Abstract
Although clinical management of melanoma has changed considerably in recent years, intrinsic treatment resistance remains a severe problem and strategies to design personal treatment regimens are highly warranted. We have applied a three-dimensional (3D) ex vivo drug efficacy assay, exposing disaggregated cells from 38 freshly harvested melanoma lymph node metastases and 21 patient derived xenografts (PDXs) to clinical relevant drugs for 7 days, and examined its potential to evaluate therapy response. A strong association between Vemurafenib response and BRAF mutation status was achieved (P < .0001), while enhanced viability was seen in some NRAS mutated tumors. BRAF and NRAS mutated tumors responded comparably to the MEK inhibitor Cobimetinib. Based on the ex vivo results, two tumors diagnosed as BRAF wild-type by routine pathology examinations had to be re-evaluated; one was subsequently found to have a complex V600E mutation, the other a double BRAF mutation (V600E/K601 N). No BRAF inhibitor resistance mechanisms were identified, but PIK3CA and NF1 mutations were identified in two highly responsive tumors. Concordance between ex vivo drug responses using tissue from PDXs and corresponding patient tumors demonstrate that PDX models represent an indefinite source of tumor material that may allow ex vivo evaluation of numerous drugs and combinations, as well as studies of underlying molecular mechanisms. In conclusion, we have established a rapid and low cost ex vivo drug efficacy assay applicable on tumor tissue from patient biopsies. The 3D/spheroid format, limiting the influence from normal adjacent cells and allowing assessment of drug sensitivity to numerous drugs in one week, confirms its potential as a supplement to guide clinical decision, in particular in identifying non-responding patients.
Introduction
Clinical management of melanomas has changed noticeably in recent years due to development of small-molecular inhibitors (BRAFi) targeting the BRAFV600E mutated protein and the use of immunotherapy [1]. Unfortunately, whereas initial responses are frequently observed in patients eligible to BRAFi treatment, nearly all relapse within one year [2], [3]. Intrinsic BRAFi resistance is seen in approximately 20% of the patients and is associated with overexpression of cyclin D1 and COT, loss of PTEN and NF1, stromal expression of hepatocyte growth factor and RAC1 and HOXD8 mutations [4]. Reports have also indicated co-existence of clones harboring either BRAF or NRAS mutation [5], [6] or BRAF/NRAS double-mutations within the same cells [7]. The majority of mechanisms of acquired BRAFi resistance include NRAS and MEK1/2 mutations, BRAFV600E amplification and alternative splicing of BRAF. In addition, dysregulation of PI3-kinase/Akt signaling and overexpression of receptor tyrosine kinases have been shown to have an impact [3]. To overcome acquired resistance, patients have been offered BRAFi in combination with MEK inhibitors (MEKi). Although progression-free survival is improved, most patients will, however, eventually experience disease progression [2], [8], [9].
Tumor cell lines grown as monolayer cultures (2D) have traditionally been used as a first step to evaluate the efficacy of anticancer therapies. This approach does, however, not adequately recapitulate the complex biology of the tumors [10], [11], [12], [13]. To date, the use of patient derived xenograft (PDX) models have been recognized as the cornerstone for evaluating the potential of novel anti-cancer therapy [14], [15] and several studies have demonstrated a strong correlation between treatment responses in PDXs and patient outcome [14], [16], [17]. The use of PDX models has, however, its limitations and is not well suited as routine assays of response prediction in individual patients. Most importantly, variability in engraftment and latency time clearly exceed what can be accepted in a clinical setting. Likewise, loss of human tumor environment and immune responses, costs and ethical considerations, limit extensive use of PDXs in routine diagnostics [18], [19].
As a compromise between 2D-cultures and PDXs, several studies have demonstrated that growth as 3D-cultures more accurately mimic tumor tissue architecture, development of hypoxia, and expression of genes associated with tumorigenesis and therapy response [13], [20], [21] and thus outperform drug response predictions in 2D assays. One example is the use of organoids, established from patient tumor tissue, which has emerged as promising preclinical models to study drug efficacy, in particular in cancers of epithelial origin [22], [23], [24]. In melanomas, the use of human cell lines grown in 2D or 3D cultures [22], [25], [26], as well as animal models, have been the standard assays to evaluate the performance of novel drugs, and to our knowledge, no assays have been developed where patient tumor cells are utilized for drug sensitivity assessments (review in [27]). In the present study, we have developed and demonstrated clinical feasibility of an ex vivo drug sensitivity assay using fresh tumor tissue from melanoma lymph node metastases. The cells were kept in 3D, avoiding influences from stromal cells, and drug responses were evaluated after one-week exposure. Proof-of-principles was demonstrated by evaluating the sensitivity to BRAF-MEK–ERK inhibitors, and comparing the output with molecular data. Based on data from the drug sensitivity test, two tumors were found misclassified as BRAFwt according to routine diagnostic examinations. Upon subsequent NGS, both tumors were confirmed to have less common BRAF mutations. In conclusion, we have demonstrated that the ex vivo drug sensitivity assay is a fast and low-cost method showing potential to provide functional information that can supplement the molecular data. Ultimately this may enhance the diagnostic precision and assist in clinical decision-making.
Materials and Methods
Patients
Randomly collected treatment naïve melanoma lymph node metastases, resected at the Norwegian Radium Hospital, Oslo University Hospital were included. The study was approved by the Regional Committee for Medical Research Ethics of South-East Norway (2014/2208, 2015/2434). Informed consent was obtained from all patients according to national guidelines.
Ex vivo Drug Response Assay
Patient tumor tissue and PDXs were mechanically disaggregated and treated with collagenase (125 U/ml) and 2.5 mg/ml DNase (Sigma Aldrich, St. Louis, MO, USA) for one hour. To remove debris and large cell clumps the suspensions were filtered through 100 μm nylon Cell Strainer (BD Falcon, Franklin Lakes, NJ, US) and washed in ice-cold PBS. If required, red blood cells were removed by ACK lysis buffer (Lonza, Verviers, Belgium). The cells were washed in cold PBS and re-suspended in RPMI-1640 medium (Lonza) supplemented with 5% fetal calf serum (FCS) (Sigma Aldrich), 2 mM L-glutamine, and penicillin/streptavidin (50 U/ml of each) (Lonza). To analyze for drug response, approximately 20,000 viable cells (assessed by Trypan Blue exclusion), resuspended in RPMI-1640 containing 5% FCS and antibiotics, were plated per well in 96-wells round bottom low adhesion plates, allowing spheroids to form (Nunc A/S, Roskilde, Denmark). Drugs were added immediately after seeding. After 5 days of treatment with Vemurafenib (Selleck Chemicals, Houston, TX, USA) and/or Cobimetinib (Selleck Chemicals), effect on viability was assessed using the CellTiter-Glo® Luminescent Cell Viability Assay (Promega, Madison, WI, USA) and reported as percentage viable cells in treated as compared to untreated control samples. For each patient sample, three technical replicates were analyzed. Several drug concentrations were initially applied, (data not shown), and 2 μM Vemurafenib and 50 nM Cobimetinib, were chosen as standard. Of the obtained tumor tissue, approximately 30% had to be discarded due to lack of viable cells in the biopsy, little material or lack of viability in control cells after analyses. However, PDX models could still be made from some of the latter and used in the ex vivo drug efficacy assay.
Molecular Analyses
DNA was extracted from 21 melanoma lymph node metastases and one PDX by the AllPrep DNA/RNA Mini Kit and AllPrep DNA/RNA/miRNA Universal kit (Qiagen, Hilden, Germany). Prior to extraction, cryo-sections were made and stained with hematoxylin/eosin. Only samples with >10% tumor cells were subjected to molecular analysis. The Ion Torrent PGM™ was used for sequencing with two different panels: the Ion AmpliSeq™ Cancer Hotspot Panel v2 covering ~2800 hotspot mutations in 50 genes, and the Oncomine Comprehensive Panel covering hotspot mutations in 73 genes, full exon coverage of 26 genes and copy number aberrations in 49 genes (Thermo Fisher Scientific, Inc., San Francisco, CA, USA).
The Torrent Suite Variant Caller version 5.0 was run using panel-optimized parameters from AmpliSeq.com for Ion AmpliSeq Cancer Hotspot Panel v2. Using hg19 as reference, single nucleotide substitutions that exceeded a 5% variant allele frequency threshold were identified. The variants were functionally annotated with ANNOVAR, using RefSeq as the underlying gene model [28] and using information from the 1000 Genomes Project [1000genomes.org] and the Catalogue of Somatic Mutations in Cancer [cancer.sanger.ac.uk/cosmic]. Detected mutations were in addition checked using the Integrative Genomics Viewer (IGV) [29]. BRAFV600E/K and NRAS mutation status were additionally established for all samples by an in-house PCR based assay used in routine diagnostics. Data supporting the findings are stored at Services for sensitive data (TSD) – University of Oslo. Access can be arranged by contacting the corresponding author (VAF) upon request.
In vivo Studies
To establish PDX models, melanoma lymph node metastases obtained from surgery were implanted subcutaneously into NOD SCID gamma mice (success rate 77%). Briefly, tumor tissue was cut into pieces of about 2 mm3 and implanted subcutaneously in the flanks of >6 months old female mice. The first passage was named P0. Totally 21 PDX models have been established, of which 16 were from patient tumors analyzed for drug effects ex vivo.
