACAGAGGGCTACAA-3′) were purchased from Invitrogen. Amplification was performed in triplicate reactions in two different runs at 95°C for 15 min, followed by 94°C for 15 s and 40 cycles of 55°C (30 s) and 72°C (30 s) on a CFX96 Thermal Cycler (Bio-Rad). The cDNA concentrations were adjusted to Cq (threshold cycle) values of HPRT1 control gene to ensure equal amplification efficiencies. Cq values were determined using CFX Manager (Bio-Rad). For the analysis, the triplicates of each run were taken together. Relative gene expression levels were calculated by the 2−ΔΔCq method compared to HPRT1 and untreated or GD2neg cells used as the reference.
ChIP-Seq Dataset and Analysis
A dataset of ChIP-seq against EZH2 using the EwS cell line SK-N-MC was generated in the laboratory of Bradley Bernstein (available from the Encyclopedia of DNA Elements [ENCODE]: ENCSR113GDB; https://www.encodeproject.org/experiments/).57 Signal fold change over control (ENCODE: ENCFF642OLV) and pseudoreplicated irreproducible discovery rate (IDR)-thresholded peak (ENCODE: ENCFF674XUJ) files after standard processing pipeline were downloaded and viewed in the UCSC Genome Browser (http://genome.ucsc.edu).58 Screenshots were produced by using the PDF/PS utility of the Genome Browser.
CRISPR/Cas9 Knockout
The single guide RNA (sgRNA) used for ST8SIA1 (NCBI Gene ID: 6489; GD3 synthase) knockout was designed using the website https://zlab.bio/guide-design-resources. The sgRNA (5′-CACCGCCATTGAAGAAATGCGCGG-3′) was cloned into the BsmBI sites of the lentiviral vector lentiCRISPR_v2 (Addgene 52961).59 To produce lentiviral supernatant for the knockout, HEK293T cells were cotransfected with lentiCRISPR_v2 and the helper plasmids pMD2.G (Addgene plasmid 12259) and psPAX2 (Addgene plasmid 12260), both a gift from Didier Trono, containing gagpol and vesicular stomatitis virus G glycoprotein (VSV-G) sequences. Lentiviral supernatant was harvested after 48 h and used to transduce the tumor cells in 10-cm plates, using 6 μg/mL Sequabrene (Sigma) overnight. After puromycin selection for 1 week, GD3S gene knockout was analyzed by staining for GD2 surface expression as described above. For wild-type (mock) control cells, the lentivirus was produced with an empty lentiCRISPR_v2 vector.
Expansion and Transduction of Human T Cells
The use of blood samples from healthy donors was approved by the University of Münster Ethical Board. Peripheral blood mononuclear cells (PBMCs) seeded at 1.5 × 106/well were stimulated for 48 h using 24-well tissue-culture plates precoated with anti-CD3 and anti-CD28 antibodies (1 μg/mL each). Culture medium consisted of RPMI 1640 or equal portions of RPMI 1640 and AIM V (Invitrogen, 12055-083) with 10% FCS, supplemented with 50 IU/mL recombinant human interleukin-2 (rhIL-2). After 48 h, the T cells were harvested and transferred to 24-well non-tissue-culture-treated plates coated with Retronectin and coincubated with viral supernatant for 48 h, as described.60 For expansion, 5 × 106 transduced or non-transduced T cells were then transferred to gas-permeable culture devices with 50 mL capacity (Wilson Wolf Manufacturing, 80040S) in 35 mL culture medium for 14–16 days.
Constructs
The GD2-specific CAR GD2-BBζ was previously described.19 Briefly, it contains the single-chain antibody domain (single-chain variable fragment [scFv]) of the mAb 14.G2a,38 the hinge domain of human IgG1, followed by the transmembrane domain of CD28 and the signaling domains derived from 4-1BB and CD3ζ. The CAR gene was codon optimized and then subcloned into the AgeI and XhoI sites of the retroviral vector SFG.60 Generation of stable retroviral producer cell lines and production of recombinant retrovirus for transduction of T cells were performed as described.60
Intracellular Cytokine Assay
5 × 105 T cells on day 13–14 after transduction and 5 × 105 target cells after 14 days of EZH2 inhibitor treatment were seeded in tubes in a total volume of 200 μL medium and co-incubated for 2 h at 37°C and 5% CO2. To block cytokine secretion, 10 μg Brefeldin A in 40 μL PBS (Sigma-Aldrich, Germany) was added overnight. After washing with washing buffer (PBS + 0.5% BSA), the CF488-conjugated anti-idiotype antibody ganglidiomab and a CD3-specific antibody were added in 100 μL washing buffer and incubated for 15 min at RT. Following another washing step, cells were incubated for 10 min at RT with 1 mL FACS Lysing Solution (BD Biosciences, Germany), diluted 1:10 in dH2O, then washed and treated for another 10 min at RT with 500 μL FACS Permeabilizing Solution (Becton Dickinson, Germany) diluted 1:10 in dH2O. Cells were washed again, resuspended in 100 μL washing buffer containing antibodies against IFN-γ and TNF-α, and then incubated for another 30 min at RT. After a final washing step, cells were fixed in 250 μL 1% paraformaldehyde (PFA), then analyzed by flow cytometry. T cells incubated with medium alone or with DMSO-treated EwS cells served as controls.
CD107a Degranulation Assay
T cells (3 × 105/well) on days 13–14 after transduction were incubated with an equal number of target cells treated with the EZH2 inhibitor for 14 days in a total volume of 200 μL, in Eppendorf tubes in the presence of Monensin (eBioscience, Germany) (1 μL/mL) and CD107a-PE antibody (BioLegend, Germany) (100 ng/mL) for 3 h at 37°C and 5% CO2. The cells were washed, then incubated with CF488-conjugated GD2 anti-idiotype antibody ganglidiomab and a CD3-specific antibody for 15 min, fixed in 1% PFA, and analyzed by flow cytometry. T cells incubated with medium alone or with DMSO-treated EwS cells served as controls.
Cytotoxicity Assay
The lytic activity of CAR T cells was tested using a calcein-acetyoxymethyl (AM) release assay.61 EwS target cells treated with 4 μM GSK126 for 14 days were washed twice with PBS and then resuspended in PBS at a final concentration of 2 × 106 cells/mL, and they were incubated with 10 μM calcein-AM (Thermo Fisher Scientific, Germany) for 30 min at 37°C with occasional shaking. After two washes in medium (RPMI and 10% FCS), cells were adjusted to 105 cells/mL. The test was performed in flat-bottom 96-well microtiter plates (Thermo Scientific, Germany). CAR T cells at effector-to-target (E:T) cell ratios from 40:1 to 10:1 were seeded in triplicates together with 1 × 104 calcein-AM-labeled target cells, with additional triplicate wells for spontaneous (only target cells in complete medium) and maximum release (only target cells in medium plus 9% Triton X-100). After 4 h at 37°C in 5% CO2, the plates were centrifuged for 5 min and then 75 μL supernatant was harvested and transferred into new black-walled 96-well microtiter plates (Greiner Bio-One, Germany). Samples were measured using a GloMax Discover multi-mode microplate reader (Promega, Germany) (excitation 475 nm and emission 500–550 nm). Data were expressed as arbitrary fluorescent units (AFUs). Specific lysis was calculated according to the following formula: [(test release − spontaneous release)/(maximum release − spontaneous release)] × 100. Non-transduced T cells and DMSO-treated EwS cells served as controls.
Statistics
The paired Student’s t test was used with paired samples and the unpaired Student’s t test with independent samples to test whether the means in each set of parametric distributed values differed significantly, and the rank-sum test was used to compare nonparametric mean values. Tests used are indicated in the figure legends; p values of less than 0.05 were considered to indicate statistically significant differences. Statistical analyses were performed using Sigmaplot 11 (Systat Software, San Jose, CA).
Author Contributions
S.K., B.A., and C.R. designed the study, supervised experiments, analyzed the data, and wrote the manuscript, with input from S.B., S.L., C.G., J.K., J.-H.M., S.S., S.J., J.M., N.F., L.G., M.F., and W.H., who performed experiments or procedures. K.K., H.N.L., N.S., I.M., and H.W. provided study materials. All authors approved the manuscript.
Conflicts of Interest
Westfälische Wilhelms-Universität Münster (WWU) has filed a patent with S.K., B.A., and C.R. as inventors composing GD2 upregulation by EZH2 inhibition in cancer.
Acknowledgments
We thank Natalia Moreno, Claudia Lanvers-Kaminsky, Marc Hotfilder, Annegret Rosemann, Anne Kruchen, Kerstin Cornils, Daniela Schwammbach, and Vijay Bhaskar Reddy for helpful contributions. This work was funded by grant RO 2402/6-1 from the Deutsche Forschungsgemeinschaft (DFG), Germany (to C.R.) and by Kinderkrebshilfe Münster e.V.
Footnotes
Supplemental Information can be found with this article online at https://doi.org/10.1016/j.ymthe.2019.02.014.
Supplemental Information
References
- 1.Eshhar Z., Waks T., Gross G., Schindler D.G. Specific activation and targeting of cytotoxic lymphocytes through chimeric single chains consisting of antibody-binding domains and the gamma or zeta subunits of the immunoglobulin and T-cell receptors. Proc. Natl. Acad. Sci. USA. 1993;90:720–724. doi: 10.1073/pnas.90.2.720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Maude S.L., Laetsch T.W., Buechner J., Rives S., Boyer M., Bittencourt H., Bader P., Verneris M.R., Stefanski H.E., Myers G.D. Tisagenlecleucel in Children and Young Adults with B-Cell Lymphoblastic Leukemia. N. Engl. J. Med. 2018;378:439–448. doi: 10.1056/NEJMoa1709866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee D.W., Kochenderfer J.N., Stetler-Stevenson M., Cui Y.K., Delbrook C., Feldman S.A., Fry T.J., Orentas R., Sabatino M., Shah N.N. T cells expressing CD19 chimeric antigen receptors for acute lymphoblastic leukaemia in children and young adults: a phase 1 dose-escalation trial. Lancet. 2015;385:517–528. doi: 10.1016/S0140-6736(14)61403-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gardner R.A., Finney O., Annesley C., Brakke H., Summers C., Leger K., Bleakley M., Brown C., Mgebroff S., Kelly-Spratt K.S. Intent-to-treat leukemia remission by CD19 CAR T cells of defined formulation and dose in children and young adults. Blood. 2017;129:3322–3331. doi: 10.1182/blood-2017-02-769208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schuster S.J., Bishop M.R., Tam C.S., Waller E.K., Borchmann P., McGuirk J.P., Jäger U., Jaglowski S., Andreadis C., Westin J.R., JULIET Investigators Tisagenlecleucel in Adult Relapsed or Refractory Diffuse Large B-Cell Lymphoma. N. Engl. J. Med. 2019;380:45–56. doi: 10.1056/NEJMoa1804980. [DOI] [PubMed] [Google Scholar]
- 6.Neelapu S.S., Locke F.L., Bartlett N.L., Lekakis L.J., Miklos D.B., Jacobson C.A., Braunschweig I., Oluwole O.O., Siddiqi T., Lin Y. Axicabtagene Ciloleucel CAR T-Cell Therapy in Refractory Large B-Cell Lymphoma. N. Engl. J. Med. 2017;377:2531–2544. doi: 10.1056/NEJMoa1707447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ladenstein R., Pötschger U., Le Deley M.C., Whelan J., Paulussen M., Oberlin O., van den Berg H., Dirksen U., Hjorth L., Michon J. Primary disseminated multifocal Ewing sarcoma: results of the Euro-EWING 99 trial. J. Clin. Oncol. 2010;28:3284–3291. doi: 10.1200/JCO.2009.22.9864. [DOI] [PubMed] [Google Scholar]
- 8.Delattre O., Zucman J., Plougastel B., Desmaze C., Melot T., Peter M., Kovar H., Joubert I., de Jong P., Rouleau G. Gene fusion with an ETS DNA-binding domain caused by chromosome translocation in human tumours. Nature. 1992;359:162–165. doi: 10.1038/359162a0. [DOI] [PubMed] [Google Scholar]
- 9.Long A.H., Highfill S.L., Cui Y., Smith J.P., Walker A.J., Ramakrishna S., El-Etriby R., Galli S., Tsokos M.G., Orentas R.J., Mackall C.L. Reduction of MDSCs with All-trans Retinoic Acid Improves CAR Therapy Efficacy for Sarcomas. Cancer Immunol. Res. 2016;4:869–880. doi: 10.1158/2326-6066.CIR-15-0230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kailayangiri S., Altvater B., Spurny C., Jamitzky S., Schelhaas S., Jacobs A.H., Wiek C., Roellecke K., Hanenberg H., Hartmann W. Targeting Ewing sarcoma with activated and GD2-specific chimeric antigen receptor-engineered human NK cells induces upregulation of immune-inhibitory HLA-G. OncoImmunology. 2016;6:e1250050. doi: 10.1080/2162402X.2016.1250050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martinez C., Hofmann T.J., Marino R., Dominici M., Horwitz E.M. Human bone marrow mesenchymal stromal cells express the neural ganglioside GD2: a novel surface marker for the identification of MSCs. Blood. 2007;109:4245–4248. doi: 10.1182/blood-2006-08-039347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Suzuki K. The pattern of mammalian brain gangliosides. II. Evaluation of the extraction procedures, postmortem changes and the effect of formalin preservation. J. Neurochem. 1965;12:629–638. doi: 10.1111/j.1471-4159.1965.tb04256.x. [DOI] [PubMed] [Google Scholar]
- 13.Rossig C., Kailayangiri S., Jamitzky S., Altvater B. Carbohydrate Targets for CAR T Cells in Solid Childhood Cancers. Front. Oncol. 2018;8:513. doi: 10.3389/fonc.2018.00513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yu A.L., Gilman A.L., Ozkaynak M.F., London W.B., Kreissman S.G., Chen H.X., Smith M., Anderson B., Villablanca J.G., Matthay K.K., Children’s Oncology Group Anti-GD2 antibody with GM-CSF, interleukin-2, and isotretinoin for neuroblastoma. N. Engl. J. Med. 2010;363:1324–1334. doi: 10.1056/NEJMoa0911123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ladenstein R., Pötschger U., Valteau-Couanet D., Luksch R., Castel V., Yaniv I., Laureys G., Brock P., Michon J.M., Owens C. Interleukin 2 with anti-GD2 antibody ch14.18/CHO (dinutuximab beta) in patients with high-risk neuroblastoma (HR-NBL1/SIOPEN): a multicentre, randomised, phase 3 trial. Lancet Oncol. 2018;19:1617–1629. doi: 10.1016/S1470-2045(18)30578-3. [DOI] [PubMed] [Google Scholar]
- 16.Heczey A., Louis C.U., Savoldo B., Dakhova O., Durett A., Grilley B., Liu H., Wu M.F., Mei Z., Gee A. CAR T Cells Administered in Combination with Lymphodepletion and PD-1 Inhibition to Patients with Neuroblastoma. Mol. Ther. 2017;25:2214–2224. doi: 10.1016/j.ymthe.2017.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pule M.A., Savoldo B., Myers G.D., Rossig C., Russell H.V., Dotti G., Huls M.H., Liu E., Gee A.P., Mei Z. Virus-specific T cells engineered to coexpress tumor-specific receptors: persistence and antitumor activity in individuals with neuroblastoma. Nat. Med. 2008;14:1264–1270. doi: 10.1038/nm.1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Louis C.U., Savoldo B., Dotti G., Pule M., Yvon E., Myers G.D., Rossig C., Russell H.V., Diouf O., Liu E. Antitumor activity and long-term fate of chimeric antigen receptor-positive T cells in patients with neuroblastoma. Blood. 2011;118:6050–6056. doi: 10.1182/blood-2011-05-354449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kailayangiri S., Altvater B., Meltzer J., Pscherer S., Luecke A., Dierkes C., Titze U., Leuchte K., Landmeier S., Hotfilder M. The ganglioside antigen G(D2) is surface-expressed in Ewing sarcoma and allows for MHC-independent immune targeting. Br. J. Cancer. 2012;106:1123–1133. doi: 10.1038/bjc.2012.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liebsch L., Kailayangiri S., Beck L., Altvater B., Koch R., Dierkes C., Hotfilder M., Nagelmann N., Faber C., Kooijman H. Ewing sarcoma dissemination and response to T-cell therapy in mice assessed by whole-body magnetic resonance imaging. Br. J. Cancer. 2013;109:658–666. doi: 10.1038/bjc.2013.356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fry T.J., Shah N.N., Orentas R.J., Stetler-Stevenson M., Yuan C.M., Ramakrishna S., Wolters P., Martin S., Delbrook C., Yates B. CD22-targeted CAR T cells induce remission in B-ALL that is naive or resistant to CD19-targeted CAR immunotherapy. Nat. Med. 2018;24:20–28. doi: 10.1038/nm.4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Walker A.J., Majzner R.G., Zhang L., Wanhainen K., Long A.H., Nguyen S.M., Lopomo P., Vigny M., Fry T.J., Orentas R.J., Mackall C.L. Tumor Antigen and Receptor Densities Regulate Efficacy of a Chimeric Antigen Receptor Targeting Anaplastic Lymphoma Kinase. Mol. Ther. 2017;25:2189–2201. doi: 10.1016/j.ymthe.2017.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Suzuki Y., Yanagisawa M., Ariga T., Yu R.K. Histone acetylation-mediated glycosyltransferase gene regulation in mouse brain during development. J. Neurochem. 2011;116:874–880. doi: 10.1111/j.1471-4159.2010.07042.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Richter G.H., Plehm S., Fasan A., Rössler S., Unland R., Bennani-Baiti I.M., Hotfilder M., Löwel D., von Luettichau I., Mossbrugger I. EZH2 is a mediator of EWS/FLI1 driven tumor growth and metastasis blocking endothelial and neuro-ectodermal differentiation. Proc. Natl. Acad. Sci. USA. 2009;106:5324–5329. doi: 10.1073/pnas.0810759106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Riggi N., Knoechel B., Gillespie S.M., Rheinbay E., Boulay G., Suvà M.L., Rossetti N.E., Boonseng W.E., Oksuz O., Cook E.B. EWS-FLI1 utilizes divergent chromatin remodeling mechanisms to directly activate or repress enhancer elements in Ewing sarcoma. Cancer Cell. 2014;26:668–681. doi: 10.1016/j.ccell.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sankar S., Theisen E.R., Bearss J., Mulvihill T., Hoffman L.M., Sorna V., Beckerle M.C., Sharma S., Lessnick S.L. Reversible LSD1 inhibition interferes with global EWS/ETS transcriptional activity and impedes Ewing sarcoma tumor growth. Clin. Cancer Res. 2014;20:4584–4597. doi: 10.1158/1078-0432.CCR-14-0072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sheffield N.C., Pierron G., Klughammer J., Datlinger P., Schönegger A., Schuster M., Hadler J., Surdez D., Guillemot D., Lapouble E. DNA methylation heterogeneity defines a disease spectrum in Ewing sarcoma. Nat. Med. 2017;23:386–395. doi: 10.1038/nm.4273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Svoboda L.K., Harris A., Bailey N.J., Schwentner R., Tomazou E., von Levetzow C., Magnuson B., Ljungman M., Kovar H., Lawlor E.R. Overexpression of HOX genes is prevalent in Ewing sarcoma and is associated with altered epigenetic regulation of developmental transcription programs. Epigenetics. 2014;9:1613–1625. doi: 10.4161/15592294.2014.988048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tomazou E.M., Sheffield N.C., Schmidl C., Schuster M., Schönegger A., Datlinger P., Kubicek S., Bock C., Kovar H. Epigenome mapping reveals distinct modes of gene regulation and widespread enhancer reprogramming by the oncogenic fusion protein EWS-FLI1. Cell Rep. 2015;10:1082–1095. doi: 10.1016/j.celrep.2015.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Riggi N., Suvà M.L., Suvà D., Cironi L., Provero P., Tercier S., Joseph J.M., Stehle J.C., Baumer K., Kindler V., Stamenkovic I. EWS-FLI-1 expression triggers a Ewing’s sarcoma initiation program in primary human mesenchymal stem cells. Cancer Res. 2008;68:2176–2185. doi: 10.1158/0008-5472.CAN-07-1761. [DOI] [PubMed] [Google Scholar]
- 31.Comet I., Riising E.M., Leblanc B., Helin K. Maintaining cell identity: PRC2-mediated regulation of transcription and cancer. Nat. Rev. Cancer. 2016;16:803–810. doi: 10.1038/nrc.2016.83. [DOI] [PubMed] [Google Scholar]
- 32.Krook M.A., Hawkins A.G., Patel R.M., Lucas D.R., Van Noord R., Chugh R., Lawlor E.R. A bivalent promoter contributes to stress-induced plasticity of CXCR4 in Ewing sarcoma. Oncotarget. 2016;7:61775–61788. doi: 10.18632/oncotarget.11240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Göttlich C., Müller L.C., Kunz M., Schmitt F., Walles H., Walles T., Dandekar T., Dandekar G., Nietzer S.L. A Combined 3D Tissue Engineered In Vitro/In Silico Lung Tumor Model for Predicting Drug Effectiveness in Specific Mutational Backgrounds. J. Vis. Exp. 2016;(110):e53885. doi: 10.3791/53885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Italiano A., Soria J.C., Toulmonde M., Michot J.M., Lucchesi C., Varga A., Coindre J.M., Blakemore S.J., Clawson A., Suttle B. Tazemetostat, an EZH2 inhibitor, in relapsed or refractory B-cell non-Hodgkin lymphoma and advanced solid tumours: a first-in-human, open-label, phase 1 study. Lancet Oncol. 2018;19:649–659. doi: 10.1016/S1470-2045(18)30145-1. [DOI] [PubMed] [Google Scholar]
- 35.Ngamukote S., Yanagisawa M., Ariga T., Ando S., Yu R.K. Developmental changes of glycosphingolipids and expression of glycogenes in mouse brains. J. Neurochem. 2007;103:2327–2341. doi: 10.1111/j.1471-4159.2007.04910.x. [DOI] [PubMed] [Google Scholar]
- 36.Battula V.L., Shi Y., Evans K.W., Wang R.Y., Spaeth E.L., Jacamo R.O., Guerra R., Sahin A.A., Marini F.C., Hortobagyi G. Ganglioside GD2 identifies breast cancer stem cells and promotes tumorigenesis. J. Clin. Invest. 2012;122:2066–2078. doi: 10.1172/JCI59735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liang Y.J., Wang C.Y., Wang I.A., Chen Y.W., Li L.T., Lin C.Y., Ho M.Y., Chou T.L., Wang Y.H., Chiou S.P. Interaction of glycosphingolipids GD3 and GD2 with growth factor receptors maintains breast cancer stem cell phenotype. Oncotarget. 2017;8:47454–47473. doi: 10.18632/oncotarget.17665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rossig C., Bollard C.M., Nuchtern J.G., Merchant D.A., Brenner M.K. Targeting of G(D2)-positive tumor cells by human T lymphocytes engineered to express chimeric T-cell receptor genes. Int. J. Cancer. 2001;94:228–236. doi: 10.1002/ijc.1457. [DOI] [PubMed] [Google Scholar]
- 39.Krause A., Guo H.F., Latouche J.B., Tan C., Cheung N.K., Sadelain M. Antigen-dependent CD28 signaling selectively enhances survival and proliferation in genetically modified activated human primary T lymphocytes. J. Exp. Med. 1998;188:619–626. doi: 10.1084/jem.188.4.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roth M., Linkowski M., Tarim J., Piperdi S., Sowers R., Geller D., Gill J., Gorlick R. Ganglioside GD2 as a therapeutic target for antibody-mediated therapy in patients with osteosarcoma. Cancer. 2014;120:548–554. doi: 10.1002/cncr.28461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Abiko K., Matsumura N., Hamanishi J., Horikawa N., Murakami R., Yamaguchi K., Yoshioka Y., Baba T., Konishi I., Mandai M. IFN-γ from lymphocytes induces PD-L1 expression and promotes progression of ovarian cancer. Br. J. Cancer. 2015;112:1501–1509. doi: 10.1038/bjc.2015.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Spurny C., Kailayangiri S., Altvater B., Jamitzky S., Hartmann W., Wardelmann E., Ranft A., Dirksen U., Amler S., Hardes J. s. Oncotarget. 2017;9:6536–6549. doi: 10.18632/oncotarget.23815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rasini V., Dominici M., Kluba T., Siegel G., Lusenti G., Northoff H., Horwitz E.M., Schäfer R. Mesenchymal stromal/stem cells markers in the human bone marrow. Cytotherapy. 2013;15:292–306. doi: 10.1016/j.jcyt.2012.11.009. [DOI] [PubMed] [Google Scholar]
- 44.Jin H.J., Nam H.Y., Bae Y.K., Kim S.Y., Im I.R., Oh W., Yang Y.S., Choi S.J., Kim S.W. GD2 expression is closely associated with neuronal differentiation of human umbilical cord blood-derived mesenchymal stem cells. Cell. Mol. Life Sci. 2010;67:1845–1858. doi: 10.1007/s00018-010-0292-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xu J., Liao W., Gu D., Liang L., Liu M., Du W., Liu P., Zhang L., Lu S., Dong C. Neural ganglioside GD2 identifies a subpopulation of mesenchymal stem cells in umbilical cord. Cell. Physiol. Biochem. 2009;23:415–424. doi: 10.1159/000218188. [DOI] [PubMed] [Google Scholar]
- 46.Shi B., Liang J., Yang X., Wang Y., Zhao Y., Wu H., Sun L., Zhang Y., Chen Y., Li R. Integration of estrogen and Wnt signaling circuits by the polycomb group protein EZH2 in breast cancer cells. Mol. Cell. Biol. 2007;27:5105–5119. doi: 10.1128/MCB.00162-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kroesen M., Büll C., Gielen P.R., Brok I.C., Armandari I., Wassink M., Looman M.W., Boon L., den Brok M.H., Hoogerbrugge P.M., Adema G.J. Anti-GD2 mAb and Vorinostat synergize in the treatment of neuroblastoma. OncoImmunology. 2016;5:e1164919. doi: 10.1080/2162402X.2016.1164919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liang Y.J., Ding Y., Levery S.B., Lobaton M., Handa K., Hakomori S.I. Differential expression profiles of glycosphingolipids in human breast cancer stem cells vs. cancer non-stem cells. Proc. Natl. Acad. Sci. USA. 2013;110:4968–4973. doi: 10.1073/pnas.1302825110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kurmasheva R.T., Sammons M., Favours E., Wu J., Kurmashev D., Cosmopoulos K., Keilhack H., Klaus C.R., Houghton P.J., Smith M.A. Initial testing (stage 1) of tazemetostat (EPZ-6438), a novel EZH2 inhibitor, by the Pediatric Preclinical Testing Program. Pediatr. Blood Cancer. 2017;64:e26218. doi: 10.1002/pbc.26218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Goswami S., Apostolou I., Zhang J., Skepner J., Anandhan S., Zhang X., Xiong L., Trojer P., Aparicio A., Subudhi S.K. Modulation of EZH2 expression in T cells improves efficacy of anti-CTLA-4 therapy. J. Clin. Invest. 2018;128:3813–3818. doi: 10.1172/JCI99760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Peng D., Kryczek I., Nagarsheth N., Zhao L., Wei S., Wang W., Sun Y., Zhao E., Vatan L., Szeliga W. Epigenetic silencing of TH1-type chemokines shapes tumour immunity and immunotherapy. Nature. 2015;527:249–253. doi: 10.1038/nature15520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Leuchte K., Altvater B., Hoffschlag S., Potratz J., Meltzer J., Clemens D., Luecke A., Hardes J., Dirksen U., Juergens H. Anchorage-independent growth of Ewing sarcoma cells under serum-free conditions is not associated with stem-cell like phenotype and function. Oncol. Rep. 2014;32:845–852. doi: 10.3892/or.2014.3269. [DOI] [PubMed] [Google Scholar]
- 53.Unland R., Clemens D., Heinicke U., Potratz J.C., Hotfilder M., Fulda S., Wardelmann E., Frühwald M.C., Dirksen U. Suberoylanilide hydroxamic acid synergistically enhances the antitumor activity of etoposide in Ewing sarcoma cell lines. Anticancer Drugs. 2015;26:843–851. doi: 10.1097/CAD.0000000000000256. [DOI] [PubMed] [Google Scholar]
- 54.Gieseke F., Kruchen A., Tzaribachev N., Bentzien F., Dominici M., Müller I. Proinflammatory stimuli induce galectin-9 in human mesenchymal stromal cells to suppress T-cell proliferation. Eur. J. Immunol. 2013;43:2741–2749. doi: 10.1002/eji.201343335. [DOI] [PubMed] [Google Scholar]
- 55.Altvater B., Pscherer S., Landmeier S., Niggemeier V., Juergens H., Vormoor J., Rossig C. CD28 co-stimulation via tumour-specific chimaeric receptors induces an incomplete activation response in Epstein-Barr virus-specific effector memory T cells. Clin. Exp. Immunol. 2006;144:447–457. doi: 10.1111/j.1365-2249.2006.03095.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lode H.N., Schmidt M., Seidel D., Huebener N., Brackrock D., Bleeke M., Reker D., Brandt S., Mueller H.P., Helm C., Siebert N. Vaccination with anti-idiotype antibody ganglidiomab mediates a GD(2)-specific anti-neuroblastoma immune response. Cancer Immunol. Immunother. 2013;62:999–1010. doi: 10.1007/s00262-013-1413-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sloan C.A., Chan E.T., Davidson J.M., Malladi V.S., Strattan J.S., Hitz B.C., Gabdank I., Narayanan A.K., Ho M., Lee B.T. ENCODE data at the ENCODE portal. Nucleic Acids Res. 2016;44(D1):D726–D732. doi: 10.1093/nar/gkv1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kent W.J., Sugnet C.W., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sanjana N.E., Shalem O., Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods. 2014;11:783–784. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Altvater B., Landmeier S., Pscherer S., Temme J., Schweer K., Kailayangiri S., Campana D., Juergens H., Pule M., Rossig C. 2B4 (CD244) signaling by recombinant antigen-specific chimeric receptors costimulates natural killer cell activation to leukemia and neuroblastoma cells. Clin. Cancer Res. 2009;15:4857–4866. doi: 10.1158/1078-0432.CCR-08-2810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Neri S., Mariani E., Meneghetti A., Cattini L., Facchini A. Calcein-acetyoxymethyl cytotoxicity assay: standardization of a method allowing additional analyses on recovered effector cells and supernatants. Clin. Diagn. Lab. Immunol. 2001;8:1131–1135. doi: 10.1128/CDLI.8.6.1131-1135.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.