Abstract
Metastatic breast cancer is the leading cause of cancer-associated death in women. The progression of this fatal disease is associated with inflammatory responses that promote cancer cell growth and dissemination, eventually leading to a reduction of overall survival. However, the mechanism(s) of the inflammation-boosted cancer progression remains unclear. In this study, we found for the first time that an extracellular cytokine, S100A8/A9, accelerates breast cancer growth and metastasis upon binding to a cell surface receptor, melanoma cell adhesion molecule (MCAM). Our molecular analyses revealed an important role of ETS translocation variant 4 (ETV4), which is significantly activated in the region downstream of MCAM upon S100A8/A9 stimulation, in breast cancer progression in vitro as well as in vivo. The MCAM-mediated activation of ETV4 induced a mobile phenotype called epithelial-mesenchymal transition (EMT) in cells, since we found that ETV4 transcriptionally upregulates ZEB1, a strong EMT inducer, at a very high level. In contrast, downregulation of either MCAM or ETV4 repressed EMT, resulting in greatly weakened tumor growth and lung metastasis. Overall, our results revealed that ETV4 is a novel transcription factor regulated by the S100A8/A9-MCAM axis, which leads to EMT through ZEB1 and thereby to metastasis in breast cancer cells. Thus, therapeutic strategies based on our findings might improve patient outcomes.
Introduction
Metastatic breast cancer classified as stage IV is the most serious cancer-related disease in women. More than 90% of such patients die of metastasis, a process by which cancer cells depart from their tumor of origin, spread systemically, and colonize at distant organs [1]. Even with the current advanced therapies, it is difficult to effectively treat stage IV breast cancer, and the establishment of an effective inhibitory treatment for breast cancer metastasis is needed. Elucidation of the complex mechanisms of breast cancer metastasis at the molecular level is needed for the establishment of such treatment.
Breast cancer cells seem to prefer the lung, liver, bone and brain as their metastatic sites. This organ-tropic metastasis is known as the “seed and soil” theory, and was first proposed by Paget et al. [2]. As to the molecular-level mechanisms underpinning the theory, accumulating evidence has revealed that cancer-mediated S100A8/A9 in the lung plays a critical role in lung tropic metastasis and the subsequent growth of cancer cells in the lung. In addition, Eisenblaetter et al. recently reported that the process by which extracellular S100A8/A9 is increased in the lung is distant cancer-mediated enrichment of a Ccl2 chemokine and CCR2highCX3CL1low monocyte population, and subsequent interaction of ligand Ccl2 with its receptor CCR2 in the abundant monocytes in the breast cancer-bearing mouse lung [3]. That process leads to an increase in immunosuppressive Treg cells, and eventually to the formation of a pre-metastatic niche in the lung, which is designated as the “soil” in the “seed and soil” model. Hiratsuka et al. clearly demonstrated in a melanoma lung metastasis model that lung S100A8/A9 functions as a strong chemokine to attract distant cancer cells through its receptor toll-like receptor 4 (TLR4) [4]. However, the receptors of S100A8/A9 are not restricted to TLR4. The receptor for advanced glycation endproducts (RAGE) also acts as an S100A8/A9 receptor. Moreover, in addition to TLR4 and RAGE, we previously revealed the presence of other important candidate receptors, MCAM, ALCAM, EMMPRIN and NPTN (isoforms α and β), which we collectively named the S100 soil sensor receptors (SSSRs). Hence, S100A8/A9 plays a crucial role in lung tropic cancer metastasis by helping to establish an immunosuppressive metastatic niche to which it then attracts remote cancer cells by stimulating various receptors on the cancer cell surface as chemokine-like ligands.
Here, we attempt to determine the main receptors functioning in metastatic breast cancer cells. Among the receptors mentioned above, we recently found that MCAM was remarkably overexpressed in metastatic breast cancer cells, which was also supported by the previous reports [5], [6], [7]. However, the MCAM downstream signal upon S100A8/A9 binding in breast cancer cells that produces the driving force for distant metastasis has remained unclear. In this study, we therefore aimed to clarify that mechanism(s) at the molecular level.
Results
Highly Up-Regulated Cellular Motility, Invasiveness and MCAM Expression in Triple-Negative Malignant MDA-MB-231 Cells
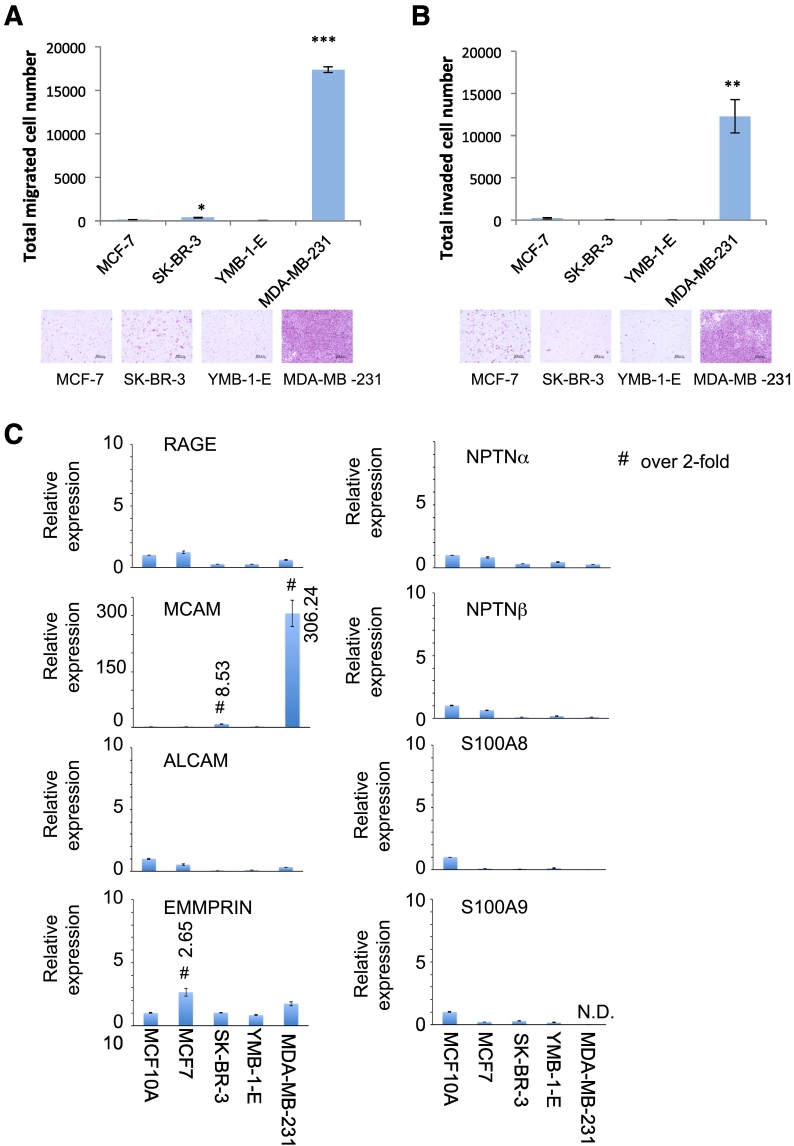
Figure 1, A and B show that MDA-MB-231 cells had the highest levels of activity for both migration and invasion among the breast cancer cells we examined. We previously reported that the S100A8/A9-SSSRs axis plays an important role in these cellular behaviors, so in our present experiments we attempted to determine the expression profile of the SSSRs in these breast cancer cell lines [8], [9], [10]. Quantitative real-time PCR analysis showed that MCAM expression was highly elevated in MDA-MB-231 cells in a specific manner (Figure 1C). By using publicly available data sets (http://watson.compbio.iupui.edu/chirayu/proggene/database/index.php), we found that the rates of both overall survival and metastasis-free survival were significantly lower in patients with MCAM-abundant tumors than in patients with MCAM-scant tumors (Figure S1A and Figure S1B). These results imply an unusual role of MCAM in the metastatic aggressiveness of breast cancer diseases. Accumulating evidence has indicated that MCAM overexpression is involved in tumorigenesis and subsequent aggressive progression in not only breast cancer cells but also various cancer species such as melanoma [11], hepatocellular carcinoma [12] and small-cell lung cancer (SCLC) [13] and that abundant MCAM in tumors is linked to poor survival of these tumor-bearing patients. We previously demonstrated that MCAM is highly increased in metastatic melanoma and functions as an S100A8/A9 receptor. The binding of S100A8/A9 derived from the remote lung region activates the melanoma and attracts melanoma cells to the S100A8/A9-enriched lung area through the cell surface MCAM sensor, resulting in melanoma lung tropic metastasis [10]. Hence, a similar mechanism may be adopted in melanoma and other breast cancer cells.
Figure 1.
Highly upregulated cellular motility, invasiveness and MCAM expression in triple-negative malignant MDA-MB-231 cells. A and B, Cellular migration and invasion were monitored according to the Boyden chamber method. The indicated cells were placed on the top surface of non-sealed inserts (A) or Matrigel-sealed inserts (B). Quantified results are displayed in the upper panel and representative images of migrating cells are shown in the bottom panel. C, Quantitative real-time PCR analysis was carried out in the indicated cells for RAGE, MCAM, ALCAM, EMMPPRIN, NPTNα, NPTNβ, S100A8 and S100A9 genes. The relative expression level of each sample is shown after calibration with the level of TBP (a suitable housekeeping gene). Data are means ± SD, *P < .05, **P < .01 and ***P < .001. N.D.: not detected.