Prior to in vivo treatment, the PDXs were re-implanted in the flanks of 6–8-week-old female atymic nude foxn1nu mice and underwent two additional passages before treatment was initiated with bilateral implantation into new mice. After four weeks, the mice were randomized into a control (6 mice) and a treatment group (8 mice) each having an average tumor –volume distribution of 135 mm3. The latter group was given 50 mg/kg Vemurafenib twice daily by oral gavage for 14 days. Controls were given 10% DMSO in 0,5% methylcellulose orally for the duration of the treatment. Tumor diameters were measured twice a week by digital calipers and tumor volume calculated by the formula 0.5 x length x width2. Data is presented as average tumor volume ± standard error of the mean (S.E.M.). All mice were bred at the Department of Comparative Medicine, The Norwegian Radium Hospital and kept according to regulations of the Norwegian Welfare Act. Experiments involving animals were approved by the Norwegian Animal Research Authority (FOTS application number 8554).
Statistical Analysis
Statistical significance was determined by the Student's two-tailed t-test using GraphPad Prism version 7.0 (GraphPad Software, San Diego, CA, USA).
Results
Ex Vivo Assessment of Patients Own Tumor for Response to Vemurafenib Reveals A Close Correlation to Known BRAF/NRAS Mutation Status in Metastatic Melanoma
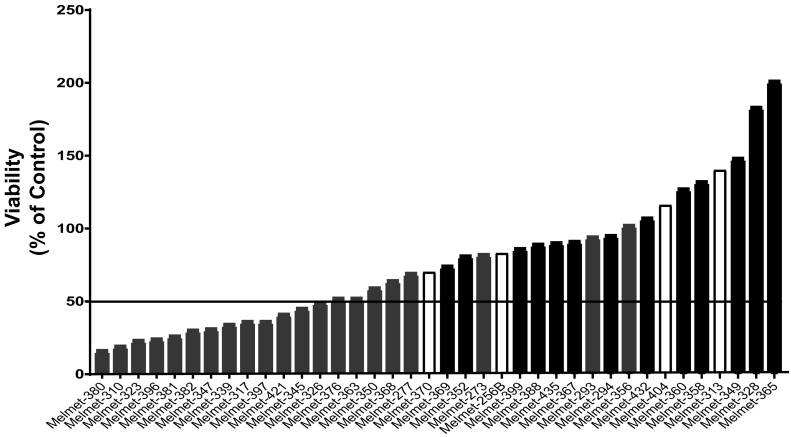
Here we aimed to establish a 3D ex vivo drug efficacy assay using freshly harvested melanoma tissue samples. As a proof of concept, the BRAFi Vemurafenib was chosen as test drug and the results correlated to BRAF mutation status. Tissue from 38 treatment naïve melanoma lymph node metastases were disaggregated and cells plated as spheroids in the presence or without 2 μM Vemurafenib for five days before viability was assessed. 50% reduction in viability was chosen as a stringent cutoff to discriminate between responders/non-responders. As shown in Figure 1, the assay verified a strong association between response and diagnostically detected BRAF mutation status (P < .0001). Of the 21 BRAFV600E mutated tumors, 12 (57%) were clearly responsive, whereas three were borderline responsive (Melmets-326, −376, −363), and six did not respond to the treatment. These numbers are in agreement with clinical experiences demonstrating an objective response to BRAF inhibition in approximately 50% of patients with metastatic melanoma [30]. None of the BRAF wild-type tumors responded to Vemurafenib while several of the BRAFwt/NRASmut tumors (in particular Melmets-328, −349, −365,), showed a marked increase in viability when tested in the ex vivo assay (Figure 1).
Figure 1.
Viability of patient derived melanoma samples analyzed ex vivo for response to Vemurafenib. Lymph node metastases from 38 patients were disaggregated and cells plated and exposed to 2 μM Vemurafenib for 5 days as described in “Materials and Methods”. Viability was assessed using CellTiter-Glo® Luminescent Cell viability assay and results presented as percentage viable cells compared to untreated controls and correlated to BRAF mutation status (P < 0.0001, Student’s two-tailed t-test). 50% reduction in viability was chosen as cutoff for response/non-response. Gray bars; BRAF mutated, black bars; NRAS mutated, white bars; Double wild-type.
It is not expected that BRAF wild-type and NRAS mutated tumors will benefit from BRAFi treatment. Therefore we also tested the effect of the MEKi Cobimetinib. The effect of Vemurafenib and Cobimetinib was overall comparable in the BRAF mutated tumors (10 cases), but two tumors (Melmet-363, Melmet-376) that were borderline responsive to Vemurafenib, responded to Cobimetinib. Of the NRAS mutated tumors, four of seven clearly responded to Cobimetinib. Surprisingly, in two NRAS mutated (Melmel-388, Melmet-432), and to a minor extent in one BRAF mutated tumor (Melmet-397), the effect of MEKi was reversed when combined with BRAFi (Table 1).
Table 1.
Relative viability of melanoma lymph node metastases analyzed ex vivo after treatment with Vemurafenib and/or Cobimetinib
| Patient No. | Mutation | Relative viability (% of control) 1 |
||
|---|---|---|---|---|
| Vemurafenib |
Cobimetinib |
Vemurafenib/Cobimetinib |
||
| (2 μM) | (50 nM) | (2 μM + 50 nM) | ||
| Melmet-339 | BRAF | 34 | 38 | n.a.* |
| Melmet-347 | BRAF | 31 | 29 | n.a. |
| Melmet-363 | BRAF | 52 | 39 | n.a. |
| Melmet-368 | BRAF | 64 | 52 | n.a. |
| Melmet-376 | BRAF | 52 | 40 | n.a. |
| Melmet-380 | BRAF | 16 | 16 | 14 |
| Melmet-381 | BRAF | 26 | 10 | 16 |
| Melmet-382 | BRAF | 30 | 35 | 29 |
| Melmet-396 | BRAF | 24 | 17 | 20 |
| Melmet-397 | BRAF | 36 | 32 | 48 |
| Melmet-352 | NRAS | 81 | 78 | n.a. |
| Melmet-360 | NRAS | 127 | 55 | n.a. |
| Melmet-367 | NRAS | 91 | 117 | n.a. |
| Melmet-369 | NRAS | 74 | 40 | n.a. |
| Melmet-388 | NRAS | 89 | 48 | 83 |
| Melmet-399 | NRAS | 86 | 49 | 55 |
| Melmet-432 | NRAS | 108 | 42 | 73 |
| Melmet-370 | Wt/Wt | 70 | 92 | n.a. |
1 Percentage survival.
* n.a. = Not analyzed.
Comparable Ex Vivo Treatment Responses in Patient Derived Tumor Cells And Corresponding PDXs
It has been previously documented that melanoma PDXs reliably reflect treatment responses seen in patients [14], [31]. We therefore aimed to examine whether therapy effects using patient tumor cells and cells derived from the corresponding PDXs (n = 16) were comparable in the ex vivo assay. In addition, five PDXs where patient tumors had not been analyzed were included. The PDX tumors were handled and exposed to treatment ex vivo as the patient samples. Despite minor variations, a good concordance was maintained throughout PDX-passages (Table 2 and data not shown). For some PDXs, however, later passages seemed to respond more similar to cells derived directly from the patient's tumor (Melmet-347, Melmet-381). Furthermore, in two cases (Melmets-350 and -356) several PDX passages showed no sign of viability in the controls when cultivated ex vivo.
Table 2.
Viability of melanoma lymph node metastases and PDXs analyzed ex vivo after treatment with Vemurafenib (2 μM)
| Patient No. | Mutation | Relative viability (% of control)1 |
||||
|---|---|---|---|---|---|---|
| Lymph node | PDX passage2,3 |
|||||
| Lowest | Highest | |||||
| Melmet-334 | BRAF | n.a.* | 52 | (4) | - | (n.a.)** |
| Melmet-347 | BRAF | 31 | 64 | (1) | 13 | (3) |
| Melmet-350 | BRAF | 59 | - | (0) | - | (1) |
| Melmet-351 | BRAF | n.a. | 57 | (2) | 56 | (6) |
| Melmet-356 | BRAF | 102 | - | (0) | - | (4) |
| Melmet-363 | BRAF | 52 | 43 | (7) | 28 | (8) |
| Melmet-376 | BRAF | 52 | 33 | (2) | 43 | (6) |
| Melmet-380 | BRAF | 16 | 26 | (0) | 42 | (3) |
| Melmet-381 | BRAF | 30 | 87 | (4) | 11 | (7) |
| Melmet-382 | BRAF | 35 | 17 | (2) | 18 | (5) |
| Melmet-389 | BRAF | n.a. | 61 | (0) | 12 | (6) |
| Melmet-393 | BRAF | n.a. | 30 | (3) | 20 | (6) |
| Melmet-358 | NRAS | 132 | 86 | (0) | 107 | (5) |
| Melmet-365 | NRAS | 201 | 122 | (1) | 116 | (5) |
| Melmet-367 | NRAS | 91 | 118 | (7) | n.a. | (n.a.) |
| Melmet-369 | NRAS | 74 | 169 | (0) | 271 | (3) |
| Melmet-388 | NRAS | 89 | 67 | (0) | 125 | (7) |
| Melmet-256 | Wt/Wt | 83 | 86 | (0) | 80 | (7) |
| Melmet-370 | Wt/Wt | 70 | 98 | (1) | 103 | (10) |
| Melmet-374 | Wt/Wt | n.a. | 79 | (3) | 103 | (5) |
| Melmet-404 | Wt/Wt | 116 | 77 | (0) | 103 | (1) |
1 Percentage survival.
2 Number of passages in parentheses.
3 PDX for Melmet-350, -356 not analyzed due to control sample not growing.
* n.a. = Not analyzed due to limited tumor material available.
