Abstract
As the only widely used live lentiviral vaccine, the equine infectious anima virus (EIAV) attenuated vaccine was developed by in vitro passaging of a virulent strain for 121 generations. In our previous study, we observed that the attenuated vaccine was gradually selected under increased environmental pressure at the population level (termed a quasispecies). To further elucidate the potential correlation between viral quasispecies evolution and pathogenesis, a systematic study was performed by sequencing env using several methods. Some key mutations were identified within Env, and we observed that increased percentages of these mutations were accompanied by an increased passage number and attenuated virulence. Phylogenetic analysis revealed that env mutations related to the loss of virulence might have occurred evolutionarily. Among these mutations, deletion of amino acid 236 in the V4 region of Env resulted in the loss of one N-glycosylation site that was crucial for virulence. Notably, the 236-deleted sequence represented a “vaccine-specific” mutation that was also found in wild EIAVLN40 strains based on single genome amplification (SGA) analysis. Therefore, our results suggest that the EIAV attenuated vaccine may originate from a branch of quasispecies of EIAVLN40. Generally, the presented results may increase our understanding of the attenuation mechanism of the EIAV vaccine and provide more information about the evolution of other lentiviruses.
Keywords: EIAV, SGA (single genome amplification), quasispecies, virulence
1. Introduction
As the simplest member of the lentivirus family, equine infectious anemia virus (EIAV) shares similar features with other lentiviruses, including its genomic structure, life cycle, cell tropism and antigen evolution [1,2]. The uniqueness of EIAV lies in the fact that most infected horses become asymptomatic carriers after recurrent febrile episodes, which are associated with a high viral load and anemia, and thus the horses achieve natural immunologic control [3]. Notably, an attenuated vaccine strain has been developed that successfully controls EIAV prevalence in China. The vaccine was generated by serial passage of a virulent EIAV strain (EIAVLN40) for 16 generations in vivo, followed by passaging for as many as 121 passages in vitro [4,5]. Due to these peculiarities, the EIAV attenuation system may provide an ideal model for elucidating the correlates of genomic evolution and immunologic control.
For RNA viruses, mutation swarms are generated rapidly through error-prone viral replication due to inaccuracy of the RNA polymerase [6]. This mutant swarm, which is also termed a viral quasispecies, appears as a whole population with a mutation-selection balance and is significantly involved in viral pathogenesis [7,8]. Viral quasispecies are also essential for viral survival because they yield beneficial viral phenotypes under in vivo selection. Interestingly, the mutation spectra are higher after long-term passage in vitro, which causes a change in virulence [9,10,11]. Hence, the relationship between RNA viral quasispecies evolution and viral pathogenesis is an intriguing problem.
As discussed previously, multi-position mutations located in the LTR and in env appeared during the EIAV attenuation process in vitro, especially in the viral env gene, which developed eight hyper variable regions located in the V3 and V4 regions [1,12,13,14]. Further analysis showed that these variations were related to viral pathogenesis during the EIAV attenuation process. A virulence-correlated parallel in env variation was also observed during the EIAV attenuation process [4]. In another study, we observed that the LTR showed a similar pattern at the population level [15]. However, the LTR is a noncoding region which could not reflect the predominant antigen gene of the EIAV evolutionary pattern. Hence, env was chosen as the target of evolutionary selection in this study to address this important question. The aim of this study was to characterize env quasispecies evolution and further investigate the related mechanism between virulence attenuation and env quasispecies. Our results will be of great interest for understanding the evolutionary mechanism of the EIAV attenuated vaccine and defining vaccine development strategies for other lentiviruses.
2. Materials and Methods
2.1. Study Subjects
All samples were stored at the Harbin Veterinary Research Institute, Chinese Academy of Agricultural Sciences (CAAS). These samples included a virulent strain (EIAVLN40) that caused an infection in horses with 100% mortality at a dose of 1 × 105 TCID50; this strain was initially isolated from an EIA positive horse and passed for 16 generations in horses, resulting in three representative strains (EIAVDLV34, EIAVDLV62, and EIAVDLV92) with 100%, 100%, and 9.1% mortality that stemmed from EIAV passage in vitro for 34, 62, and 92 generations, respectively (Figure S1) [16]. Additionally, we included a vaccine strain (EIAVDLV121) that provided 85% protection against EIAVLN40 challenge and a full-length infectious molecular clone (pLGFD3-8) constructed from a vaccine strain and then passaged in fetal donkey dermal (FDD) cells for 3 generations. Animal experiments showed that this infectious clone was avirulent [17].