Significance of the S100A8/A9-MCAM Axis in Cancer Mortality
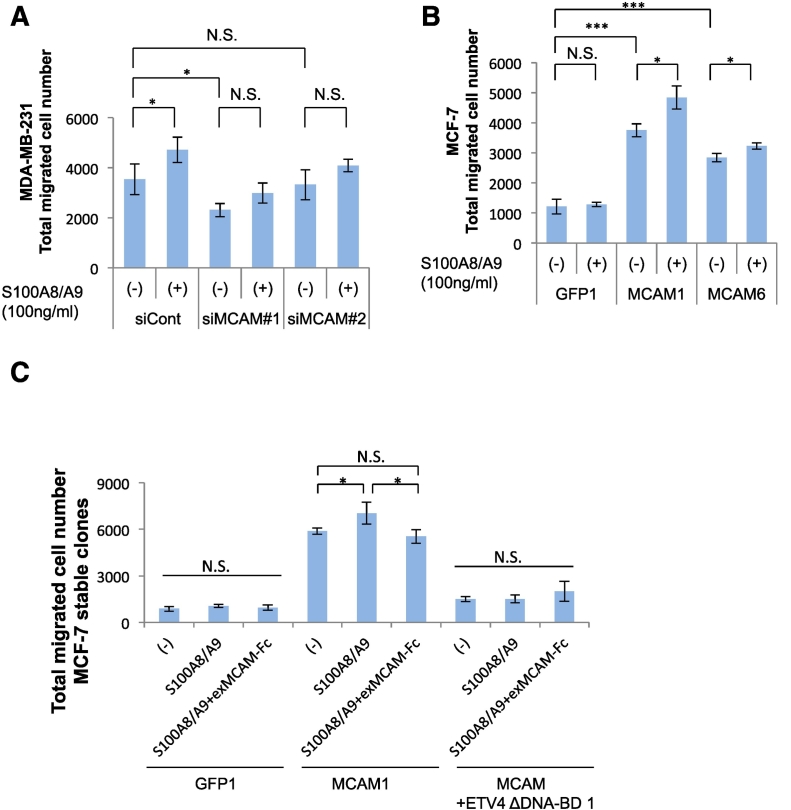
We next attempted to determine the significance of the S100A8/A9-MCAM axis in the metastatic mortality of MDA-MB-231 cells. We used the siRNA technique to attenuate the intrinsic MCAM function. We confirmed the knockdown effects of the siRNAs used and we found that the efficacy of siMCAM #1 was higher than that of siMCAM #2 (Figure S2). As shown in Figure 2A, we found that the two siRNAs induced downregulation of the migration ability of MDA-MB-231 cells not only under the basal condition but also in response to stimulation with S100A8/A9. Conversely, we then evaluated the effect of MCAM overexpression. Since the expression of endogenous MCAM in MCF-7 cells is relatively low, we forced MCF-7 cells to express foreign MCAM at a higher level in a sustained manner, resulting in the establishment of MCF-7-derived stable sublines (Figure S3). We randomly chose four GFP (Figure S3A, left), four MCAM (Figure S3B, top left), and four MCAM+ETV4 ΔDNA-BD clones (Figure S3B, top right). All of the selected GFP clones showed lower migration activity (Figure S3A, right), while the MCAM clones showed a tendency for increased basal migration ability (Figure S3B, bottom). The migration ability was further upregulated by S100A8/A9 stimulation in MCAM clones (#1 and #6) but not in the GFP clone (#1) (Figure 2B). To further understand the importance of S100A8/A9-MCAM interaction in breast cancer migration, we treated the MCAM-overexpressed MCF-7 cells with an exMCAM-Fc decoy that prevents interaction between S100A8/A9 and cellular MCAM in a ligand S100A8/A9-absorptive manner. As expected, the decoy almost completely abrogated the elevation of S100A8/A9-mediated migration (Figure 2C).
Figure 2.
Significance of the S100A8/A9-MCAM axis in cancer mortality. A, MDA-MB-231 cells were transfected with either MCAM siRNAs (siMCAM #1, siMCAM #2) or a negative control scrambled siRNA (siCont.) and then assessed for their migration abilities in response to extracellular S100A8/A9 (100 ng/ml). B, MCF-7-derived stable sublines, GFP #1, MCAM #1 and MCAM #6 (see Figure S3), were assessed for their migration abilities in response to extracellular S100A8/A9 (100 ng/ml). C, MCF-7-derived stable sublines, GFP #1, MCAM #1 and MCAM+ETV4 ΔDNA-BD #1 (see Figure S3), were assessed for their migration abilities in response to extracellular S100A8/A9 (100 ng/ml) in the presence or absence of a decoy exMCAM-Fc recombinant protein (1000 ng/ml). Data are means ± SD, N.S.: not significant, *P < .05 and ***P < .001.
Identification of the S100A8/A9-MCAM-ETV4-ZEB1 Pathway
Our previous search using protein/DNA array I (Thermo Fisher Scientific) for transcription factors that could generate the metastatic driving force and could be regulated by an MCAM downstream signal provided four potential candidates, ETS, GATA, NFI and PPAR (Supplementary Table S1, marked in red color) among the MCAM-induced several transcription factors [14]. Among these, we focused on the significance of ETS, since an ETS decoy oligonucleotide in the four candidates’ decoys (Supplementary Table S2) had a preventive effect on the migration activity in MDA-MB-231 cells (Figure S4A). That spurred us to try to identify the ETS protein producing the metastatic force in breast cancer cells from ETS family proteins. Quantitative real-time PCR analysis showed that ETV1, ETV4, ETV5 and ELK3 were expressed in MDA-MB-231 cells at very high levels and in a cell-specific manner (Figure S4B). When we forced MCF-7 cells to overexpress these proteins, we found that the expression of ETV1, ETV4 and ETV5, but not that of ELK3, resulted in the acquisition of much higher cell-migration activity (Figure S5A). Since epithelial-mesenchymal transition (EMT) facilitates metastasis of cancer cells, we investigated the induction of EMT markers by ETV1, ETV4, ETV5 and ELK3 transcription factors. As shown in Figure S5B, we found that ETV4 alone has the ability to induce expression of several EMT markers, including N-cadherin, TWIST1 and ZEB1, at the highest levels. To determine which EMT markers are regulated by ETV4 with the help of MCAM, we transfected MCF-7 cells again with an empty vector, MCAM, ETV4 wt or ETV4 ΔDNA-BD vector (deletion of C-terminal DNA-binding domain: 342-484 aa). This approach showed that ZEB1 is the most suitable EMT molecule for regulation by MCAM and subsequent ETV4 activation (Figure S6A). ETV4, also called E1AF or PEA3, has been shown to be closely involved in breast cancer progression [15]. Due to its protein function as a transcription factor, ETV4 has many target genes associated with cancer promotion. Jiand et al. reported that ETV4 enhances cell cycle progression via the upregulation of cyclin D3 transcription [16]. MMP2, the renowned invasion-associated matrix metalloprotease, is also positively regulated by ETV4 [17]. We also newly found that ZEB1, a strong EMT inducer, is a potential target gene of ETV4.
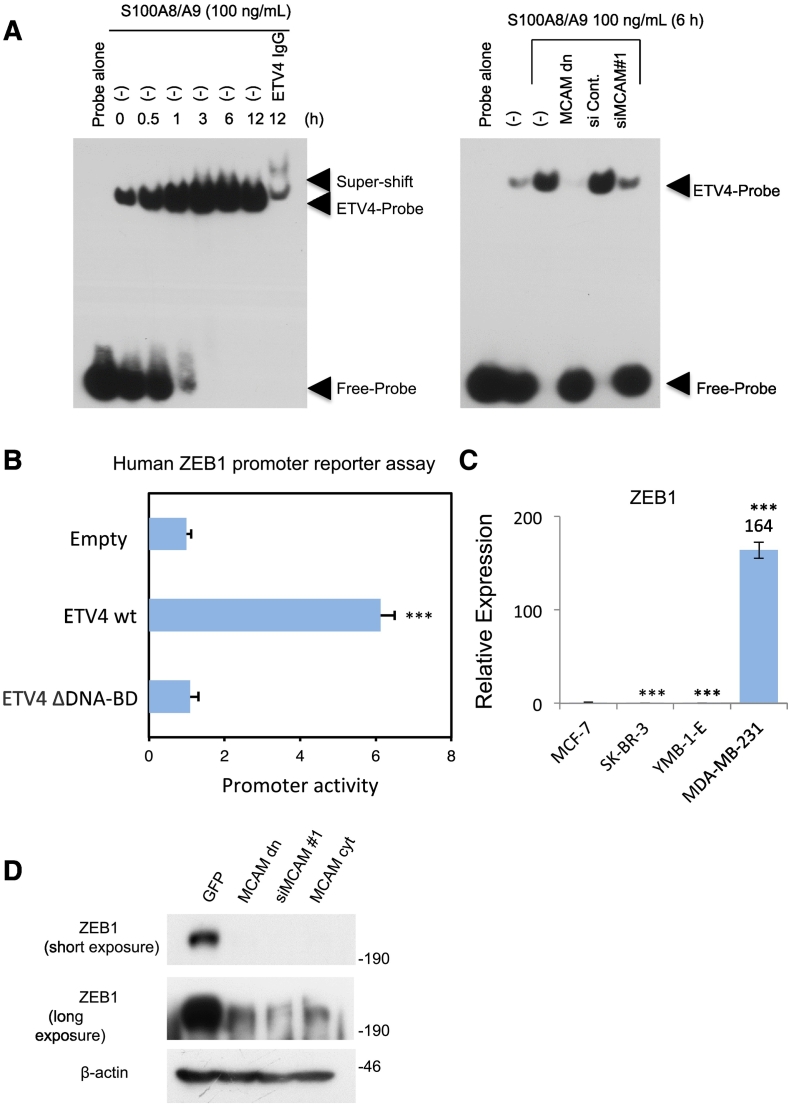
To further clarify S100A8/A9-MCAM-ETV4-ZEB1, the signaling pathway that appeared to be responsible for the acquisition of metastatic force by breast cancer cells, we first performed an electrophoretic mobility shift assay (EMSA) to detect the activation of ETV4 through MCAM upon S100A8/A9 binding. EMSA analysis revealed time-dependent activation of intrinsic ETV4 after stimulation with S100A8/A9 (Figure 3A, left). In addition, the S100A8/A9-mediated activation of ETV4 was remarkably downregulated by procedures for the prevention of endogenous MCAM expression by using either forced expression of MCAM dn (deletion of cytoplasmic tail: D 584-646 aa) or siMCAM#1 transduction into cells (Figure 3A, right). The activation of ETV4 is closely linked to MCAM-mediated elevation of migration ability, since ETV4 ΔDNA-BD overexpression effectively reduced cellular migration even under an abundant MCAM condition (Figure S6B). These results indicate that ETV4 is positively regulated by MCAM via S100A8/A9 binding and that such activated ETV4 plays a critical role in the enhancement of cellular migration. Thus, ETV4-mediated cancer migration may be a critical step for breast cancer metastasis and may greatly affect patient survival. Yuan et al. reported that a highly elevated level of ETV4 protein in triple-negative breast cancer was associated with a high risk of distant metastasis in their patients [18]. In addition, our web-based search (http://watson.compbio.iupui.edu/chirayu/proggene/database/index.php) of the literature indicated a marked reduction in both overall (Figure S7A) and relapse-free (Figure S7B) survival of breast cancer patients with elevated levels of ETV4 protein.
Figure 3.