** Only one PDX passage analyzed.
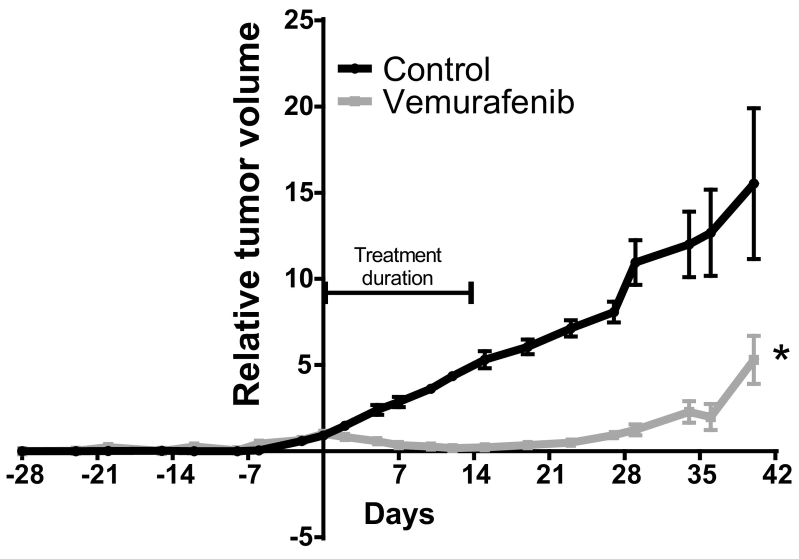
As a final confirmation step, PDX of Melmet-382 (passage 4) was examined in vivo for response to Vemurafenib. As was observed in the ex vivo assay performed on patient- and PDX-derived material, a strong significant response was achieved (Figure 2).
Figure 2.
Antitumor efficacy of Vemurafenib in vivo. Melmet-382 PDX was treated with Vemurafenib (50 mg/kg) given twice daily by oral gavage for 14 days. Control mice were given 10% DMSO in 0,5% methylcellulose orally. Tumor volume was measured twice a week and results presented as relative volume related to tumor volume at initiation of the treatment. Error bars represent ±S.E.M.
Targeted Sequencing of Patient Tumor Samples Combined with Ex Vivo Drug Sensitivity Assessment Provide Precise Diagnostic Information
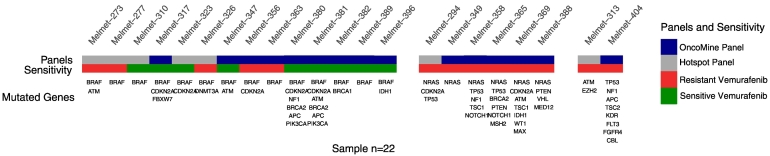
In attempt to reveal molecular mechanisms of treatment response, targeted sequencing (IonTorrent™ Oncomine and/or Cancer Hotspot Panel) was performed on 21 of the patient samples and one PDX. The sequence data were filtered against databases for common mutations (1000 Genomes) and known cancer mutations (COSMIC). Following variant calling, non-synonymous mutations were reported (Figure 3 and Table S1 for complete list of mutations). As expected, BRAFV600E (62%) was the most common mutation followed by CDKN2A (33%) and NRAS (29%). It appeared that the mutation load was higher in NRAS compared to BRAF mutated tumors. The Melmet-323 tumor, found highly responsive to BRAFi (viability, 25% of control) was shown to have a rare dinucleotide BRAF mutation yielding a complex V600E variant (c.1799_1800TG > AA). This tumor had previously been diagnosed as BRAF/NRAS wild-type by a PCR based assay routinely used for diagnostic evaluation. Likewise, the PDX from Melmet-389 (patient tissue not analyzed) diagnosed as BRAF wild-type showed remarkable response to Vemurafenib when analyzed ex vivo. In this case, targeted sequencing revealed a double BRAF mutation (V600E and K601 N). Two other tumors carrying double BRAF mutations, BRAFV600E/K601E (Melmet-363) and BRAFV600E/S605R (Melmet-273), were found borderline sensitive and resistant, respectively.
Figure 3.
Distribution of mutations in 21 melanoma lymph node metastases and one PDX (melmet 389). Sequencing was performed using IonTorrent Oncomine (blue and cancer Hotspot (gray) panels. Mutation analysis was performed using the Torrent Suite Variant Caller version 5.9 and annotated using ANNOVAR as described in Materials and Methods. Response to Vemurafenib is shown in bottom bar; sensitive (green), resistant (red).
Interestingly, despite that aberrations in the PI3K/Akt pathway and NF1 mutations have been associated with BRAFi resistance, two highly responsive BRAFV600E mutated tumors (Melmet-380, Melmet-381) were shown to have a PIK3CA (p.H1047R) or NF1 mutation (Melmet-380). Other candidate genes with suggested impact on treatment response [32], [33] and found affected included three TP53, two IDH1 and one EZH2 mutations (Figure 3 and Table S1).
Discussion
Despite promising molecular anti-cancer targets, lack of model systems and/or biomarkers identifying responders have clearly limited the success of targeted therapy. Melanoma is one of the most heterogeneous cancer forms and differences in BRAF mutation status have been observed between primary tumors and corresponding metastases, between different metastases as well as intra-tumorally [34], [35]. This makes it difficult to identify patients likely to benefit from targeting therapy based solely on molecular screening of a single biopsy. During the last decades, various 3D-culture systems [36], [37] and organoid models, the latter in particular from epithelial derived cancers [23], have been developed to assess response to anti-cancer treatment. However, no ex vivo assay based on the patient's own tumor cells has, to the best of our knowledge, so far been established in routine diagnostics [23], [38]. In the current study, we applied a modified version of the ATP-based tumor sensitivity- [39] and extreme drug resistance assays [40] that we previously successfully have used to predict primary platinum resistance in ovarian cancer patients [41]. Here, when melanoma lymph node metastases were analyzed for response to Vemurafenib ex vivo, two important observations were made. First, a strong correlation between response and verified BRAF status were achieved in the sense that all patients that responded to the treatment harbored a BRAF mutation, whereas this was not the case for any of the NRAS or BRAF wild-type tumors. Second, and in agreement with intrinsic resistance seen in the clinic, not all BRAF mutated tumors responded to the treatment. Furthermore, ex vivo analysis of tumor material harvested from various passages of corresponding PDXs retained the response profile seen in the matching patient tumor samples, as was also seen when treating the PDX in vivo.
Numerous studies have concluded that permanent cancer cell lines grown as adherent 2D-cultures poorly reflect the complexity of a solid tumor [12]. Furthermore, most melanoma cell lines have been derived from highly proliferative tumors [42], exposed to high selection pressure due to BRAF or NRAS mutations and loss of CDKN2A [43],. This may partly explain the high failure rate of novel targeted therapy since the test system usually has been based on the use of such cell lines. During the course of this study, we aimed to establish adherent in vitro cell lines from some of the tumors and PDXs. In cases where we successfully were able to establish permanent cell lines, they all seemed to be highly proliferative (personal observation) and to harbor BRAF or NRAS mutations. In addition, we experienced, as also has been reported by others [42], that the primary cultures were easily over-grown by fibroblasts. In contract, stromal cells will not grow anchorage-independently making the 3D assay superior to the more time consuming establishment of stably growing cell cultures in 2D.
Studies have suggested that assay-guided therapy more accurately identifies ineffective than effective drugs [44], [45]. Using the stringent 50% reduction in viability as cutoff to discriminate between responders and non-responders [46], all NRAS mutated or wild-type tumors were resistant to Vemurafenib, and some showed increased viability as compared to controls. The latter is in accordance with studies showing paradoxical reactivation of MAPK signaling and increased proliferation when wild-type or NRAS mutated tumors are treated with BRAFi [47].
In accordance with clinical observations [48], approximately 60% of the BRAF mutated tumors responded in the 3D assay. In agreement with a recent study, in which melanoma tissue was cultured as micro tumor fragments [49], complete loss of viability following BRAF or MEK inhibition was, however, not achieved, a finding that may be explained by intra-tumor heterogeneity and/or the presence of normal cell infiltration.