2.2. Viral RNA Extraction and cDNA Synthesis
Viral RNA was extracted from 140 μL of the virus samples using the Viral RNA Mini Kit (QIAamp, Dusseldorf, Germany) according to the manufacturer’s protocol. Reverse transcription of RNA to single-stranded cDNA was performed using the SuperScriptTM IV Reverse Transcriptase System (Life Technologies, Carlsbad, USA). First, 1 μg of RNA, dNTPs (0.5 mM each), and 0.5 μM of the env NR primer (5’-CAGCTACAATGGCAGCTATTATAGCAG-3’; nucleotides (nt) 6702 to 6676 of the EIAV sequence) were incubated for 5 min at 65 °C to denature the RNA secondary structure. First-strand cDNA synthesis was carried out in 20 μL reaction mixtures with 5× SSIV buffer, 5 mM DTT, 2 U/μL of an RNase inhibitor (RNaseOUT) (Life Technologies, Carlsbad, USA), and 10 U/μL of SuperScriptTM IV (Life Technologies, Carlsbad, USA). The reaction mixture was incubated at 55°C for 15 min and heated to 80°C for 10 min to complete the reverse transcription reaction, followed by RNase H (Life Technologies, Carlsbad, USA) digestion at 37 °C for 20 min. The synthesized cDNA was used immediately for PCR or stored at −80 °C.
2.3. Bulk PCR
The full-length hypervariable region of the env cassette was amplified by nested PCR from the viral cDNA. The specific method was as follows. First, 0.6 μL of bulk cDNA was used for the first-round PCR in a 20 μL volume. The PCR was performed using the KOD FX (Toyobo, Osaka, Japan) system, which included 2× PCR buffer, 0.4 mM dNTPs, and 0.1 μM of the env NF (5’-GAAGGCCATCAGGGAGGGAAG-3’; (nt) 5240 to 5260) and env NR primers. The cycling conditions were as follows: 94 °C for 2 min, followed by 35 cycles of 94 °C for 30 s, 58 °C for 30 s, and 68 °C for 1 min 30 s, and a final extension of 68 °C for 2 min. The second-round PCR was performed using 0.6 μL of the first-round PCR product and the V3-1 (5’-ACAAACATATACAGGACATCT-3’; (nt) 5808 to 5828) and V4-1 (5’-CTCCAATATTCCAAGAAATAC-3’; (nt) 6218 to 6198) primers under the same conditions used for the first-round PCR. The final PCR products were analyzed by 1% agarose gel electrophoresis. The recycled products were ligated into pMD18-T (TaKaRa, Dalian, China). The constructed plasmids were transformed into E. coli DH5α (TaKaRa, Dalian, China) and spread onto a plate. A total of 98 positive clones identified by PCR were sequenced by COME (Jilin, China).
2.4. Probe Design and Real-Time PCR
Two specific probes and primers were synthesized by General Biosystems (Anhui, China). The first probe was the 6-carboxy-fluorescein (FAM) probe (probe-FAM: 5’-TGCAGCAAAGTAAAAACACTTGGAT-3’; (nt) 6010 to 6034), which targeted the env-Δ236D-phenotype sequence, and the second was the hexachloride fluorescein (HEX) probe (probe-HEX: 5’-TGCAGCAAAGCGATAATAACACTTG-3’; (nt) 6010 to 6034), which targeted the env-236D-phenotype sequence. The env-Δ236D-phenotype and env-236D-phenotype sequences were named depending on the presence of the 236D deletion in the EIAV Env. The DNA fragments of env-Δ236D-phenotype and env-236D-phenotype were amplified with primer F (5’-ATAGGAGGTAGACTAAATGGTTCAGG-3’; (nt) 5634-5668) and primer R (5’-AAACAAAAAGAATGGAGGTTGGACA-3’; (nt) 6143-6119) and ligated into the pMD-18T (TaKaRa, Dalian, China) vector. Then, the two plasmids were serially diluted to 102 to 109 copies/μL as standards to construct real-time PCR standard curves. The experiments were performed using 1 μM of each primer, 10 μL of premix Ex Taq (TaKaRa, Dalian, China), 0.25 μM of the HEX-labeled or FAM-labeled probe, 2 μL of the cDNA samples and RNase-free water to a final volume of 20 μL. The samples were amplified using TaqMan-based real-time PCR on the ABI Prism 7500 sequence detection system (Applied Biosystems GmbH, Thermo Fisher Scientific, Wilmington, USA) with one cycle at 95 °C (5 min), followed by 40 cycles at 95 °C (15 s), 58 °C (30 s), and 72 °C (20 s). The calculated efficiencies for all primers were determined by dilution experiments and ranged from 97% to 98%; thus, the target sequences were amplified with similar efficiencies. All samples were run with at least three duplicates.
2.5. SGA (Single Genome Amplification)
According to the Poisson distribution [18], the DNA dilution yielding positive PCR products in no more than 30% of the wells will contain one amplicon resulting from single molecule amplification in over 80% of these wells. The viral cDNA was serially diluted to yield a single copy by nested PCR as described for the bulk PCR. All products derived from cDNA dilutions conforming to the above description were sequenced with the V3-1 and V4-1 primers by GENEWIZ (Suzhou, China).
2.6. Data Processing and Analysis
The sequences generated in this study have been submitted to GenBank (GenBank acc. no. MK268242 to MK268339 for bulk PCR and MK278920 to MK279296 for SGA). A preliminary sequence analysis was conducted using the SeqMan and MegAlign programs from the Lasergene DNA & Protein analysis software (version 7.0, DNASTAR Inc., Madison, WI, USA), the BioEdit Sequence Alignment Editor (version 7.2.5), and ClustalX (version 2.1). A maximum likelihood tree was constructed using MEGA 7.0 [19]. The amino acid alignment was automatically generated by MargFreq (version 1.02).
