Abstract
Probiotic bacteria exopolysaccharides (EPS) have been recognized as molecules that regulate immune development and have anti-inflammation and anticancer effects. Yet, these bioactivities are of interspecies diversity; thus, examining the gene clusters of EPS and biosynthesis pathways are essential for selecting the better application of specific EPS. In this study, we isolated a new Bifidobacterium strain, named B. breve lw01. A complete genome of B. breve lw01 was sequenced revealing a circular 2,313,172 bp chromosome. Furthermore, a deep excavation of genome sequence from different database based on the comparison-selected results was performed to explore the gene cluster responsible for EPS synthesis. We found that B. breve lw01 harbors a new EPS-encoding cluster with 14 predicted genes, which could be divided into three groups according to the biosynthesis pathway hypothesis. Using tertiary purification, high purity EPS were obtained. EPS is composed of rhamnose (Rha), arabinose (Ara), galactose (Gal), glucose (Glc), and mannose (Man) in a molar ratio of 0.35:0.44:1.38:0.67:1.65. With reference to its bioactivity, it showed to possess anticancer activity against Head and Neck Squamous Cell Carcinoma cell line by regulating cell cycle arrest and cell apoptosis promotion. To sum up, this study examined the biosynthesis and bioactivity of EPS using a new isolated B. breve strain, which could be used to clarify its further application in functional food or drug industry.
Keywords: Bifidobacterium, exopolysaccharide, genome, biosynthesis, anticancer, probiotic, functional food
Introduction
Microbial EPS are carbohydrate polymers surrounding the envelope of most bacteria. The bacterial EPS has initially gained lots of attention from researchers for its effect on biofilm or capsular polysaccharides formation that act as virulence factors in infectious diseases (Gonzalez-Escobedo et al., 2011; Yamanaka et al., 2011). Recently, bacterial EPS have re-gained attention for their implication on human health, especially since bacteria with some probiotic traits and their EPS could contribute to the host health maintenance. Among these, Lactobacillus and Bifidobacterium are mostly used in the formulation of probiotic foods or supplements for human consumption.
Bifidobacterium, which is one of the dominant early colonization bacteria in human intestinal tract, was proven to regulate immune development (Arboleya et al., 2012), immune modulation (Fanning et al., 2012), and to possess an anti-inflammation (Strisciuglio et al., 2015) and anticancer (Wang et al., 2017) activity. The immune response is stain-specific (Lopez et al., 2010) and one of the mechanisms driving the immune functions is regulated by bifido-EPS. Bifido-EPS may facilitate Bifidobacterial-host interaction through immune modulation (Fanning et al., 2012) and provide pathogen protection through a physical barrier, also known as “Bifidobacterial biofilm” (Ruas-Madiedo et al., 2010). In addition, bifido-EPS has shown to possess an antioxidant activity, which may further reduce tissue damage (Li et al., 2014). In addition, EPS can inhibit DNA synthesis (You et al., 2004) and the expression of gene involved in angiogenesis. Yet, these bioactivities, which rely on the related physicochemical characteristic, are of interspecies diversity. Thus, for better application of specific bifido-EPS, gene clusters, chemical composition and its hypothetical biosynthesis pathways prediction need to be carefully examined.
With regards to chemical composition and its synthesis method, HePS, which are type of the EPS polymers reported to be the unique form in Bifidobacterium, compared to the HoPS and HePS types in Lactobacillus strain (Ruas-Madiedo, 2014). Leivers et al. (2011) have reported the hypothetical biosynthesis pathway based on one kind of EPS-unit of HMW-EPS in strain B. animalis subsp. lactis IPLA-R1, which mainly included three steps: activated precursor’s synthesis and EPS-unit formation, export-polymerization process and chain length determination.
In this study, we isolated a new probiotic and EPS-producing Bifidobaccterium, named B. breve lw01. After genome sequencing and in silico analysis, a new EPS cluster and the hypothetical synthesis pathway were discovered. The extraction of EPS, purity determination and scanning electron microscopy observation confirmed its morphology, thus determining the composition of this EPS. Finally, we focused on its anti-tumor properties and preliminary investigated its mechanism. This study provided genetic information on EPS cluster from new isolated EPS-producing Bifidobacterium, connecting it with biosynthesis pathway and exploring its anti-tumor activities. Biochemical evidence was shown to explain its anti-tumor activity. Our research provided a theoretical basis for further application of this bifido-EPS in the probiotic field.
Materials and Methods
Isolation of Bacteria Stains
Fresh infant fecal sample was collected and incubated in a modified MRS-C liquid medium (MRS medium plus 0.5% cysteine • HCl) for 24 h. The medium was then transferred into a modified MRS-C agar plate and cultured at 37°C in anaerobic conditions for 48–72 h. Next, the different morphologies were picked up, and viscous colonies were separately inoculated for pure culture. Staining and morphological characteristics of the isolated bacteria were tested by microscopy. The protocol was approved by the Biomedical Ethics Committee of Peking University School and Hospital of Stomatology. Written informed consent was obtained from parents of the infant.
Cell Lines and Culture Conditions
Human Head and Neck Squamous Cell Carcinoma cell line SCC15 (ATCC® CRL-1623TM), CAL 27 (ATCC® CRL-2095TM) and WSU-HN6 (obtained from Central Laboratory of Peking University School and Hospital of Stomatology) were used in this study. SCC15 were cultured in a 1:1 mixture of DMEM and Ham’s F12 medium (Life technology, Carlsbad, CA, United States) containing 10% FBS (Invitrogen, Waltham, MA, United States), 0.2% hydrocortisone (Selleck Chemicals, Houston, TX, United States) and 1% Penicillin/Streptomycin (Gibco, Waltham, MA, United States). CAL27 and WSU-HN6 were cultured in DMEM medium containing 10% FBS and 1% Penicillin/Streptomycin. All the cells were kept in a humidified atmosphere containing 95% air/5% CO2 at 37°C. For the anaerobic culture, we used the chamber with 93% N2/5% CO2/2% O2.
Genomic DNA Isolation, Sequencing and Analysis of EPS Cluster in Bifidobacterium
Genome DNA was isolated using QIAamp DNA Mini Kit (Cat. 51304, QIAGEN, MD, United States) according the manufacturer’s instructions. 16s rRNA PCR was used to verify the purified strain type. The whole genome was sequenced on Illumina Hiseq 2000 platform (2 × 100 bp) second-generation sequencing platform. Sequence blast searches were performed on National Center for Biotechnology Information website1. The high-quality reads were assembled by SOAPdenovo. Based on the comparison-selected results, the relative abundance of different functional levels and EPS cluster searchers were investigated using KEGG (Kyoto Encyclopedia of genes and genomes), RAST (Rapid Annotation Subsystem Technology) and COG database. The genomes and EPS cluster information available in the GenBank database were used for the comparative analysis among B. breve lw01 and several Bifidobacterium strains.
Extraction and Purification of EPS
For EPS extraction, purified bacteria were anaerobically cultured in 1 L of 10% skimmed milk at 37°C for 48 h. To isolate the crude EPS from the culture broth, 1 L of culture samples were centrifuged at 12,000 ×g for 15 min at 4°C. After obtaining the supernatant, TCA was added to a final concentration of 10% for 12 h at 4°C. Precipitated proteins were then removed by centrifugation at 4000 ×g for 20 min at 4°C. The EPS was precipitated from the supernatant with 3 volumes of cold ethanol followed by an overnight incubation at 4°C. After centrifugation at 6000 ×g for 30 min at 4°C, the pellet containing EPS was resuspended in 2 mL of distilled water and dialyzed (molecular weight cut-off: 6000–8000 Da) against 1 L of distilled water for 2 days with three water changes per 8 h.
Ion exchange chromatography of DEAE Sepharose Fast Flow (16 × 25 mm, GE Healthcare, Amersham, Uppsala, Sweden) was used for further EPS purification. Distilled water was applied to keep the pH of the ion exchange column neutral, eluted with 0.1, 0.2, 0.3, 0.4, and 0.5 M NaCl. The loading volume was 1 mL and the flow rate was 1 mL/min. Stepwise collection of 5 mL was applied for each tube. Then, the purified EPS from ion exchange chromatography was further purified by gel filtration chromatography of Sepharose CL-6B (GE Healthcare, Amersham, Uppsala, Sweden). Elution was performed with distilled water at a flow rate of 1 mL/min. The final eluted EPS was freeze-dried and stored at 4°C until analysis.
A HPLC (Agilent, Santa Clara, CA, United States) equipped with a NH2 column (Agilent, Santa Clara, CA, United States) was used for EPS further purification. The mobile phase consisted of 80% ammonium acetate solution and 20% acetonitrile at a flow rate of 1.0 mL/min, and the column temperature was kept on 30°C. The refractive index detector (RID) was used to detect the EPS.
The amount of EPS was determined using the phenol/sulfuric acid method. Glucose was used as the standard sugar. After the color reaction of the glucose standard solution with phenol/sulfuric acid, its absorbance was measured at 490 nm, and the OD490-standard sugar concentration curve was drawn. The diluted sample of EPS solution was examined using the same color reaction. The 490 nm absorbance value was measured, and the corresponding sugar concentration was found on the standard curve. The sugar concentration of the sample to be tested was calculated.
Scanning Electron Microscopy
Scanning electron microscope was used to examine the morphology of the EPS. Briefly, the freeze-dried EPS powder was mounted on specimens slide, sputter coated with gold and examined using SEM (SEM; Hitachi, S-4700, Hitachi Ltd., Tokyo, Japan) at accelerating voltages of 10 and 15 kV, respectively.
Monomer Composition of EPS
The monosaccharide standards dried to constant weight were weighed separately, and L-Rha, L-Ara, D-Gal, D-Glc, D-Man, and D-Fru prepared at different concentrations (0.2, 0.5, 1.0, 4.0, and 10.0 μg/mL) were mixed to standard solution. Standard curve for each monosaccharide was drew with monosaccharide concentration (μg/mL) as the abscissa (x) and peak area (nC × min) as the ordinate (y). 200 μL concentrated H2SO4 was added to 5 mg EPS and was left to stand still. Then 800 distilled water was added and hydrolyzed at 100°C for 2.5 h. After cooling, add to 2 mL and neutralize with solid BaCO3. Everything was centrifuged at 5000 r/min for 10 min to remove impurities, and remove the supernatant. The supernatant was diluted 20 times, filtered through a 0.20 μm microporous membrane and subjected to test.
High pH anion exchange chromatography with pulsed amperometric detection (HAPEC-PAD) was used to determine monosaccharide composition on a DIONEX-2500 ion chromatograph system, equipped with on-line automatic degassing GS50 quaternary gradient pump, ED50A electrochemical detector (pulse amperometry detection), Au working electrode and Ag/AgCl reference electrode. Chromatographic conditions were set as follows: sugar analysis column (CarboPacTM PA10, 4 × 250 mm); sugar protection column (CarboPac PA10, 4 × 50 mm); mobile phase: A. H2O, B. 200 mmol/L NaOH, A:B = 91:9; injection volume: 25 μL; flow rate: 1.0 mL/min; column temperature: 30°C. Chromeleon 6.5 chromatography was used as workstation.
Cell Proliferation and Cytotoxic Analysis
The EPS effect on cell proliferation and cytotoxicity were examined using CCK-8 assay and DAPI staining. Appropriate number of cells in 100 μL suspension were plated into 96-well plate and incubated in normoxic or hypoxic environment descripted above. After each time point, 10 μL of the CCK-8 solution (Bimake, Houston, TX, United States) was added to each well and incubated for another 4 h at 37°C. The absorbance at 450 nm was determined using a microplate reader. Cell nuclear morphology was stained by DAPI (Zhongshan Golden Bridge Biotechnology Co., Beijing, China). Each sample had three replicates and each experiment was run in triplicate.
Western Blot
After treating cells with EPS, cells were harvested and lysed in RIPA buffer (Applygen Technology, Beijing, China) containing proteinase inhibitors and phosphatase inhibitors (ROCHE, Basel, Switzerland). BCA kit (Thermo Fisher Scientific, Rockford, IL, United States) was used to measure the protein concentration. Total of 70 μg amounts of protein samples were separated by 10% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to polyvinylidene difluoride (PVDF) membranes by wet blotting. The membranes were blocked in 10% non-fat dry milk for 1 h and then probed with antibodies against MCM2 (1:1,000, ABclonal, Boston, MA, United States), Caspases-3 (1:1,000, CST, Boston, MA, United States), PARP (1:1,000, CST, Boston, MA, United States), cleaved-PARP (1:1,000, CST, Boston, MA, United States) and RPS18 (1:1,000, ABclonal, Boston, MA, United States) separately at 4°C overnight. Followed by incubation with peroxidase-linked secondary antibodies (1:10,000, CST, Boston, MA, United States) for 1 h at room temperature, the enhanced chemiluminescent (ECL) reagent (Thermo Fisher Scientific, Rockford, IL, United States) was used to visualize the immunoreactive proteins.
Statistical Analysis
The absorbance value in CCK-8 assay and western blot results were expressed as means ± standard deviation (SD). One-way ANOVA was used for comparison among different concentration group and Fisher’s least significant difference (LSD) method was used for multiple comparisons in CCK-8 assay. Independent sample t-test was used to compare the treatment group and control group in western blot assay. All calculations and analyses were performed using SPSS 20.0 software (SPSS Inc., Chicago, IL, United States). A P-value < 0.05 was considered statistically significant.
Results
Isolation of Bifidobacterium
A total of 653 strains bacteria were isolated from the infant fecal samples. Among those, 23 viscous strains were isolated and transferred in a modified MRS-C medium. One strain was typically rod-shaped or bifurcated, non-spore-forming. Consequently, this clone was analyzed using 16S rRNA PCR (16S rRNA-F: 5′-AGAGTTTGATCMTGGCTCAG-3′ and 16S rRNA-R: 5′-TACGGYTACCTTGTTACGACTT-3′). The 16S rRNA sequence analyses showed 99% identity with B. breve DSM20213 (Supplementary Figure S1).
Genome Information of B. breve lw01
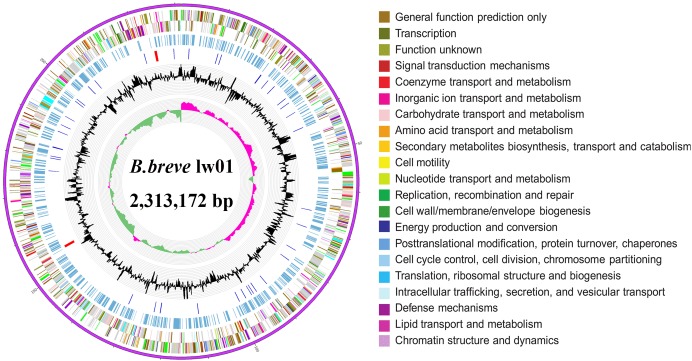
The complete genome sequence of our new isolated B. breve strain was submitted to GenBank under the accession Number CP034192. The complete genome of this strain is composed of one circular chromosome of 2,313,172 bp with a GC content of 58.7%, which is similar to other reported B. breve strains (GC content of 58.5–62.8%). In addition, the new strain contains a total of 63 RNAs including 54 tRNA genes, 2 rRNA, and 3 ncRNAs. A total of 1862 coding genes out of 2077 total genes were classified into 23 COG categories (Tables 1, 2 and Figure 1). The genome comparative analyses among B. breve lw01 and other seven Bifidobacterium strains are presented in Supplementary Table S1. Consequently, this strain, named B. breve lw01 was deposited in China Center of Industrial Culture Collection (CICC) under the number CICC 24633.
Table 1.
Features of B. breve lw01 genome.
| Features | Chromosome |
|---|---|
| Genome size (bp) | 2,313,172 |
| GC content (%) | 58.7 |
| RNAs | 63 |
| rRNAs | 2, 2, 2 (5S, 16S, 23S) |
| tRNAs | 54 |
| ncRNAs | 3 |
| CDSs (with protein) | 1862 |
Table 2.
COG categories of coding proteins in B. breve lw01 genome.
| COG | Name | Count | Percent (%) |
|---|---|---|---|
| A | RNA processing and modification | 2 | 0.10 |
| C | Energy production and conversion | 53 | 2.71 |
| D | Cell cycle control, cell division, chromosome partitioning | 31 | 1.59 |
| E | Amino acid transport and metabolism | 209 | 10.69 |
| F | Nucleotide transport and metabolism | 80 | 4.09 |
| G | carbohydrate transport and metabolism | 253 | 12.94 |
| H | Coenzyme transport and metabolism | 77 | 3.94 |
| I | Lipid transport and metabolism | 54 | 2.76 |
| J | Translation, ribosomal structure and biogenesis | 167 | 8.54 |
| K | Transcription | 153 | 7.83 |
| L | Replication, recombination and repair | 90 | 4.60 |
| M | Cell wall/membrane/envelope biogenesis | 106 | 5.42 |
| N | Cell motility | 12 | 0.61 |
| O | Posttranslational modification, protein turnover, chaperones | 74 | 3.79 |
| P | Inorganic ion transport and metabolism | 94 | 4.81 |
| Q | Secondary metabolites biosynthesis, transport and catabolism | 21 | 1.07 |
| R | General function prediction only | 153 | 7.83 |
| S | Function unknown | 93 | 4.76 |
| T | Signal transduction mechanisms | 92 | 4.71 |
| U | Intracellular trafficking, secretion, and vesicular transport | 15 | 0.77 |
| V | Defense mechanisms | 67 | 3.43 |
| W | Extracellular structures | 4 | 0.20 |
| X | Mobilome: prophages, transposons | 55 | 2.81 |
FIGURE 1.
Circle genome map of B. breve lw01. Genome sequences (ring 1). COG Annotated coding sequences (ring 2 and 3), KEGG enzymes (ring 4), RNA genes (ring 5), GC content (ring 6), and GC skew (ring 7) are shown. Very short features were enlarged to enhance the visibility. Clustered genes, such as several rRNA genes, may appear as one-line due to space limitations. Image was created by the software Circos.
In silico Analysis of Bifido-EPS Cluster
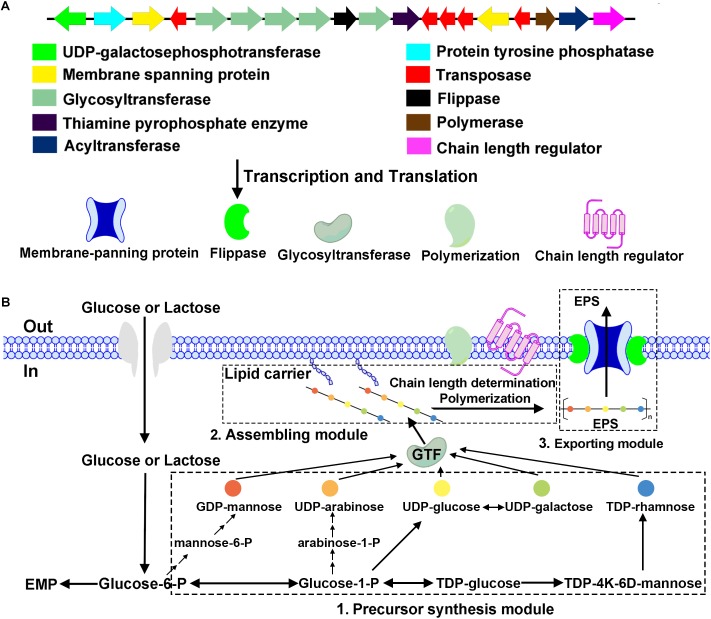
Bifidobacterium breve lw01 harbors an EPS-encoding cluster with 14 predicted genes and 5 transposase coding genes. Priming-GTF (p-gtf) is the enzyme that catalyzes the initial step of EPS-unit synthesis; consequently, we first found this enzyme: UDP-galactosephosphotransferase (EH245_02110) coding for proteins of 576 amino acids. Five genes were coding for the GTFs (EH245_02130, EH245_02135, EH245_02140, EH245_02145, EH245_02155). Two types of genes could be responsible for transporting of the repeat EPS-units across the cytoplasmic membrane: flippase (EH245_02150) and membrane spanning protein (EH245_02120, EH245_02180). The chain length regulation and polymerization system were also found in this EPS-unit, which included the polymerase (EH245_02190), chain length regulator (EH245_02200) and the protein tyrosine phosphatase (EH245_02115). Another discovered feature was the presence of transposase (EH245_02125, EH245_02165, EH245_02170, EH245_02175, EH245_02185), an enzyme participated putative horizontal transfer of EPS among different taxa. There were also several genes coding for the precursor involved in the biosynthesis of EPS-unit: thiamine pyrophosphate enzyme (EH245_02160) and acyltransferase (EH245_02195) (Figure 2A).
FIGURE 2.
Physical map of the putative EPS cluster of B. breve lw01 and a schematic diagram of EPS biosynthesis in this strain. (A) The genes were categorized according to their potential functions, which are indicated with the colored arrows. (B) Hypothetical EPS biosynthesis pathway in strain B. breve lw01 based on its homology analysis of the EPS cluster.
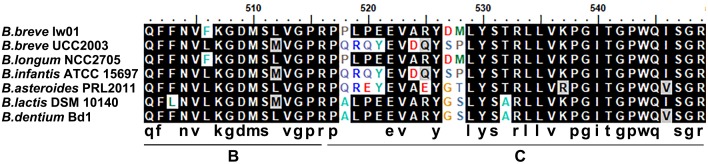
We compared the EPS cluster information with other seven Bifidobacteium strains (Supplementary Table S1). Interestingly, the EPS cluster region has a G+C DNA content (49.7%) markedly lower than the B. breve strains (GC content of 58.5–62.8%), which inferred that it may be acquired by horizontal DNA transfer. The comparison of amino acid sequences of p-gtf showed that it was quite similar to the strain B. longum NCC2705 (with 98% identity). Partial amino acid identity of p-gtf among B. breve lw01 and other seven Bifidobacterium stains is shown in Figure 3. The results showed that they process the similar sequences particularly in the carboxy terminus regions.
FIGURE 3.
Alignment of an fragment of amino acid sequence of priming-GTF in B. breve lw01 and other Bifidobacterium strains. B at the bottom represented blocks involved in interaction with lipid carrier and C represented blocks involved in sugar-specific recognition.
Hypothetical Pathway of EPS Biosynthesis in B. breve lw01
The hypothetical pathway of EPS biosynthesis in B. breve lw01 was divided into three modules. First module was the precursor synthesis. After glycose and lactose fluxed into the cytoplasm, activated NDP-sugar precursors modules were formed from the intermediate molecules of glycolysis. Second module was the assembling module. EPS-unit was catalyzed by p-gtf enzyme (UDP-galactosephosphotransferase). With the help of p-gtf (UDP-galactosephosphotransferase), one sugar-1-phosphate was transferred to the lipophilic carrier molecule and anchored to the cell membrane. Other five glycosyltransferases (GTFs) catalyzed the formation of glycosidic bonds by transferring new sugar moieties from a donor nucleotide sugar to the initial monosaccharide of the unit. A chain length regulator and polymerization system then assembled the EPS-lipophilic carrier unit and formed the finished EPS-unit. In the third exporting module, flippase and membrane spanning protein helped this finished EPS-unit to cross the cytoplasmic membrane to the extracellular face and build the final EPS polymers (Figure 2B).
Extraction, Purification and Analysis of EPS
Crude EPS extracted from 1 L of 10% skimmed milk was about 80 mg. Only one kind of EPS fraction was obtained from Ion exchange chromatography with 0.1 mol/L NaCl solution, which appeared at 8–18 tubes (about 50 mL). When the elution tubes exceeded 50 tubes and the concentration of NaCl was above 0.3 M, no obvious polysaccharide component was eluted. After Ion exchange chromatography, we yielded about 26 mg EPS. After gel filtration chromatography and HPLC purification, 5 and 1 mg EPS were obtained, respectively. The single and symmetric peak showed that the EPS was purified with high quality (Figure 4).
FIGURE 4.
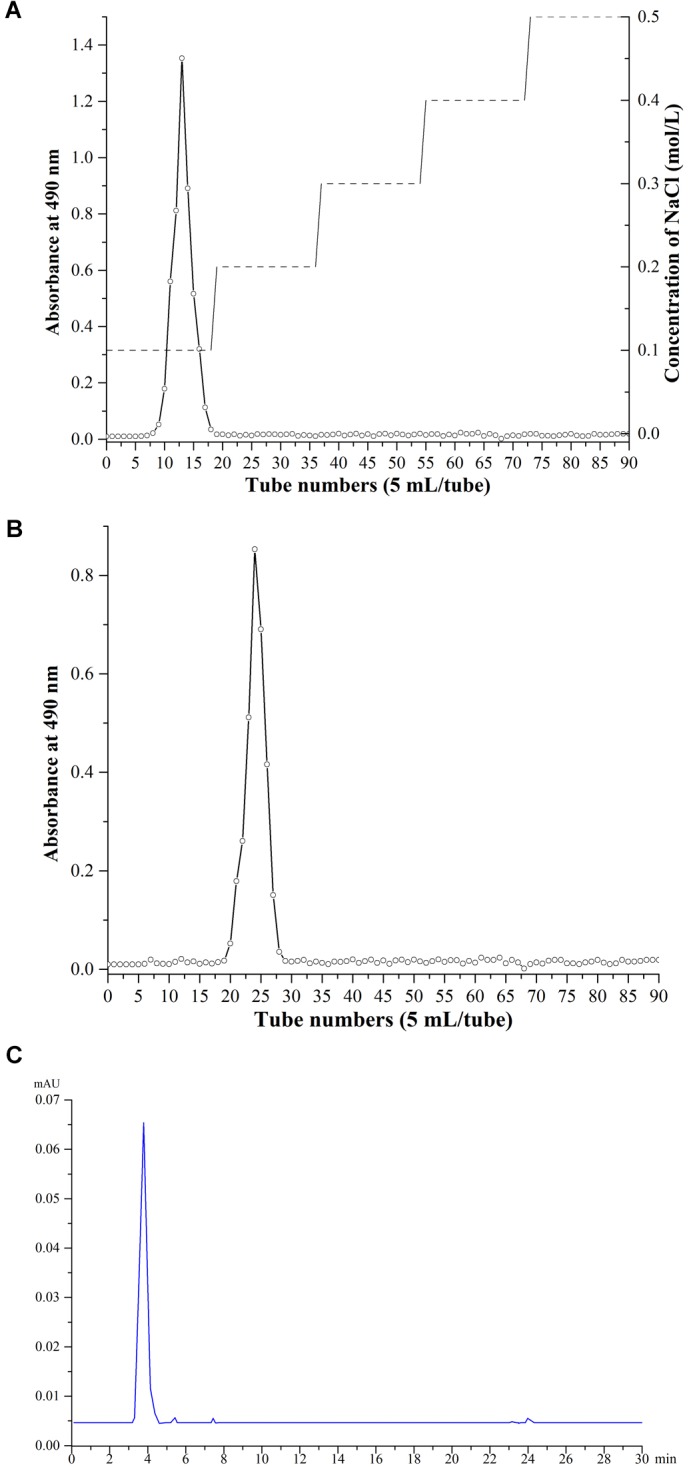
Purification and analysis of EPS. (A) Ion exchange chromatography. (B) gel filtration chromatography purification. (C) HPLC purification.
Scanning Electron Microscopy Images of EPS From B. breve lw01
Scanning electron microscope is a useful method to investigate the surface and three-dimensional morphology of biomacromeolecules. The morphology of EPS observed by SEM showed that EPS was mainly composed of freely distributed irregular spherical bodies, flakes and nets. EPS was relatively rough, and was composed of depressions and voids, which may be due to the nature and structure of EPS, and the formation of branches. The use of different methods in the process of extraction and purification of EPS could cause different ultrastructure (Figure 5).
FIGURE 5.
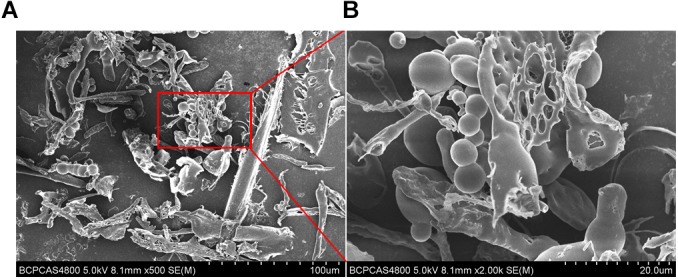
Scanning electron microscope images of purified EPS from B. breve lw01. (A) Magnification 500×. (B) In part magnified of A (magnification 2000×).
Chemical Composition of B. breve lw01’s EPS
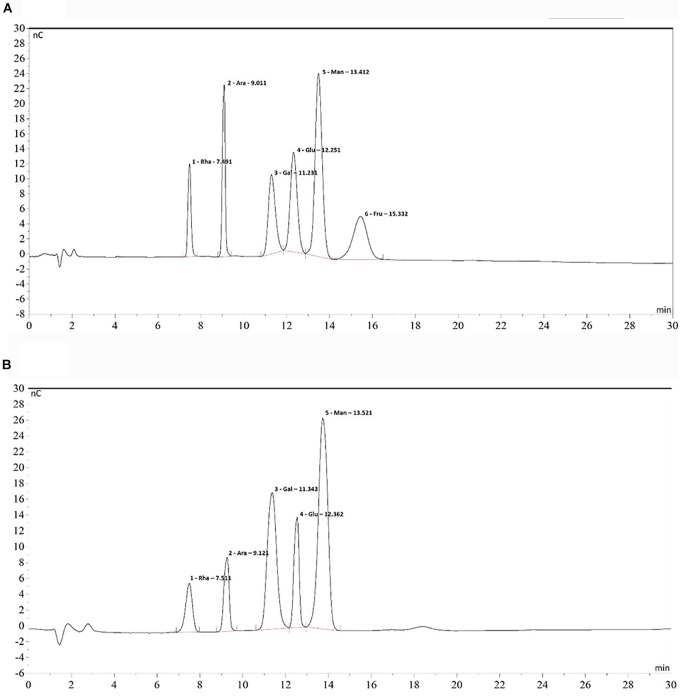
The monomer composition of B. breve lw01’s EPS was determined by HPAEC-PAD. The results of monomer analysis indicated that EPS was a heteropolysaccharide mainly composed of rhamnose, arabinose, galactose, glucose and mannose in a molar ratio of 0.35:0.44:1.38:0.67:1.65 compared with the HPAEC spectra of standard monosaccharides (Figure 6). The constitution of our EPS was similar to the EPS composition produced by B. animalis RH (Shang et al., 2013).
FIGURE 6.
Chromatogram of standard monosaccharide (A) and EPS monosaccharide compositions (B) on HPAEC-PAD. From left to right: 1. Rhamnose 2. Arabinose 3. Galactose 4. Glucose 5. Mannose 6. Fructose.
Anticancer Property of B. breve lw01’s EPS on HNSCC Cell Lines
Anti-proliferation Effect in Aerobic and Anaerobic Environment
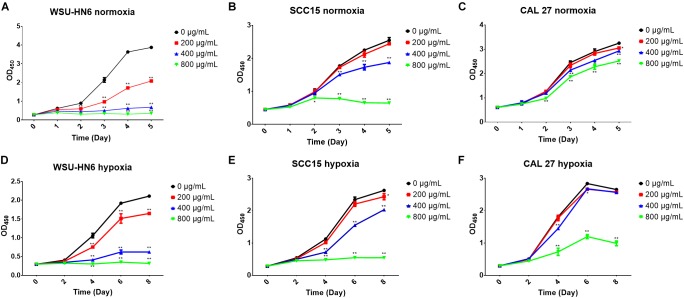
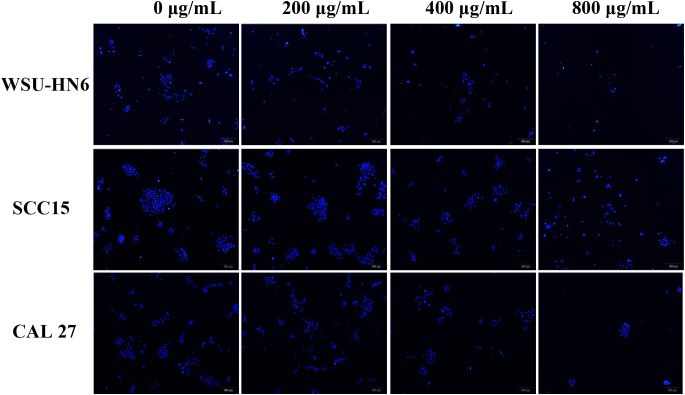
CCK-8 assay and DAPI staining were used to examine the anti-tumor effect of EPS on HNSCC cell lines. CCK-8 results showed that EPS could inhibit cell proliferation in a concentration dependent manner. However, different cell line showed different sensitivity to EPS. The highest effect was observed in WSU-HN6, followed by SCC15 and CAL 27. On day 5, EPS inhibited the growth of WSU-HN6 by 46% at 200 μg/mL in aerobic environment. For CAL 27 cell line, in normoxia condition, 800 μg/mL EPS could inhibit CAL 27 cell line proliferation slightly, while the proliferation rate drastically decreased when culturing cells under hypoxic condition. This strengthened effect could be observed in the other cell lines as well (Figure 7). Furthermore, DAPI staining of cell nuclear with different treatment concentration presented the same tendency when cell lines were cultured in normoxia condition (Figure 8).
FIGURE 7.
Cell proliferation inhibition of EPS from B. breve lw01 by CCK-8 assay. (A–C) Three HNSCC cell lines were cultured in normoxia condition and treated by gradient concentration of EPS from B. breve lw01. (D–F) Three HNSCC cell lines were cultured in hypoxia condition. All the experiments were run in triplicate. ∗p < 0.05; ∗p < 0.01.
FIGURE 8.
DAPI staining images of cell nuclear after treated with gradient concentration of EPS. All the cells were cultured in normoxia condition. magnification 4×.
Cell Cycle and Apoptosis-Associated Protein Expression
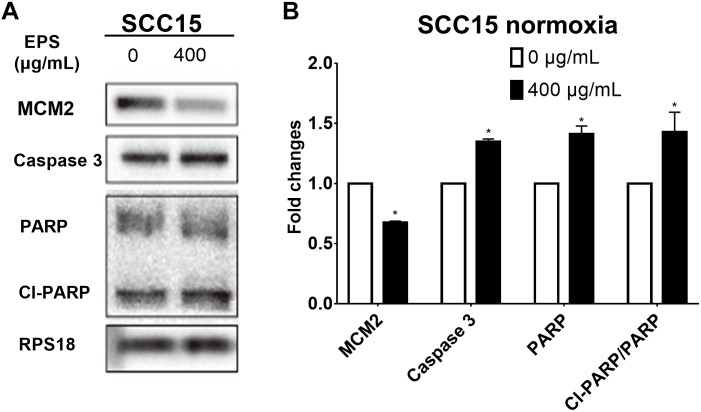
In order to find the detailed mechanism on the anti-proliferation effect of EPS, we tested the expression of cell-cycle and apoptosis associated protein by western bolt assay in a represented cell line SCC15 in normoxia condition. Decreased expression of MCM2 suggested that the cell cycle arrest occurs at G1-S phase. Increased expression of cell apoptosis protein caspase 3, PARP and the proportion of Cl-PARP/PARP inferred that EPS could promote the cell apoptosis (Figure 9).
FIGURE 9.
Cell cycle and apoptosis protein expression after treating SCC15 cell line with 400 μg/mL EPS. (A) Western blot assay. (B) Relative fold changes by calculating relative gray value using software Quantity One. ∗p < 0.05.
Discussion
Currently, EPS obtained from probiotic bacteria Bifidobacterium have been gaining increasing attention, mainly due to their role in immune system and gut microbiota modulation. In the present study, we isolated a new B. breve strain from infant feces. After sequencing its genome, we predicted its EPS cluster and hypothetic synthesis pathway. Based on the EPS extraction and high-quality purification, we determined its chemical composition. Finally, we focused our attention on its anticancer activity and provided biochemical evidence to explain it. Thus, this study provided a better understanding of the function-genome cluster relation and biosynthesis in bifido-EPS, which could facilitate genetic and metabolic engineering indispensable for production of tailor-made EPS used in food or drug industry.
Extraction and purification were necessary to examine the biological activity of B. breve lw01’s EPS. The yield of crude EPS released to culture medium can be influenced by environmental and nutritional conditions, such as the medium composition, pH, temperature, oxygen and the growth phase (Leroy and De Vuyst, 2016; Bengoa et al., 2018). In our study, slimmed milk was chosen for the initial cultivation to avoid sugar in the medium that interferes with the EPS extraction. Crude bifido-EPS was followed by the further purification (Ion exchange chromatography and gel filtration chromatography), which yielded high purity bifido-EPS, thus providing good research basis for the biological activity. Multiple purification induced the poor yield, which was affected by many factors such as EPS sample loading volume, eluent and elution rate. In the course of the research, we observed that the low purification efficiency affected our further research. Therefore, future studies should focus on the elevation of the purification efficiency of EPS isolation.
Previous studies have indicated that genes responsible for the biosynthesis of EPS are presented in the form of clusters in Bifidobacterium genome (Leivers et al., 2011; Fanning et al., 2012; Hidalgo-Cantabrana et al., 2014). Due to the high interspecies variability existence, and deep excavation of genome sequences from different database, the comparison-selected results were used to examine the gene cluster responsible for EPS synthesis. We find complete EPS gene cluster was involved in the biosynthesis of EPS and the gene cluster could be divided into three following groups: the gene responsible for repeated unit synthesis, polymerization/chain length determination and exportation. No transcriptional regulator was observed as in other Bifidobacterium (Ruas-Madiedo, 2014). The EPS biosynthesis pathway was similar but not identical to that reported in B. breve UCC2003. In B. breve UCC2003, the EPS synthesis was bidirectional and responsible for production of one or two polymers (Fanning et al., 2012). For the further confirmation of the exact function of each gene involved in our bifdo-EPS gene cluster, mutants by deletion part of the EPS clusters could be used by comparing to their counterparts.
Among the biosynthesis pathway, repeated unit synthesis was the key step to determine the final EPS-unit constitution and GTF was the key enzyme involved in this part. The amino acid identity of p-gtf among B. breve lw01 and other seven Bifidobacterium stains was lower. The similar sequences were observed, particularly in the carboxy terminus regions, due to the presence of regions involved in interaction with lipid carrier (region B) and sugar specificity (region C) (Provencher et al., 2003; Yan et al., 2017). However, the composition and the structure of bifido-EPS have not yet been fully illustrated for any of the strains containing completed genomes (Hidalgo-Cantabrana et al., 2014). Therefore, although the numbers and specificities of GTFs could contribute to the different EPS-unit constitution and structures, it is still not possible to fully establish the association between the genetics and physico-chemical characteristics (Hidalgo-Cantabrana et al., 2014). Moreover, in this work, we did not focus on the structure of EPS. Accordingly, future studies should focus on investigating the genetics and physico-chemical correlation based on EPS genome-composition-structure characters.
In our previous study, we already verified that the strain B. breve possess an anti-tumor effect on HNSCC tumor-bearing model (Wang et al., 2017). Yet, the exact mechanism was not fully explored. Existing studies have shown that the presence of bacterial molecules structure (DNA, proteins, EPS or metabolites) could have a specific effect on the immune or inflammation system (Menard et al., 2010; Hevia et al., 2014; Hidalgo-Cantabrana et al., 2014). In the present study, we further examined whether the new extracted bifido-EPS may be responsible for the anticancer bioactivity.
Currently, there are only few studies on the anticancer effect of EPS from Bifidobacterium strains. In vitro experiments have shown that polysaccharide fraction extracted from B. bifidum BGN4 could inhibit the growth of colon cancer cell lines HT-29 and HCT-116, but had no effect on Caco-2 cell line. Previous study has reported that the inhibition rate on HT-29 cell line was 50.5 ± 3.6% at 80 μg/mL by Trypan blue exclusion assay. It is speculated that the anticancer effect may be related to the inhibition of DNA synthesis and reduction of cell proliferation capacity (You et al., 2004). The research from Nam Joo Ha’s group showed that butanol extract of B. adolescentis SPM0212 could inhibit the growth of Caco-2, HT-29, and SW480 cells by 70, 30, and 40%, respectively, at 200 μg/mL. The mechanism may rely on inducing macrophage activation and increasing the production TNF-α and NO (Lee et al., 2008). In the present study, we investigated anticancer effect of bifido-EPS on HNSCC cell line, which exerted proliferation inhibition effect in dose-dependent manner. In addition, different cells presented different sensitivity to bifido-EPS.
As for the mechanisms of the anti-cancer property of EPS, except for the inhibition of DNA synthesis and immune modulation reported above, EPS has anti-oxidative properties and can inhibit the expression of gene involved in angiogenesis (Deepak et al., 2016). Cell proliferative activity is closely related to the cell cycle, which is regulated by the DNA replication initiation factor. MCM 2, known as DNA replication licensing factor, is involved in pre-replicative complex formation during G1 phase and it allows for the DNA replication initiation in the subsequent S-phase (Gambus et al., 2011). Downregulation of MCM2 could increase cell death when the cells encounter replication stress during S-phase (Ibarra et al., 2008). MCM2 has also been applied as a proliferation marker in many types of cancer (Tan et al., 2001; Yousef et al., 2017; Zhou et al., 2018). In addition, its downregulation is considered as an attractive target in tumor chemotherapy (Simon and Schwacha, 2014). In this study, the expression of MCM2 was downregulated, which suggested that EPS could arrest cell cycle in G1-S phase in HNSCC cell line. PARP is activated when cells sustained DNA damage and it is the main target of caspase 3, whose activation could mediate a latent specific proteolytic cleavage in apoptosis (Brauns et al., 2005; Yu et al., 2006). In our study, EPS increased the expression of caspase 3 and the cleaved-PARP proportion, while inferred cell apoptosis occurred in treatment group.
The chemical composition of our EPS, which included rhamnose, arabinose, galactose, glucose, and mannose, could explain its anti-tumor effect to some extent. It is quite interesting that mannose accounted for the largest proportion of the bifido-EPS, which have been reported to have a significant anti-tumor activity. Mannose could impair tumor growth by inhibiting the proliferation while promoting the apoptosis of cancer cell. It also revealed chemo-sensitization character. Administration of doxorubicin with mannose showed an obviously enhanced anti-tumor effect (Gonzalez et al., 2018). Moreover, the level of mannose in serum is associated with the prognosis of esophageal adenocarcinoma patients. The patients with low level of d-mannose have higher risk of recurrence and death (Gu et al., 2017). Besides, rhamnose has also been reported to have anti-tumor activity, since it can enhance the immunogenicity of melanoma-associated antigen A3, which stimulates antitumor immune responses (Zhang et al., 2016). The galactose is widely used backbone of the nanocarrier in cancer therapy, beneficial for targeted therapy of tumors (Jain et al., 2018). When conjugated with platinum (II), the complex has well therapeutic and target effect (Wu et al., 2016). In our next study, we plan to further investigate the effect of our bifido-EPS in vivo.
To sum up, the current work isolated one new probiotic strain B. breve lw01 and sequenced its genome, which led to discovery of a new EPS biosynthesis cluster and prediction of its biosynthesis pathway. After extraction and purification, high purity of our EPS showed to possess an anticancer activity in HNSCC cell lines. In addition, the composition of bifido-EPS showed that mannose accounted for the largest proportion. The preliminary mechanisms could rely on its cell cycle arrest and promote cell apoptosis effect. We believe that this bifido-EPS could be used to facilitate genetic and metabolic engineering and to produce tailor-made EPS for further application in functional food or drug industry.
Ethics Statement
The protocol was approved by the Biomedical Ethics Committee of Peking University School and Hospital of Stomatology. Written informed consent was obtained from parents of the infant.
Author Contributions
GL and CG contributed the conception and design of the study. LW and KT organized the database and analysis EPS cluster information. YW, QL, and LX carried out the study. YW performed the statistical analysis and wrote section of the manuscript. LW wrote the first draft of the manuscript. All authors contributed to manuscript revision, read and approved the submitted version.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank team members from Beijing Engineering and Technology Research Center of Food Additives to provide machine and technical support in EPS purification and composition determination.
Abbreviations
- bifido-EPS
Bifidobacterium-EPS
- COG
clusters of orthologous groups
- EPS
exopolysaccharide
- HAPEC-PAD
High pH anion exchange chromatography with pulsed amperometric detection
- HePS
heteropolysaccharides
- HNSCC
head and neck squamous cell carcinoma
- HoPS
homopolysaccharides
- HPLC
high-performance liquid chromatography
- MCM2
mini-chromosome maintenance 2
- p-gtf
priming-GTF
- SEM
scanning electron microscope
- TCA
trichloroacetic acid
Funding. This study was supported by the National Natural Science Foundation of China (Nos. 81470707 and 81802699), as well as Peking University School and Hospital of Stomatology Youth Research Fund (No. PKUSS20180118). This research was also supported by the fund of the Beijing Engineering and Technology Research Center of Food Additives, Beijing Technology and Business University (BTBU).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.01044/full#supplementary-material
References
- Arboleya S., Binetti A., Salazar N., Fernandez N., Solis G., Hernandez-Barranco A., et al. (2012). Establishment and development of intestinal microbiota in preterm neonates. FEMS Microbiol. Ecol. 79 763–772. 10.1111/j.1574-6941.2011.01261.x [DOI] [PubMed] [Google Scholar]
- Bengoa A. A., Llamas M. G., Iraporda C., Duenas M. T., Abraham A. G., Garrote G. L. (2018). Impact of growth temperature on exopolysaccharide production and probiotic properties of Lactobacillus paracasei strains isolated from kefir grains. Food Microbiol. 69 212–218. 10.1016/j.fm.2017.08.012 [DOI] [PubMed] [Google Scholar]
- Brauns S. C., Dealtry G., Milne P., Naude R., Van de Venter M. (2005). Caspase-3 activation and induction of PARP cleavage by cyclic dipeptide cyclo(Phe-Pro) in HT-29 cells. Anticancer Res. 254197–4202. [PubMed] [Google Scholar]
- Deepak V., Ramachandran S., Balahmar R. M., Pandian S. R., Sivasubramaniam S. D., Nellaiah H., et al. (2016). In vitro evaluation of anticancer properties of exopolysaccharides from Lactobacillus acidophilus in colon cancer cell lines. In Vitro Cell Dev. Biol. Anim. 52 163–173. 10.1007/s11626-015-9970-3 [DOI] [PubMed] [Google Scholar]
- Fanning S., Hall L. J., Cronin M., Zomer A., MacSharry J., Goulding D., et al. (2012). Bifidobacterial surface-exopolysaccharide facilitates commensal-host interaction through immune modulation and pathogen protection. Proc. Natl. Acad. Sci. U.S.A. 109 2108–2113. 10.1073/pnas.1115621109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gambus A., Khoudoli G. A., Jones R. C., Blow J. J. (2011). MCM2-7 form double hexamers at licensed origins in Xenopus egg extract. J. Biol. Chem. 286 11855–11864. 10.1074/jbc.M110.199521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez P. S., O’Prey J., Cardaci S., Barthet V. J. A., Sakamaki J.-i., Beaumatin F., et al. (2018). Mannose impairs tumour growth and enhances chemotherapy. Nature 563 719–723. 10.1038/s41586-018-0729-3 [DOI] [PubMed] [Google Scholar]
- Gonzalez-Escobedo G., Marshall J. M., Gunn J. S. (2011). Chronic and acute infection of the gall bladder by Salmonella Typhi: understanding the carrier state. Nat. Rev. Microbiol. 9 9–14. 10.1038/nrmicro2490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu J., Liang D., Pierzynski J. A., Zheng L., Ye Y., Zhang J., et al. (2017). D-mannose: a novel prognostic biomarker for patients with esophageal adenocarcinoma. Carcinogenesis 38 162–167. 10.1093/carcin/bgw207 [DOI] [PubMed] [Google Scholar]
- Hevia A., Lopez P., Suarez A., Jacquot C., Urdaci M. C., Margolles A., et al. (2014). Association of levels of antibodies from patients with inflammatory bowel disease with extracellular proteins of food and probiotic bacteria. Biomed. Res. Int. 2014:351204. 10.1155/2014/351204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hidalgo-Cantabrana C., Sanchez B., Milani C., Ventura M., Margolles A., Ruas-Madiedo P. (2014). Genomic overview and biological functions of exopolysaccharide biosynthesis in Bifidobacterium spp. Appl. Environ. Microbiol. 80 9–18. 10.1128/AEM.02977-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibarra A., Schwob E., Mendez J. (2008). Excess MCM proteins protect human cells from replicative stress by licensing backup origins of replication. Proc. Natl. Acad. Sci. U.S.A. 105 8956–8961. 10.1073/pnas.0803978105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain A., Jain A., Parajuli P., Mishra V., Ghoshal G., Singh B., et al. (2018). Recent advances in galactose-engineered nanocarriers for the site-specific delivery of siRNA and anticancer drugs. Drug Discov. Today 23 960–973. 10.1016/j.drudis.2017.11.003 [DOI] [PubMed] [Google Scholar]
- Lee D. K., Jang S., Kim M. J., Kim J. H., Chung M. J., Kim K. J., et al. (2008). Anti-proliferative effects of Bifidobacterium adolescentis SPM0212 extract on human colon cancer cell lines. BMC Cancer 8:310. 10.1186/1471-2407-8-310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivers S., Hidalgo-Cantabrana C., Robinson G., Margolles A., Ruas-Madiedo P., Laws A. P. (2011). Structure of the high molecular weight exopolysaccharide produced by Bifidobacterium animalis subsp. lactis IPLA-R1 and sequence analysis of its putative eps cluster. Carbohydr. Res. 346 2710–2717. 10.1016/j.carres.2011.09.010 [DOI] [PubMed] [Google Scholar]
- Leroy F., De Vuyst L. (2016). Advances in production and simplified methods for recovery and quantification of exopolysaccharides for applications in food and health. J. Dairy Sci. 99 3229–3238. 10.3168/jds.2015-9936 [DOI] [PubMed] [Google Scholar]
- Li S., Huang R., Shah N. P., Tao X., Xiong Y., Wei H. (2014). Antioxidant and antibacterial activities of exopolysaccharides from bifidobacterium bifidum WBIN03 and Lactobacillus plantarum R315. J. Dairy Sci. 97 7334–7343. 10.3168/jds.2014-7912 [DOI] [PubMed] [Google Scholar]
- Lopez P., Gueimonde M., Margolles A., Suarez A. (2010). Distinct bifidobacterium strains drive different immune responses in vitro. Int. J. Food Microbiol. 138 157–165. 10.1016/j.ijfoodmicro.2009.12.023 [DOI] [PubMed] [Google Scholar]
- Menard O., Gafa V., Kapel N., Rodriguez B., Butel M. J., Waligora-Dupriet A. J. (2010). Characterization of immunostimulatory CpG-rich sequences from different Bifidobacterium species. Appl. Environ. Microbiol. 76 2846–2855. 10.1128/AEM.01714-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Provencher C., LaPointe G., Sirois S., Van Calsteren M.-R., Roy D. (2003). Consensus-degenerate hybrid oligonucleotide primers for amplification of priming glycosyltransferase genes of the exopolysaccharide locus in strains of the Lactobacillus casei group. Appl. Environ. Microbiol. 69 3299–3307. 10.1128/AEM.69.6.3299-3307.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruas-Madiedo P. (2014). “Biosynthesis and bioactivity of exopolysaccharidesproduced by probiotic bacteria,” in Food Oligosaccharides: Production, Analysis and Bioactivity, eds Moreno F. J., Sanz M. L. (Hoboken, NJ: JohnWiley & Sons Ltd; ), 118–1133. [Google Scholar]
- Ruas-Madiedo P., Medrano M., Salazar N., De Los Reyes-Gavilan C. G., Perez P. F., Abraham A. G. (2010). Exopolysaccharides produced by Lactobacillus and Bifidobacterium strains abrogate in vitro the cytotoxic effect of bacterial toxins on eukaryotic cells. J. Appl. Microbiol. 109 2079–2086. 10.1111/j.1365-2672.2010.04839.x [DOI] [PubMed] [Google Scholar]
- Shang N., Xu R., Li P. (2013). Structure characterization of an exopolysaccharide produced by Bifidobacterium animalis RH. Carbohydr. Polym. 91 128–134. 10.1016/j.carbpol.2012.08.012 [DOI] [PubMed] [Google Scholar]
- Simon N. E., Schwacha A. (2014). The Mcm2-7 replicative helicase: a promising chemotherapeutic target. Biomed. Res. Int. 2014:549719. 10.1155/2014/549719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strisciuglio C., Miele E., Giugliano F. P., Vitale S., Andreozzi M., Vitale A., et al. (2015). Bifidobacteria enhance antigen sampling and processing by dendritic cells in pediatric inflammatory bowel disease. Inflamm. Bowel Dis. 21 1491–1498. 10.1097/MIB.0000000000000389 [DOI] [PubMed] [Google Scholar]
- Tan D. F., Huberman J. A., Hyland A., Loewen G. M., Brooks J. S., Beck A. F., et al. (2001). MCM2–a promising marker for premalignant lesions of the lung: a cohort study. BMC Cancer 1:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Vuletic I., Deng D., Crielaard W., Xie Z., Zhou K., et al. (2017). Bifidobacterium breve as a delivery vector of IL-24 gene therapy for head and neck squamous cell carcinoma in vivo. Gene Ther. 24 699–705. 10.1038/gt.2017.74 [DOI] [PubMed] [Google Scholar]
- Wu M., Li H., Liu R., Gao X., Zhang M., Liu P., et al. (2016). Galactose conjugated platinum(II) complex targeting the Warburg effect for treatment of non-small cell lung cancer and colon cancer. Eur. J. Med. Chem. 110 32–42. 10.1016/j.ejmech.2016.01.016 [DOI] [PubMed] [Google Scholar]
- Yamanaka T., Yamane K., Furukawa T., Matsumoto-Mashimo C., Sugimori C., Nambu T., et al. (2011). Comparison of the virulence of exopolysaccharide-producing Prevotella intermedia to exopolysaccharide non-producing periodontopathic organisms. BMC Infect. Dis. 11:228. 10.1186/1471-2334-11-228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan S., Zhao G., Liu X., Zhao J., Zhang H., Chen W. (2017). Production of exopolysaccharide by Bifidobacterium longum isolated from elderly and infant feces and analysis of priming glycosyltransferase genes. RSC Adv. 7 31736–31744. 10.1039/c7ra03925e [DOI] [Google Scholar]
- You H. J., Oh D. K., Ji G. E. (2004). Anticancerogenic effect of a novel chiroinositol-containing polysaccharide from Bifidobacterium bifidum BGN4. FEMS Microbiol. Lett. 240 131–136. 10.1016/j.femsle.2004.09.020 [DOI] [PubMed] [Google Scholar]
- Yousef E. M., Furrer D., Laperriere D. L., Tahir M. R., Mader S., Diorio C., et al. (2017). MCM2: an alternative to Ki-67 for measuring breast cancer cell proliferation. Mod. Pathol. 30 682–697. 10.1038/modpathol.2016.231 [DOI] [PubMed] [Google Scholar]
- Yu S. W., Andrabi S. A., Wang H., Kim N. S., Poirier G. G., Dawson T. M., et al. (2006). Apoptosis-inducing factor mediates poly(ADP-ribose) (PAR) polymer-induced cell death. Proc. Natl. Acad. Sci. U.S.A. 103 18314–18319. 10.1073/pnas.0606528103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Wang B., Ma Z., Wei M., Liu J., Li D., et al. (2016). L-rhamnose enhances the immunogenicity of melanoma-associated antigen A3 for stimulating antitumor immune responses. Bioconjug. Chem. 27 1112–1118. 10.1021/acs.bioconjchem.6b00081 [DOI] [PubMed] [Google Scholar]
- Zhou H., Qin Y., Ji S., Ling J., Fu J., Zhuang Z., et al. (2018). SOX9 activity is induced by oncogenic Kras to affect MDC1 and MCMs expression in pancreatic cancer. Oncogene 37 912–923. 10.1038/onc.2017.393 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.