Both pre-clinical and clinical studies have demonstrated that combined BRAF and MEK inhibition may be beneficial for patients with BRAF mutated tumors. Moreover, selective MEK inhibition has shown efficacy in NRAS mutated melanoma (reviewed in [49]). In the current study, response of BRAF mutated tumors to Vemurafenib and/or Cobimetinib was in most cases comparable, and in accordance with previous studies [50], half of the NRAS mutated tumors responded to MEK inhibition. Response to MEK inhibition was, however, less pronounced in NRAS mutated tumors than response to BRAF inhibition in BRAF mutated tumors. Of particular note, in a recent study [51], BRAFi were shown to amplify the effect of MEKi in NRAS mutated melanomas whereas in another study [49], an antagonistic effect of combining MEK and mutated BRAF inhibition was observed. In support of the latter, in three cases (two NRAS and one BRAF mutated) the combined treatment was less efficient than the mono-treatments. Taken together this clearly demonstrates that there is a need to extend the current molecular examinations with functional tests reporting on drug sensitivity to provide precise diagnostics for guidance of clinical treatment decisions.
Two tumors, originally diagnosed as BRAF wild-type by PCR-based in-house routine pathology examination, showed excellent response to Vemurafenib in the ex vivo assay. Based on this, targeted sequencing was performed and both were found to be BRAF mutated. Several reasons may explain the discrepancy such as the sensitivity of the molecular analyses or intra-tumoral heterogeneity. In support of the former, one tumor was found to harbor a complex BRAF mutation that was not analyzed for in the diagnostic assay. Furthermore, in support of heterogeneity, a study by Saint-Jean et al. [52] demonstrated that seven percent of melanomas diagnosed as BRAF wild-type by the first biopsy examination, revealed BRAF mutations following analysis of repeated biopsies. Likewise, a recent meta-analysis revealed intra-tumoral discrepancy in BRAF status among patients with metastatic melanoma [53]. The current cohort of samples consisted exclusively of stage III lymph node metastases that were not offered treatment besides removal of the malignant lesion. Some, however, developed distant metastases (stage IV) and five of these (two sensitive and three resistant from the ex vivo assay) received BRAFi treatment. In contrast to responses observed in the ex vivo assay the general clinical response was in all cases poor. For three patients a mixed response was observed; some metastases declined whereas some grew progressively, a finding strongly supporting melanoma heterogeneity. Together, these results strongly suggest more thorough molecular analysis of cases where discrepancy between ex vivo viability results and clinical diagnosis is observed and underscores the necessity, in a diagnostic setting, to examine multiple biopsies from each tumor [34]. The ex vivo assay will, however, to some extent account for intra-tumor heterogeneity as a larger fraction of the lesion is disaggregated and examined for drug sensitivity. It should be mentioned also that a meta-analysis comprising more than 15,000 tumors demonstrated that drug resistance could be foreseeable with high accuracy using various assays, whereas sensitivity, on the other hand, was less predictable [38], as also supported by our findings.
Except for mutated NRAS being strongly associated with BRAFi resistance, no other mechanisms of resistance were revealed. Aberrations in the PI3K/Akt pathway as well as NF1 mutations have been associated with BRAFi resistance [4]. This is in contrast to our findings demonstrating co-existence of PIK3CA mutations and one NF1 mutation in two of the most BRAFi responsive tumors. In agreement with our findings, however, it has been claimed that oncogenic PIK3CA mutation does not play a major role in Vemurafenib resistance [54], and a study by Krauthammer et al. suggested that loss of NF1 not necessarily is associated with BRAFi resistance [55].
The high success rate of establishing melanoma PDX models, and their ability to reliably recapitulate patient tumor architecture, genotype and response to treatment, have made them powerful tools to develop new and improved therapeutic strategies [14], [56], [57]. In a study by Einarsdottir et al. [58], PDX models in passage three were claimed to develop fast enough to guide treatment decisions. Although the use of PDXs in routine diagnostic may not be a realistic goal due to variability in engraftment, latency period, number of animals required and costs [14], they may provide an unlimited resource of tumor cells for both small-scale as well as large-scale ex vivo drug screening. Here we demonstrated that PDXs assessed for treatment responses using the ex vivo assay show a high degree of concordance with results observed when analyzing the corresponding patient tumors directly, or when treating PDXs in vivo, supporting previous observations that early PDX passages resemble the original tumor [59]. In agreement with our findings, short-time ex vivo cultures of breast cancer PDXs were recently found to predict in vivo drug responses [60]. When analyzing several PDX passages for treatment response ex vivo, concordance was in most cases achieved, indicating PDX stability [61]. Notably, although cells from the parental tumors in general were easy to cultivate ex vivo, serial PDX-passages from two of the tumors showed no sign of viability, suggesting dependence of factors provided by the host or tumor stromal cells. However, in general, in cases where the amount of tumor material is scarce, PDX models may be established and used as an indefinite source of tumor material for ex vivo drug testing.
In conclusion, the presented data strongly support the potential of the ex vivo assay to provide valuable functional information in the patient tumor. The fast and reliable analyses, combined with the low cost, make the assay attractive to supplement molecular data in clinical decisions. Furthermore, the findings underscore the importance of considering intra-tumor heterogeneity as well as heterogeneity between various metastases in the individual patients when analyzing drug effects ex vivo. Finally, we hypothesize that analyzing drug effects ex vivo will be of particular importance in pinpointing patients that are not likely to respond to targeted therapy.
The following is the supplementary data related to this article.
Mutations identified in 21 melanoma lymph node metastases and one PDX by IonTorrent Cancer Hotspot and Oncomine Comprehensive Panel.
Footnotes
Acknowledgements: This work was supported by South-Eastern Norway Regional Health Authority (KFK, EM, PW) and Research Council of Norway under the program from Publicly Initiated Clinical Trial Studies (grant number 218325VN, IRB) and the Cancer Society of Norway (IRB).
References
- 1.Shah DJ, Dronca RS. Latest advances in chemotherapeutic, targeted, and immune approaches in the treatment of metastatic melanoma. Mayo Clin Proc. 2014;89(4):504–519. doi: 10.1016/j.mayocp.2014.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]; DJ Shah and RS Dronca, Latest advances in chemotherapeutic, targeted, and immune approaches in the treatment of metastatic melanoma. Mayo Clin Proc, 2014. 89(4): p. 504-519. [DOI] [PMC free article] [PubMed]
- 2.Long GV, Stroyakovskiy D, Gogas H, Levchenko E, de Braud F, Larkin J, Garbe C, Jouary T, Hauschild A, Grob JJ. Combined BRAF and MEK inhibition versus BRAF inhibition alone in melanoma. N Engl J Med. 2014;371(20):1877–1888. doi: 10.1056/NEJMoa1406037. [DOI] [PubMed] [Google Scholar]; GV Long, D Stroyakovskiy, H Gogas, E Levchenko, F de Braud, J Larkin, C Garbe, T Jouary, A Hauschild, JJ Grob, et al., Combined BRAF and MEK inhibition versus BRAF inhibition alone in melanoma. N Engl J Med, 2014. 371(20): p. 1877-1888. [DOI] [PubMed]
- 3.Sullivan RJ, Flaherty KT. Resistance to BRAF-targeted therapy in melanoma. Eur J Cancer. 2013;49(6):1297–1304. doi: 10.1016/j.ejca.2012.11.019. [DOI] [PubMed] [Google Scholar]; RJ Sullivan and KT Flaherty, Resistance to BRAF-targeted therapy in melanoma. Eur J Cancer, 2013. 49(6): p. 1297-1304. [DOI] [PubMed]
- 4.Manzano JL, Layos L, Buges C, de Los Llanos Gil M, Vila L, Martinez-Balibrea E, Martinez-Cardus A. Resistant mechanisms to BRAF inhibitors in melanoma. Ann Transl Med. 2016;4(12):237. doi: 10.21037/atm.2016.06.07. [DOI] [PMC free article] [PubMed] [Google Scholar]; JL Manzano, L Layos, C Buges, M de Los Llanos Gil, L Vila, E Martinez-Balibrea and A Martinez-Cardus, Resistant mechanisms to BRAF inhibitors in melanoma. Ann Transl Med, 2016. 4(12): p. 237. [DOI] [PMC free article] [PubMed]
- 5.Jovanovic B, Egyhazi S, Eskandarpour M, Ghiorzo P, Palmer JM, Bianchi Scarra G, Hayward NK, Hansson J. Coexisting NRAS and BRAF mutations in primary familial melanomas with specific CDKN2A germline alterations. J Invest Dermatol. 2010;130(2):618–620. doi: 10.1038/jid.2009.287. [DOI] [PMC free article] [PubMed] [Google Scholar]; B Jovanovic, S Egyhazi, M Eskandarpour, P Ghiorzo, JM Palmer, G Bianchi Scarra, NK Hayward and J Hansson, Coexisting NRAS and BRAF mutations in primary familial melanomas with specific CDKN2A germline alterations. J Invest Dermatol, 2010. 130(2): p. 618-620. [DOI] [PMC free article] [PubMed]
- 6.Sensi M, Nicolini G, Petti C, Bersani I, Lozupone F, Molla A, Vegetti C, Nonaka D, Mortarini R, Parmiani G. Mutually exclusive NRASQ61R and BRAFV600E mutations at the single-cell level in the same human melanoma. Oncogene. 2006;25(24):3357–3364. doi: 10.1038/sj.onc.1209379. [DOI] [PubMed] [Google Scholar]; M Sensi, G Nicolini, C Petti, I Bersani, F Lozupone, A Molla, C Vegetti, D Nonaka, R Mortarini, G Parmiani, et al., Mutually exclusive NRASQ61R and BRAFV600E mutations at the single-cell level in the same human melanoma. Oncogene, 2006. 25(24): p. 3357-3364. [DOI] [PubMed]
- 7.Raaijmakers MI, Widmer DS, Narechania A, Eichhoff O, Freiberger SN, Wenzina J, Cheng PF, Mihic-Probst D, Desalle R, Dummer R. Co-existence of BRAF and NRAS driver mutations in the same melanoma cells results in heterogeneity of targeted therapy resistance. Oncotarget. 2016;7(47):77163–77174. doi: 10.18632/oncotarget.12848. [DOI] [PMC free article] [PubMed] [Google Scholar]; MI Raaijmakers, DS Widmer, A Narechania, O Eichhoff, SN Freiberger, J Wenzina, PF Cheng, D Mihic-Probst, R Desalle, R Dummer, et al., Co-existence of BRAF and NRAS driver mutations in the same melanoma cells results in heterogeneity of targeted therapy resistance. Oncotarget, 2016. 7(47): p. 77163-77174. [DOI] [PMC free article] [PubMed]
- 8.Johnson DB, Flaherty KT, Weber JS, Infante JR, Kim KB, Kefford RF, Hamid O, Schuchter L, Cebon J, Sharfman WH. Combined BRAF (Dabrafenib) and MEK inhibition (Trametinib) in patients with BRAFV600-mutant melanoma experiencing progression with single-agent BRAF inhibitor. J Clin Oncol. 2014;32(33):3697–3704. doi: 10.1200/JCO.2014.57.3535. [DOI] [PMC free article] [PubMed] [Google Scholar]; DB Johnson, KT Flaherty, JS Weber, JR Infante, KB Kim, RF Kefford, O Hamid, L Schuchter, J Cebon, WH Sharfman, et al., Combined BRAF (Dabrafenib) and MEK inhibition (Trametinib) in patients with BRAFV600-mutant melanoma experiencing progression with single-agent BRAF inhibitor. J Clin Oncol, 2014. 32(33): p. 3697-3704. [DOI] [PMC free article] [PubMed]
- 9.Welsh SJ, Rizos H, Scolyer RA, Long GV. Resistance to combination BRAF and MEK inhibition in metastatic melanoma: Where to next? Eur J Cancer. 2016;62:76–85. doi: 10.1016/j.ejca.2016.04.005. [DOI] [PubMed] [Google Scholar]; SJ Welsh, H Rizos, RA Scolyer and GV Long, Resistance to combination BRAF and MEK inhibition in metastatic melanoma: Where to next? Eur J Cancer, 2016. 62: p. 76-85. [DOI] [PubMed]
- 10.Das V, Bruzzese F, Konecny P, Iannelli F, Budillon A, Hajduch M. Pathophysiologically relevant in vitro tumor models for drug screening. Drug Discov Today. 2015;20(7):848–855. doi: 10.1016/j.drudis.2015.04.004. [DOI] [PubMed] [Google Scholar]; V Das, F Bruzzese, P Konecny, F Iannelli, A Budillon and M Hajduch, Pathophysiologically relevant in vitro tumor models for drug screening. Drug Discov Today, 2015. 20(7): p. 848-855. [DOI] [PubMed]
- 11.Gillet JP, Calcagno AM, Varma S, Marino M, Green LJ, Vora MI, Patel C, Orina JN, Eliseeva TA, Singal V. Redefining the relevance of established cancer cell lines to the study of mechanisms of clinical anti-cancer drug resistance. Proc Natl Acad Sci U S A. 2011;108(46):18708–18713. doi: 10.1073/pnas.1111840108. [DOI] [PMC free article] [PubMed] [Google Scholar]; JP Gillet, AM Calcagno, S Varma, M Marino, LJ Green, MI Vora, C Patel, JN Orina, TA Eliseeva, V Singal, et al., Redefining the relevance of established cancer cell lines to the study of mechanisms of clinical anti-cancer drug resistance. Proc Natl Acad Sci U S A, 2011. 108(46): p. 18708-18713. [DOI] [PMC free article] [PubMed]
- 12.Mitra A, Mishra L, Li S. Technologies for deriving primary tumor cells for use in personalized cancer therapy. Trends Biotechnol. 2013;31(6):347–354. doi: 10.1016/j.tibtech.2013.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]; A Mitra, L Mishra and S Li, Technologies for deriving primary tumor cells for use in personalized cancer therapy. Trends Biotechnol, 2013. 31(6): p. 347-354. [DOI] [PMC free article] [PubMed]
- 13.Zeeberg K, Cardone RA, Greco MR, Saccomano M, Nohr-Nielsen A, Alves F, Pedersen SF, Reshkin SJ. Assessment of different 3D culture systems to study tumor phenotype and chemosensitivity in pancreatic ductal adenocarcinoma. Int J Oncol. 2016;49(1):243–252. doi: 10.3892/ijo.2016.3513. [DOI] [PubMed] [Google Scholar]; K Zeeberg, RA Cardone, MR Greco, M Saccomano, A Nohr-Nielsen, F Alves, SF Pedersen and SJ Reshkin, Assessment of different 3D culture systems to study tumor phenotype and chemosensitivity in pancreatic ductal adenocarcinoma. Int J Oncol, 2016. 49(1): p. 243-252. [DOI] [PubMed]
- 14.Byrne AT, Alferez DG, Amant F, Annibali D, Arribas J, Biankin AV, Bruna A, Budinska E, Caldas C, Chang DK. Interrogating open issues in cancer medicine with patient-derived xenografts. Nat Rev Cancer. 2017;17(10):632. doi: 10.1038/nrc.2017.85. [DOI] [PubMed] [Google Scholar]; AT Byrne, DG Alferez, F Amant, D Annibali, J Arribas, AV Biankin, A Bruna, E Budinska, C Caldas, DK Chang, et al., Interrogating open issues in cancer medicine with patient-derived xenografts. Nat Rev Cancer, 2017. 17(10): p. 632. [DOI] [PubMed]
- 15.Cassidy JW, Caldas C, Bruna A. Maintaining Tumor Heterogeneity in Patient-Derived Tumor Xenografts. Cancer Res. 2015;75(15):2963–2968. doi: 10.1158/0008-5472.CAN-15-0727. [DOI] [PMC free article] [PubMed] [Google Scholar]; JW Cassidy, C Caldas and A Bruna, Maintaining Tumor Heterogeneity in Patient-Derived Tumor Xenografts. Cancer Res, 2015. 75(15): p. 2963-2968. [DOI] [PMC free article] [PubMed]
- 16.Kemper K, Krijgsman O, Cornelissen-Steijger P, Shahrabi A, Weeber F, Song JY, Kuilman T, Vis DJ, Wessels LF, Voest EE. Intra- and inter-tumor heterogeneity in a vemurafenib-resistant melanoma patient and derived xenografts. EMBO Mol Med. 2015;7(9):1104–1118. doi: 10.15252/emmm.201404914. [DOI] [PMC free article] [PubMed] [Google Scholar]; K Kemper, O Krijgsman, P Cornelissen-Steijger, A Shahrabi, F Weeber, JY Song, T Kuilman, DJ Vis, LF Wessels, EE Voest, et al., Intra- and inter-tumor heterogeneity in a vemurafenib-resistant melanoma patient and derived xenografts. EMBO Mol Med, 2015. 7(9): p. 1104-1118. [DOI] [PMC free article] [PubMed]