3. Results
3.1. General Characteristics of EIAV env Variability during the Course of EIAV Attenuation Using Bulk PCR
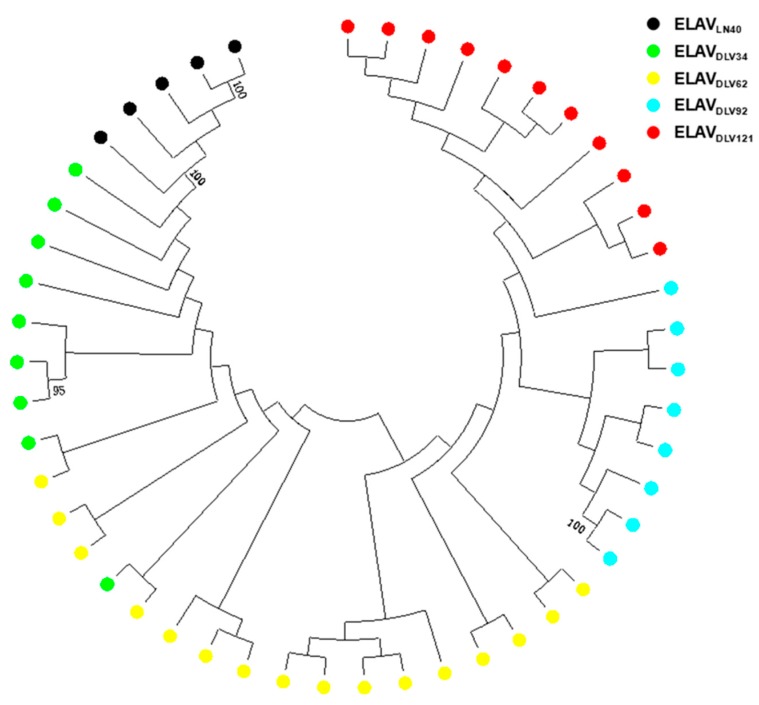
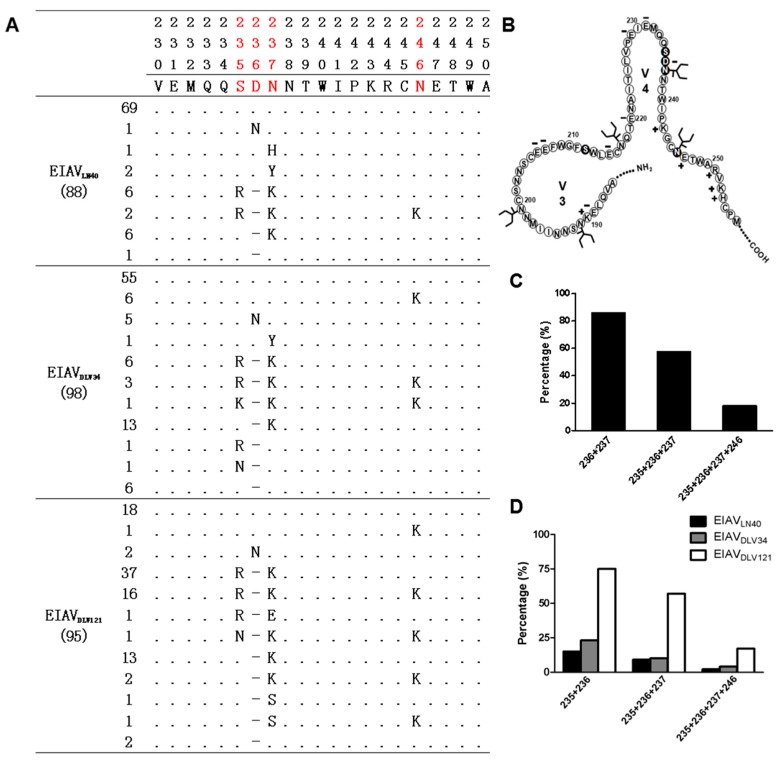
To verify the characteristics of EIAV env variation, some env genes derived from EIAV strains during the critical stages of the attenuation process were analyzed using bulk PCR [20,21]. A phylogenetic tree was constructed based on 98 env sequences amplified from EIAV strains with differing virulence (EIAVLN40, EIAVDLV34, EIAVDLV62, EIAVDLV92, and EIAVDLV121) (the morbidity of all strains showed in Figure S1). The general features of env variability are shown in Figure 1. Obviously, individual clusters accompanied by a decrease in virulence were generated. In other words, the evolutionary pattern of the env gene was consistent with the loss of virulence. Notably, the sequences derived from EIAVDLV62 displayed a separated distribution between EIAVLN40 and EIAVDLV121. This result suggested that EIAVDLV62 was an important viral evolutionary state that was associated with EIAV pathogenicity. The details of the sequences that also displayed several mutations in each cluster are presented in Table 1. The isolates from each generation displayed the same mutation types (insertions and deletions) but had different mutation frequencies and sites. We observed that the proportion of strains carrying this mutation gradually increased with the serial passage number in vitro (EIAVDLV34, EIAVDLV62, EIAVDLV92, and EIAVDLV121 = 6.25%, 18.5%, 42.1%, and 100%, respectively). We also noted that most mutations were random except for a successful amino acid deletion (236D) located in the V4 domain of Env compared to the reference strain sequence (EIAVLN40), as shown in Figure 2. This mutation was also stable in horses inoculated with EIAVDLV121 [22]. Thus, we speculated that this site might represent a vaccine-specific mutation.