Identification of the S100A8/A9-MCAM-ETV4-ZEB1 pathway. A, MDA-MB-231 cells were treated with S100A8/A9 (100 ng/ml) for regular time intervals, and the nuclei of the treated cells were collected at different time points, lysed and subjected to EMSA analysis. To clarify the ETV4-mediated band, an ETV4 antibody was used for a super shift experiment (left). MDA-MB-231 cells were transfected with MCAM dn plasmid, siCont., or siMCAM #1 and then treated with S100A8/A9 (100 ng/ml). ETV4 activity was monitored again by EMSA analysis for the treated cell nuclear extracts (right). B, A luciferase (luc)-based ZEB1 promoter reporter assay was performed in HEK293 cells after transfection with the reporter vector in combination with a control empty vector, ETV4 wt or ETV4 ΔDNA-BD expression vector. C, Quantitative real-time PCR analysis was carried out in the indicated cells for the ZEB-1 gene. The relative expression level of each sample is shown after calibration with the TBP value. D, MDA-MB-231 cells were transfected with GFP, MCAM dn, siMCAM #1 or MCAM cyt expression vector. The transfected cells were lysed and subjected to WB analysis to examine endogenous ZEB1 conditions. Data are means ± SD, ***P < .001.
At this point, we need to address how ETV4 is activated upon MCAM-S100A8/A9 binding. It is known that ERK1/2-mediated phosphorylation of ETV4 has a crucial role in the transcriptional activation of ETV4. We also found that ERK1/2 is highly activated by MCAM stimulation with S100A8/A9 (Figure S8). We therefore speculate that ERK1/2 activation under MCAM is at least partly responsible for triggering the ETV4 activation.
Our next interest was in ZEB1 activation through the S100A8/A9-MCAM-ETV4 axis. ETV4 and ZEB1 have long been independently recognized for their unusual roles in cancer aggressiveness [19], [20]. However, it has not been determined whether their roles are linked in some way. To determine whether ETV4 can lead to changes in the expression of target genes, we performed a ZEB1 promoter reporter assay. The results showed that the ETV4 wt, but not ETV4 ΔDNA-BD, induced promoter activation of ZEB1 at a much higher level than that induced by the control empty vector (Figure 3B). Interestingly, ZEB1 expression was markedly elevated in MDA-MB-231 cells (Figure 3C), which also showed critical expression levels of intrinsic MCAM (Figure 1C) and ETV4 (Figure S4B). These molecular patterns were also linked to cellular metastatic phenotypes (Figure 1, A and B). When we inhibited MCAM function using three different methods—i.e., MCAM dn-forced expression, siMCAM transduction and MCAM cyt-forced expression (a competitor of the MCAM signal)—we found that all three procedures inhibited ZEB1 expression in MDA-MB-231 cells (Fig. 3D). The identified pathway is likely to universally arise in triple-negative breast cancer cells. In this line of investigation, it also turned out that forced expression of ETV4 ΔDNA-BD effectively mitigated the S100A8/A9-induced migration of another triple-negative breast cancer cell line (Figure S9A). Concomitant with this, the S100A8/A9-induced upregulation of ETV4 activity (Figure S9B) and subsequent ZEB1 induction (Figure S9C) were both pronouncedly reduced by the forced expression of ETV4 ΔDNA-BD as well as MCAM knock down. Thus, we revealed for the first time that the MCAM-ETV4 signal is a crucial regulator of the ZEB1 gene that consistently occurs in triple-negative breast cancer cells.
EMT Phenotype Expression Through the MCAM-ETV4 Axis
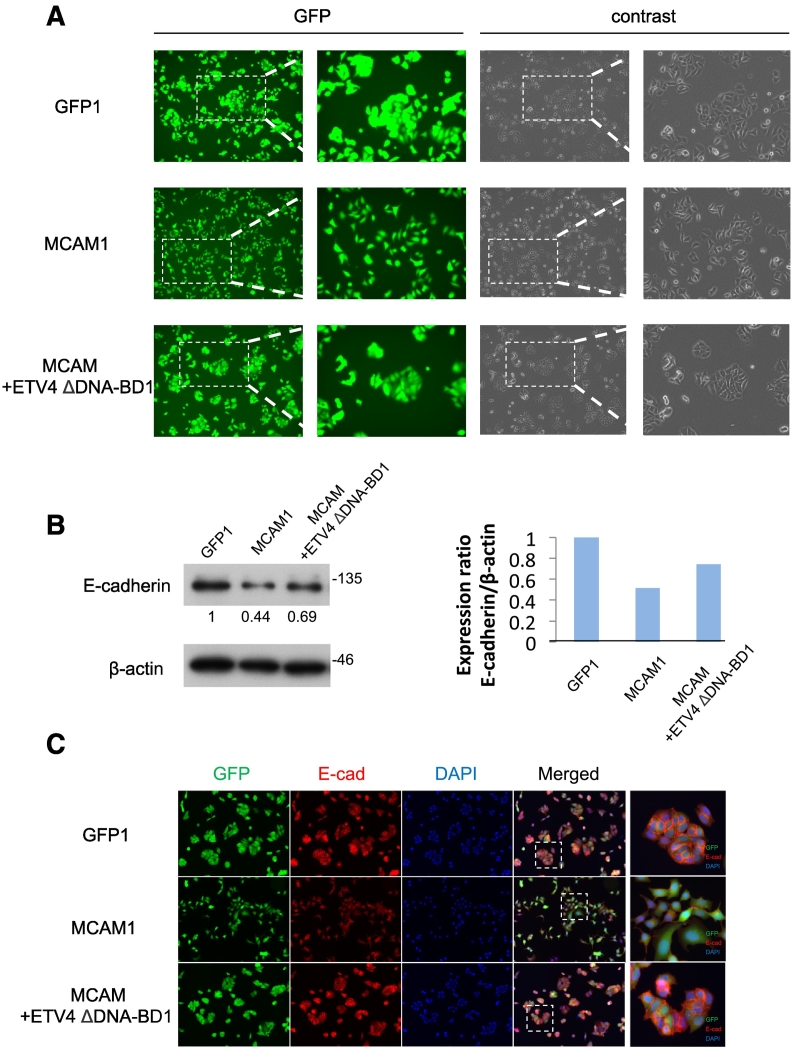
In order to investigate whether a biological connection truly exists between the identified pathway and the EMT phenomenon, we further established MCAM+ETV4 ΔDNA-BD-overexpressed stable sublines (#1, #2, #3 and #6) from MCF-7 cells (Figure S3B). The clones all showed relatively low levels of migration activity even at a higher level of foreign MCAM expression (Figure S3B, right) compared to those of the clones overexpressing MCAM alone (Figure S3B, left). When we closely examined the appearance of the individual cells in these established cell clones (GFP #1, MCAM #1, and MCAM+ETV4 ΔDNA-BD #1), we found that only the MCAM #1 clone was sparsely distributed in a culture space with a mesenchymal phenotype, while ETV4 ΔDNA-BD (MCAM+ETV4 ΔDNA-BD #1) never showed this activity, resulting in a phenotype similar to that of the GFP #1 clone, which shows an epithelial phenotype with tightly attached together (Figure 4A). We observed these same characteristics in all the clones that we established (data not shown). Since E-cadherin is a reliable epithelial marker that functions in cell-cell adhesion, we further examined the E-cadherin level in each of the clones as indicated in Figure 4A. In a reflection of the cellular phenotypes, we found that MCAM overexpression (MCAM #1 clone) reduced E-cadherin expression and that ETV4 ΔDNA-BD (MCAM+ETV4 ΔDNA-BD #1 clone) rescued E-cadherin from its own MCAM-mediated reduction (Figure 4, B and C). These results indicated that the S100A8/A9-MCAM-ETV4-ZEB1 pathway plays a major role in breast cancer migration concomitant with EMT, which may contribute to breast cancer malignancy. In fact, Jang et al. reported that increased expression of ZEB1 was correlated with abundant MCAM in triple-negative breast cancer cells, and was associated with poor clinical outcomes in patients with triple-negative breast cancer [21].
Figure 4.
EMT phenotype expression through the MCAM-ETV4 axis. A, Cell distributions in the MCF-7-derived clones, GFP #1, MCAM #1 and MCAM+ETV4 ΔDNA-BD #1 (see Figure S3), were monitored for their GFP expression by using a fluorescence microscope. B, The cell sublines were collected, lysed, and subjected to WB analysis to detect endogenous E-cadherin levels (left). The quantification of E-cadherin band intensities is shown after calibration with b-actin bands as an internal control. C, Immunofluorescence staining was carried out to detect E-cadherin.
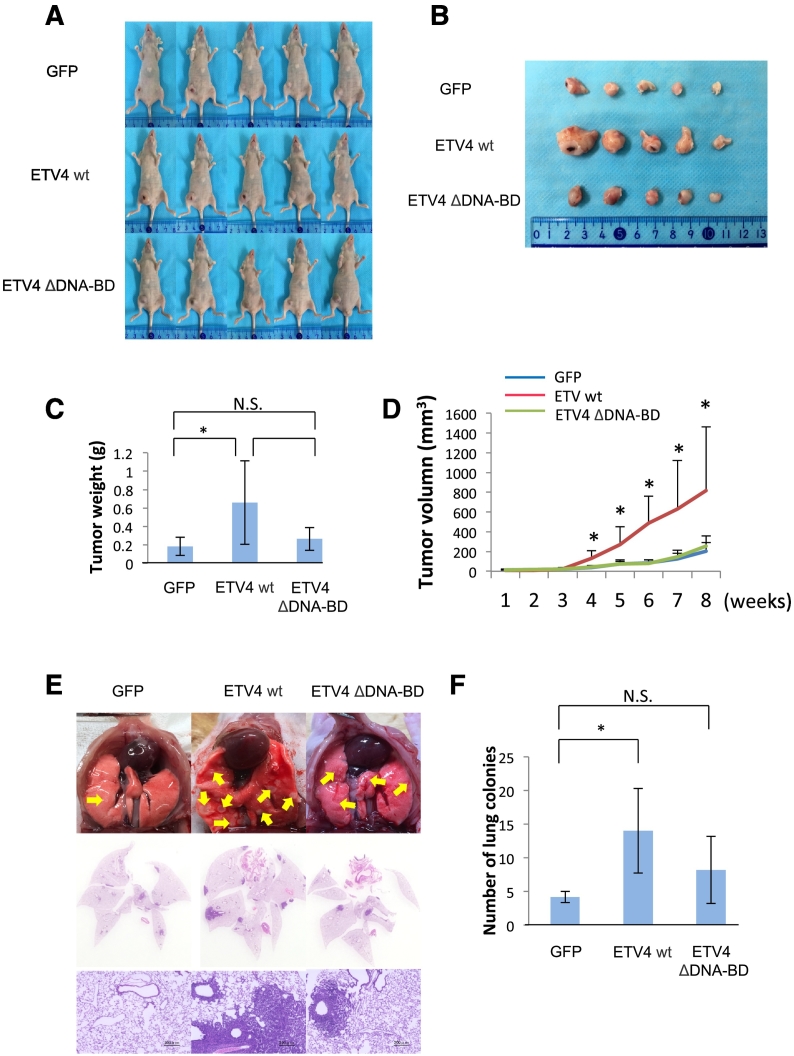
ETV4 is Essential for the Growth and Metastasis of Breast Cancer In Vivo
The biological role of ETV4 in breast cancer progression was further assessed in an in vivo setting using an orthotopic xenograft model (Figure 5). Overexpression of the ETV4 wt in MDA-MB-231 cells (ETV4 wt #1 clone) significantly promoted tumor growth (Figure 5, A and B) as evidenced by the larger tumor weight (Figure 5C) and faster growth kinetics (Figure 5D) compared to the GFP control group (GFP #1 clone), who were treated by mammary fat pad injection (Figure 5). These results support the potential positive correlation between ETV4 expression and worse clinical outcomes in breast cancer patients (Figure S7). On the other hand, ETV4 ΔDNA-BD (ETV4 ΔDNA-BD #1 clone) had no positive effect on tumor progression. To investigate whether ETV4 confers an increased metastatic potential other than tumor growth, we performed tail vein injection of MDA-MB-231 stable clones and assessed their lung colonization. Two months after injection, both macro images (Figure 5E, top panels) and hematoxylin-eosin staining (Figure 5E, middle and bottom panels) of the dissected lungs showed a much larger number of metastatic nodules in the ETV4 wt group compared with the GFP control or ETV4 ΔDNA-BD group (Figure 5F). These results indicate that ETV4 plays an important role in both tumor growth and metastasis in breast cancers. Overall, the results support the idea that the newly identified axis S100A8/A9-MCAM-ETV4 greatly contributes to the progression of breast cancers by inducing ETV4 target genes, including ZEB1, that enhance tumor growth and metastasis, and this axis may be linked to the poor survival of patients with breast cancers in which MCAM (Figure S1) and ETV4 (Figure S7) are jointly produced.
Figure 5.
ETV4 is essential for growth and metastasis of breast cancer in vivo. A, Representative imaging of mice bearing MDA-MB-231 stable clones (GFP #1, ETV4 wt #1 and ETV4 ΔDNA-BD #1) after mammary fat pad injection. n=5. B, Photograph of the dissected tumor. C, Weight of the dissected tumor. D, A tumor growth curve was established after injection, and the tumor diameter was measured every week. E, Each representative clone (5 x 105 cells) was injected into mice through the tail vein. Lung metastatic nodules were quantified after 2 months. Lung tumor nodules were observed macroscopically (top) and their sections were also observed by H&E staining (middle and bottom). n=5. F, Number of lung colonies in E. Data are means ± SD, N.S: not significant, *P < .05, **P < .01 and ***P < .001.
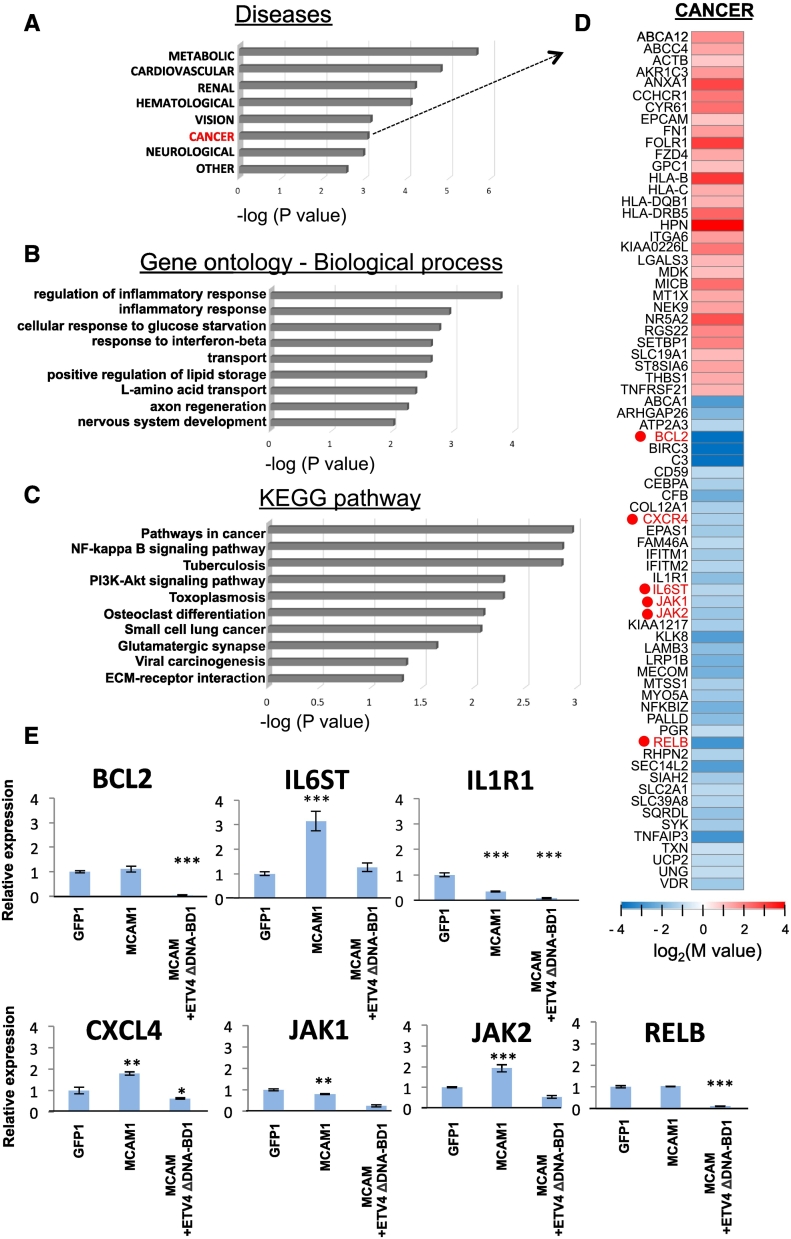
ETV4 Leads to a Global Reprogramming of Gene Expression
Finally, due to the critical function of ETV4 as a transcription factor in tumor progression, we attempted to gather information on unidentified genes targeted by ETV4 (other than ZEB1) in order to better understand the cancer-associated role of ETV4, which may also contribute to breast cancer aggressiveness coordinately with ZEB1. To identify other MCAM-ETV4 target genes relevant to breast cancer aggressiveness, we carried out a comprehensive analysis of gene expression in an MCF-7-derived stable clone, MCAM+ETV4 ΔDNA-BD #1, in comparison to that in the MCAM #1 clone (Figure 6). By this approach, we found that genes whose expressions were either increased or decreased in ETV4-preventive cells (MCAM+ETV4 ΔDNA-BD #1) were associated with cancer disease (Figure 6A, marked in red color), and more often associated with metabolic disease than with any other conditions (Figure 6A). When we focused on the genes for which expression was downregulated by ETV4 ΔDNA-BD overexpression, we found that many genes were associated with inflammation and glucose metabolism (Figure 6B). Since cancer cells frequently change their own metabolic pathway from aerobic to anabolic using the glycolytic system in response to characteristic alterations of the cancer-surrounding environments [22], ETV4 may play a significant role in the regulation of cancer metabolisms. Analysis of the altered genes in expression on the KEGG pathway also yielded the important finding that a number of cancer-related pathways, including NFκB and Akt, and leading to small lung cancer and viral carcinogenesis, were all associated with ETV4 (Figure 6C). Thus, the reason for the critical role of ETV4 in breast cancer progression may be that ETV4 induces a large number of cancer-associated molecules (Figure 6D and 6E) and establishes the pathways involving them, and these pathways in turn may function coordinately.
Figure 6.
RNA-seq analysis. A, Functional enrichment analysis (P < .05) was performed for protein-coding RNAs in GAD_DISEASE_CLASS with upregulated and downregulated genes in the MCF-7-derived MCAM+ETV4 ΔDNA-BD #1 clone in comparison to those in the MCAM #1 clone. B and C, For the downregulated genes, functional enrichment analysis (P < .05) was also performed in GO (B) and KEGG (C) analysis. D, Heat maps are displayed for the selected genes that are upregulated and downregulated in the cancer category of the disease clustering as indicated in (A). These analyses were performed using the database for annotation, visualization and integrated discovery (DAVID) v6.8 (http://david.ncifcr.gov/). E, Quantitative real-time PCR analysis was carried out in the indicated cells for the genes shown in (D). The relative expression level of each sample is shown after calibration with the TBP (a suitable housekeeping gene) value. Data are means ± SD, *P < .05, **P < .01 and ***P < .001. ND: not detected.
Discussion
Breast cancer is a major cause of cancer-related mortality in women worldwide. The vast majority of deaths are attributed to metastasis. The key factors in breast cancer tumorigenesis and metastasis are cytokines in the tumor microenvironment. In normal tissue, cytokines have a growth inhibitory role in the epithelium [23]. However, in breast cancer, through a series of genetic and epigenetic changes, and autocrine and paracrine signals, the tumor microenvironment is activated, losing its protective effects and becoming the major factor in the initiation and the progression of the tumors [24], [25], [26], [27]. In our study, we demonstrated that the cytokine S100A8/A9 is a novel key regulator that binds to MCAM, leading to significant activation of the oncogenetic transcription factor ETV4, and eventually promoting activation of the EMT marker ZEB1. We found that induction of a remarkable level of ETV4 was a cause of the onset of breast cancer dissemination and subsequent distant lung metastasis.
MCAM (or CD146, Mel-CAM, MUC18, S-endo1), also known as the melanoma cell adhesion molecule, was first described in malignant melanomas, where it was expressed at high levels and found to be associated with poor prognosis [28], [29]. More recently, high-level expression of MCAM has been associated with the metastatic progression in hepatocellular carcinoma [12], [30] and ovarian cancer [31]. The presence of several protein kinase recognition motifs in the cytoplasmic domain suggests the involvement of MCAM in downstream cell signaling [32]. In a recent study, we showed that one of the mitogen-activated protein kinase kinase kinases (MAP3Ks), which directly binds to MCAM, plays a key role in the activation of malignant melanoma metastasis. This finding revealed a distinctive function of MCAM other than adhesion. To determine the mechanism(s) by which MCAM modulates metastasis, we applied three different techniques to inhibit the function of MCAM. We found that each of these inactivated forms of MCAM—i.e., MCAM inactivated by transduction with a dominant negative vector, siRNA or MCAM cytoplasmic tail vector—abrogated the S100A8/A9-induced expression of ETV4 (Figure 3A, right), probably in an ERK1/2-dependent manner (Figure S8). Since robust ERK1/2 activation was observed in S100A8/A9-MCAM-induced breast cancer metastasis, we postulated that ETV4, a positive regulator of ERK/MAPK, is involved in the migration of cancer cells. This speculation was also supported by the results of a recent study. Namely, Fung reported that a novel signal pathway, ERK-MAPK-ETV4-MMP-E-cadherin, drove the malignance of esophageal carcinoma [33]. Consistent with their findings, we clearly demonstrated that MCAM exacerbated breast cancer migration, and this effect was associated with enhanced ERK1/2-ETV4 activation.
Our observations indicate that ETV4 transcription factor that is positively regulated by MCAM may be a key nuclear effector of oncogenic MAPK signaling. Indeed, the results of a prior study suggested that ETV4 may activate a RAS and MAPK transcriptional program in the absence of the MAPK pathway [34]. ETV4 is known to be involved in events participating in tumor development and progression, indicating that ETV4 has a function similar to that of MCAM. In this study, we demonstrated that ETV4 is activated by MCAM, a conclusion that appears to be supported by the analogous overall survival of breast cancer patients with MCAM-activated ETV4 from public databases (Figure S1 and Figure S7). ETV4 is also known to be a significant mediator of tumorigenesis through the activation of several downstream pathways that are associated with migration, invasion and stemness [35], [36]. For instance, inhibition of ETV4 in gastric carcinoma and pancreatic carcinoma significantly impairs the invasive capacity of cancer cells [20], [37]. Our data demonstrated that ZEB1 expression at the mRNA level is positively correlated with ETV4 expression levels and ectopic overexpression of the ETV4 wt in MDA-MB-231 cells further promoted their cancer progression in an orthotopic mouse model (Figure 5). As an epithelial-to-mesenchymal transition (EMT) marker, the importance of ZEB1 has been well documented. However, the role of EMT in activating metastasis has recently been challenged [38], [39], with the Snail and Twist EMT transcription factors increasingly thought to play key roles in breast and pancreatic cancer. In contrast, our results clearly described that ZEB1 stimulates breast cancer lung metastasis based on ETV4 activation.
One of the most striking consequences of our findings concerns the critical function of ETV4. The regulatory potential of ETV4 is not limited to effects on a few crucial downstream target genes but rather leads to a global reprogramming of gene expression patterns, and controls not only EMT but also other programs and pathways. The results of our RNA-seq analysis (Figure 6) brought many genes of interest to our attention (Figure 6D). Next, therefore, we examined the expression profiles of key genes in each of the clones (Figure 6E). Due to the strong anti-apoptotic function of BCL2 and survival function of RELB, a component of NFκB, both BCL2 and RELB may confer the survival ability to breast cancer cells through ETV4. For the acquisition of metastatic ability, it has long been recognized that the Cxcl12 receptor, C-X-C chemokine receptor 4 (CXCR4), promotes breast cancer cell metastasis to the lung. In addition, IL6ST, JAK1 and JAK2 are also interesting, since these molecules play a critical role in STAT3 activation. Accumulating evidence has indicated that the transcription factor STAT3 promotes breast cancer aggressiveness [40]. The predicted ETV4-targeted genes, which are positively regulated by the MCAM-ETV4 pathway, may cooperate with ZEB1 to induce breast cancer progression. Further studies are required to determine the association between these molecules in the MCAM-ETV4 pathway and breast cancer progression.
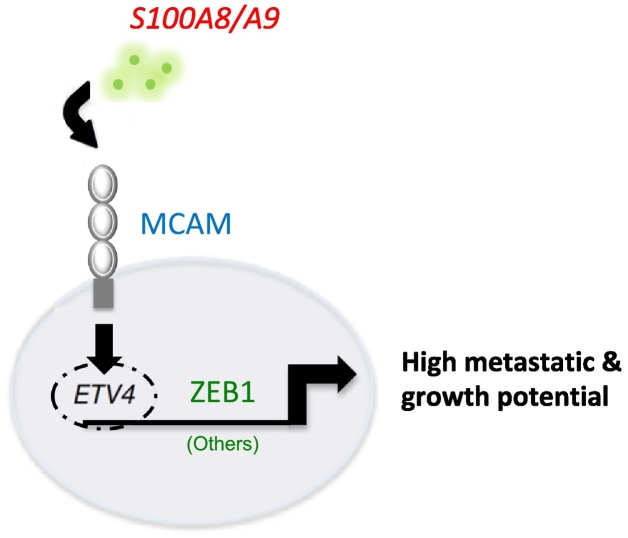
Taken together, the results of our study revealed a critical role of MCAM in breast cancer upon S100A8/A9 binding extracellularly, indicating an essential function of inflammatory cytokines in cancer-associated microenvironments. Downstream signaling analysis showed that MCAM promotes activation of ETV4, which further modulates ZEB1 and others transcriptionally (Figure 7). Inhibition of this pathway would be a promising therapeutic alternative in breast cancer treatment.
Figure 7.
Graphical schematic of breast cancer metastasis based on the S100A8/A9-MCAM-ETV4-ZEB1 axis.
Conclusions
Here we showed that the S100A8/A9-MCAM-mediated signal pathway leads to an aggressive metastasis of breast cancer to the lung in a tumor-bearing mouse. That the identified molecules were activated one after another in response to MCAM stimulation via S100A8/A9 was a novel finding, and the revealed signaling pathway showed that MCAM promotes activation of ETV4, which further modulates ZEB1 (Figure 7). We hope that our findings will lead to a better understanding of the physiological and pathological mechanisms in breast cancer metastasis, and suppression of the identified pathway might be an effective approach for prevention of metastasis by targeting the S100A8/A9-MCAM axis in breast cancer and probably other cancers.
Materials and Methods
Cell Lines
The following cell lines were used in this study: HEK293T (a human embryonic kidney cell line stably expressing the SV40 large T antigen; RIKEN BioResource Center, Tsukuba, Japan), MCF-10A (a non-tumorigenic human breast epithelial cell line; ATCC, Rockville, MD), MCF-7 (a human breast cancer cell line; ATCC), SK-BR-3 (a human breast cancer cell line; ATCC), YMB-1-E (a human breast cancer cell line; JCRB Cell Bank, Tokyo), and MDA-MB-231 (a human breast cancer cell line; ATCC). MCF-10A cells were cultured in DMEM/Ham's F-12 (D/F) medium (Thermo Fisher Scientific, Waltham, MA) supplemented with 100 ng/ml cholera toxin, 20 ng/ml epidermal growth factor (EGF), 0.01 mg/ml insulin, 500 ng/ml hydrocortisone and 5% FBS (Intergen, Purchase, NY). HEK293T, MCF-7, SK-BR-3, YMB-1-E and MDA-MB-231 cells were all cultivated in D/F medium (Thermo Fisher Scientific) supplemented with 10% FBS (Intergen).
Recombinant Proteins
Human S100A8/A9 recombinant protein was prepared from the FreeStyle 293 Expression System (Thermo Fisher Scientific) as previously reported [10]. The exMCAM-Fc decoy protein was prepared from its transduced CHO-S stable clone conditioned media. The CHO clone was established by a convenient electroporation gene delivery method using an improved plasmid named pSAKA-1B [9] based on the pIDT-SMART (C-TSC) vector [41]. After collection of the serum-free conditioned medium from large-scale culture of the established CHO-S clone, exMCAM-Fc was purified by protein-G affinity chromatography according to the manufacturer’s instructions.
Plasmids
The pIDT-SMART (C-TSC) vector, abbreviated as pCMViR-TSC, was used for temporal overexpression of foreign genes. The cDNAs inserted into the vector were as follows: human cDNAs encoding MCAM, MCAM dn (deletion of cytoplasmic tail: D 584-646 aa), MCAM cyt (cytoplasmic tail: 584-646 aa), ETS family genes (ETV1, ETV4, ETV5, ELK3), and ETV4 ΔDNA-BD (deletion of C-terminal DNA-binding domain: D 342-484 aa). MCAM, MCAM dn and MCAM cyt were designed for expression as C-terminal 3xHA-6His-tagged forms. ETS family genes and ETV4 ΔDNA-BD were designed for expression as C-terminal 3xMyc-6His-tagged forms. The cells were transiently transfected with the plasmid vectors using FuGENE-HD (Promega, Madison, WI).
To obtain stable transformants that show significantly high expression levels of the transduced genes in a sustained manner, we used another improved vector based on pIDT-SMART (C-TSC), named pSAKA-4B, which is useful for a broad range of human cell lines but not rodent CHO cells. MCAM-3xHA-6His and ETV4 ΔDNA-BD -3xMyc-6His were inserted into pSAKA-4B [42]. A series of clones (see Figure S3) was established by a convenient electroporation gene delivery method and subsequent selection with puromycin at 20 μg/ml.
siRNA
Human MCAM siRNA (#1, ID No. s8571; #2, ID No. s8572) and control siRNA (Silencer Negative control siRNA #1) were purchased from Thermo Fisher Scientific. The siRNAs were transfected using Lipofectamin RNAiMAX reagent (Thermo Fisher Scientific).
RNA-seq Based Analysis
Total RNA was extracted from cells with ISOGEN (Nippon Gene, Tokyo) according to the manufacturer’s instructions. The isolated RNA was subjected to RNA-seq-based analysis of gene expression (Bioengineering Lab, Kanagawa, Japan).
Quantitative Real-Time PCR
Real-time RT-PCR was performed in a LightCycler rapid thermal cycler system (ABI 7900HT; Applied Biosystems, Foster City, CA) using a LightCycler 480 SYBR Green I Master Kit (Roche Diagnostics, Indianapolis, IN) according to the manufacturer’s instructions. The forward and reverse primer pairs used (5’ to 3’) are shown in Supplementary Table S3.
Western Blot Analysis
Western blotting was performed under conventional conditions. The antibodies used were as follows: rabbit anti-ZEB1 antibody (Proteintech Japan, Tokyo), mouse anti-E-cadherin antibody (BD Biosciences, Bedford, MA), mouse anti-HA tag antibody (Cell Signaling Technology, Beverly, MA), mouse anti-Myc antibody (Cell Signaling Technology) and mouse anti-human β-actin antibody (Sigma-Aldrich). The secondary antibody was horseradish peroxidase-conjugated anti-mouse or anti-rabbit IgG antibody (Cell Signaling Technology).
Luciferase Assay
The human ZEB1 (3391 bp with SacI and HindIII sites from the translation start site) promoter was amplified with genomic DNA from HepG2 cells by PCR and cloned into the luciferase reporter vector pGL4.11 (Promega). The primers used to amplify the promoter were 5′-caaggataaaataagataaaatcagc-3′ and 5′-aaagccacatcagcaacagc-3′.
pGL4.11 carrying the human ZEB1 promoter and pGL4.74 encoding Renilla luciferase regulated under the HSV-TK promoter as an internal control were transfected into HEK293T cells with polyethyleneimine Max (Polyscience, Warrington, PA) as a transfection reagent. For co-transfection, pCMViR-TSC vectors carrying ETV4 expression plasmids (wt and ΔDNA-BD) were used for monitoring the ZEB1 promoter. After 48 h, transfected cells were measured using a Dual-Glo Luciferase Assay System (Promega).
Electrophoretic Mobility Shift Assay (EMSA)
Cell nuclear extracts were prepared according to the manufacturer’s instructions by using NE-PER nuclear and cytoplasmic extraction reagents (Thermo Fisher Scientific), and EMSA was performed using a LightShift Chemiluminescent EMSA kit (Thermo Fisher Scientific). A 5′-biotin-labeled double-stranded probe (forward: 5′-agtacCGGAAGTaca-3′; reverse: 3′-tgtACTTCCGgtact-5′) was purchased from Sigma-Aldrich. Rabbit anti-human PEA3/ETV4 antibody (Novus, Littleton, CO) was used for super shift analysis. After the reactions, all samples were fractionated by 7% PAGE and blotted onto a Biodyne B nylon membrane (Pall, Tokyo).
Cell Migration and Invasion
Migration and invasion assays were performed by the Boyden chamber method. For the invasion assay, 8-μm pore filters set in Transwell culture inserts (BD Biosciences) were coated with Matrigel. Cells were placed in the upper chamber in a low-serum medium, D/F (0.5% FBS), and the lower chamber was filled with high-serum medium, D/F (10% FBS), in the presence or absence of S100A8/A9 recombinant protein (100 ng/ml). After 24 hours (for MDA-MB-231 cells) and after 48 hours (for MCF-7 cells), cells that had passed through the membrane were stained with hematoxylin eosin (H&E) (Muto Pure Chemicals, Tokyo). Each transwell insert was microscopically imaged in five distinct regions at 10× in triplicate. The numbers of cells that had migrated to the five distinct regions were counted using the software (BZ-analysis application; Keyence) and summed as the total cell number.
Animal Experiments
Experimental protocols were approved by the Animal Experiment Committee of Okayama University (approval no. OKU-2014011). All of the mouse procedures and euthanasia, including cell transplantations, were done painlessly or under anesthesia according to the strict guidelines of the Experimental Animal Committee of Okayama University. To examine tumor metastasis in vivo, MDA-MB-231 cells and the clones from which they originated were used in animal experiments. The established clones (5x105 cells) were injected into the mammary fad pads of Balb/c nude mice (Charles River Laboratories, Yokohama, Japan) at the age of 6 weeks. Two months later, the mice were sacrificed and the tumors were dissected out and examined. As a lung metastasis model, 5x105 cells re-suspended in 100 μl of sterile PBS were injected into the tail veins of nude mice. Lung colonization was monitored at day 60. The distribution of the metastasized cancer cells in the whole lung was observed after HE staining.
Statistical Analysis
All values are expressed as means ± SD. All data were analyzed by unpaired Student's t-test for significant differences between the mean values of each group.
Acknowledgments
Acknowledgement
This research was supported in part by JSPS KAKENHI Grant Number 17H03577 to M.S., and by funds to M.S. from the Smoking Research Foundation, the Terumo Life Science Foundation, and the Takeda Science Foundation.
Authors’ contributions
Conception: Masakiyo Sakaguchi
Funding acquisition: Masakiyo Sakaguchi
Study supervision: Masakiyo Sakaguchi
Design of strategy: Youyi Chen, Toshihiko Hibino and Masakiyo Sakaguchi
Establishment of key cell sublines: Youyi Chen
Acquisition of key data through studies: Youyi Chen
Contribution to in vitro data collection: Youyi Chen, I Wayan Sumardika, Nahoko Tomonobu, Rie Kinoshita, Yusuke Inoue, Yosuke Mitsui, I Made Winarsa Ruma and Junichi Soh
Contribution to in vivo data collection: Youyi Chen, I Wayan Sumardika, Hitoshi Murata, Ken-ichi Yamamoto, Miyoko Kubo, Endy Widya Putranto, Masahiro Nishibori and Shinichi Toyooka
Data analysis: Youyi Chen, Nahoko Tomonobu, Rie Kinoshita, Yosuke Mitsui, I Made Winarsa Ruma, Akira Yamauchi, Hitoshi Murata, Shuta Tomida, Kazuhiko Shien and Endy Widya Putranto
Methodology: Youyi Chen, Nahoko Tomonobu, Rie Kinoshita, I Made Winarsa Ruma, Hidekazu Iioka, Ken Saito, Eisaku Kondo Hiroki Sato, Akira Yamauchi, Hitoshi Murata, Shuta Tomida, Kazuhiko Shien, Hiromasa Yamamoto, Junichi Soh, Junichiro Futami, Miyoko Kubo, Endy Widya Putranto, Ming Liu, Masahiro Nishibori, Shinichi Toyooka and Masakiyo Sakaguchi
Project administration: Youyi Chen, Rie Kinoshita, Yusuke Inoue, Hitoshi Murata, Hiromasa Yamamoto, Ming Liu, Masahiro Nishibori, Shinichi Toyooka and Masakiyo Sakaguchi
Experimental Resources: Youyi Chen, I Wayan Sumardika, Rie Kinoshita, Hitoshi Murata, Ken-ichi Yamamoto, Kazuhiko Shien, Junichiro Futami, Endy Widya Putranto, Ming Liu, Masahiro Nishibori, Shinichi Toyooka and Masakiyo Sakaguchi
Luc-expressing cell lines: Takashi Murakami
Software: Youyi Chen
Validation evaluation: Youyi Chen and Masakiyo Sakaguchi
Visualization: Youyi Chen, Masahiro Nishibori and Shinichi Toyooka
Writing: Youyi Chen and Masakiyo Sakaguchi
Conflict-of-interest disclosure statement
The authors declare that they have no competing interest.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.neo.2019.04.006.
Appendix A. Supplementary Data
Supplementary tables
Fig. S1. Survival curves of patients bearing breast cancers with MCAM at a high or low level. A and B, Overall (A) and metastasis-free (B) survival plots according to MCAM expression levels were obtained from a publicly available web site (http://watson.compbio.iupui.edu/chirayu/proggene/database/index.php).
Fig. S2. Validation of the knockdown effect of MCAM siRNA. MDA-MB-231 cells were transfected with either MCAM siRNAs (siMCAM#1, siMCAM#2) or a negative control scrambled siRNA (siCont.), and cellular MCAM protein was detected by WB.
Fig. S3. Establishment of MCF-7-derived sublines that stably express foreign GFP, MCAM or MCAM+ETV4 ΔDNA-BD. A and B, Selected clones resistant to puromycin selection were examined for the transduced foreign genes including GFP alone (A, left), MCAM alone (B, top left) and MCAM+ETV4 ΔDNA-BD (B, top right). Four of each of the clones that were positive for the transduced genes were randomly chosen as indicated by arrowheads, resulting in selection of GFP (#1, #2, #3 and #8), MCAM (#1, #2, #3 and #6) and MCAM+ETV4 ΔDNA-BD (#1, #2, #3 and #6) clones. The selected clones were all examined for their migration ability (A, right; B, bottom). Data are means ± SD, *P < .05, **P < .01 and ***P < .001.
Fig. S4. Screening of a candidate transcription factor(s) that is important in breast cancer migration through MCAM. A, MDA-MB-231 cells were transfected with the indicated decoy DNA oligonucleotides that compete with host DNA cis-elements via binding with their target transcription factors. B, Quantitative real-time PCR analysis was carried out in the indicated cells for ETS family genes. The relative expression level of each sample is shown after calibration with TBP (a suitable housekeeping gene) value. Data are means ± SD, *P < .05, **P < .01 and ***P < .001. ND: not detected.
Fig. S5. Identification of the gene(s) to induce MCAM-mediated migration from the four selected ETS candidates (ETV1, ETV4, ETV5 and ELK3). A and B, An MCF-7-derived MCAM-overexpressed clone, MCAM #1, was temporarily transfected with the four selected ETS candidates (ETV1, ETV4, ETV5 and ELK3), and then its migration (A) and expression of EMT-relevant genes (B) were examined by the Boyden chamber assay and quantitative real-time PCR analysis, respectively. Data are means ± SD, *P < .05, **P < .01 and ***P < .001.
Fig. S6. Identification of an EMT-relevant gene, ZEB1, that is regulated by the MCAM-ETV4 pathway. A, Quantitative real-time PCR analysis was carried out for the CDH2 (N-cadherin), TWIST1 and ZEB1 genes in MCF-7 cells after transfection with an empty vector (EV), and with the MCAM, ETV4 wt and ETV4 ΔDNA-BD expression vectors. The relative expression level of each sample is shown after calibration with the TBP value. B, MDA-MB-231 cells were temporarily transfected with the GFP, MCAM dn and MCAM+ETV4 ΔDNA-BD expression vectors, respectively. The migration ability of the transduced cells was evaluated by Boyden chamber assay. Data are means ± SD, *P < .05, **P < .01 and ***P < .001.
Fig. S7. Survival curves of patients bearing breast cancers with ETV4 expression at a high or low level. A and B, Overall (A) and relapse-free (B) survival plots according to the ETV4 expression levels were obtained from a publicly available web site (http://watson.compbio.iupui.edu/chirayu/proggene/database/index.php).
Fig. S8. Down-regulation of MCAM inhibits ERK1/2 phosphorylation. A, MDA-MB-231 cells were transfected with either siMCAM or a negative control scrambled siCont, and cellular MCAM, p-ERK1/2 and t-ERK1/2 in the treated cells were detected by WB.
Fig. S9. The S100A8/A9-MCAM-ETV4-ZEB1 pathway in triple-negative breast cancer cell lines. A, Three luciferase (Luc)-expressed triple-negative breast cancer cell lines, MDA-MD-231-Luc, Hs578T-Luc and BT-549-Luc, were obtained from the JCRB Cell Bank (Tokyo, Japan). Cellular migration was monitored according to the Boyden chamber method. These Luc-expressed cell lines were transiently transfected with either control GFP or ETV4 ΔDNA-BD plasmid, and the transfected cells were placed at the top of a transwell chamber. Then they were stimulated by S100A8/A9 (100 ng/ml) for 24 h at the bottom of the chamber. Data are means ± SD, *P < .05 and **P < .01, N.S.: not significant. B, The four prepared cell lines (MDA-MB-231, MDA-MD-231-Luc, Hs578T-Luc and BT-549-Luc) were transfected with siCont., siMCAM #1, GFP plasmid or ETV4 ΔDNA-BD plasmid and then they were treated with S100A8/A9 (100 ng/ml) for 6 h (top) or 12 h (bottom). The ETV4 activities in the prepared nuclear extracts were monitored by EMSA analysis. C, Cells prepared by a method similar to that described in B (bottom) were also subjected to WB analysis of their whole cell lysates to examine endogenous ZEB1 levels.
References
- 1.DeSantis CE, Ma J, Goding Sauer A, Newman LA, Jemal A. Breast cancer statistics, 2017, racial disparity in mortality by state. CA Cancer J Clin. 2017;67(6):439–448. doi: 10.3322/caac.21412. [DOI] [PubMed] [Google Scholar]
- 2.Paget S. The distribution of secondary growths in cancer of the breast. 1889. Cancer Metastasis Rev. 1989;8(2):98–101. [PubMed] [Google Scholar]
- 3.Eisenblaetter M, Flores-Borja F, Lee JJ, Wefers C, Smith H, Hueting R, Cooper MS, Blower PJ, Patel D, Rodriguez-Justo M. Visualization of Tumor-Immune Interaction - Target-Specific Imaging of S100A8/A9 Reveals Pre-Metastatic Niche Establishment. Theranostics. 2017;7(9):2392–2401. doi: 10.7150/thno.17138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hiratsuka S, Watanabe A, Sakurai Y, Akashi-Takamura S, Ishibashi S, Miyake K, Shibuya M, Akira S, Aburatani H, Maru Y. The S100A8-serum amyloid A3-TLR4 paracrine cascade establishes a pre-metastatic phase. Nat Cell Biol. 2008;10(11):1349–1355. doi: 10.1038/ncb1794. [DOI] [PubMed] [Google Scholar]
- 5.Mishra R, Thorat D, Soundararajan G, Pradhan SJ, Chakraborty G, Lohite K, Karnik S, Kundu GC. Semaphorin 3A upregulates FOXO 3a-dependent MelCAM expression leading to attenuation of breast tumor growth and angiogenesis. Oncogene. 2015;34(12):1584–1595. doi: 10.1038/onc.2014.79. [DOI] [PubMed] [Google Scholar]
- 6.Zeng Q, Zhang P, Wu Z, Xue P, Lu D, Ye Z, Zhang X, Huang Z, Feng J, Song L. Quantitative proteomics reveals ER-alpha involvement in CD146-induced epithelial-mesenchymal transition in breast cancer cells. J Proteome. 2014;103:153–169. doi: 10.1016/j.jprot.2014.03.033. [DOI] [PubMed] [Google Scholar]
- 7.Zeng Q, Li W, Lu D, Wu Z, Duan H, Luo Y, Feng J, Yang D, Fu L, Yan X. CD146, an epithelial-mesenchymal transition inducer, is associated with triple-negative breast cancer. Proc Natl Acad Sci U S A. 2012;109(4):1127–1132. doi: 10.1073/pnas.1111053108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hibino T, Sakaguchi M, Miyamoto S, Yamamoto M, Motoyama A, Hosoi J, Shimokata T, Ito T, Tsuboi R, Huh NH. S100A9 is a novel ligand of EMMPRIN that promotes melanoma metastasis. Cancer Res. 2013;73(1):172–183. doi: 10.1158/0008-5472.CAN-11-3843. [DOI] [PubMed] [Google Scholar]
- 9.Sakaguchi M, Yamamoto M, Miyai M, Maeda T, Hiruma J, Murata H, Kinoshita R, Winarsa Ruma IM, Putranto EW, Inoue Y. Identification of an S100A8 Receptor Neuroplastin-β and its Heterodimer Formation with EMMPRIN. J Invest Dermatol. 2016;136(11):2240–2250. doi: 10.1016/j.jid.2016.06.617. [DOI] [PubMed] [Google Scholar]
- 10.Ruma IM, Putranto EW, Kondo E, Murata H, Watanabe M, Huang P, Kinoshita R, Futami J, Inoue Y, Yamauchi A. MCAM, as a novel receptor for S100A8/A9, mediates progression of malignant melanoma through prominent activation of NF-κB and ROS formation upon ligand binding. Clin Exp Metastasis. 2016;33(6):609–627. doi: 10.1007/s10585-016-9801-2. [DOI] [PubMed] [Google Scholar]
- 11.Hayward NK, Wilmott JS, Waddell N, Johansson PA, Field MA, Nones K, Patch AM, Kakavand H, Alexandrov LB, Burke H. Whole-genome landscapes of major melanoma subtypes. Nature. 2017;545(7653):175–180. doi: 10.1038/nature22071. [DOI] [PubMed] [Google Scholar]
- 12.Wang J, Tang X, Weng W, Qiao Y, Lin J, Liu W, Liu R, Ma L, Yu W, Yu Y. The membrane protein melanoma cell adhesion molecule (MCAM) is a novel tumor marker that stimulates tumorigenesis in hepatocellular carcinoma. Oncogene. 2015;34(47):5781–5795. doi: 10.1038/onc.2015.36. [DOI] [PubMed] [Google Scholar]
- 13.Tripathi SC, Fahrmann JF, Celiktas M, Aguilar M, Marini KD, Jolly MK, Katayama H, Wang H, Murage EN, Dennison JB. MCAM Mediates Chemoresistance in Small-Cell Lung Cancer via the PI3K/AKT/SOX2 Signaling Pathway. Cancer Res. 2017;77(16):4414–4425. doi: 10.1158/0008-5472.CAN-16-2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen Y, Sumardika IW, Tomonobu N, Winarsa Ruma IM, Kinoshita R, Kondo E, Inoue Y, Sato H, Yamauchi A, Murata H. Melanoma cell adhesion molecule is the driving force behind the dissemination of melanoma upon S100A8/A9 binding in the original skin lesion. Cancer Lett. 2019;452:178–190. doi: 10.1016/j.canlet.2019.03.023. [DOI] [PubMed] [Google Scholar]
- 15.Yuen HF, Chan YK, Grills C, McCrudden CM, Gunasekharan V, Shi Z, Wong AS, Lappin TR, Chan KW, Fennell DA. Polyomavirus enhancer activator 3 protein promotes breast cancer metastatic progression through Snail-induced epithelial-mesenchymal transition. J Pathol. 2011;224(1):78–89. doi: 10.1002/path.2859. [DOI] [PubMed] [Google Scholar]
- 16.Jiang J, Wei Y, Liu D, Zhou J, Shen J, Chen X, Zhang S, Kong X, Gu J. E1AF promotes breast cancer cell cycle progression via upregulation of Cyclin D3 transcription. Biochem Biophys Res Commun. 2007;358(1):53–58. doi: 10.1016/j.bbrc.2007.04.043. [DOI] [PubMed] [Google Scholar]
- 17.Bieche I, Tozlu S, Girault I, Onody P, Driouch K, Vidaud M, Lidereau R. Expression of PEA3/E1AF/ETV4, an Ets-related transcription factor, in breast tumors: positive links to MMP2, NRG1 and CGB expression. Carcinogenesis. 2004;25(3):405–411. doi: 10.1093/carcin/bgh024. [DOI] [PubMed] [Google Scholar]
- 18.Yuan ZY, Dai T, Wang SS, Peng RJ, Li XH, Qin T, Song LB, Wang X. Overexpression of ETV4 protein in triple-negative breast cancer is associated with a higher risk of distant metastasis. Onco Targets Ther. 2014;7:1733–1742. doi: 10.2147/OTT.S66692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Keld R, Guo B, Downey P, Cummins R, Gulmann C, Ang YS, Sharrocks AD. PEA3/ETV4-related transcription factors coupled with active ERK signalling are associated with poor prognosis in gastric adenocarcinoma. Br J Cancer. 2011;105(1):124–130. doi: 10.1038/bjc.2011.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krebs AM, Mitschke J, Lasierra Losada M, Schmalhofer O, Boerries M, Busch H, Boettcher M, Mougiakakos D, Reichardt W, Bronsert P. The EMT-activator Zeb1 is a key factor for cell plasticity and promotes metastasis in pancreatic cancer. Nat Cell Biol. 2017;19(5):518–529. doi: 10.1038/ncb3513. [DOI] [PubMed] [Google Scholar]
- 21.Jang MH, Kim HJ, Kim EJ, Chung YR, Park SY. Expression of epithelial-mesenchymal transition-related markers in triple-negative breast cancer: ZEB1 as a potential biomarker for poor clinical outcome. Hum Pathol. 2015;46(9):1267–1274. doi: 10.1016/j.humpath.2015.05.010. [DOI] [PubMed] [Google Scholar]
- 22.Lehuede C, Dupuy F, Rabinovitch R, Jones RG, Siegel PM. Metabolic plasticity as a determinant of tumor growth and metastasis. Cancer Res. 2016;76(18):5201–5208. doi: 10.1158/0008-5472.CAN-16-0266. [DOI] [PubMed] [Google Scholar]
- 23.Kaukonen R, Mai A, Georgiadou M, Saari M, De Franceschi N, Betz T, Sihto H, Ventela S, Elo L, Jokitalo E. Normal stroma suppresses cancer cell proliferation via mechanosensitive regulation of JMJD1a-mediated transcription. Nat Commun. 2016;7(12237) doi: 10.1038/ncomms12237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kojima Y, Acar A, Eaton EN, Mellody KT, Scheel C, Ben-Porath I, Onder TT, Wang ZC, Richardson AL, Weinberg RA. Autocrine TGF-beta and stromal cell-derived factor-1 (SDF-1) signaling drives the evolution of tumor-promoting mammary stromal myofibroblasts. Proc Natl Acad Sci U S A. 2010;107(46):20009–20014. doi: 10.1073/pnas.1013805107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nakamura T, Matsumoto K, Kiritoshi A, Tano Y, Nakamura T. Induction of hepatocyte growth factor in fibroblasts by tumor-derived factors affects invasive growth of tumor cells: in vitro analysis of tumor-stromal interactions. Cancer Res. 1997;57(15):3305–3313. [PubMed] [Google Scholar]
- 26.Kurose K, Gilley K, Matsumoto S, Watson PH, Zhou XP, Eng C. Frequent somatic mutations in PTEN and TP53 are mutually exclusive in the stroma of breast carcinomas. Nat Genet. 2002;32(3):355–357. doi: 10.1038/ng1013. [DOI] [PubMed] [Google Scholar]
- 27.Hu M, Yao J, Cai L, Bachman KE, van den Brule F, Velculescu V, Polyak K. Distinct epigenetic changes in the stromal cells of breast cancers. Nat Genet. 2005;37(8):899–905. doi: 10.1038/ng1596. [DOI] [PubMed] [Google Scholar]
- 28.Lehmann JM, Riethmuller G, Johnson JP. MUC18, a marker of tumor progression in human melanoma, shows sequence similarity to the neural cell adhesion molecules of the immunoglobulin superfamily. Proc Natl Acad Sci U S A. 1989;86(24):9891–9895. doi: 10.1073/pnas.86.24.9891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shih IM, Elder DE, Hsu MY, Herlyn M. Regulation of Mel-CAM/MUC18 expression on melanocytes of different stages of tumor progression by normal keratinocytes. Am J Pathol. 1994;145(4):837–845. [PMC free article] [PubMed] [Google Scholar]
- 30.Jiang G, Zhang L, Zhu Q, Bai D, Zhang C, Wang X. CD146 promotes metastasis and predicts poor prognosis of hepatocellular carcinoma. J Exp Clin Cancer Res. 2016;35(38) doi: 10.1186/s13046-016-0313-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ma Y, Zhang H, Xiong C, Liu Z, Xu Q, Feng J, Zhang J, Wang Z, Yan X. CD146 mediates an E-cadherin-to-N-cadherin switch during TGF-beta signaling-induced epithelial-mesenchymal transition. Cancer Lett. 2018;430:201–214. doi: 10.1016/j.canlet.2018.05.016. [DOI] [PubMed] [Google Scholar]
- 32.Sers C, Kirsch K, Rothbacher U, Riethmuller G, Johnson JP. Genomic organization of the melanoma-associated glycoprotein MUC18: implications for the evolution of the immunoglobulin domains. Proc Natl Acad Sci U S A. 1993;90(18):8514–8518. doi: 10.1073/pnas.90.18.8514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fung TM, Ng KY, Tong M, Chen JN, Chai S, Chan KT, Law S, Lee NP, Choi MY, Li B. Neuropilin-2 promotes tumourigenicity and metastasis in oesophageal squamous cell carcinoma through ERK-MAPK-ETV4-MMP-E-cadherin deregulation. J Pathol. 2016;239(3):309–319. doi: 10.1002/path.4728. [DOI] [PubMed] [Google Scholar]
- 34.Hollenhorst PC, Ferris MW, Hull MA, Chae H, Kim S, Graves BJ. Oncogenic ETS proteins mimic activated RAS/MAPK signaling in prostate cells. Genes Dev. 2011;25(20):2147–2157. doi: 10.1101/gad.17546311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dumortier M, Ladam F, Damour I, Vacher S, Bieche I, Marchand N, de Launoit Y, Tulasne D, Chotteau-Lelievre A. ETV4 transcription factor and MMP13 metalloprotease are interplaying actors of breast tumorigenesis. Breast Cancer Res. 2018;20(1):73. doi: 10.1186/s13058-018-0992-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kurpios NA, MacNeil L, Shepherd TG, Gludish DW, Giacomelli AO, Hassell JA. The Pea3 Ets transcription factor regulates differentiation of multipotent progenitor cells during mammary gland development. Dev Biol. 2009;325(1):106–121. doi: 10.1016/j.ydbio.2008.09.033. [DOI] [PubMed] [Google Scholar]
- 37.Lu YX, Ju HQ, Liu ZX, Chen DL, Wang Y, Zhao Q, Wu QN, Zeng ZL, Qiu HB, Hu PS. ME1 regulates NADPH homeostasis to promote gastric cancer growth and metastasis. Cancer Res. 2018;78(8):1972–1985. doi: 10.1158/0008-5472.CAN-17-3155. [DOI] [PubMed] [Google Scholar]
- 38.Fischer KR, Durrans A, Lee S, Sheng J, Li F, Wong ST, Choi H, El Rayes T, Ryu S, Troeger J. Epithelial-to-mesenchymal transition is not required for lung metastasis but contributes to chemoresistance. Nature. 2015;527(7579):472–476. doi: 10.1038/nature15748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zheng X, Carstens JL, Kim J, Scheible M, Kaye J, Sugimoto H, Wu CC, LeBleu VS, Kalluri R. Epithelial-to-mesenchymal transition is dispensable for metastasis but induces chemoresistance in pancreatic cancer. Nature. 2015;527(7579):525–530. doi: 10.1038/nature16064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yates LR, Knappskog S, Wedge D, Farmery JHR, Gonzalez S, Martincorena I, Alexandrov LB, Van Loo P, Haugland HK, Lilleng PK. Genomic evolution of breast cancer metastasis and relapse. Cancer Cell. 2017;32(2) doi: 10.1016/j.ccell.2017.07.005. [169-184 e167] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sakaguchi M, Watanabe M, Kinoshita R, Kaku H, Ueki H, Futami J, Murata H, Inoue Y, Li SA, Huang P. Dramatic increase in expression of a transgene by insertion of promoters downstream of the cargo gene. Mol Biotechnol. 2014;56(7):621–630. doi: 10.1007/s12033-014-9738-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sumardika IW, Youyi C, Kondo E, Inoue Y, Ruma IMW, Murata H, Kinoshita R, Yamamoto KI, Tomida S, Shien K. beta-1,3-Galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase 3 increases MCAM stability, which enhances S100A8/A9-mediated cancer motility. Oncol Res. 2018;26(3):431–444. doi: 10.3727/096504017X15031557924123. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary tables
Fig. S1. Survival curves of patients bearing breast cancers with MCAM at a high or low level. A and B, Overall (A) and metastasis-free (B) survival plots according to MCAM expression levels were obtained from a publicly available web site (http://watson.compbio.iupui.edu/chirayu/proggene/database/index.php).
Fig. S2. Validation of the knockdown effect of MCAM siRNA. MDA-MB-231 cells were transfected with either MCAM siRNAs (siMCAM#1, siMCAM#2) or a negative control scrambled siRNA (siCont.), and cellular MCAM protein was detected by WB.
Fig. S3. Establishment of MCF-7-derived sublines that stably express foreign GFP, MCAM or MCAM+ETV4 ΔDNA-BD. A and B, Selected clones resistant to puromycin selection were examined for the transduced foreign genes including GFP alone (A, left), MCAM alone (B, top left) and MCAM+ETV4 ΔDNA-BD (B, top right). Four of each of the clones that were positive for the transduced genes were randomly chosen as indicated by arrowheads, resulting in selection of GFP (#1, #2, #3 and #8), MCAM (#1, #2, #3 and #6) and MCAM+ETV4 ΔDNA-BD (#1, #2, #3 and #6) clones. The selected clones were all examined for their migration ability (A, right; B, bottom). Data are means ± SD, *P < .05, **P < .01 and ***P < .001.
Fig. S4. Screening of a candidate transcription factor(s) that is important in breast cancer migration through MCAM. A, MDA-MB-231 cells were transfected with the indicated decoy DNA oligonucleotides that compete with host DNA cis-elements via binding with their target transcription factors. B, Quantitative real-time PCR analysis was carried out in the indicated cells for ETS family genes. The relative expression level of each sample is shown after calibration with TBP (a suitable housekeeping gene) value. Data are means ± SD, *P < .05, **P < .01 and ***P < .001. ND: not detected.
Fig. S5. Identification of the gene(s) to induce MCAM-mediated migration from the four selected ETS candidates (ETV1, ETV4, ETV5 and ELK3). A and B, An MCF-7-derived MCAM-overexpressed clone, MCAM #1, was temporarily transfected with the four selected ETS candidates (ETV1, ETV4, ETV5 and ELK3), and then its migration (A) and expression of EMT-relevant genes (B) were examined by the Boyden chamber assay and quantitative real-time PCR analysis, respectively. Data are means ± SD, *P < .05, **P < .01 and ***P < .001.
Fig. S6. Identification of an EMT-relevant gene, ZEB1, that is regulated by the MCAM-ETV4 pathway. A, Quantitative real-time PCR analysis was carried out for the CDH2 (N-cadherin), TWIST1 and ZEB1 genes in MCF-7 cells after transfection with an empty vector (EV), and with the MCAM, ETV4 wt and ETV4 ΔDNA-BD expression vectors. The relative expression level of each sample is shown after calibration with the TBP value. B, MDA-MB-231 cells were temporarily transfected with the GFP, MCAM dn and MCAM+ETV4 ΔDNA-BD expression vectors, respectively. The migration ability of the transduced cells was evaluated by Boyden chamber assay. Data are means ± SD, *P < .05, **P < .01 and ***P < .001.
Fig. S7. Survival curves of patients bearing breast cancers with ETV4 expression at a high or low level. A and B, Overall (A) and relapse-free (B) survival plots according to the ETV4 expression levels were obtained from a publicly available web site (http://watson.compbio.iupui.edu/chirayu/proggene/database/index.php).
Fig. S8. Down-regulation of MCAM inhibits ERK1/2 phosphorylation. A, MDA-MB-231 cells were transfected with either siMCAM or a negative control scrambled siCont, and cellular MCAM, p-ERK1/2 and t-ERK1/2 in the treated cells were detected by WB.
Fig. S9. The S100A8/A9-MCAM-ETV4-ZEB1 pathway in triple-negative breast cancer cell lines. A, Three luciferase (Luc)-expressed triple-negative breast cancer cell lines, MDA-MD-231-Luc, Hs578T-Luc and BT-549-Luc, were obtained from the JCRB Cell Bank (Tokyo, Japan). Cellular migration was monitored according to the Boyden chamber method. These Luc-expressed cell lines were transiently transfected with either control GFP or ETV4 ΔDNA-BD plasmid, and the transfected cells were placed at the top of a transwell chamber. Then they were stimulated by S100A8/A9 (100 ng/ml) for 24 h at the bottom of the chamber. Data are means ± SD, *P < .05 and **P < .01, N.S.: not significant. B, The four prepared cell lines (MDA-MB-231, MDA-MD-231-Luc, Hs578T-Luc and BT-549-Luc) were transfected with siCont., siMCAM #1, GFP plasmid or ETV4 ΔDNA-BD plasmid and then they were treated with S100A8/A9 (100 ng/ml) for 6 h (top) or 12 h (bottom). The ETV4 activities in the prepared nuclear extracts were monitored by EMSA analysis. C, Cells prepared by a method similar to that described in B (bottom) were also subjected to WB analysis of their whole cell lysates to examine endogenous ZEB1 levels.