- 17.Kemper K, Krijgsman O, Kong X, Cornelissen-Steijger P, Shahrabi A, Weeber F, van der Velden DL, Bleijerveld OB, Kuilman T, Kluin RJC. BRAF(V600E) Kinase Domain Duplication Identified in Therapy-Refractory Melanoma Patient-Derived Xenografts. Cell Rep. 2016;16(1):263–277. doi: 10.1016/j.celrep.2016.05.064. [DOI] [PMC free article] [PubMed] [Google Scholar]; K Kemper, O Krijgsman, X Kong, P Cornelissen-Steijger, A Shahrabi, F Weeber, DL van der Velden, OB Bleijerveld, T Kuilman, RJC Kluin, et al., BRAF(V600E) Kinase Domain Duplication Identified in Therapy-Refractory Melanoma Patient-Derived Xenografts. Cell Rep, 2016. 16(1): p. 263-277. [DOI] [PMC free article] [PubMed]
- 18.Aparicio S, Hidalgo M, Kung AL. Examining the utility of patient-derived xenograft mouse models. Nat Rev Cancer. 2015;15(5):311–316. doi: 10.1038/nrc3944. [DOI] [PubMed] [Google Scholar]; S Aparicio, M Hidalgo and AL Kung, Examining the utility of patient-derived xenograft mouse models. Nat Rev Cancer, 2015. 15(5): p. 311-316. [DOI] [PubMed]
- 19.Hidalgo M, Amant F, Biankin AV, Budinska E, Byrne AT, Caldas C, Clarke RB, de Jong S, Jonkers J, Maelandsmo GM. Patient-derived xenograft models: an emerging platform for translational cancer research. Cancer Discov. 2014;4(9):998–1013. doi: 10.1158/2159-8290.CD-14-0001. [DOI] [PMC free article] [PubMed] [Google Scholar]; M Hidalgo, F Amant, AV Biankin, E Budinska, AT Byrne, C Caldas, RB Clarke, S de Jong, J Jonkers, GM Maelandsmo, et al., Patient-derived xenograft models: an emerging platform for translational cancer research. Cancer Discov, 2014. 4(9): p. 998-1013. [DOI] [PMC free article] [PubMed]
- 20.Nath S, Devi GR. Three-dimensional culture systems in cancer research: Focus on tumor spheroid model. Pharmacol Ther. 2016;163:94–108. doi: 10.1016/j.pharmthera.2016.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]; S Nath and GR Devi, Three-dimensional culture systems in cancer research: Focus on tumor spheroid model. Pharmacol Ther, 2016. 163: p. 94-108. [DOI] [PMC free article] [PubMed]
- 21.Onion D, Argent RH, Reece-Smith AM, Craze ML, Pineda RG, Clarke PA, Ratan HL, Parsons SL, Lobo DN, Duffy JP. 3-Dimensional Patient-Derived Lung Cancer Assays Reveal Resistance to Standards-of-Care Promoted by Stromal Cells but Sensitivity to Histone Deacetylase Inhibitors. Mol Cancer Ther. 2016;15(4):753–763. doi: 10.1158/1535-7163.MCT-15-0598. [DOI] [PubMed] [Google Scholar]; D Onion, RH Argent, AM Reece-Smith, ML Craze, RG Pineda, PA Clarke, HL Ratan, SL Parsons, DN Lobo, JP Duffy, et al., 3-Dimensional Patient-Derived Lung Cancer Assays Reveal Resistance to Standards-of-Care Promoted by Stromal Cells but Sensitivity to Histone Deacetylase Inhibitors. Mol Cancer Ther, 2016. 15(4): p. 753-763. [DOI] [PubMed]
- 22.Beaumont KA, Anfosso A, Ahmed F, Weninger W, Haass NK. Imaging- and Flow Cytometry-based Analysis of Cell Position and the Cell Cycle in 3D Melanoma Spheroids. J Vis Exp. 2015;106 doi: 10.3791/53486. [DOI] [PMC free article] [PubMed] [Google Scholar]; KA Beaumont, A Anfosso, F Ahmed, W Weninger and NK Haass, Imaging- and Flow Cytometry-based Analysis of Cell Position and the Cell Cycle in 3D Melanoma Spheroids. J Vis Exp, 2015(106): p. e53486. [DOI] [PMC free article] [PubMed]
- 23.Weeber F, Ooft SN, Dijkstra KK, Voest EE. Tumor Organoids as a Pre-clinical Cancer Model for Drug Discovery. Cell Chem Biol. 2017;24(9):1092–1100. doi: 10.1016/j.chembiol.2017.06.012. [DOI] [PubMed] [Google Scholar]; F Weeber, SN Ooft, KK Dijkstra and EE Voest, Tumor Organoids as a Pre-clinical Cancer Model for Drug Discovery. Cell Chem Biol, 2017. 24(9): p. 1092-1100. [DOI] [PubMed]
- 24.Xu H, Lyu X, Yi M, Zhao W, Song Y, Wu K. Organoid technology and applications in cancer research. J Hematol Oncol. 2018;11(1):116. doi: 10.1186/s13045-018-0662-9. [DOI] [PMC free article] [PubMed] [Google Scholar]; H Xu, X Lyu, M Yi, W Zhao, Y Song and K Wu, Organoid technology and applications in cancer research. J Hematol Oncol, 2018. 11(1): p. 116. [DOI] [PMC free article] [PubMed]
- 25.Spoerri L, Beaumont KA, Anfosso A, Haass NK. Real-Time Cell Cycle Imaging in a 3D Cell Culture Model of Melanoma. Methods Mol Biol. 2017;1612:401–416. doi: 10.1007/978-1-4939-7021-6_29. [DOI] [PubMed] [Google Scholar]; L Spoerri, KA Beaumont, A Anfosso and NK Haass, Real-Time Cell Cycle Imaging in a 3D Cell Culture Model of Melanoma. Methods Mol Biol, 2017. 1612: p. 401-416. [DOI] [PubMed]
- 26.Haass NK, Sproesser K, Nguyen TK, Contractor R, Medina CA, Nathanson KL, Herlyn M, Smalley KS. The mitogen-activated protein/extracellular signal-regulated kinase kinase inhibitor AZD6244 (ARRY-142886) induces growth arrest in melanoma cells and tumor regression when combined with docetaxel. Clin Cancer Res. 2008;14(1):230–239. doi: 10.1158/1078-0432.CCR-07-1440. [DOI] [PubMed] [Google Scholar]; NK Haass, K Sproesser, TK Nguyen, R Contractor, CA Medina, KL Nathanson, M Herlyn and KS Smalley, The mitogen-activated protein/extracellular signal-regulated kinase kinase inhibitor AZD6244 (ARRY-142886) induces growth arrest in melanoma cells and tumor regression when combined with docetaxel. Clin Cancer Res, 2008. 14(1): p. 230-239. [DOI] [PubMed]
- 27.Marconi A, Quadri M, Saltari A, Pincelli C. Progress in melanoma modelling in vitro. Exp Dermatol. 2018;27(5):578–586. doi: 10.1111/exd.13670. [DOI] [PubMed] [Google Scholar]; A Marconi, M Quadri, A Saltari and C Pincelli, Progress in melanoma modelling in vitro. Exp Dermatol, 2018. 27(5): p. 578-586. [DOI] [PubMed]
- 28.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38(16):e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]; K Wang, M Li and H Hakonarson, ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res, 2010. 38(16): p. e164. [DOI] [PMC free article] [PubMed]
- 29.Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. Integrative genomics viewer. Nat Biotechnol. 2011;29(1):24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]; JT Robinson, H Thorvaldsdottir, W Winckler, M Guttman, ES Lander, G Getz and JP Mesirov, Integrative genomics viewer. Nat Biotechnol, 2011. 29(1): p. 24-26. [DOI] [PMC free article] [PubMed]
- 30.Chapman PB, Hauschild A, Robert C, Haanen JB, Ascierto P, Larkin J, Dummer R, Garbe C, Testori A, Maio M. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med. 2011;364(26):2507–2516. doi: 10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed] [Google Scholar]; PB Chapman, A Hauschild, C Robert, JB Haanen, P Ascierto, J Larkin, R Dummer, C Garbe, A Testori, M Maio, et al., Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med, 2011. 364(26): p. 2507-2516. [DOI] [PMC free article] [PubMed]
- 31.Gao H, Korn JM, Ferretti S, Monahan JE, Wang Y, Singh M, Zhang C, Schnell C, Yang G, Zhang Y. High-throughput screening using patient-derived tumor xenografts to predict clinical trial drug response. Nat Med. 2015;21(11):1318–1325. doi: 10.1038/nm.3954. [DOI] [PubMed] [Google Scholar]; H Gao, JM Korn, S Ferretti, JE Monahan, Y Wang, M Singh, C Zhang, C Schnell, G Yang, Y Zhang, et al., High-throughput screening using patient-derived tumor xenografts to predict clinical trial drug response. Nat Med, 2015. 21(11): p. 1318-1325. [DOI] [PubMed]
- 32.Kunz M, Holzel M. The impact of melanoma genetics on treatment response and resistance in clinical and experimental studies. Cancer Metastasis Rev. 2017;36(1):53–75. doi: 10.1007/s10555-017-9657-1. [DOI] [PubMed] [Google Scholar]; M Kunz and M Holzel, The impact of melanoma genetics on treatment response and resistance in clinical and experimental studies. Cancer Metastasis Rev, 2017. 36(1): p. 53-75. [DOI] [PubMed]
- 33.Mahmoud F, Shields B, Makhoul I, Hutchins LF, Shalin SC, Tackett AJ. Role of EZH2 histone methyltrasferase in melanoma progression and metastasis. Cancer Biol Ther. 2016;17(6):579–591. doi: 10.1080/15384047.2016.1167291. [DOI] [PMC free article] [PubMed] [Google Scholar]; F Mahmoud, B Shields, I Makhoul, LF Hutchins, SC Shalin and AJ Tackett, Role of EZH2 histone methyltrasferase in melanoma progression and metastasis. Cancer Biol Ther, 2016. 17(6): p. 579-591. [DOI] [PMC free article] [PubMed]
- 34.Grzywa TM, Paskal W, Wlodarski PK. Intratumor and Intertumor Heterogeneity in Melanoma. Transl Oncol. 2017;10(6):956–975. doi: 10.1016/j.tranon.2017.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]; TM Grzywa, W Paskal and PK Wlodarski, Intratumor and Intertumor Heterogeneity in Melanoma. Transl Oncol, 2017. 10(6): p. 956-975. [DOI] [PMC free article] [PubMed]
- 35.Yancovitz M, Litterman A, Yoon J, Ng E, Shapiro RL, Berman RS, Pavlick AC, Darvishian F, Christos P, Mazumdar M. Intra- and inter-tumor heterogeneity of BRAF(V600E)mutations in primary and metastatic melanoma. PLoS One. 2012;7(1) doi: 10.1371/journal.pone.0029336. [DOI] [PMC free article] [PubMed] [Google Scholar]; M Yancovitz, A Litterman, J Yoon, E Ng, RL Shapiro, RS Berman, AC Pavlick, F Darvishian, P Christos, M Mazumdar, et al., Intra- and inter-tumor heterogeneity of BRAF(V600E))mutations in primary and metastatic melanoma. PLoS One, 2012. 7(1): p. e29336. [DOI] [PMC free article] [PubMed]
- 36.Fang Y, Eglen RM. Three-Dimensional Cell Cultures in Drug Discovery and Development. SLAS Discov. 2017;22(5):456–472. doi: 10.1177/1087057117696795. [DOI] [PMC free article] [PubMed] [Google Scholar]; Y Fang and RM Eglen, Three-Dimensional Cell Cultures in Drug Discovery and Development. SLAS Discov, 2017. 22(5): p. 456-472. [DOI] [PMC free article] [PubMed]
- 37.Friedrich J, Seidel C, Ebner R, Kunz-Schughart LA. Spheroid-based drug screen: considerations and practical approach. Nat Protoc. 2009;4(3):309–324. doi: 10.1038/nprot.2008.226. [DOI] [PubMed] [Google Scholar]; J Friedrich, C Seidel, R Ebner and LA Kunz-Schughart, Spheroid-based drug screen: considerations and practical approach. Nat Protoc, 2009. 4(3): p. 309-324. [DOI] [PubMed]
- 38.Volm M, Efferth T. Prediction of Cancer Drug Resistance and Implications for Personalized Medicine. Front Oncol. 2015;5:282. doi: 10.3389/fonc.2015.00282. [DOI] [PMC free article] [PubMed] [Google Scholar]; M Volm and T Efferth, Prediction of Cancer Drug Resistance and Implications for Personalized Medicine. Front Oncol, 2015. 5: p. 282. [DOI] [PMC free article] [PubMed]
- 39.Konecny G, Crohns C, Pegram M, Felber M, Lude S, Kurbacher C, Cree IA, Hepp H, Untch M. Correlation of drug response with the ATP tumorchemosensitivity assay in primary FIGO stage III ovarian cancer. Gynecol Oncol. 2000;77(2):258–263. doi: 10.1006/gyno.2000.5728. [DOI] [PubMed] [Google Scholar]; G Konecny, C Crohns, M Pegram, M Felber, S Lude, C Kurbacher, IA Cree, H Hepp and M Untch, Correlation of drug response with the ATP tumorchemosensitivity assay in primary FIGO stage III ovarian cancer. Gynecol Oncol, 2000. 77(2): p. 258-263. [DOI] [PubMed]
- 40.Kim HS, Kim TJ, Chung HH, Kim JW, Kim BG, Park NH, Song YS, Bae DS, Kang SB. In vitro extreme drug resistance assay to taxanes or platinum compounds for the prediction of clinical outcomes in epithelial ovarian cancer: a prospective cohort study. J Cancer Res Clin Oncol. 2009;135(11):1513–1520. doi: 10.1007/s00432-009-0598-0. [DOI] [PubMed] [Google Scholar]; HS Kim, TJ Kim, HH Chung, JW Kim, BG Kim, NH Park, YS Song, DS Bae and SB Kang, In vitro extreme drug resistance assay to taxanes or platinum compounds for the prediction of clinical outcomes in epithelial ovarian cancer: a prospective cohort study. J Cancer Res Clin Oncol, 2009. 135(11): p. 1513-1520. [DOI] [PubMed]
- 41.Hetland TE, Kaern J, Skrede M, Sandstad B, Trope C, Davidson B, Florenes VA. Predicting platinum resistance in primary advanced ovarian cancer patients with an in vitro resistance index. Cancer Chemother Pharmacol. 2012;69(5):1307–1314. doi: 10.1007/s00280-012-1835-9. [DOI] [PubMed] [Google Scholar]; TE Hetland, J Kaern, M Skrede, B Sandstad, C Trope, B Davidson and VA Florenes, Predicting platinum resistance in primary advanced ovarian cancer patients with an in vitro resistance index. Cancer Chemother Pharmacol, 2012. 69(5): p. 1307-1314. [DOI] [PubMed]
- 42.Raaijmakers MI, Widmer DS, Maudrich M, Koch T, Langer A, Flace A, Schnyder C, Dummer R, Levesque MP. A new live-cell biobank workflow efficiently recovers heterogeneous melanoma cells from native biopsies. Exp Dermatol. 2015;24(5):377–380. doi: 10.1111/exd.12683. [DOI] [PubMed] [Google Scholar]; MI Raaijmakers, DS Widmer, M Maudrich, T Koch, A Langer, A Flace, C Schnyder, R Dummer and MP Levesque, A new live-cell biobank workflow efficiently recovers heterogeneous melanoma cells from native biopsies. Exp Dermatol, 2015. 24(5): p. 377-380. [DOI] [PubMed]
- 43.Garman B, Anastopoulos IN, Krepler C, Brafford P, Sproesser K, Jiang Y, Wubbenhorst B, Amaravadi R, Bennett J, Beqiri M. Genetic and Genomic Characterization of 462 Melanoma Patient-Derived Xenografts, Tumor Biopsies, and Cell Lines. Cell Rep. 2017;21(7):1936–1952. doi: 10.1016/j.celrep.2017.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]; B Garman, IN Anastopoulos, C Krepler, P Brafford, K Sproesser, Y Jiang, B Wubbenhorst, R Amaravadi, J Bennett, M Beqiri, et al., Genetic and Genomic Characterization of 462 Melanoma Patient-Derived Xenografts, Tumor Biopsies, and Cell Lines. Cell Rep, 2017. 21(7): p. 1936-1952. [DOI] [PMC free article] [PubMed]
- 44.Fruehauf JP, Alberts DS. In vitro drug resistance versus chemosensitivity: two sides of different coins. J Clin Oncol. 2005;23(15):3641–3643. doi: 10.1200/JCO.2005.05.281. author reply 3646-3648. [DOI] [PubMed] [Google Scholar]; JP Fruehauf and DS Alberts, In vitro drug resistance versus chemosensitivity: two sides of different coins. J Clin Oncol, 2005. 23(15): p. 3641-3643; author reply 3646-3648. [DOI] [PubMed]
- 45.Kreahling JM, Altiok S. Special Technologies for Ex Vivo Analysis of Cancer. Cancer Control. 2015;22(2):226–231. doi: 10.1177/107327481502200215. [DOI] [PubMed] [Google Scholar]; JM Kreahling and S Altiok, Special Technologies for Ex Vivo Analysis of Cancer. Cancer Control, 2015. 22(2): p. 226-231. [DOI] [PubMed]
- 46.Santo VE, Rebelo SP, Estrada MF, Alves PM, Boghaert E, Brito C. Drug screening in 3D in vitro tumor models: overcoming current pitfalls of efficacy read-outs. Biotechnol J. 2017;12(1) doi: 10.1002/biot.201600505. [DOI] [PubMed] [Google Scholar]; VE Santo, SP Rebelo, MF Estrada, PM Alves, E Boghaert and C Brito, Drug screening in 3D in vitro tumor models: overcoming current pitfalls of efficacy read-outs. Biotechnol J, 2017. 12(1). [DOI] [PubMed]
- 47.Vu HL, Aplin AE. Targeting mutant NRAS signaling pathways in melanoma. Pharmacol Res. 2016;107:111–116. doi: 10.1016/j.phrs.2016.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]; HL Vu and AE Aplin, Targeting mutant NRAS signaling pathways in melanoma. Pharmacol Res, 2016. 107: p. 111-116. [DOI] [PMC free article] [PubMed]
- 48.Eroglu Z, Ribas A. Combination therapy with BRAF and MEK inhibitors for melanoma: latest evidence and place in therapy. Ther Adv Med Oncol. 2016;8(1):48–56. doi: 10.1177/1758834015616934. [DOI] [PMC free article] [PubMed] [Google Scholar]; Z Eroglu and A Ribas, Combination therapy with BRAF and MEK inhibitors for melanoma: latest evidence and place in therapy. Ther Adv Med Oncol, 2016. 8(1): p. 48-56. [DOI] [PMC free article] [PubMed]
- 49.Seidel D, Rothe R, Kirsten M, Jahnke HG, Dumann K, Ziemer M, Simon JC, Robitzki AA. A multidimensional impedance platform for the real-time analysis of single and combination drug pharmacology in patient-derived viable melanoma models. Biosens Bioelectron. 2019;123:185–194. doi: 10.1016/j.bios.2018.08.049. [DOI] [PubMed] [Google Scholar]; D Seidel, R Rothe, M Kirsten, HG Jahnke, K Dumann, M Ziemer, JC Simon and AA Robitzki, A multidimensional impedance platform for the real-time analysis of single and combination drug pharmacology in patient-derived viable melanoma models. Biosens Bioelectron, 2019. 123: p. 185-194. [DOI] [PubMed]
- 50.Solit DB, Garraway LA, Pratilas CA, Sawai A, Getz G, Basso A, Ye Q, Lobo JM, She Y, Osman I. BRAF mutation predicts sensitivity to MEK inhibition. Nature. 2006;439(7074):358–362. doi: 10.1038/nature04304. [DOI] [PMC free article] [PubMed] [Google Scholar]; DB Solit, LA Garraway, CA Pratilas, A Sawai, G Getz, A Basso, Q Ye, JM Lobo, Y She, I Osman, et al., BRAF mutation predicts sensitivity to MEK inhibition. Nature, 2006. 439(7074): p. 358-362. [DOI] [PMC free article] [PubMed]
- 51.Niessner H, Sinnberg T, Kosnopfel C, Smalley KSM, Beck D, Praetorius C, Mai M, Beissert S, Kulms D, Schaller M. BRAF Inhibitors Amplify the Proapoptotic Activity of MEK Inhibitors by Inducing ER Stress in NRAS-Mutant Melanoma. Clin Cancer Res. 2017;23(20):6203–6214. doi: 10.1158/1078-0432.CCR-17-0098. [DOI] [PMC free article] [PubMed] [Google Scholar]; H Niessner, T Sinnberg, C Kosnopfel, KSM Smalley, D Beck, C Praetorius, M Mai, S Beissert, D Kulms, M Schaller, et al., BRAF Inhibitors Amplify the Proapoptotic Activity of MEK Inhibitors by Inducing ER Stress in NRAS-Mutant Melanoma. Clin Cancer Res, 2017. 23(20): p. 6203-6214. [DOI] [PMC free article] [PubMed]
- 52.Saint-Jean M, Quereux G, Nguyen JM, Peuvrel L, Brocard A, Vallee A, Knol AC, Khammari A, Denis MG, Dreno B. Is a single BRAF wild-type test sufficient to exclude melanoma patients from vemurafenib therapy? J Invest Dermatol. 2014;134(5):1468–1470. doi: 10.1038/jid.2013.378. [DOI] [PubMed] [Google Scholar]; M Saint-Jean, G Quereux, JM Nguyen, L Peuvrel, A Brocard, A Vallee, AC Knol, A Khammari, MG Denis and B Dreno, Is a single BRAF wild-type test sufficient to exclude melanoma patients from vemurafenib therapy? J Invest Dermatol, 2014. 134(5): p. 1468-1470. [DOI] [PubMed]
- 53.Valachis A, Ullenhag GJ. Discrepancy in BRAF status among patients with metastatic malignant melanoma: A meta-analysis. Eur J Cancer. 2017;81:106–115. doi: 10.1016/j.ejca.2017.05.015. [DOI] [PubMed] [Google Scholar]; A Valachis and GJ Ullenhag, Discrepancy in BRAF status among patients with metastatic malignant melanoma: A meta-analysis. Eur J Cancer, 2017. 81: p. 106-115. [DOI] [PubMed]
- 54.Romano E, Pradervand S, Paillusson A, Weber J, Harshman K, Muehlethaler K, Speiser D, Peters S, Rimoldi D, Michielin O. Identification of multiple mechanisms of resistance to vemurafenib in a patient with BRAFV600E-mutated cutaneous melanoma successfully rechallenged after progression. Clin Cancer Res. 2013;19(20):5749–5757. doi: 10.1158/1078-0432.CCR-13-0661. [DOI] [PubMed] [Google Scholar]; E Romano, S Pradervand, A Paillusson, J Weber, K Harshman, K Muehlethaler, D Speiser, S Peters, D Rimoldi and O Michielin, Identification of multiple mechanisms of resistance to vemurafenib in a patient with BRAFV600E-mutated cutaneous melanoma successfully rechallenged after progression. Clin Cancer Res, 2013. 19(20): p. 5749-5757. [DOI] [PubMed]
- 55.Krauthammer M, Kong Y, Bacchiocchi A, Evans P, N Pornputtapong C Wu, McCusker JP, Ma S, Cheng E, Straub R. Exome sequencing identifies recurrent mutations in NF1 and RASopathy genes in sun-exposed melanomas. Nat Genet. 2015;47(9):996–1002. doi: 10.1038/ng.3361. [DOI] [PMC free article] [PubMed] [Google Scholar]; M Krauthammer, Y Kong, A Bacchiocchi, P Evans, N Pornputtapong, C Wu, JP McCusker, S Ma, E Cheng, R Straub, et al., Exome sequencing identifies recurrent mutations in NF1 and RASopathy genes in sun-exposed melanomas. Nat Genet, 2015. 47(9): p. 996-1002. [DOI] [PMC free article] [PubMed]
- 56.Krepler C, Sproesser K, Brafford P, Beqiri M, Garman B, Xiao M, Shannan B, Watters A, Perego M, Zhang G. A Comprehensive Patient-Derived Xenograft Collection Representing the Heterogeneity of Melanoma. Cell Rep. 2017;21(7):1953–1967. doi: 10.1016/j.celrep.2017.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]; C Krepler, K Sproesser, P Brafford, M Beqiri, B Garman, M Xiao, B Shannan, A Watters, M Perego, G Zhang, et al., A Comprehensive Patient-Derived Xenograft Collection Representing the Heterogeneity of Melanoma. Cell Rep, 2017. 21(7): p. 1953-1967. [DOI] [PMC free article] [PubMed]
- 57.Krepler C, Xiao M, Sproesser K, Brafford PA, Shannan B, Beqiri M, Q Liu W Xu, Garman B, Nathanson KL. Personalized Preclinical Trials in BRAF Inhibitor-Resistant Patient-Derived Xenograft Models Identify Second-Line Combination Therapies. Clin Cancer Res. 2016;22(7):1592–1602. doi: 10.1158/1078-0432.CCR-15-1762. [DOI] [PMC free article] [PubMed] [Google Scholar]; C Krepler, M Xiao, K Sproesser, PA Brafford, B Shannan, M Beqiri, Q Liu, W Xu, B Garman, KL Nathanson, et al., Personalized Preclinical Trials in BRAF Inhibitor-Resistant Patient-Derived Xenograft Models Identify Second-Line Combination Therapies. Clin Cancer Res, 2016. 22(7): p. 1592-1602. [DOI] [PMC free article] [PubMed]
- 58.Einarsdottir BO, Bagge RO, Bhadury J, Jespersen H, Mattsson J, Nilsson LM, Truve K, Lopez MD, Naredi P, Nilsson O. Melanoma patient-derived xenografts accurately model the disease and develop fast enough to guide treatment decisions. Oncotarget. 2014;5(20):9609–9618. doi: 10.18632/oncotarget.2445. [DOI] [PMC free article] [PubMed] [Google Scholar]; BO Einarsdottir, RO Bagge, J Bhadury, H Jespersen, J Mattsson, LM Nilsson, K Truve, MD Lopez, P Naredi, O Nilsson, et al., Melanoma patient-derived xenografts accurately model the disease and develop fast enough to guide treatment decisions. Oncotarget, 2014. 5(20): p. 9609-9618. [DOI] [PMC free article] [PubMed]
- 59.Hartsough EJ, Aplin AE. Of Mice and Melanoma: PDX System for Modeling Personalized Medicine. Clin Cancer Res. 2016;22(7):1550–1552. doi: 10.1158/1078-0432.CCR-15-3054. [DOI] [PMC free article] [PubMed] [Google Scholar]; EJ Hartsough and AE Aplin, Of Mice and Melanoma: PDX System for Modeling Personalized Medicine. Clin Cancer Res, 2016. 22(7): p. 1550-1552. [DOI] [PMC free article] [PubMed]
- 60.Bruna A, Rueda OM, Greenwood W, Batra AS, Callari M, Batra RN, Pogrebniak K, Sandoval J, Cassidy JW, Tufegdzic-Vidakovic A. A Biobank of Breast Cancer Explants with Preserved Intra-tumor Heterogeneity to Screen Anticancer Compounds. Cell. 2016;167(1):260–274. doi: 10.1016/j.cell.2016.08.041. e222. [DOI] [PMC free article] [PubMed] [Google Scholar]; A Bruna, OM Rueda, W Greenwood, AS Batra, M Callari, RN Batra, K Pogrebniak, J Sandoval, JW Cassidy, A Tufegdzic-Vidakovic, et al., A Biobank of Breast Cancer Explants with Preserved Intra-tumor Heterogeneity to Screen Anticancer Compounds. Cell, 2016. 167(1): p. 260-274 e222. [DOI] [PMC free article] [PubMed]
- 61.Sia D, Moeini A, Labgaa I, Villanueva A. The future of patient-derived tumor xenografts in cancer treatment. Pharmacogenomics. 2015;16(14):1671–1683. doi: 10.2217/pgs.15.102. [DOI] [PubMed] [Google Scholar]; D Sia, A Moeini, I Labgaa and A Villanueva, The future of patient-derived tumor xenografts in cancer treatment. Pharmacogenomics, 2015. 16(14): p. 1671-1683. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Mutations identified in 21 melanoma lymph node metastases and one PDX by IonTorrent Cancer Hotspot and Oncomine Comprehensive Panel.