Figure 1.
The evolutionary history was inferred by applying MEGA 7.0 program based on the general time reversible model. Whole genome sequences were obtained by bulk PCR. Bootstrap values above 80% are indicated. EIAVLN40, EIAVDLV34, EIAVDLV62, EIAVDLV92, and EIAVDLV121 are presented with black circles, black triangles, gray circles, blue circles and red circles, respectively.
Table 1.
Amino acid differences in Env V4 among different Equine Infectious Anima Virus (EIAV) strains obtained using bulk PCR.
| 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | Mutation Rate (236D/-) |
Morbidity (Horses) |
|
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 5 | 5 | 5 | |||
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 0 | 1 | 2 | |||
| EIAVLN40 | N29 | A29 | I29 | T29 | I29 | L29 | V29 | P29 | E29 | V29 | E29 | M28 | Q29 | Q28 | S29 | D29 | N28 | N29 | T29 | W29 | I29 | P29 | K29 | R29 | C27 | N29 | E29 | T29 | W29 | A29 | R29 | V29 | 0.00% | 100% |
| I1 | R1 | S1 | Y2 | |||||||||||||||||||||||||||||||
| EIAVDLV34 | S9 | A16 | I15 | T15 | I16 | L16 | V16 | P16 | E16 | V10 | E16 | M16 | Q16 | Q14 | S16 | D14 | N16 | N16 | T16 | W16 | I16 | P16 | K16 | G8 | C16 | N14 | E13 | T16 | W16 | A16 | R12 | V16 | 6.25% | 100% |
| N7 | V1 | A1 | I6 | K1 | N1 | R8 | K2 | K3 | K4 | |||||||||||||||||||||||||
| R1 | -1 | |||||||||||||||||||||||||||||||||
| EIAVDLV62 | N9 | A16 | I16 | T16 | I16 | L16 | V16 | P16 | E16 | V9 | E16 | M16 | Q16 | Q14 | S16 | D9 | N16 | N16 | T16 | W16 | I16 | P16 | K16 | R11 | C16 | N13 | E13 | T16 | W16 | A16 | R12 | V16 | 18.5% | 100% |
| S7 | I7 | R2 | N4 | G5 | K3 | K2 | K4 | |||||||||||||||||||||||||||
| -3 | G1 | |||||||||||||||||||||||||||||||||
| EIAVDLV92 | N11 | A19 | I19 | T16 | I19 | L19 | V19 | P19 | E19 | V18 | E19 | M19 | Q19 | Q17 | S19 | D3 | N13 | N18 | T16 | W19 | I18 | P19 | K19 | R17 | C19 | N14 | E18 | T18 | W19 | A19 | R12 | V19 | 42.1% | 9.1% |
| S8 | A3 | I1 | R1 | N8 | K5 | D1 | A3 | V1 | G2 | K5 | K1 | A1 | K7 | |||||||||||||||||||||
| K1 | -8 | R1 | ||||||||||||||||||||||||||||||||
| EIAVDLV121 | N11 | A17 | I18 | T17 | I18 | L18 | V18 | P18 | E17 | V17 | E17 | M18 | Q18 | Q18 | R15 | -18 | K17 | N18 | T18 | W18 | I17 | P18 | K18 | R17 | C18 | K9 | E10 | T18 | W18 | A18 | K13 | V17 | 100% | 0 |
| S7 | T1 | A1 | K1 | I1 | K1 | S3 | S1 | V1 | G1 | N8 | K8 | R5 | I1 | |||||||||||||||||||||
| E1 |
The additional amino acid D, which consistently appeared in virulent EIAV strains, is marked with dark grey, and N-linked glycosylation sites are marked with light grey.
Figure 2.
Distribution of the consensus mutations located in the V3 and V4 regions and schematic structural representations. (A) Red letters are defined as critical consensus mutation sites displayed within the Env region during the EIAV attenuation process. (B) Schematic figure of the EIAVDLV121 V3 and V4 regions. N-Glycosylation sites are shown as branched lines. (C) Comparison of the proportions of several combined mutations at 235, 236, 237, and 246 for the whole mutants. (D) The percentages of these combined mutations at the key stages of vaccine development (EIAVLN40, EIAVDLV34, and EIAVDLV121).
3.2. Precise Characteristics of the EIAV env Mutation Distribution during the Attenuation Process
Once the vaccine-specific mutation was verified, we wanted to investigate whether this mutation could be detected during serial passage using a higher sensitivity method. These data will further benefit our understanding of the relationship between env quasispecies and virulence. Firstly, we adopted double-probe real-time PCR to accurately determine the proportion of strains with the env-Δ236D-phenotype and env-236D-phenotype sequences (which refer to virus sequences without and with the 236D mutation) in different EIAV generations [23,24]. An env-236D-phenotype-specific probe (HEX) and an env-Δ236D-phenotype-specific probe (FAM) were designed based on the V4 domain sequences (Figure S2). To avoid the influence of the initial viral copy number on the detection sensitivity, samples containing different initial viral copy numbers were analyzed in parallel (102 copies/μL, 103 copies/μL and 104 copies/μL). As presented in Figure S3, the env-Δ236D-phenotype sequence was detected with low copy numbers from all virulent strains (EIAVDLV34 and EIAVDLV62) except for EIAVLN40, and the proportions of strains with the env-Δ236D-phenotype gradually increased with the passage number (from 2.47% to 67.5% at the 103 copies/μL concentration), whereas the proportion of strains with the env-236D-phenotype decreased concomitantly.
Although a gradual increase in the percentage of vaccine-specific sequences was observed using double-probe real-time PCR, the distribution of this mutation in EIAVLN40 was still unclear. The SGA method can avoid the recombination events induced by the polymerase and better detect minor nucleotide changes at the same position; thus, this technique has been performed recently for viral quasispecies analysis [25,26,27]. Thus, we were interested in using SGA to resequence env to obtain more information about env quasispecies during long-term passage. Samples obtained separately from EIAVLN40 (n = 88), EIAVDLV34 (n = 98), EIAVDLV121 (n = 95) and infectious clone pLGFD3-8 (n = 100) were amplified by SGA and aligned. As expected, the 236D mutation was found in the EIAV strains derived from each generation. The proportion of sequences with the 236D mutation best matched the results derived from the bulk PCR (17.0%, 30.6%, 77.9%, and 100%, respectively) (Table 2). Importantly, we detected 236D (15/72) in the EIAVLN40 reservoir, which was not observed using bulk PCR. Additionally, the SGA method exhibited a relatively higher mutation detection rate than bulk PCR. Hence, more variants located at the same position in the EIAV population were observed; for instance, multiple substitutions (E, V, K or Q) instead of a single substitution (E) were detected at position 229 in EIAVLN40 using the SGA method. In particular, in addition to position 236, we found that three other positions, which were separately located at S235R, N237K, and N246K, presented gradual and successful mutation accumulation with the increasing passage number, as presented in Table 2. Further analysis showed that the deletion of amino acid 237D resulted in the loss of a N-glycosylation site (237NNTW240) and that the N237K/N246K mutations would also be involved in N-glycosylation site formation (Figure 2A,B). We also noted that these key mutations (235, 236, 237, and 247) were absent or present in combination; for instance, the frequency of the combined mutations at sites 236 and 237 was 85.8% (109/127). The proportions of mutation that concurred at 235, 236, and 237 was 57.5% (73/127). Meanwhile, the sequences with mutations at 235, 236, 237, and 246 showed a lower frequency than the other sequences, as shown in Figure 2C. Moreover, the proportion of these combined mutations at sites 235–236, 235–236–237 and 235–236–237–246 gradually increased as the virulence of the strains decreased, especially in 235–236 (shown in Figure 2D). Hence, we speculated the existence of a link between the sites 236 and 237. According to these results, we speculated that long-term passage in vitro not only had a significant effect on the env quasispecies compositions but also might contribute to viral attenuation.
Table 2.
Amino acid differences in Env V4 among different EIAV strains detected using single genome amplification (SGA).
| 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | Mutation Rate (236D/-) |
|
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 5 | 5 | 5 | ||
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 0 | 1 | 2 | ||
| EIAVLN40 | N86 | A88 | I88 | T88 | I88 | L88 | V88 | P88 | E85 | V87 | E88 | M88 | Q87 | Q88 | S80 | D72 | N71 | N88 | T88 | W88 | I88 | P88 | K87 | R80 | C88 | N86 | E87 | T87 | W88 | A88 | R81 | V88 | 17.0% |
| S2 | V1 | I1 | H1 | R8 | -15 | K14 | N1 | G8 | K2 | K1 | I1 | K7 | |||||||||||||||||||||
| K1 | N1 | H1 | |||||||||||||||||||||||||||||||
| Q1 | |||||||||||||||||||||||||||||||||
| EIAVDLV34 | N51 | A97 | I98 | T98 | I98 | L97 | V98 | P97 | E94 | V60 | E96 | M98 | Q98 | Q82 | S86 | D63 | N75 | N98 | T98 | W95 | I95 | P97 | K96 | R57 | C97 | N85 | E79 | T96 | W94 | A94 | R75 | V94 | 30.6% |
| S47 | T1 | V1 | L1 | K3 | I38 | K2 | K8 | R10 | -30 | K22 | S1 | S2 | L1 | Q1 | G41 | W1 | K10 | K17 | A1 | C3 | C2 | K20 | I4 | ||||||||||
| D1 | R8 | K1 | N5 | Y1 | C2 | C1 | P1 | S3 | G2 | I1 | R1 | S1 | T3 | ||||||||||||||||||||
| N1 | T1 | ||||||||||||||||||||||||||||||||
| EIAVDLV121 | N76 | A95 | I95 | T92 | I95 | L95 | V94 | P95 | E92 | I10 | E93 | M94 | Q94 | Q91 | R54 | -74 | K69 | N95 | T95 | W95 | I95 | P95 | K95 | R68 | C95 | N74 | E83 | T94 | W95 | A93 | K55 | V95 | 77.9% |
| S18 | A3 | I1 | K1 | V84 | K1 | V1 | H1 | R3 | S40 | D19 | N23 | G25 | K21 | K11 | A1 | T2 | R40 | ||||||||||||||||
| K1 | D1 | L1 | V1 | K1 | N1 | N2 | S2 | K2 | G1 | ||||||||||||||||||||||||
| Q1 | E1 | ||||||||||||||||||||||||||||||||
| pLGFD3-8 | N100 | A100 | I100 | T100 | I100 | L100 | V100 | P100 | E99 | V97 | E99 | M100 | Q98 | Q100 | S87 | -100 | K100 | N100 | T97 | W98 | I99 | P100 | K100 | G88 | C99 | K99 | K99 | T100 | W99 | A100 | R97 | V100 | 100% |
| Q1 | I3 | K1 | H1 | R13 | I3 | C2 | V1 | R12 | G1 | N1 | E1 | C1 | K2 | ||||||||||||||||||||
| R1 | M1 |
The additional amino acid D, which consistently appeared in virulent EIAV strains, is marked with dark grey, and N-linked glycosylation sites are marked with light grey.
3.3. The Relationship between EIAV Attenuation In Vitro and env Gene Quasispecies
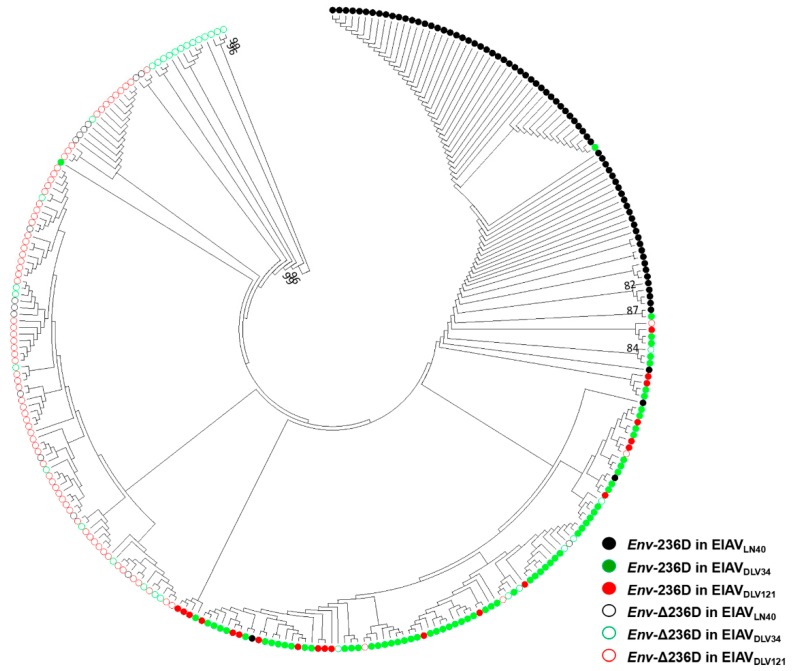
To better analyze the relationship between virulence and env quasispecies, we constructed a maximum likelihood tree using the MEGA 7.0 program based on the SGA results. As presented in Figure 3, two obvious major groups of env sequences have evolved, as represented by the env-236D-phenotype (indicated with solid circles) and env-Δ236D-phenotype (indicated with hollow circles). Then, serial passaging events continued to lead to accumulation of the env-Δ236D-phenotype in the whole population, which made the variant distribution even more complex (i.e., the variant distribution of EIAVDLV34). Over the course of 121 generations in vitro, the cluster distribution of related sequences taken from EIAVDLV121 showed a pattern similar to that of EIAVLN40 except for reversal of the predominant phenotype from env-236D to env-Δ236D at the individual population level. We observed that two typical env phenotypes displayed a gradual transition pattern. These results demonstrated that a predominant phenotype transition occurred in the natural virus population under selection, which was consistent with the LTR investigation results [15].
Figure 3.
Construction of a maximum likelihood tree using the MEGA 7.0 program based on the Tamura-Nei model. Bootstrap values above 80% are indicated. EIAVLN40, EIAVDLV34, and EIAVDLV121 are shown in black, green and red, respectively. Solid and hollow circles represent the env-236D-phenotype and env-Δ236D-phenotype sequences, respectively. All samples were collected from three virus strains (EIAVLN40, EIAVDLV34, and EIAVDLV121). A total of 381 sequences were analyzed.
4. Discussion
In contrast to the use of artificial mutagenesis to induce RNA viral attenuation, the EIAV live-attenuated vaccine was developed based on natural evolution through consecutive passaging in vitro. There is growing evidence that analyzing virulence determinants from an evolutionary perspective rather than nucleotide mutations in specific virulence genes is very important, particularly for viruses undergoing consecutive passages in vitro [28,29]. Although a quasispecies is well accepted as a key factor for viral virulence [30], the relationship between these factors remains an open question. In other words, not all viruses will lose their virulence even through serial passage in vitro, such as the CDV Rockborn strain generated in canine macrophage cells [31].
In our previous study, we observed that the EIAV attenuated vaccine also existed as a complex quasispecies [15]. Furthermore, our results suggested that the attenuated vaccine might be a result driven by selection of the dominant EIAV variant at the whole population level. However, whether a similar evolutionary pattern existed in the predominant virulence gene of EIAV was not clear. Here, we used the EIAV attenuation system to investigate the relationship between virulence and EIAV env gene variation. In the present study, we used three different methods to answer this question. Regardless of the method used, some stable mutations located in the V3 and V4 regions of Env were verified. Compared with those of the other methods, SGA showed the highest detection sensitivity for minor nucleotide changes; for instance, the mutations located at positions 235, 236, 237, 246, 248, and 250 could be verified only using the SGA method. In other words, more variants were detected at the same positions. Several key mutations, including S235R, N237K, and N246K, attracted our attention. Not only did the percentages of these mutations parallel with the increased passage number but these mutations (237NNTW240 and N237K/N246K) could also result in loss of an N-glycosylation site or formation of an adjacent N-glycosylation site. We confirmed that this position in combination with three other N-glycosylation site mutations in Env resulted in reversion of virulence and influenced the neutralizing activity [32]. Further analysis showed that inter-linkages existed among these key mutations. Based on these results, we speculated that the observed pattern of Env variation was consistent with the loss of viral virulence.
In particular, we noted that the env-Δ236D-phenotype, which could present as a vaccine-specific mutation, was present in its parental virus reservoir (EIAVLN40) at a lower level. Hence, our observations indicate that the EIAV vaccine strain (env-Δ236D-phenotype) may preexist in the EIAV population as a minority population; indeed, another study from our laboratory detected vaccine-specific LTR sequences in horses infected with virulent strains [15]. According to these results, we proposed a model describing env evolution during the attenuation process, as shown in Figure 4. In this model, the env-236D-phenotype is the predominant hypervirulent EIAV quasispecies that causes typical EIA symptoms in infected horses and induces 100% mortality. The env-Δ236D-phenotype is an avirulent quasispecies that may be a beneficial phenotype for new culture environments (equine macrophages), but the underlying reason is complicated (possibly due to its high fitness or harboring a selectable trait) [6]. To adapt to new culture conditions, the avirulent phenotype selectively accumulated [8,33]. The shape of the phylogenetic tree further reflected this phenomenon in which the env-Δ236D-phenotype gradually became predominant within the whole population but progressively lost its virulence. Correspondingly, the env-Δ236D-phenotype became dominant with ongoing passages, and thus the strain lost its virulence in animals.
Figure 4.
A schematic evolutionary model comparing the EIAV virulent strain to the vaccine strain during long-term passage in vitro. The virulent virus strains are presented with black circles, and the avirulent virus strains are presented with red circles. In this model, the env-Δ236D-phenotype sequences preexisted in EIAVLN40 as a minority population. With serial passages in vitro, sequences containing the env-Δ236D-phenotype gradually became the predominant EIAV quasispecies and generated high replicative fidelity under the new environmental selection conditions. Virulence was simultaneously attenuated as a result, as discussed in the text.
In summary, our study characterized EIAV env quasispecies and observed a virulence-correlated parallel in env variation during EIAV passage in vitro. However, this strain in the context of the ensemble of the predominant population progressively lost its virulence in vivo. Hence, our data support the view of quasispecies theory and provide a role for the env quasispecies in EIAV pathogenesis, which partly explains the attenuation-related mechanism of EIAV.
Acknowledgments
We thank Han GZ for technical assistance on evolutionary analysis.
Supplementary Materials
The following are available online at https://www.mdpi.com/1999-4915/11/4/380/s1, Figure S1: The morbidity of varied EIAV strains. Figure S2: WebLogo presentation of variability in EIAV env. Figure S3: Quantification of the env-Δ236D-phenotype and env-236D-phenotype sequences located in the V4 region of env by double-probe real-time PCR.
Author Contributions
Conceived and designed the experiments: Y.L. and X.W. Performed the experiments: C.L., X.-F.W., Y.W., and J.C. Analyzed the data: C.L. Wrote the paper: Z.Z., Y.L. and X.W.
Funding
This research was funded by National Natural Science Foundation of China (31672533, 31302066, and 31672578).
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Yuan Q., Liu C., Liang Z., Chen X., Diao D., Kong X. The comparison of genetic variation in the envelope protein between various immunodeficiency viruses and equine infectious anemia virus. Virol. Sin. 2012;27:241–247. doi: 10.1007/s12250-012-3253-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cook R.F., Leroux C., Issel C.J. Equine infectious anemia and equine infectious anemia virus in 2013: A review. Vet. Microbiol. 2013;167:181–204. doi: 10.1016/j.vetmic.2013.09.031. [DOI] [PubMed] [Google Scholar]
- 3.Ma J., Shi N., Jiang C.G., Lin Y.Z., Wang X.F., Wang S., Lv X.L., Zhao L.P., Shao Y.M., Kong X.G., et al. A proviral derivative from a reference attenuated eiav vaccine strain failed to elicit protective immunity. Virology. 2011;410:96–106. doi: 10.1016/j.virol.2010.10.032. [DOI] [PubMed] [Google Scholar]
- 4.Wang X., Wang S., Lin Y., Jiang C., Ma J., Zhao L., Lv X., Wang F., Shen R., Kong X., et al. Genomic comparison between attenuated chinese equine infectious anemia virus vaccine strains and their parental virulent strains. Arch. Virol. 2011;156:353–357. doi: 10.1007/s00705-010-0877-8. [DOI] [PubMed] [Google Scholar]
- 5.Wang X.F., Lin Y.Z., Li Q., Liu Q., Zhao W.W., Du C., Chen J., Wang X., Zhou J.H. Genetic evolution during the development of an attenuated EIAV vaccine. Retrovirology. 2016;13:9. doi: 10.1186/s12977-016-0240-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bingham R.J., Dykeman E.C., Twarock R. RNA virus evolution via a quasispecies-based model reveals a drug target with a high barrier to resistance. Viruses. 2017;9:347. doi: 10.3390/v9110347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Colizzi E.S., Hogeweg P. Evolution of functional diversification within quasispecies. Genome Biol. Evol. 2014;6:1990–2007. doi: 10.1093/gbe/evu150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Park J.M., Niestemski L.R., Deem M.W. Quasispecies theory for evolution of modularity. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 2015;91:012714. doi: 10.1103/PhysRevE.91.012714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Domingo E., Sheldon J., Perales C. Viral quasispecies evolution. Microbiol. Mol. Biol. Rev. 2012;76:159–216. doi: 10.1128/MMBR.05023-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Andino R., Domingo E. Viral quasispecies. Virology. 2015;479–480:46–51. doi: 10.1016/j.virol.2015.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Domingo E., Martin V., Perales C., Grande-Perez A., Garcia-Arriaza J., Arias A. Viruses as quasispecies: Biological implications. Curr. Top. Microbiol. 2006;299:51–82. doi: 10.1007/3-540-26397-7_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Craigo J.K., Ezzelarab C., Cook S.J., Chong L., Horohov D., Issel C.J., Montelaro R.C. Envelope determinants of equine lentiviral vaccine protection. PLoS ONE. 2013;8:e66093. doi: 10.1371/journal.pone.0066093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Du J.S., Wang X.F., Ma J., Wang J.X., Qin Y.Y., Zhu C.H., Liu F., Shao Y.M., Zhou J.H., Qiao W.T., et al. Structural and biochemical insights into the v/i505t mutation found in the eiav gp45 vaccine strain. Retrovirology. 2014;11 doi: 10.1186/1742-4690-11-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Duan L., Du J., Wang X., Zhou J., Wang X., Liu X. Structural and functional characterization of EIAV gp45 fusion peptide proximal region and asparagine-rich layer. Virology. 2016;491:64–72. doi: 10.1016/j.virol.2016.01.010. [DOI] [PubMed] [Google Scholar]
- 15.Wang X.F., Liu Q., Wang Y.H., Wang S., Chen J., Lin Y.Z., Ma J., Zhou J.H., Wang X. Characterization of equine infectious anemia virus long terminal repeat quasispecies in vitro and in vivo. J. Virol. 2018;92 doi: 10.1128/JVI.02150-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin Y.Z., Shen R.X., Zhu Z.Y., Deng X.L., Cao X.Z., Wang X.F., Ma J., Jiang C.G., Zhao L.P., Lv X.L., et al. An attenuated eiav vaccine strain induces significantly different immune responses from its pathogenic parental strain although with similar in vivo replication pattern. Antiviral Res. 2011;92:292–304. doi: 10.1016/j.antiviral.2011.08.016. [DOI] [PubMed] [Google Scholar]
- 17.Jiang C.G., Gao X., Ma J., Lin Y.Z., Wang X.F., Zhao L.P., Hua Y.P., Liu D., Zhou J.H. C-terminal truncation of the transmembrane protein of an attenuated lentiviral vaccine alters its in vitro but not in vivo replication and weakens its potential pathogenicity. Virus Res. 2011;158:235–245. doi: 10.1016/j.virusres.2011.04.007. [DOI] [PubMed] [Google Scholar]
- 18.Hayat M.J., Higgins M. Understanding poisson regression. J. Nurs. Educ. 2014;53:207–215. doi: 10.3928/01484834-20140325-04. [DOI] [PubMed] [Google Scholar]
- 19.Kumar S., Stecher G., Tamura K. Mega7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Craigo J.K., Montelaro R.C. Eiav envelope diversity: Shaping viral persistence and encumbering vaccine efficacy. Curr. HIV Res. 2010;8:81–86. doi: 10.2174/157016210790416398. [DOI] [PubMed] [Google Scholar]
- 21.Etemad B., Ghulam-Smith M., Gonzalez O., White L.F., Sagar M. Single genome amplification and standard bulk pcr yield hiv-1 envelope products with similar genotypic and phenotypic characteristics. J. Virol. Methods. 2015;214:46–53. doi: 10.1016/j.jviromet.2015.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang X.F., Wang S., