Abstract
The γ-carbonic anhydrases (CAs, EC 4.2.1.1) present in the Antarctic marine bacteria Pseudoalteromonas haloplanktis and Colwellia psychrerythraea, herein referred to as PhaCA and CpsCA, respectively, were investigated for their activation with a panel of 24 amino acids and amines. Both bacteria are considered Antarctic models for the investigation of photosynthetic and metabolic pathways in organisms adapted to live in cold seawater. PhaCA was much more sensitive to activation by these compounds compared to the genetically related enzyme CpsCA. The most effective PhaCA activators were d-Phe, l-/d-DOPA, l-Tyr and 2-pyridyl-methylamine, with the activation constant KA values of 0.72–3.27 µM. d-His, l-Trp, d-Tyr, histamine, dopamine, serotonin anddicarboxylic amino acids were also effective activators of PhaCA, with KA values of 6.48–9.85 µM. CpsCA was activated by d-Phe, d-DOPA, l-Trp, l-/d-Tyr, 4-amino-l-Phe, histamine, 2-pyridyl-methylamine and l-/d-Glu with KA values of 11.2–24.4 µM. The most effective CpsCA activator was l-DOPA (KA of 4.79 µM). Given that modulators of CAs from Antarctic bacteria have not been identified and investigated in detail for their metabolic roles to date, this research sheds some light on these poorly understood processes.
Keywords: Antarctic bacteria, carbonic anhydrase, metalloenzymes, amine, amino acid, activator, Pseudoalteromonas haloplanktis, Colwellia psychrerythraea
1. Introduction
Pseudoalteromonas haloplanktis and Colwellia psychrerythraea are bacteria adapted to live in the extreme conditions of the Antarctic Ocean. These marine heterotrophic bacteria are thought to rely on simplified metabolic strategies which evolved in order to overcome thermodynamic constraints connected toliving at low temperatures and in the dark for prolonged periods during the Antarctic winter [1,2,3,4,5,6,7]. Thus, these two bacteria are considered model organisms for studying metabolic processes and, more generally, strategies used for adaptation to harsh environmental conditions typical of the Antarctic region, which may also provide interesting insights for potential biotechnological or biomedical applications of enzymes encoded in their genomes [1,2,5].
Carbonic anhydrases (CAs, EC 4.2.1.1) are metalloproteins involved in several metabolic processes [8,9,10,11,12,13,14,15,16,17] either directly, by providing CO2/bicarbonate for carboxylating reactions [8,9,12,13], or indirectly, by modulating pH [13,14,15,16,17]. Indeed, CAs convert the neutral molecules CO2 and water to a weak base (bicarbonate) and a strong acid (hydronium ion) with very high efficacy [8,13]. For this reason, in many organisms including bacteria, CAs are the main players in pH homeostasis and related physiologic processes, which are involved in the metabolism, survival and colonization of various niches in which these organisms thrive [18,19,20,21,22,23,24,25]. These processes were mainly investigated for pathogenic microorganisms, such as bacteria [9,13,16,17], protozoa [10,11,21,22] and fungi [17,26]. However, the wide distributions of these enzymes in non-pathogenic Archaea [25] and Bacteria [3,4,6,7,27] make them of interest for understanding crucial biochemical processes connected with metabolism, adaptation to various environmental conditions, and survival in extreme environments. Indeed, we have reported CAs from extremophilic bacteria living at 110 °C in volcanic hot springs [28,29,30], as well as CAs from Antarctic organisms which thrive at very low temperatures, such as PhaCA from P. haloplanktis [3,4] and CpsCA from C. psychrerythraea [6,7]. Both these CAs belong to the γ-CA family, which is the least investigated CA family to date [9,12,13,25]. It should be mentioned that seven genetically unrelated CA families are known to date, the α-, β-, γ-, δ-, ζ-, η- and θ-CAs [12,23], and many of their inhibitors possess pharmacologic or environmental applications [14,15,31,32,33]. However, the activators of CA enzymes have been investigated in far less detail, with exception for the α-class enzymes. Indeed, activation of the human CAs, of which 15 isoforms are known to date [14,15], was recently shown to have the potential for pharmacologic applications in memory therapy and learning [34,35].
Here we report the first activation study of γ-CAs from Antarctic bacteria. We present an amino acid and amine activation study of PhaCA cloned from P. haloplanktis, and of CpsCA from C. psychrerythraea.
