Abstract
As a flavor and platform chemical, m-cresol (3-methylphenol) is a valuable industrial compound that currently is mainly synthesized by chemical methods from fossil resources. In this study, we present the first biotechnological de novo production of m-cresol from sugar in complex yeast extract-peptone medium with the yeast Saccharomyces cerevisiae. A heterologous pathway based on the decarboxylation of the polyketide 6-methylsalicylic acid (6-MSA) was introduced into a CEN.PK yeast strain. For synthesis of 6-MSA, expression of different variants of 6-MSA synthases (MSASs) were compared. Overexpression of codon-optimized MSAS from Penicillium patulum together with activating phosphopantetheinyl transferase npgA from Aspergillus nidulans resulted in up to 367 mg/L 6-MSA production. Additional genomic integration of the genes had a strongly promoting effect and 6-MSA titers reached more than 2 g/L. Simultaneous expression of 6-MSA decarboxylase patG from A. clavatus led to the complete conversion of 6-MSA and production of up to 589 mg/L m-cresol. As addition of 450–750 mg/L m-cresol to yeast cultures nearly completely inhibited growth our data suggest that the toxicity of m-cresol might be the limiting factor for higher production titers.
Keywords: 6-Methylsalicylic acid synthase, m-Cresol, 6-Methylsalicylic acid decarboxylase, Polyketide synthase, Codon-optimization, Saccharomyces cerevisiae
Abbreviations: Acyl carrier protein, ACP; Acyltransferase, AT; ketoreductase, KR; ketosynthase, KS; 6-methylsalicylic acid, 6-MSA; 6-methylsalicylic acid decarboxylase, PatG; 6-methylsalicylic acid synthase, MSAS; optical density, OD; phosphopantetheinyl transferase, PPT; polyketide synthase, PKS; thioester hydrolase, TH
Graphical abstract
Highlights
-
•
Expression of 6-methylsalicylic acid synthase (MSAS) and decarboxylase enables m-cresol production from sugars in complex medium in S. cerevisiae
-
•
6-methylsalicylic acid synthase is limiting 6-MSA and m-cresol production rates
-
•
Genomic integration of heterologous genes improves product titers
-
•
Toxicity of m-cresol to yeast cells limits increased production titers
1. Introduction
Meta-cresol (m-cresol, 3-methylphenol) is an important specialty chemical and platform compound. m-Cresol and 4-chloro-m-cresol are utilized as disinfectants and antiseptic agents because of their antibacterial and antifungal properties (Lambert et al., 1998; McDonnell and Russel, 1999; Nishimura et al., 2008; Spray and Lodge, 1943). Cresols also act as antioxidants scavenging reactive oxygen species (Yeung et al., 2002). As platform compound m-cresol is suitable for synthesis of several chemicals with high market value. The most prominent industrial example is menthol, which is chemically synthesized via alkylation of m-cresol to thymol and further hydrogenation to menthol (Yadav and Pathre, 2005). Because of its peppermint odor, L-menthol is a desired flavor in chewing gum and toothpaste (Berger, 2007).
Currently, m-cresol is mainly produced by chemical processes from fossil resources, and additional purification steps have to be applied to isolate it from o-, m-, p-cresol mixtures. Due to limitations in fossil resource reserves and environmental concerns, chemical synthesis of m-cresol is not sustainable and biotechnological production from renewable resources desirable. Many Penicillium and Aspergillus species can natively synthesize m-cresol as an intermediate in biosynthesis of the mycotoxin patulin (Puel et al., 2010).
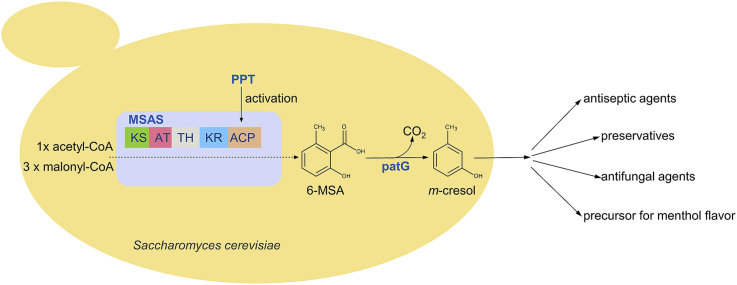
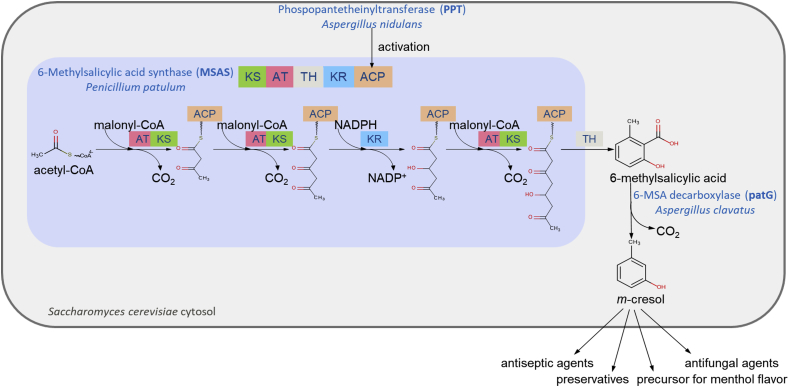
The first steps of patulin biosynthesis are catalyzed by the iterative polyketide synthase (PKS) 6-methylsalicylic acid synthase (MSAS) and the 6-methylsalicylic acid (6-MSA) decarboxylase leading to formation of m-cresol (Puel et al., 2010). MSAS contains different functional domains such as ketosynthase (KS), acyltransferase (AT), thioester hydrolase (TH), ketoreductase (KR), and acyl carrier protein (ACP) (Parascandolo et al., 2016) on 180 kDa homotetramer subunits (Spencer and Jordan, 1992) and catalyzes chain elongation and modification in an iterative fashion (Fig. 1). This PKS utilizes one acetyl-CoA and three malonyl-CoA in three decarboxylative claisen thioester condensations and one TH-mediated hydrolysis after the third elongation round to form 6-methylsalicylic acid (6-MSA) (Parascandolo et al., 2016).
Fig. 1.
Metabolic pathway for m-cresol production in S. cerevisiae via 6-methylsalicylic acid (6-MSA) synthesis. The 6-methylsalicylic acid synthase (MSAS) consists of multiple domains: the ketoacylsynthase (KS), acyltransferase (AT), thioester hydrolase (TH), ketoreductase (KR), and acyl carrier protein (ACP). MSAS must be activated by phosphopantetheinylation, and catalyzes the synthesis of 6-MSA from one acetyl-CoA and three malonyl-CoA under consumption of one NADPH. 6-MSA decarboxylase can further convert 6-MSA to m-cresol, valuable for the flavor and pharmaceutical industry. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Because of their anticancer, antibiotic, mycotoxic or cholesterol-lowering properties of polyketides including polyphenols, macrolides, polyenes, enediynes and polyethers (Hertweck, 2009), interest for high-level production of polyketides has increased. Due to difficult cultivations and genetical limitations of their native fungal hosts, heterologous production systems are promising alternatives. The MSAS gene of Penicillium patulum was already heterologously expressed in Streptomyces coelicolor, Escherichia coli and Saccharomyces cerevisiae for 6-MSA production (Bedford et al., 1995; Kealey et al., 1998; Wattanachaisaereekul et al., 2007). In E. coli and S. cerevisiae co-expression of a suitable phosphopantetheinyl transferase (PPT), for example sfp from Bacillus subtilis or npgA from Aspergillus nidulans, was required for an active holo-form of the ACP domain of MSAS (Kealey et al., 1998; Wattanachaisaereekul et al., 2007). High-level expression together with engineering of the endogenous metabolism and precursor supply resulted in 6-MSA titers up to 67 mg/L in S. coelicolor (Bedford et al., 1995), 75 mg/L in E. coli, 554 mg/L in S. cerevisiae in minimal medium (Wattanachaisaereekul et al., 2008) and 1.7 g/L in S. cerevisiae in YPD (Kealey et al., 1998). Besides MSAS, also the 6-MSA decarboxylase patG from Aspergillus clavatus has already been expressed in S. cerevisiae but was used only in biotransformation assays after supplementation of the medium with 6-MSA (Snini et al., 2014). S. cerevisiae as a heterologous host for biotechnological production processes has several advantages compared to other microorganisms. It is quite robust in harsh industrial fermentation conditions, not sensitive against phages, able to ferment sugars at low pH, and many genetic tools are available for genetic engineering (Gibson et al., 2007; Liu, 2011; Weber et al., 2010).
In this study, we established the pathway for de novo production of m-cresol from glucose in S. cerevisiae. First, we compared different MSAS variants and PPT for production of the intermediate 6-MSA in S. cerevisiae. We established functional expression of patG from A. clavatus in biotransformation assays with 6-MSA and evaluated the toxic effect of the product m-cresol on yeast. Finally, combining the heterologous expression of the most promising variants of MSAS, PPT and patG from multi-copy plasmids together with genomic integration of the expression constructs enabled de novo production of m-cresol from glucose in complex medium in yeast up to toxic levels.
2. Material and methods
2.1. Strains and plasmids
Yeast strains and plasmids used in this study are listed in Table 1. Yeast strains from freshly streaked YPD (= complex medium with glucose) plates (20 g/L peptone, 10 g/L yeast extract, 20 g/L glucose) were used for cultivations. SCD medium consisted of 1.7 g/L yeast nitrogen base without amino acids, 5 g/L ammonium sulfate and 20 g/L glucose. E. coli DH10β (Gibco BRL, Gaithersburg, MD) was utilized for subcloning of plasmids and grown in lysogeny broth (LB)-medium (10 g/L trypton, 5 g/L yeast extract, 5 g/L sodium chloride, pH 7.5). For plasmid maintenance appropriate antibiotics (200 mg/L hygromycin, 200 mg/L G418, 100 mg/L ampicillin) were added to media.
Table 1.
Plasmids and yeast strains used in the present study. Genes from Saccharomyces cerevisiae (Sc), Penicillium patulum (Pp), Aspergillus nidulans (An), Aspergillus niger (Ani) Aspergillus clavatus (Ac), codon-optimized genes (opt) or variants previously used by Wattanachaisaereekul et al. (2008) (var) are indicated by prefixes in superscript. Other abbreviations: hphNT1: hygromycin resistance; Ampr: ampicillin resistance; kanMX: kanamycin resistance. If not stated otherwise, promoters (p) were taken 1–500 bp upstream and terminators (t) 1–300 bp downstream of respective open reading frames.
| Plasmid | Plasmid based on | Relevant features | Reference |
|---|---|---|---|
| pRS42K | – | 2μ, kanMX, Ampr | Taxis and Knop (2006) |
| pRS62H | – | 2μ, hphNT1, Ampr, HXT7p−1- −392, FBA1t | Farwick et al. (2014) |
| pJHV1 | pRS42K | 2μ, kanMX, Ampr, MET25p−1- −384, CYC1t | This work |
| pJHV2 | pRS62H | 2μ, hphNT1, Ampr, HXT7p−1 -−392-AnoptnpgA-FBA1t | This work |
| pJHV5 | pJHV7 | 2μ, kanMX, Ampr, PGK1p-AniMSAS-CYC1t | This work |
| pJHV7 | pRS42K | 2μ, kanMX, Ampr, PGK1p, CYC1t | This work |
| pJHV11 | pJHV7 | 2μ, kanMX, Ampr, PGK1p-PpMSAS-CYC1t | This work |
| pJHV13 | pJHV7 | 2μ, kanMX, Ampr, PGK1p, CYC1t, FBA1p-AcoptpatG-ADH1t | This work |
| pJHV17 | pJHV7 | 2μ, kanMX, Ampr, PGK1p-PpvarMSAS-CYC1t | This work |
| pJHV20 | pRS62H | 2μ, hphNT1, Ampr, HXT7p−1- −392-AnnpgA-FBA1t | This work |
| pJHV36 | pJHV7 | 2μ, kanMX, Ampr, PGK1p-PpoptMSAS-CYC1t | This work |
| pJHV49 | pJHV36 | 2μ, kanMX, Ampr, PGK1p-PpoptMSAS-CYC1t, HXT7p−1- −392-AnoptnpgA-FBA1t | This work |
| pJHV53 | pJHV13 | 2μ, kanMX, Ampr, PGK1p-PpoptMSAS-CYC1t, HXT7p−1- −392-AnoptnpgA-FBA1t, FBA1p-AcoptpatG-ADH1t | This work |
| pRCC-K_URA3 | – | 2μ, kanMX, Ampr, ROX3p-optCas9-CYC1t, pSNR52-gRNA for URA3 | Mara Reifenrath, University of Frankfurt |
| SiHV33 | – | ConLS′-gfp-dropout-ConRE′-KanMX-URA 3′Hom-KanR-ColE1-URA3 5′Hom | Simon Harth, University of Frankfurt |
| pRS426CTMSA-PP | – | 2μ, URA3, Ampr, TEF1p-PpvarMSAS-CYC1t | Verena Siewers, Chalmers, Gothenburg |
| pDKP4832 |
– |
2μ, URA3, Ampr, GAL1p-AnnpgA |
Verena Siewers, Chalmers, Gothenburg |
|
S. cerevisiae strain |
Parent strain |
Relevant features |
Reference |
| CEN.PK2-1C | – | MATa leu2-3112 ura3-52 trp1-289 his3-Δ1 MAL2–8cSUC2 | Entian and Kötter (2007) |
| JHY162 | CEN.PK2–1C | ura3::PGK1p-PpoptMSAS-CYC1t_HXT7p−1- −392-AnoptnpgA-FBA1t_FBA1p-AcoptpatG-ADH1t | This work |
| JHY163 | CEN.PK2–1C | ura3::PGK1p-PpoptMSAS-CYC1t_HXT7p−1- −392-AnoptnpgA-FBA1t | This work |
2.2. Plasmid and strain construction
The DNA sequences of patG, PpMSAS without the intron (Beck et al., 1990) and npgA (respective GeneBank accession numbers JN698985.1, X55776.1, AF198117.1) were codon-optimized with the JCat tool (Grote et al., 2005). They as well as the native PpMSAS gene were ordered from Thermo Fischer Scientific (Germany) as one or more GeneArt Strings DNA fragments. PpvarMSAS and native npgA were received on the plasmids pRS426CTMSA-PP (Wattanachaisaereekul et al., 2008) and pDPK4832 (Wattanachaisaereekul et al., 2007) from Chalmers University of Technology. Codon-optimized sequences were deposited in GenBank under the accession numbers MK791642 (PpoptMSAS), MK791643 (PpvarMSAS), MK791644 (optnpgA) and MK791645 (optpatG). For DNA amplification of AniMSAS (GeneBank accession number XM_001402371.2) genomic DNA of A. niger was donated by Technical University of Munich. Open reading frames, promoters and terminators were amplified by PCR from genomic DNA of CEN.PK2-1C or from plasmids with 35 bp homologous overlaps using primers shown in supplementary Table S1. Plasmids were assembled in yeast via homologous recombination of overlapping PCR fragments or String DNA fragments and linearized vector backbone as described previously (Schadeweg and Boles, 2016). Yeast was transformed with DNA fragments according to Gietz and Schiestl (2007). Assembled plasmids were recovered by yeast DNA preparations and were transformed in E. coli for propagation and amplification. If only one PCR fragment and a vector backbone were assembled, Gibson assembly was used (Gibson et al., 2009). Genomic integrations into the ura3 locus of CEN.PK2–1C were performed with the CRISPR/Cas9 system described in Generoso et al. (2016) with the CRISPR/Cas9 plasmid pRCC-K_URA3 from Mara Reifenrath (University of Frankfurt) described in (Reifenrath and Boles (2018)). The donor DNA for insertion and the up to 500 bp homologous regions upstream and downstream of ura3 were amplified from plasmid pJHV53 and SIHV33 with the primers listed in Table S1. After yeast transformation cells were streaked out on selective YPD medium.
2.3. Cell cultivation
For fermentations for m-cresol and 6-MSA production precultures were cultivated in 150 mL YPD supplemented with corresponding antibiotics in 500 mL Erlenmeyer shake flasks at 180 rpm and 30 °C. Precultures were harvested in exponential phase and for main culture 25 mL YPD with respective antibiotics in 100 mL Erlenmeyer shake flasks were inoculated to an optical density (OD600 nm) of 0.1 in low-OD fermentations and 5 or higher in high-OD fermentations and shaken at 180 rpm 30 °C for 144 h. Cultures for m-cresol production were incubated in a 30 °C shaking waterbath (Memmert, Germany) under a hood to avoid m-cresol inhalation. Samples for cell density determination and HPLC analysis were taken every 0, 6, 24, 48, 72 (or in some cases 120) and 144 h.
For biotransformation experiments of 6-MSA to m-cresol CEN.PK2-1C expressing the codon-optimized patG from A. clavatus on a plasmid and an empty vector control were grown in YPD medium with corresponding antibiotic at 180 rpm and 30 °C. Twenty-five milliliter YPD with respective antibiotic was supplemented with 3.8 mg 6-MSA in 25 μL ethanol to a final concentration of 1 mM, inoculated with the preculture to an OD600 nm of 0.1 and shaken at 180 rpm and 30 °C. Optical density, 6-MSA consumption and m-cresol production were followed at 0, 3, 6, 24, 48 and 72 h by taking samples of the fermentation.
To determine the toxicity of m-cresol a preculture of CEN.PK2-1C was prepared in 150 mL YPD in 500 mL shake flasks at 180 rpm and 30 °C overnight. Fifty milliliter YPD in 300 mL shake flasks were inoculated at an optical density of 0.1, supplemented with different concentrations of m-cresol (750, 600, 450, 300, 150 and 0 mg/L) and grown at 180 rpm and 30 °C for 144 h. Cell densities were followed with the Cell Growth Quantifier (Aquila Biolabs GmbH, Germany) (Bruder et al., 2016). All experiments were carried out in two biological replicates.
2.4. Growth and metabolite analysis
For fermentations and biotransformation experiments cell growth was monitored in the spectrophotometer Ultrospec 2100 pro (GE Healthcare, USA) at an optical density of 600 nm. Culture supernatants for HPLC analysis of 6-MSA and m-cresol formation were obtained by centrifugation at 10,000 rpm for 2 min, 400 μL were treated with 100 μL acetonitrile, centrifuged again and stored at -20 °C until analysis. Samples were analysed in a HPLC (Dionex) using an Agilent Zorbax SB-C8 column (4.6 × 150 mm, 3.5 μm) at 40 °C. 6-MSA and m-cresol were separated by the following gradient of solvent A (0.1% (v/v) formic acid in ddH2O) and solvent B (0.1% (v/v) formic acid in acetonitrile) at a flow rate of 1 mL/min: 5 min 15% B, 20 min linear gradient to 40% B, 1 min linear gradient to 100% B, 4 min 100% B, 1 min linear gradient to 15% B, 4 min 15% B. 6-MSA was detected at 308 nm and m-cresol at 270 nm in an UV detector (Dionex UltiMate 3000 Variable Wavelength Detector). For quantification and calibration, 6-MSA/cresol standards were prepared in equal concentrations (w/v) in 50% (v/v) DMSO/H2O from 6-MSA purchased from Cayman Chemical Company (19199) and m-cresol purchased from Carl Roth (9269.1). For data analysis and graphing the software Prism 5 (GraphPad, USA) was utilized. Each data point represents the mean of at least two biological replicates and the error bars represent the standard deviation as calculated by the software.
3. Results & discussion
3.1. Biosynthesis of the precursor 6-methylsalicylic acid
In order to proof the principle of de novo m-cresol production from glucose in S. cerevisiae, the precursor 6-MSA is required in sufficient amounts. To produce high levels of 6-MSA, we therefore first expressed and compared native MSAS from A. niger (AniMSAS) (Fisch et al., 2009) and three MSAS variants from P. patulum, namely native (PpMSAS) (Beck et al., 1990), codon-optimized (PpoptMSAS) and a variant previously used by Wattanachaisaereekul et al. (2008) (PpvarMSAS). Compared to native PpMSAS this variant exhibits two amino acid exchanges (A699S and N1677S) and 32 silent mutations within its coding region.
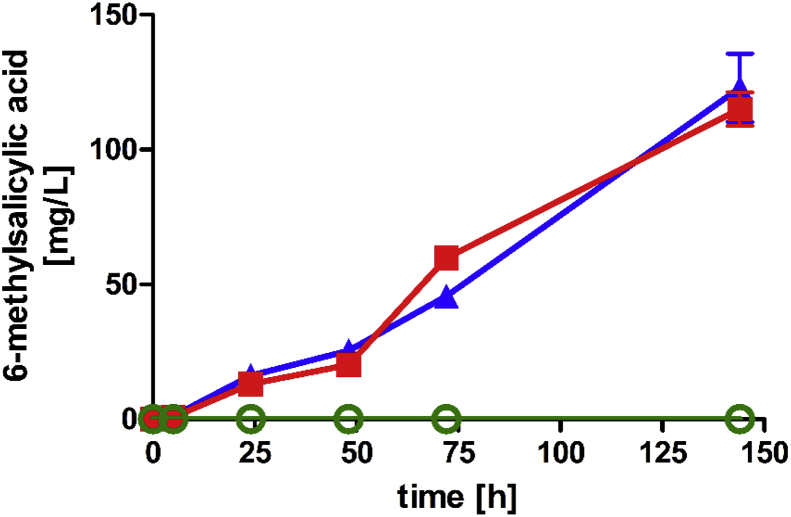
Wattanachaisaereekul et al. (2007, 2008) and Choi and Da Silva (2014) already showed 6-MSA biosynthesis in S. cerevisiae in previous work. Similarly, we transformed the S. cerevisiae strain CEN.PK2–1C with multi-copy plasmids expressing PpvarMSAS (Wattanachaisaereekul et al., 2008) and native phosphopantetheinyl transferase npgA from A. nidulans under control of the strong and constitutive promoters PGK1 and HXT7−1- −392, respectively. Low-OD fermentations of yeast transformants in YPD supplemented with G418 and hygromycin with 20 g/L glucose led to accumulation of up to 115 mg/L 6-MSA in the extracellular medium after 144 h (Fig. 2). Growth was not affected and was comparable to the strain with empty plasmids which did not produce any 6-MSA (not shown).
Fig. 2.
6-MSA formation with strain CEN.PK2–1C carrying the empty vectors pJHV7 and pRS62H as control (green circles), expressing PpvarMSAS and native npgA from multi-copy plasmids pJHV17 and pJHV20 (red squares) and expressing PpvarMSAS and codon-optimized optnpgA from multi-copy plasmids pJHV17 and pJHV2 (blue triangles). Cultures were inoculated at low OD (0.1) and cultivated for 144 h at 30 °C in 25 mL YPD supplemented with G418 and hygromycin. 6-MSA concentrations were determined in the supernatants. Error bars represent the standard deviation of biological duplicates. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
It has been shown before that codon-optimization of heterologous genes can improve protein expression (Kaishima et al., 2016) and substrate conversion in S. cerevisiae (Wiedemann and Boles, 2008). However, co-expression of a codon-optimized npgA (optnpgA) together with PpvarMSAS increased 6-MSA titers only slightly up to 123 mg/L after 144 h. Nevertheless, in all further experiments optnpgA was used. We also performed similar fermentations in defined SCD medium expressing PpvarMSAS from the plasmid pRS426CTMSA-PP and co-expressing optnpgA but reached only up to 22 mg/L 6-MSA. Interestingly, also Sydor et al. (2010) observed improved titers of resveratrol production with yeast by using complex medium – a heterologous pathway which is also dependent on a polyketide synthase-like enzyme. Because of the better results in YPD medium we decided to continue with YPD medium for further experiments.
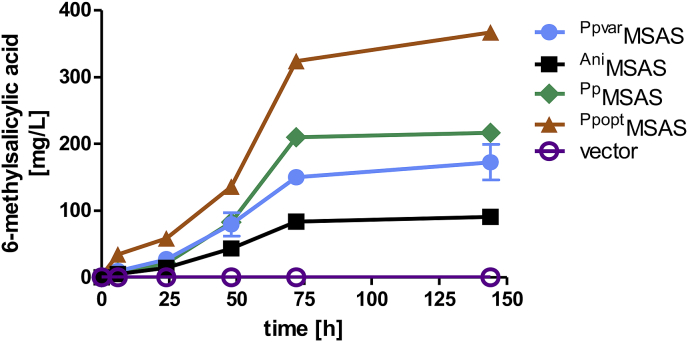
Compared to the low-OD fermentations, in high-OD fermentations starting with an OD600 of 9 the strain expressing PpvarMSAS and optnpgA even reached 6-MSA titers up to 173 mg/L (Fig. 3).
Fig. 3.
6-MSA production by different MSASs in high-OD fermentations. Yeast strain CEN.PK2–1C carrying the empty vectors pJHV7 and pRS62H as control (purple), and strains expressing optnpgA (pJHV2) and the MSAS variants PpvarMSAS (pJHV17; light blue), AniMSAS (pJHV5; black), PpMSAS (pJHV11; green) or PpoptMSAS (pJHV36; orange) from multi-copy plasmids were inoculated at an OD of 9, and cultivated for 144 h at 30 °C in 25 mL YPD supplemented with G418 and hygromycin. 6-MSA concentrations were determined in the supernatants. Error bars represent the standard deviation of biological duplicates. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Next, we tested the native MSASs from P. patulum (PpMSAS) (Beck et al., 1990) and from A. niger (AniMSAS), also in high-OD fermentations. The AniMSAS shares less than 50% identity with the PpMSAS. Compared to PpvarMSAS, yeast cells (CEN.PK2–1C) expressing AniMSAS (co-expressed with optnpgA) produced 47% less 6-MSA (91 mg/L) (Fig. 3). Despite the differences in sequence, the performance of PpMSAS and PpvarMSAS did not differ much. Compared to PpvarMSAS, PpMSAS produced slightly more 6-MSA (216 mg/L) (Fig. 3). Finally, a codon-optimized version of PpMSAS (PpoptMSAS) was expressed in CEN.PK2–1C together with optnpgA. Codon-optimization of PpMSAS led to a major increase of 6-MSA titers up to 367 mg/L (Fig. 3).
With these approaches, we could show that the yeast strain expressing the codon-optimized MSAS PpoptMSAS from P. patulum together with codon-optimized npgA from A. nidulans was the best 6-MSA producer. Moreover, high-OD fermentations were beneficial for higher levels of 6-MSA. Therefore, these variants and conditions were further used for de novo m-cresol biosynthesis in yeast.
3.2. Toxicity of the final product m-cresol
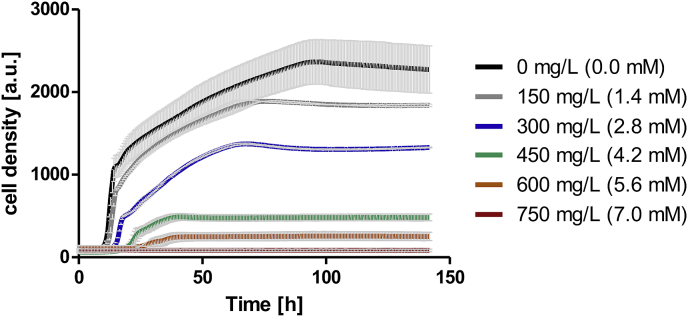
As many phenolic compounds display toxic effects on yeast cells already at low concentrations (Adeboye et al., 2014; Gottardi et al., 2017) and m-cresol has been shown to possess antifungal and antibacterial properties (Lambert et al., 1998; McDonnell and Russel, 1999), we wanted to evaluate the toxicity of m-cresol before studying its biosynthesis from glucose in S. cerevisiae. For this purpose we added different concentrations of m-cresol to cultures of the wild type strain CEN.PK2–1C and followed growth in YPD medium for 144 h. Already at the lowest tested m-cresol concentration of 150 mg/L growth of CEN.PK2–1C was slightly reduced (Fig. 4). Growth rates were strongly reduced at concentrations of 450–600 mg/L, and growth was completely prevented at 750 mg/L m-cresol (corresponding to 7 mM). Therefore, toxicity is in the same range as that of coniferyl aldehyde, ferulic acid, p-coumaric acid and vanillin, reported to impair growth at concentrations of 1.1 mM, 1.8 mM, 9.1 mM and 9.7 mM, respectively (Adeboye et al., 2014). Additionally, Wood et al. (2015) compared the toxicity of furfural, hydroxymethylfurfural, 4-hydroxy-methyl-benzaldehyde, vanillin and guaiacol with that of m-cresol. They found that m-cresol exhibited the lowest inhibitory effect on yeast growth rate but strongly influenced final cell densities. Altogether, it can be concluded that m-cresol exhibits notable toxicity on S. cerevisiae.
Fig. 4.
Toxic effects of m-cresol on growth of CEN.PK2–1C in YPD supplemented with different m-cresol concentrations. Cell densities (starting OD = 0.1) were followed over 144 h with the Cell Growth Quantifier (Aquila Biolabs GmbH) and are depicted as arbitrary units (a.u.). Growth curves represent average of two biological replicates including standard deviations (light grey bars). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.3. Biotransformation of 6-MSA into m-cresol by 6-MSA decarboxylase
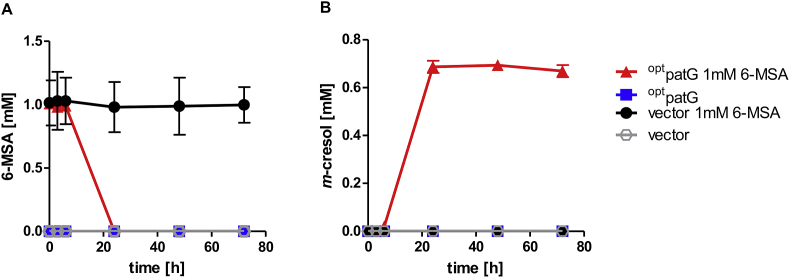
The in vivo functionality of the 6-MSA decarboxylase PatG from A. clavatus (Snini et al., 2014) expressed in yeast was first tested in biotransformation experiments with added 6-MSA. For this, CEN.PK2–1C cells transformed with the codon-optimized patG (optpatG) gene expressed under control of the strong FBA1 promoter from a multi-copy plasmid or an empty vector as control were cultivated in YPD with G418 supplemented with or without 1 mM 6-MSA. In yeast cultures with the empty vector the 6-MSA concentration did not change over 72 h, indicating that 6-MSA cannot normally be converted by yeast cells (Fig. 5A). The patG expressing strain consumed 6-MSA completely within 24 h after an initial 6-h delay, and at the same time 0.67 mM (72 mg/L) of m-cresol was found in the extracellular medium (Fig. 5B). This demonstrates that 6-MSA can be taken up by the yeast cells and is subsequently decarboxylated to m-cresol by PatG, confirming the biotransformation experiments of Snini et al. (2014) and Li et al. (2019). However, the product concentration was 30% lower than the utilized substrate concentration, suggesting either slightly limited secretion of m-cresol, partial loss by evaporation or further conversion. We investigated the volatility of m-cresol at 700 mg/L in YPD medium under fermentation conditions but without yeast cells, and found that less than 5% m-cresol had evaporated even after 144 h.
Fig. 5.
6-MSA uptake and conversion. A) 6-MSA consumption and B) m-cresol production of CEN.PK2–1C expressing 6-MSA decarboxylase optpatG from multi-copy plasmid pJHV13 or carrying empty vector pJHV7 as reference. Strains were cultivated for 72 h in YPD plus G418 with and without supplementation of 1 mM 6-MSA with an initial OD of 0.2.6-MSA and m-cresol concentrations were determined in the supernatants. Error bars represent standard deviation of biological duplicates. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.4. De novo production of m-cresol from glucose in complex medium
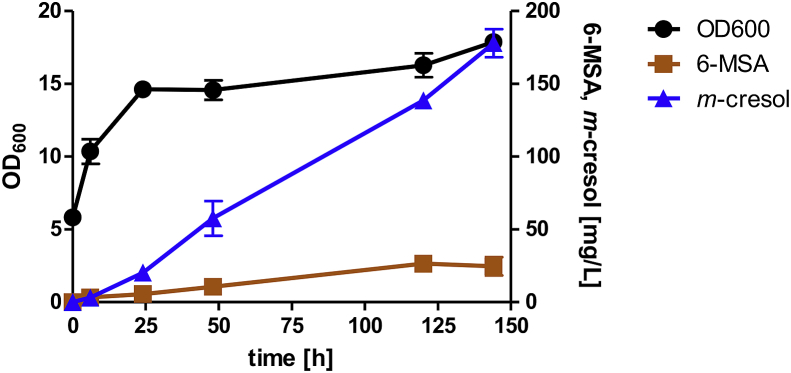
The previous experiments demonstrated that the precursor 6-MSA is provided in high amounts when PpoptMSAS and optnpgA are expressed in S. cerevisiae. Furthermore, 6-MSA can be transported in both directions across the plasma membrane and should be available for intracellular m-cresol production. Additionally, PatG can in vivo decarboxylate 6-MSA to m-cresol. Now, we expressed PpoptMSAS, optnpgA and the codon-optimized optpatG together under control of the PGK1, HXT7−1- −392 and FBA1 promoters, respectively, from a multi-copy plasmid in CEN.PK2–1C, and performed high-OD fermentations (starting OD600 = 5). The yeast cells produced up to 178 mg/L m-cresol from 20 g/L glucose after 144 h in YPD supplemented with G418 (Fig. 6). Nevertheless, the intermediate 6-MSA was accumulating in the extracellular medium up to 26 mg/L, indicating a bottleneck in the PatG reaction. Final cell densities of the m-cresol production strain were slightly lower than those of the strain carrying only an empty vector (OD600 of 18 versus 21, respectively), probably reflecting the inhibitory effects of m-cresol.
Fig. 6.
Production of the intermediate 6-MSA (orange), final product m-cresol (blue) and growth (black) of CEN.PK2–1C expressing PpoptMSAS,optnpgA and optpatG from multi-copy plasmid pJHV53. Fermentations (starting OD = 5) were performed in biological duplicates at 30 °C in YPD supplemented with G418.6-MSA and m-cresol concentrations were determined in the supernatants. Error bars represent standard deviation of biological duplicates. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.5. Genomic integration of the heterologous pathway genes increased 6-MSA and m-cresol production
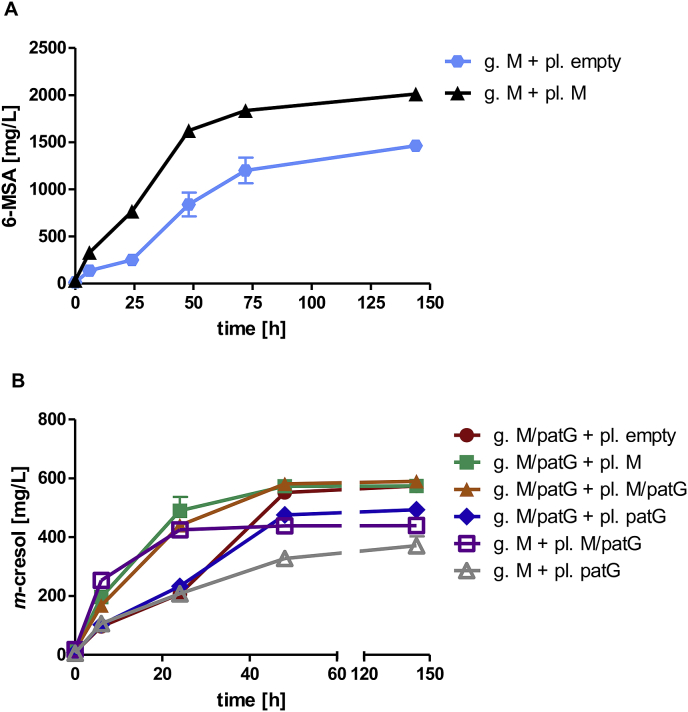
To stabilize expression of pathway genes and prevent plasmid burden and heterogeneity issues by expression from multi-copy plasmids (Krivoruchko et al., 2013; De Jong et al., 2015; Schadeweg and Boles, 2016), the heterologous m-cresol pathway genes PpoptMSAS, optnpgA and optpatG under control of the PGK1, HXT7−1- −392 and FBA1 promoters, respectively, were genomically integrated into the ura3 locus of CEN.PK2–1C. In high-OD fermentations (starting OD = 6), compared to the plasmid-based production strains, genomic integration of PpoptMSAS and optnpgA (without patG) led to a 4-fold increase of 6-MSA production up to 1461 mg/L 6-MSA after 144 h in YPD (Fig. 8A). Additional genomic expression of optpatG under control of FBA1 promoter resulted in a 3-fold increase of m-cresol production (580 mg/L) (Fig. 7C), compared to the plasmid-based strain. Compared to the empty vector control strain, both m-cresol producing strains reached lower final cell densities reflecting the toxic effects of m-cresol (Fig. 7A). Moreover, the genomic integration nearly completely abolished accumulation of the intermediate 6-MSA (Fig. 7B), suggesting that the bottleneck in the plasmid-based approach was a plasmid-born or heterogeneity problem or an unbalanced expression ratio between PpoptMSAS, optnpgA and optpatG.
Fig. 8.
Determination of the limiting factors for 6-MSA and m-cresol production. A) 6-MSA titers produced by strain JHY163 (ura3::PpoptMSAS-optnpgA) expressing additionally PpoptMSAS and optnpgA from multi-copy plasmid pJHV49 (pl. M; black) or as control the empty plasmid pRS42K (pl. Empty; light blue). B) m-cresol titers produced by strain JHY162 (ura3::PpoptMSAS-optnpgA-optpatG) carrying additionally as a control the empty plasmid pRS42K (pl. Empty; red), or expressing PpoptMSAS,optnpgA,optpatG from pJHV53 (pl. M/patG; orange) or optpatG from pJHV13 (pl. patG; blue) and strain JHY163 (ura3::PpoptMSAS-optnpgA) (g. M) expressing additionally PpoptMSAS,optnpgA,optpatG from pJHV53 (pl. M/patG; purple) or optpatG from pJHV13 (pl. patG; grey). High-OD fermentations (starting OD = 5) were performed in biological duplicates at 30 °C in YPD supplemented with G418 (error bars represent standard deviations). g. M. indicates genomic expression of MSAS/npgA, g. M/patG indicates genomic expression of MSAS/npgA and patG, + pl. indicates additional overexpression from multi-copy plasmids. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
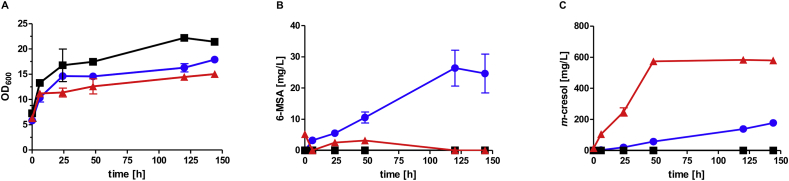
Fig. 7.
Increase in m-cresol production by genomic integration of the pathway genes. Growth (A) and production of 6-MSA (B) and m-cresol (C) of CEN.PK2–1C expressing PpoptMSAS,optnpgA and optpatG from multi-copy plasmid pJHV53 (blue) or from genome (strain JHY162; red). As control, the empty vector pRS42K was transformed into CEN.PK2–1C (black). High-OD fermentations (starting OD = 6) were performed in biological duplicates at 30 °C in YPD supplemented with G418 for plasmid maintenance (error bars represent standard deviations). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.6. Evaluating limitations in the heterologous m-cresol pathway
Because by expression of the m-cresol pathway from a multi-copy plasmid the intermediate 6-MSA was accumulating, the question remained whether expression of the cresol pathway was not balanced or whether a bottleneck in the pathway was limiting m-cresol production. To explore this in more detail, in addition to the genomic expression cassette for PpoptMSAS, optnpgA and/or optpatG, PpoptMSAS/optnpgA and/or optpatG were additionally expressed from a multi-copy plasmid in different combinations.
When in addition to the genomic PpoptMSAS/optnpgA copy, PpoptMSAS/optnpgA was expressed from a plasmid this led to strong increases in the 6-MSA production rate and final 6-MSA titers (2009 mg/L compared to 1461 mg/L with only the genomic copy and an empty vector as control) (Fig. 8A), indicating that MSAS activity was limiting 6-MSA production.
The limiting role of PpoptMSAS/optnpgA with only the genomic expression copy was also reflected in the m-cresol production rates. In all tested combinations, additional overexpresssion of PpoptMSAS/optnpgA from a plasmid always strongly increased m-cresol production rates, independent of optpatG copy numbers (Fig. 8B).
On the other hand, (additional) overexpression of optpatG from a multi-copy plasmid had no positive effect on the m-cresol production rates and did not increase the final m-cresol titers. In contrast, surprisingly, overexpression from the multi-copy plasmid without simultaneous overexpression of PpoptMSAS/optnpgA or from only the plasmid copy had even a slightly negative effect on m-cresol titers (493 mg/L with optpatG from plasmid additionally to the genomic copy compared to 573 mg/L with only an empty vector, and 440 mg/L or 370 mg/L with optpatG only from plasmid with or without PpoptMSAS/optnpgA). The results indicate that MSAS activity is limiting the m-cresol production rate while a balanced expression of patG is necessary for optimal m-cresol titers. However, it must also be considered that the reached m-cresol titers are already very toxic to the cells (compare with Fig. 4), and this toxicity might limit higher m-cresol production titers.
4. Conclusions
This is the first reported de novo production of m-cresol from glucose in S. cerevisiae. By engineering various aspects of the heterologous m-cresol pathway, we could progressively increase 6-MSA and m-cresol titers. Testing different enzyme variants for the MSAS reaction, changing media and fermentation conditions, and performing biotransformation assays were useful for the establishment of a functional pathway and revealed that genes codon-optimized for S. cerevisiae and high-OD fermentations in complex medium were beneficial for high-level production of 6-MSA and m-cresol. Another important aspect was the stable integration of the expression cassettes into the genome. Genomic expression of the pathway was clearly superior to expression from multi-copy plasmids. Nevertheless, simultaneous expression of the heterologous genes from genomic cassettes and multi-copy plasmids revealed MSAS activity as a bottleneck in the m-cresol formation rate. It remains to be investigated how additional engineering of precursor supply of acetyl-CoA, malonyl-CoA and NADPH (Choi and Da Silva, 2014; Kildegaard et al., 2016; Shiba et al., 2007; Wattanachaisaereekul et al., 2008) will further influence m-cresol production. Moreover, the clarification of the stimulating effect on 6-MSA production of complex yeast extract-peptone medium compared to synthetic defined medium still needs further investigations. However, toxicity of m-cresol will remain the biggest challenge for future optimizations of m-cresol production with yeast. To solve this problem, in situ product extraction through e.g. biphasic fermentations with dodecane overlay, scale-up and fed-batch bioreactors (Mehrer et al., 2018) might be promising approaches. Finally, as many Penicillium and Aspergillus species can natively synthesize m-cresol as an intermediate in biosynthesis of the mycotoxin patulin (Puel et al., 2010), it could also be interesting to investigate m-cresol production and tolerance with these ascomycetes by e.g. inactivating late stage patulin biosynthesis genes or by heterolous expression of missing genes.
Acknowledgements
We thank Verena Siewers for the plasmids pRS426CTMSA-PP and pDKP4832, Mara Reifenrath for plasmid pRCC-K_URA3, Simon Harth for plasmid SiHV33 and Phillip Benz for the donation of genomic DNA from A. niger. We thank Mislav Oreb and Martin Grininger (Goethe-University Frankfurt) for fruitful discussions. This work has been financially supported by the Hessen State Ministry of Higher Education, Research and the Arts as part of the LOEWE research initiative MegaSyn.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.mec.2019.e00093.
Contributor Information
Julia Hitschler, Email: j.hitschler@bio.uni-frankfurt.de.
Eckhard Boles, Email: e.boles@bio.uni-frankfurt.de.
Competing interests
The authors declare that they have no competing interests.
Authors’ contribution
JH and EB contributed equally in the design of the study. JH performed the experimental work. JH and EB wrote the paper. All authors have read and approved the submission of the manuscript.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- Adeboye P.T., Bettiga M., Olsson L. The chemical nature of phenolic compounds determines their toxicity and induces distinct physiological responses in Saccharomyces cerevisiae in lignocellulose hydrolysates. Amb. Express. 2014;4:1–10. doi: 10.1186/s13568-014-0046-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck J., Ripka S., Siegner A., Schiltz E., Schweizer E. The multifunctional 6-methylsalicylic acid synthase gene of Penicillium patulum. Eur. J. Biochem. 1990;192:487–498. doi: 10.1111/j.1432-1033.1990.tb19252.x. [DOI] [PubMed] [Google Scholar]
- Bedford D.J., Schweizer E., Hopwood D.A., Khosla C. Expression of a functional fungal polyketide synthase in the bacterium Streptomyces coelicolor A3 ( 2 ) J. Bacteriol. 1995;177:4544–4548. doi: 10.1128/jb.177.15.4544-4548.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger R.G. Springer Berlin Heidelberg; 2007. Flavours and Fragrances Chemistry, Bioprocessing and Sustainability. [Google Scholar]
- Bruder S., Reifenrath M., Thomik T., Boles E., Herzog K. Parallelised online biomass monitoring in shake flasks enables efficient strain and carbon source dependent growth characterisation of Saccharomyces cerevisiae. Microb. Cell Factories. 2016;15:127. doi: 10.1186/s12934-016-0526-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J.W., Da Silva N.A. Improving polyketide and fatty acid synthesis by engineering of the yeast acetyl-CoA carboxylase. J. Biotechnol. 2014;187:56–59. doi: 10.1016/j.jbiotec.2014.07.430. [DOI] [PubMed] [Google Scholar]
- De Jong B.W., Shi S., Valle-Rodríguez J.O., Siewers V., Nielsen J. Metabolic pathway engineering for fatty acid ethyl ester production in Saccharomyces cerevisiae using stable chromosomal integration. Ind. Microbiol. Biotechnol. 2015;42:477–486. doi: 10.1007/s10295-014-1540-2. [DOI] [PubMed] [Google Scholar]
- Entian K.-D., Kötter P. Yeast genetic strain and plasmid collections. Methods Microbiol. 2007;36:629–666. doi: 10.1016/S0580-9517(06)36025-4. [DOI] [Google Scholar]
- Farwick A., Bruder S., Schadeweg V., Oreb M., Boles E. Engineering of yeast hexose transporters to transport D -xylose without inhibition by D -glucose. Proc. Natl. Acad. Sci. U.S.A. 2014;111:5159–5164. doi: 10.1073/pnas.1323464111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisch K.M., Gillaspy A.F., Gipson M., Henrikson J.C., Hoover A.R., Jackson L., Najar F.Z., Wägele H., Cichewicz R.H. Chemical induction of silent biosynthetic pathway transcription in Aspergillus Niger. Ind. Microbiol. Biotechnol. 2009;36:1199–1213. doi: 10.1007/s10295-009-0601-4. [DOI] [PubMed] [Google Scholar]
- Generoso W.C., Gottardi M., Oreb M., Boles E. Simplified CRISPR-Cas genome editing for Saccharomyces cerevisiae. J. Microbiol. Methods. 2016;127:203–205. doi: 10.1016/j.mimet.2016.06.020. [DOI] [PubMed] [Google Scholar]
- Gibson B.R., Lawrence S.J., Leclaire J.P.R., Powell C.D., Smart K.A. Yeast responses to stresses associated with industrial brewery handling. FEMS Microbiol. Rev. 2007;31:535–569. doi: 10.1111/j.1574-6976.2007.00076.x. [DOI] [PubMed] [Google Scholar]
- Gibson D.G., Young L., Chuang R.-Y., Venter J.C., Hutchison C.A., III, Smith H.O. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods. 2009;6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- Gietz R.D., Schiestl R.H. Quick and easy yeast transformation using the LiAc/ss carrier DNA/PEG method. Nat. Protoc. 2007;2:35–37. doi: 10.1038/nprot.2007.14. [DOI] [PubMed] [Google Scholar]
- Gottardi M., Knudsen J.D., Prado L., Oreb M., Branduardi P., Boles E. De novo biosynthesis of trans -cinnamic acid derivatives in Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 2017;10:4883–4893. doi: 10.1007/s00253-017-8220-x. [DOI] [PubMed] [Google Scholar]
- Grote A., Hiller K., Scheer M., Münch R., Nörtemann B., Hempel D.C., Jahn D. JCat : a novel tool to adapt codon usage of a target gene to its potential expression host. Nucleic Acids Res. 2005;33:526–531. doi: 10.1093/nar/gki376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertweck C. The biosynthetic logic of polyketide diversity. Angew. Chem. Int. Ed. 2009;48:4688–4716. doi: 10.1002/anie.200806121. [DOI] [PubMed] [Google Scholar]
- Kaishima M., Ishii J., Matsuno T., Fukuda N., Kondo A. Expression of varied GFPs in Saccharomyces cerevisiae : codon optimization yields stronger than expected expression and fluorescence intensity. Sci. Rep. 2016;6:35932. doi: 10.1038/srep35932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kealey J.T., Liu L., Santi D.V., Betlach M.C., Barr P.J. Production of a polyketide natural product in nonpolyketide-producing prokaryotic and eukaryotic hosts. Proc. Natl. Acad. Sci. U.S.A. 1998;95:505–509. doi: 10.1073/pnas.95.2.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kildegaard K.R., Jensen N.B., Schneider K., Czarnotta E., Özdemir E., Klein T., Maury J., Ebert B.E., Christensen H.B., Chen Y., Kim I.K., Herrgård M.J., Blank L.M., Forster J., Nielsen J., Borodina I. Engineering and systems-level analysis of Saccharomyces cerevisiae for production of 3-hydroxypropionic acid via malonyl-CoA reductase-dependent pathway. Microb. Cell Factories. 2016;15:53. doi: 10.1186/s12934-016-0451-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krivoruchko A., Serrano-Amatriain C., Chen Y., Siewers V., Nielsen J. Improving biobutanol production in engineered Saccharomyces cerevisiae by manipulation of acetyl-CoA metabolism. Ind. Microbiol. Biotechnol. 2013;40:1051–1056. doi: 10.1007/s10295-013-1296-0. [DOI] [PubMed] [Google Scholar]
- Lambert R.J., Johnston M.D., Simons E.A. Disinfectant testing : use of the Bioscreen Microbiological Growth Analyser for laboratory biocide screening. Lett. Appl. Microbiol. 1998;26:288–292. doi: 10.1046/j.1472-765x.1998.00334.x. [DOI] [PubMed] [Google Scholar]
- Li B., Chen Y., Zong Y., Shang Y., Zhang Z., Xu X., Wang X., Long M., Tian S. Dissection of patulin biosynthesis, spatial control and regulation mechanism in Penicillium expansum. Environ. Microbiol. 2019:1462–2920. doi: 10.1111/1462-2920.14542. [DOI] [PubMed] [Google Scholar]
- Liu Z.L. Molecular mechanisms of yeast tolerance and in situ detoxification of lignocellulose hydrolysates. Appl. Microbiol. Biotechnol. 2011;90:809–825. doi: 10.1007/s00253-011-3167-9. [DOI] [PubMed] [Google Scholar]
- McDonnell G., Russel A.D. Antiseptics and disinfectants: activity , action , and resistance. Clin. Microbiol. Rev. 1999;12:147–179. doi: 10.1128/cmr.12.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehrer C.R., Incha M.R., Politz M.C., Pfleger B.F. Anaerobic production of medium-chain fatty alcohols via a β-reduction pathway. Metab. Eng. 2018;48:63–71. doi: 10.1016/j.ymben.2018.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura H., Higo Y., Ohno M., Tsutsui T.W., Tsutsui T. Ability of root canal antiseptics used in dental practice to induce chromosome aberrations in human dental pulp cells. Mutat. Res. 2008;649:45–53. doi: 10.1016/j.mrgentox.2007.07.007. [DOI] [PubMed] [Google Scholar]
- Parascandolo J.S., Havemann J., Potter H.K., Huang F., Riva E., Connolly J., Wilkening I., Song L., Leadlay P.F., Tosin M. Insights into 6-methylsalicylic acid bio-assembly by using chemical probes. Angew. Chem. Int. Ed. 2016;55:3463–3467. doi: 10.1002/anie.201509038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puel O., Galtier P., Oswald I.P. Biosynthesis and toxicological effects of patulin. Toxins. 2010;2:613–631. doi: 10.3390/toxins2040613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reifenrath M., Boles E. Engineering of hydroxymandelate synthases and the aromatic amino acid pathway enables de novo biosynthesis of mandelic and 4-hydroxymandelic acid with Saccharomyces cerevisiae. Metab. Eng. 2018;45:246–254. doi: 10.1016/j.ymben.2018.01.001. [DOI] [PubMed] [Google Scholar]
- Schadeweg V., Boles E. n-Butanol production in Saccharomyces cerevisiae is limited by the availability of coenzyme A and cytosolic acetyl-CoA. Biotechnol. Biofuels. 2016;9:44. doi: 10.1186/s13068-016-0456-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiba Y., Paradise E.M., Kirby J., Ro D., Keasling J.D. Engineering of the pyruvate dehydrogenase bypass in Saccharomyces cerevisiae for high-level production of isoprenoids. Metab. Eng. 2007;9:160–168. doi: 10.1016/j.ymben.2006.10.005. [DOI] [PubMed] [Google Scholar]
- Snini S.P., Tadrist S., Laffitte J., Jamin E.L., Oswald I.P., Puel O. The gene patG involved in the biosynthesis pathway of patulin, a food-borne mycotoxin, encodes a 6-methylsalicylic acid decarboxylase. Int. J. Food Microbiol. 2014;171:77–83. doi: 10.1016/j.ijfoodmicro.2013.11.020. [DOI] [PubMed] [Google Scholar]
- Spencer J.B., Jordan P.M. Purification and properties of 6-methylsalicylic acid synthase from Penicillium patulum. Biochem. J. 1992;288:839–846. doi: 10.1042/bj2880839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spray G., Lodge R.M. The effects of resorcinol and of m-cresol on the growth of Bact. lactis aerogenes. Trans. Faraday Soc. 1943;39:424–431. [Google Scholar]
- Sydor T., Schaffer S., Boles E. Considerable increase in resveratrol production by recombinant industrial yeast strains with use of rich medium. Appl. Environ. Microbiol. 2010;76:3361–3363. doi: 10.1128/AEM.02796-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taxis C., Knop M. System of centromeric , episomal , and integrative vectors based on drug resistance markers for Saccharomyces cerevisiae. Biotechniques. 2006;40:73–77. doi: 10.2144/000112040. [DOI] [PubMed] [Google Scholar]
- Wattanachaisaereekul S., Lantz A., Nielsen M., Andresson O., Nielsen J. Optimization of heterologous production of the polyketide 6-MSA in Saccharomyces cerevisiae. Biotechnol. Bioeng. 2007;97:893–900. doi: 10.1002/bit. [DOI] [PubMed] [Google Scholar]
- Wattanachaisaereekul S., Lantz A.E., Nielsen M.L., Nielsen J. Production of the polyketide 6-MSA in yeast engineered for increased malonyl-CoA supply. Metab. Eng. 2008;10:246–254. doi: 10.1016/j.ymben.2008.04.005. [DOI] [PubMed] [Google Scholar]
- Weber C., Farwick A., Benisch F., Brat D., Dietz H., Subtil T., Boles E. Trends and challenges in the microbial production of lignocellulosic bioalcohol fuels. Appl. Microbiol. Biotechnol. 2010;87:1303–1315. doi: 10.1007/s00253-010-2707-z. [DOI] [PubMed] [Google Scholar]
- Wiedemann B., Boles E. Codon-optimized bacterial genes improve L -arabinose fermentation in recombinant Saccharomyces cerevisiae. Appl. Environ. Microbiol. 2008;74:2043–2050. doi: 10.1128/AEM.02395-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood J.A., Orr V.C.A., Luque L. High-throughput screening of inhibitory compounds on growth and ethanol production of Saccharomyces cerevisiae. Bioenergy Res. 2015;8:423–430. doi: 10.1007/s12155-014-9535-4. [DOI] [Google Scholar]
- Yadav G.D., Pathre G.S. Novel mesoporous solid superacidic catalysts : activity and selectivity in the synthesis of thymol by isopropylation of m -cresol with 2-propanol over UDCaT-4 , -5 , and -6. J. Phys. Chem. A. 2005;109:11080–11088. doi: 10.1021/jp052335e. [DOI] [PubMed] [Google Scholar]
- Yeung S.Y., Lan W.H., Huang C.S., Lin C.P., Chan C.P., Chang M.C., Jeng J.H. Scavenging property of three cresol isomers against H2O2, hypochlorite, superoxide and hydroxyl radicals. Food Chem. Toxicol. 2002;40:1403–1413. doi: 10.1016/S0278-6915(02)00102-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.