2. Results and Discussion
The catalytic activity of the recombinant γ-CAs from P. haloplanktis and C. psychrerythraea for the CO2 hydration reaction was reported earlier by our group [3,4,6,7]. Both enzymes showed a significant catalytic activity for the physiologic CO2 hydration reaction, with the following kinetic parameters: kcat = 1.4 × 105 s−1 and kcat/Km = 1.9 × 106 M−1·s−1 for PhaCA, and kcat = 6.0 × 105 s−1 and kcat/Km = 4.7 × 106 M−1·s−1 for CpsCA. Considering that these enzymes belong to the γ-CA class, which has the lowest CO2 hydrase activity of all CAs, the measured activities are indeed rather significant, especially when compared to those of other CAs (such as the human isoforms hCA I and II) used in our experiments as controls (Table 1). The activity of the enzymes PhaCA and CpsCA were also inhibited by acetazolamide (AZA, 5-acetamido-1,3,4-thiadiazole-2-sulfonamide), a clinically used sulfonamide inhibitor with KI values ranging between 403 and 502 nM [3,4,6,7].
Table 1.
Activation of human carbonic anhydrase (hCA) isozymes I, II, PhaCA and CpsCA with l-Trp, at 25 °C, for the CO2 hydration reaction [42].
| Isozyme | kcat 1 (s−1) | KM 1 (mM) | (kcat) l-Trp 2 (s−1) | KA 3 (μM) l-Trp |
|---|---|---|---|---|
| hCA I a | 2.0 × 105 | 4.0 | 3.4 × 105 | 44.0 |
| hCA II a | 1.4 × 106 | 9.3 | 4.9 × 106 | 27.0 |
| PhaCA b | 1.4 × 105 | 7.3 | 7.6 × 105 | 7.12 |
| CpsCA b | 6.0 × 105 | 12.7 | 9.9 × 105 | 21.3 |
1 Observed catalytic rate without activator. KM values in the presence and the absence of activators were the same for the various carbonic anhydrases (CAs) (data not shown). 2 Observed catalytic rate in the presence of 10 μM activator. 3 The activation constant (KA) for each enzyme was obtained by fitting the observed catalytic enhancements as a function of the activator concentration [41]. The mean was obtained from at least three determinations by a stopped-flow CO2 hydrase method [42]. Standard errors were in the range of 5–10% of the reported values (data not shown). a Human recombinant isozymes, from [32]; b Antarctic bacteria recombinant enzyme, this work.
As for all CA classes investigated so far, the activators of the γ-CAs participate in the catalytic cycle [35,36,37,38,39,40,41,42], most probably by facilitating the rate-determining step of the catalytic cycle (shown schematically by the ping-pong Reactions (1) and (2) below). In fact, the rate-determining step for many CAs is the generation of the nucleophilic species of the enzyme (EZnOH), depicted by Equation (2):
| (1) |
| (2) |
In all CAs, the step shown in Equation (2) is assisted by amino acid residues from the active site [35], which perform the function of shuttling protons between the catalytic active site and the solvent media. Thus, in the presence of sufficiently high concentrations of CA activators CAAs, the intermolecular proton transfer reaction (Equation (2)), becomes an intramolecular step within the enzyme–activator complex, which is favored thermodynamically. In this way, the activators (CAAs) participate in the rate-determining step of the catalytic cycle, which is represented by Equation (3):
| (3) |
The enzyme–activator complexes (shown as E–A, where E stands for the enzyme and A for the activator) favor the intramolecular proton transfer step. In this way, the zinc-coordinated water loses a proton to the environment (in this intramolecular step), and promotes the formation of the catalytically active nucleophilic species of the enzyme, with the hydroxide ion coordinating the zinc ion [35]. In all X-ray crystal structures available so far for CAs complexed to activators, these molecules were observed to be bound within the active site crevice of the enzyme [35,36,37,38,39,40,41] in a region of the cavity that is distinct from the inhibitor binding site(s). However, to date, only the α-class CAs have been investigated in detail by this technique. Indeed, the activator binding sites for α-CAs such as hCA I and II (h = human isoform) were identified to be at the entrance of the active site cavity, near the amino acid residue His64 (hCA II numbering [35]), which is also the natural proton shuttle residue for enzymes belonging to the α-class [35,36,37,38,39,40,41]. It should be mentioned that the nature of the amino acid residue(s) involved in the proton shuttling for the γ-CAs is for the moment rather controversial, although several proposals were reported, such as for example two Glu residues (for CAM, the first such enzyme investigated in detail by Ferry’s group from the archaeon Methanosarcina thermophila [25]) situated at the entrance of the cavity.
We measured the kinetic constants (kcat and KM) of the two γ-CAs (for the physiologic CO2 hydration reaction) investigated here in the presence of 10 μM l-Trp as an activator in order to see whether the activation mechanism is different from that of the α-class enzymes investigated earlier [35]. The data of Table 1 show that the presence of l-Trp does not change the value of KM for both of the two enzymes belonging to the α-class (hCA I/II) as well as for the two bacterial Antarctic enzymes PhaCA and CpsCA. In fact, the activator has an effect only on the kcat, which at a 10 µM concentration of the activator leads to an enhancement of 5.42 times of the kinetic constant for PhaCA, and of 1.65 times for CpsCA (Table 1). The same type of enhancement of the kcat with no influence on Km was observed for hCA I and II [35], indicating that the activation mechanisms of the two enzyme classes must be rather similar.
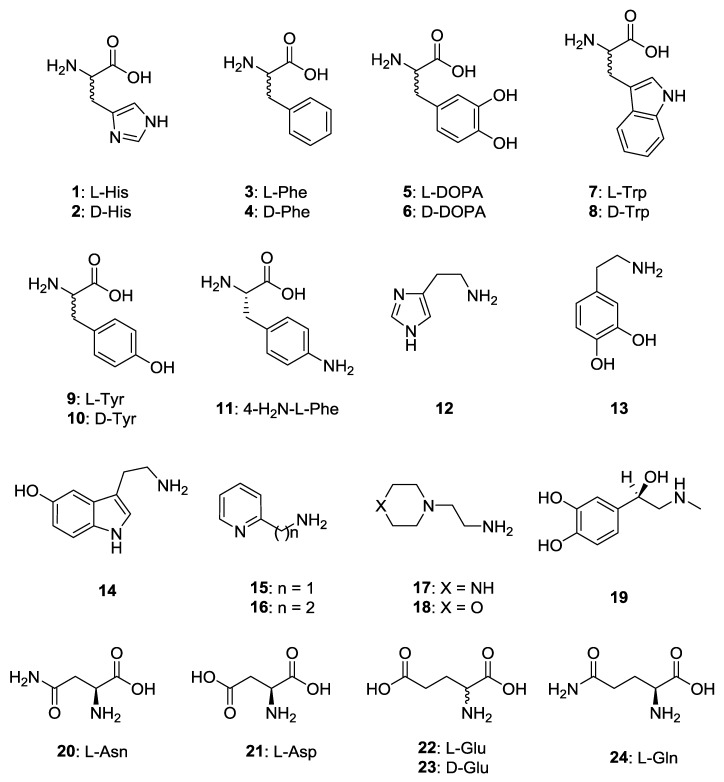
We report the activation profiles of PhaCA and CpsCA with a wide range of amino acid and amine activators of types 1–24 (Figure 1), which were obtained by measuring the dose response curves of the activation of the two enzymes in the presence of increasing concentrations of activators. It should be mentioned that in order to act as CAAs, the amino and COOH moiety of the inhibitor should generally not be derivatized [35]. Small modifications such as N-methylation or the transformation of the COOH to COOMe groups are tolerated and do not significantly change the CA activating properties, as demonstrated earlier by us [35,40,41]. This is the reason why we include amines and amino acids 1–24 in our study and not some of their derivatives. In this way, the KA values of the 24 compounds were determined for the two enzymes (see Materials and Methods for details). We included in our study the amino acids and amines that were investigated as activators of CAs belonging to various classes of enzymes present in diverse organisms, both eukaryotic and prokaryotic [32,33,34,35,36,37,40,41,42].
Figure 1.
CA activators CAAs of types 1–24 used in the present study.
The structure–activity relationship (SAR) for the activation of PhaCA/CpsCA with compounds 1–24 revealed the following observations (Table 2):
-
(i)
Firstly, PhaCA is much more sensitive to activation with amines and amino acids 1–24, compared to CpsCA, although both enzymes are prokaryotic, belonging to the same class. Indeed, the activation constants of these amines ranged between 0.72 and 32.4 µM for PhaCA, whereas they were only in the range of 4.79 to 100 µM for CpsCA.
-
(ii)
The most effective PhaCA activators were d-Phe, l-/d-DOPA, l-Tyr and amine 15, which showed KA values ranging between 0.72 and 3.27 µM. Compounds such as d-His, l-Trp, d-Tyr, histamine, dopamine, serotonin and 20–23 were also effective activators, with KA values ranging between 6.48 and 9.85 µM. Medium potency activators were l-His, l-Phe, d-Trp, amines 16–19 and l-Gln, which showed KA values ranging between 10.1 and 32.4 µM (Table 1). Thus, the SAR was rather complex, but generally the d-amino acids were better activators than their l-enantiomers (except for the enantiomer pairs l-/d-Trp and l-/d-Tyr). In some cases, the amines (histamine) were more effective activators than the structurally related amino acids (l- and d-His), whereas in other cases (l-/d-DOPA) the amino acids were more effective CAAs compared to the structurally related amine (dopamine). No major differences were observed between carboxylate/carboxamide derivatives in some cases (l-Asp/l-Asn), whereas for l-Glu, the carboxamide (l-Gln) was more than three times less effective as an activator.
-
(iii)
For CpsCA, l-Phe and l-Gln were devoid of activating effects up to a 100 µM concentration of compound in the assay system. Weak activators were also l-/d-His, d-Trp, dopamine, serotonin, amines 16–19, as well as l-Asn and l-Asp (KA values ranging between 27.3 and 79.8 µM). On the other hand, better activators of CpsCA were d-Phe, d-DOPA, l-Trp, l-/d-Tyr, 4-amino-l-Phe, histamine, 2-pyridyl-methylamine and l-/d-Glu, with KA values ranging between 11.2 and 24.4 µM. The most effective CpsCA activator was l-DOPA, with a KA of 4.79 µM. It is obvious that very small structural changes in the molecule of the activator have drastic effects on the enzyme activating effects. For example, the two enantiomers of DOPA have values of KA which differ by a factor of 2.33, with the l-enantiomer being the most active. However, the amino acid in which one of the two OH moieties of DOPA is missing, l-Tyr, was 4 times less effective compared to DOPA, whereas l-Phe was more than 20 times less effective as an activator of this isoform (the compound lacking both phenolic OH groups present in DOPA).
-
(iv)
As no X-ray crystal structure for adducts of activators with γ-CAs are available so far, we cannot rationalize our SAR data in detail. However, all the observations reported above concur with the fact that these compounds bind within the enzyme active site and facilitate the generation of the nucleophilic zinc hydroxide species of the enzyme.
Table 2.
Activation constants of hCA I, hCA II and the bacterial enzymes PhaCA (Pseudoalteromonas haloplanktis) and CpsCA (Colwellia psychrerythraea) with amino acids and amines 1–24, by a stopped-flow CO2 hydrase assay [42].
| No. | Compound | KA (μM) * | |||
|---|---|---|---|---|---|
| hCA I a | hCA II a | PhaCA b | CpsCA b | ||
| 1 | l-His | 0.03 | 10.9 | 12.6 | 47.5 |
| 2 | d-His | 0.09 | 43 | 9.41 | 35.9 |
| 3 | l-Phe | 0.07 | 0.013 | 15.8 | >100 |
| 4 | d-Phe | 86 | 0.035 | 3.19 | 15.4 |
| 5 | l-DOPA | 3.1 | 11.4 | 1.08 | 4.79 |
| 6 | d-DOPA | 4.9 | 7.8 | 0.72 | 11.2 |
| 7 | l-Trp | 44 | 27 | 7.12 | 21.3 |
| 8 | d-Trp | 41 | 12 | 13.9 | 36.8 |
| 9 | l-Tyr | 0.02 | 0.011 | 1.02 | 19.5 |
| 10 | d-Tyr | 0.04 | 0.013 | 7.35 | 18.4 |
| 11 | 4-H2N-l-Phe | 0.24 | 0.15 | 3.27 | 17.2 |
| 12 | Histamine | 2.1 | 125 | 6.48 | 20.6 |
| 13 | Dopamine | 13.5 | 9.2 | 8.70 | 32.1 |
| 14 | Serotonin | 45 | 50 | 9.05 | 34.8 |
| 15 | 2-Pyridyl-methylamine | 26 | 34 | 2.39 | 21.5 |
| 16 | 2-(2-Aminoethyl)pyridine | 13 | 15 | 18.7 | 38.2 |
| 17 | 1-(2-Aminoethyl)-piperazine | 7.4 | 2.3 | 15.1 | 33.0 |
| 18 | 4-(2-Aminoethyl)-morpholine | 0.14 | 0.19 | 10.1 | 34.3 |
| 19 | l-Adrenaline | 0.09 | 96.0 | 17.5 | 79.8 |
| 20 | l-Asn | 11.3 | >100 | 9.80 | 27.9 |
| 21 | l-Asp | 5.20 | >100 | 9.85 | 27.3 |
| 22 | l-Glu | 6.43 | >100 | 9.01 | 24.4 |
| 23 | d-Glu | 10.7 | >100 | 4.72 | 12.0 |
| 24 | l-Gln | >100 | >50 | 32.4 | >100 |
3. Materials and Methods
The protocol described in [3,4,6,7] was used to obtain purified recombinant Pseudoalteromonas haloplanktis (PhaCA) and Colwellia psychrerythraea (CpsCA) enzymes.
CA Activity/Activation Measurements
An Sx.18Mv-R Applied Photophysics (Oxford, UK) stopped-flow instrument was used to assay the catalytic activity of various CA isozymes for CO2 hydration reaction [42]. Phenol red (at a concentration of 0.2 mM) was used as an indicator, working at the absorbance maximum of 557 nm, with 10 mM Hepes (pH 7.5, for α-CAs) or TRIS (pH 8.3, for γ-CAs) as buffers, and 0.1 M NaClO4 (for maintaining constant ionic strength), following the CA-catalyzed CO2 hydration reaction for a period of 10 s at 25 °C. The CO2 concentrations ranged from 1.7 to 17 mM for the determination of the kinetic parameters and inhibition constants. For each activator, at least six traces of the initial 5–10% of the reaction were used to determine the initial velocity. The uncatalyzed rates were determined in the same manner and subtracted from the total observed rates. Stock solutions of activators (at 0.1 mM) were prepared in distilled-deionized water and dilutions up to 1 nM were made thereafter with the assay buffer. Enzyme and activator solutions were pre-incubated together for 15 min prior to the assay, in order to allow for the formation of the enzyme–activator complexes. The activation constant (KA), defined similarly as the inhibition constant KI, can be obtained by considering the classical Michaelis–Menten equation (Equation (4)), which was fitted by non-linear least squares by using PRISM 3:
| (4) |
where [A]f is the free concentration of activator.
Working at substrate concentrations considerably lower than KM ([S] << KM), and considering that [A]f can be represented in the form of the total concentration of the enzyme ([E]t) and activator ([A]t), the obtained competitive steady-state equation for determining the activationconstant is given by Equation (5):
| (5) |
where v0 represents the initial velocity of the enzyme-catalyzed reaction in the absence of an activator [43,44,45]. The equilibrium constants measured using this approach are in excellent agreement with those obtained from other methods, including native mass spectrometry and fluorescence spectroscopy [46].
4. Conclusions
γ-CAs are present in bacteria, archaea and plants, constituting probably the most archaic class of such enzymes [9]. In the Antarctic marine bacteria Pseudoalteromonas haloplanktis and Colwellia psychrerythraea, two such enzymes—PhaCA and CpsCA, respectively—were recently reported. However, their role in the life cycle of these prokaryotes is poorly investigated. However, both these bacteria are considered model Antarctic organisms for the investigation of photosynthetic and metabolic pathways in organisms adapted to live in extreme conditions that are characterized by cold seawater and absence of light during the Antarctic winter. A panel of 24 amino acid and amines were investigated as activators of the two enzymes. PhaCA was much more sensitive to activation by these compounds compared to the genetically related enzyme CpsCA. The best PhaCA activators were d-Phe, l-/d-DOPA, l-Tyr and 2-pyridyl-methylamine, with KA values of 0.72–3.27 µM. d-His, l-Trp, d-Tyr, histamine, dopamine, serotonin and dicarboxylic amino acids were also effective activators, with KA values of 6.48–9.85 µM. CpsCA was activated by d-Phe, d-DOPA, l-Trp, l-/d-Tyr, 4-amino-l-Phe, histamine, 2-pyridyl-methylamine and l-/d-Glu, with KA values of 11.2–24.4 µM. The most effective CpsCA activator was l-DOPA (KA of 4.79 µM). Sincemodulators of CAs from Antarctic bacteria have not yet been investigated in detail for their metabolic roles, these data provide valuable insights into these understudied processes.
Author Contributions
A.A. measured the kinetics and activation; S.D.P. performed the cloning, expression and purification of the recombinant coral enzymes; S.M.O. and Z.A. performed kinetic data analysis; W.A.D., C.C. and C.T.S. wrote and edited the manuscript. C.T.S. supervised the project.
Funding
This research was financed in part by a Distinguished Scientists Fellowship Programme (DSFP) of King Saud University, Riyadh, Saudi Arabia, to Z.A. and C.T.S. W.A.D. and C.T.S. thank the Australian Research Council for funding.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Mocali S., Chiellini C., Fabiani A., Decuzzi S., de Pascale D., Parrilli E., Tutino M.L., Perrin E., Bosi E., Fondi M., et al. Ecology of cold environments: New insights of bacterial metabolic adaptation through an integrated genomic-phenomic approach. Sci. Rep. 2017;7:839. doi: 10.1038/s41598-017-00876-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fondi M., Bosi E., Presta L., Natoli D., Fani R. Modelling microbial metabolic rewiring during growth in a complex medium. BMC Genom. 2016;17:970. doi: 10.1186/s12864-016-3311-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.De Luca V., Vullo D., Del Prete S., Carginale V., Scozzafava A., Osman S.M., AlOthman Z., Supuran C.T., Capasso C. Cloning, characterization and anion inhibition studies of a new γ-carbonic anhydrase from the Antarctic bacterium Pseudoalteromonas haloplanktis. Bioorg. Med. Chem. 2015;23:4405–4409. doi: 10.1016/j.bmc.2015.06.021. [DOI] [PubMed] [Google Scholar]
- 4.Vullo D., De Luca V., Del Prete S., Carginale V., Scozzafava A., Capasso C., Supuran C.T. Sulfonamide inhibition studies of the γ-carbonic anhydrase from the Antarctic bacterium Pseudoalteromonas haloplanktis. Bioorg. Med. Chem. Lett. 2015;25:3550–3555. doi: 10.1016/j.bmcl.2015.06.079. [DOI] [PubMed] [Google Scholar]
- 5.Czajka J.J., Abernathy M.H., Benites V.T., Baidoo E.E.K., Deming J.W., Tang Y.J. Model metabolic strategy for heterotrophic bacteria in the cold ocean based on Colwellia psychrerythraea 34H. Proc. Natl. Acad. Sci. USA. 2018;115:12507–12512. doi: 10.1073/pnas.1807804115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vullo D., De Luca V., Del Prete S., Carginale V., Scozzafava A., Osman S.M., AlOthman Z., Capasso C., Supuran C.T. Sulfonamide inhibition studies of the γ-carbonic anhydrase from the Antarctic bacterium Colwellia psychrerythraea. Bioorg. Med. Chem. Lett. 2016;26:1253–1259. doi: 10.1016/j.bmcl.2016.01.023. [DOI] [PubMed] [Google Scholar]
- 7.De Luca V., Vullo D., Del Prete S., Carginale V., Osman S.M., AlOthman Z., Supuran C.T., Capasso C. Cloning, characterization and anion inhibition studies of a γ-carbonic anhydrase from the Antarctic bacterium Colwellia psychrerythraea. Bioorg. Med. Chem. 2016;24:835–840. doi: 10.1016/j.bmc.2016.01.005. [DOI] [PubMed] [Google Scholar]
- 8.Supuran C.T. Carbonic Anhydrases and Metabolism. Metabolites. 2018;8:25. doi: 10.3390/metabo8020025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Capasso C., Supuran C.T. An overview of the alpha-, beta-and gamma-carbonic anhydrases from Bacteria: Can bacterial carbonic anhydrases shed new light on evolution of bacteria? J. Enzyme Inhib. Med. Chem. 2015;30:325–332. doi: 10.3109/14756366.2014.910202. [DOI] [PubMed] [Google Scholar]
- 10.Bua S., Haapanen S., Kuuslahti M., Parkkila S., Supuran C.T. Sulfonamide Inhibition Studies of a New β-Carbonic Anhydrase from the Pathogenic Protozoan Entamoebahistolytica. Int. J. Mol. Sci. 2018;19:3946. doi: 10.3390/ijms19123946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haapanen S., Bua S., Kuuslahti M., Parkkila S., Supuran C.T. Cloning, Characterization and Anion Inhibition Studies of a β-Carbonic Anhydrase from the Pathogenic Protozoan Entamoeba histolytica. Molecules. 2018;23:3112. doi: 10.3390/molecules23123112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Supuran C.T. Structure and function of carbonic anhydrases. Biochem. J. 2016;473:2023–2032. doi: 10.1042/BCJ20160115. [DOI] [PubMed] [Google Scholar]
- 13.Supuran C.T., Capasso C. An Overview of the Bacterial Carbonic Anhydrases. Metabolites. 2017;7:56. doi: 10.3390/metabo7040056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Supuran C.T. Carbonic anhydrases: Novel therapeutic applications for inhibitors and activators. Nat. Rev. Drug Discov. 2008;7:168–181. doi: 10.1038/nrd2467. [DOI] [PubMed] [Google Scholar]
- 15.Neri D., Supuran C.T. Interfering with pH regulation in tumours as a therapeutic strategy. Nat. Rev. Drug Discov. 2011;10:767–777. doi: 10.1038/nrd3554. [DOI] [PubMed] [Google Scholar]
- 16.Supuran C.T., Capasso C. New light on bacterial carbonic anhydrases phylogeny based on the analysis of signal peptide sequences. J. Enzyme Inhib. Med. Chem. 2016;31:1254–1260. doi: 10.1080/14756366.2016.1201479. [DOI] [PubMed] [Google Scholar]
- 17.Supuran C.T., Capasso C. Biomedical applications of prokaryotic carbonic anhydrases. Expert Opin. Ther. Pat. 2018;28:745–754. doi: 10.1080/13543776.2018.1497161. [DOI] [PubMed] [Google Scholar]
- 18.Supuran C.T. Carbonic Anhydrase Inhibition and the Management of Hypoxic Tumors. Metabolites. 2017;7:48. doi: 10.3390/metabo7030048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Supuran C.T. Advances in structure-based drug discovery of carbonic anhydrase inhibitors. Expert Opin. Drug Discov. 2017;12:61–88. doi: 10.1080/17460441.2017.1253677. [DOI] [PubMed] [Google Scholar]
- 20.Nishimori I., Onishi S., Takeuchi H., Supuran C.T. The α and β-Classes Carbonic Anhydrases from Helicobacter pylori as Novel Drug Targets. Curr. Pharm. Des. 2008;14:622–630. doi: 10.2174/138161208783877875. [DOI] [PubMed] [Google Scholar]
- 21.da Silva Cardoso V., Vermelho A.B., Ricci Junior E., Almeida Rodrigues I., Mazotto A.M., Supuran C.T. Antileishmanial activity of sulphonamidenanoemulsions targeting the β-carbonic anhydrase from Leishmania species. J. Enzyme Inhib. Med. Chem. 2018;33:850–857. doi: 10.1080/14756366.2018.1463221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vermelho A.B., da Silva Cardoso V., Ricci Junior E., Dos Santos E.P., Supuran C.T. Nanoemulsions of sulfonamide carbonic anhydrase inhibitors strongly inhibit the growth of Trypanosomacruzi. J. Enzyme Inhib. Med. Chem. 2018;33:139–146. doi: 10.1080/14756366.2017.1405264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Supuran C.T. How many carbonic anhydrase inhibition mechanisms exist? J. Enzyme Inhib. Med. Chem. 2016;31:345–360. doi: 10.3109/14756366.2015.1122001. [DOI] [PubMed] [Google Scholar]
- 24.Murray A.B., Aggarwal M., Pinard M., Vullo D., Patrauchan M., Supuran C.T., McKenna R. Structural Mapping of Anion Inhibitors to β-Carbonic Anhydrase psCA3 from Pseudomonas aeruginosa. Chem. Med. Chem. 2018;13:2024–2029. doi: 10.1002/cmdc.201800375. [DOI] [PubMed] [Google Scholar]
- 25.Zimmerman S.A., Ferry J.G., Supuran C.T. Inhibition of the archaeal beta-class (Cab) and gamma-class (Cam) carbonic anhydrases. Curr. Top. Med. Chem. 2007;7:901–908. doi: 10.2174/156802607780636753. [DOI] [PubMed] [Google Scholar]
- 26.Nocentini A., Vullo D., Del Prete S., Osman S.M., Alasmary F.A.S., AlOthman Z., Capasso C., Carta F., Gratteri P., Supuran C.T. Inhibition of the β-carbonic anhydrase from the dandruff-producing fungus Malasseziaglobosa with monothiocarbamates. J. Enzyme Inhib. Med. Chem. 2017;32:1064–1070. doi: 10.1080/14756366.2017.1355307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.De Simone G., Supuran C.T. (In)organic anions as carbonic anhydrase inhibitors. J. Inorg. Biochem. 2012;111:117–129. doi: 10.1016/j.jinorgbio.2011.11.017. [DOI] [PubMed] [Google Scholar]
- 28.Di Fiore A., Capasso C., De Luca V., Monti S.M., Carginale V., Supuran C.T., Scozzafava A., Pedone C., Rossi M., De Simone G. X-ray structure of the first “extremo-α-carbonic anhydrase”, a dimeric enzyme from the thermophilic bacterium Sulfurihydrogenibium yellowstonense YO3AOP1. Acta Crystallogr. Sect. D Biol. Crystallogr. 2013;69:1150–1159. doi: 10.1107/S0907444913007208. [DOI] [PubMed] [Google Scholar]
- 29.Akdemir A., Vullo D., De Luca V., Scozzafava A., Carginale V., Rossi M., Supuran C.T., Capasso C. The extremo-α-carbonic anhydrase (CA) from Sulfurihydrogenibium azorense, the fastest CA known, is highly activated by amino acids and amines. Bioorg. Med. Chem. Lett. 2013;23:1087–1090. doi: 10.1016/j.bmcl.2012.12.009. [DOI] [PubMed] [Google Scholar]
- 30.Vullo D., De Luca V., Scozzafava A., Carginale V., Rossi M., Supuran C.T., Capasso C. The first activation study of a bacterial carbonic anhydrase (CA). The thermostable α-CA from Sulfurihydrogenibium yellowstonense YO3AOP1 is highly activated by amino acids and amines. Bioorg. Med. Chem. Lett. 2012;22:6324–6327. doi: 10.1016/j.bmcl.2012.08.088. [DOI] [PubMed] [Google Scholar]
- 31.Supuran C.T. Applications of carbonic anhydrases inhibitors in renal and central nervous system diseases. Expert Opin. Ther. Pat. 2018;28:713–721. doi: 10.1080/13543776.2018.1519023. [DOI] [PubMed] [Google Scholar]
- 32.Supuran C.T. Carbonic anhydrase inhibitors and their potential in a range of therapeutic areas. Expert Opin. Ther. Pat. 2018;28:709–712. doi: 10.1080/13543776.2018.1523897. [DOI] [PubMed] [Google Scholar]
- 33.Nocentini A., Supuran C.T. Carbonic anhydrase inhibitors as antitumor/antimetastaticagents: A patent review (2008–2018) Expert Opin. Ther. Pat. 2018;28:729–740. doi: 10.1080/13543776.2018.1508453. [DOI] [PubMed] [Google Scholar]
- 34.Canto de Souza L., Provensi G., Vullo D., Carta F., Scozzafava A., Costa A., Schmidt S.D., Passani M.B., Supuran C.T., Blandina P. Carbonic anhydrase activation enhances object recognition memory in mice through phosphorylation of the extracellular signal-regulated kinase in the cortex and the hippocampus. Neuropharmacology. 2017;118:148–156. doi: 10.1016/j.neuropharm.2017.03.009. [DOI] [PubMed] [Google Scholar]
- 35.Supuran C.T. Carbonic anhydrase activators. Future Med. Chem. 2018;10:561–573. doi: 10.4155/fmc-2017-0223. [DOI] [PubMed] [Google Scholar]
- 36.Angeli A., Kuuslahti M., Parkkila S., Supuran C.T. Activation studies with amines and amino acids of the α-carbonic anhydrase from the pathogenic protozoan Trypanosoma cruzi. Bioorg. Med. Chem. 2018;26:4187–4190. doi: 10.1016/j.bmc.2018.07.011. [DOI] [PubMed] [Google Scholar]
- 37.Angeli A., Del Prete S., Alasmary F.A.S., Alqahtani L.S., AlOthman Z., Donald W.A., Capasso C., Supuran C.T. The first activation studies of the η-carbonic anhydrase from the malaria parasite Plasmodium falciparum with amines and amino acids. Bioorg. Chem. 2018;80:94–98. doi: 10.1016/j.bioorg.2018.06.002. [DOI] [PubMed] [Google Scholar]
- 38.Stefanucci A., Angeli A., Dimmito M.P., Luisi G., Del Prete S., Capasso C., Donald W.A., Mollica A., Supuran C.T. Activation of β- and γ-carbonic anhydrases from pathogenic bacteria with tripeptides. J. Enzyme Inhib. Med. Chem. 2018;33:945–950. doi: 10.1080/14756366.2018.1468530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Angeli A., Alasmary F.A.S., Del Prete S., Osman S.M., AlOthman Z., Donald W.A., Capasso C., Supuran C.T. The first activation study of a δ-carbonic anhydrase: TweCAδ from the diatom Thalassiosiraweissflogii is effectively activated by amines and amino acids. J. Enzyme Inhib. Med. Chem. 2018;33:680–685. doi: 10.1080/14756366.2018.1447570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Briganti F., Mangani S., Orioli P., Scozzafava A., Vernaglione G., Supuran C.T. Carbonic anhydrase activators: X-ray crystallographic and spectroscopic investigations for the interaction of isozymes I and II with histamine. Biochemistry. 1997;36:10384–10392. doi: 10.1021/bi970760v. [DOI] [PubMed] [Google Scholar]
- 41.Clare B.W., Supuran C.T. Carbonic anhydrase activators. 3: Structure-activity correlations for a series of isozyme II activators. J. Pharm. Sci. 1994;83:768–773. doi: 10.1002/jps.2600830603. [DOI] [PubMed] [Google Scholar]
- 42.Khalifah R.G. The carbon dioxide hydration activity of carbonic anhydrase. I. Stop-flow kinetic studies on the native human isoenzymes B and C. J. Biol. Chem. 1971;246:2561–2573. [PubMed] [Google Scholar]
- 43.Temperini C., Scozzafava A., Vullo D., Supuran C.T. Carbonic anhydrase activators. Activation of isoforms I, II, IV, VA, VII, and XIV with l- and d-phenylalanine and crystallographic analysis of their adducts with isozymeII: Stereospecific recognition within the active site of an enzyme and its consequences for the drug design. J. Med. Chem. 2006;49:3019–3027. doi: 10.1021/jm0603320. [DOI] [PubMed] [Google Scholar]
- 44.Angeli A., Donald W.A., Parkkila S., Supuran C.T. Activation studies with amines and amino acids of the β-carbonic anhydrase from the pathogenic protozoan Leishmania donovanichagasi. Bioorg. Chem. 2018;78:406–410. doi: 10.1016/j.bioorg.2018.04.010. [DOI] [PubMed] [Google Scholar]
- 45.Rami M., Winum J.Y., Supuran C.T., Melnyk P., Yous S. (Hetero)aryl substituted thiazol-2,4-yl scaffold as human carbonic anhydrase I, II, VII and XIV activators. J. Enzyme Inhib. Med. Chem. 2019;34:224–229. doi: 10.1080/14756366.2018.1543292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nguyen G.T.H., Tran T.N., Podgorski M.N., Bell S.G., Supuran C.T., Donald W.A. Nanoscale ion emitters in native mass spectrometry for measuring ligand-protein binding affinities. ACS Cent. Sci. 2019;5:308–318. doi: 10.1021/acscentsci.8b00787. [DOI] [PMC free article] [PubMed] [Google Scholar]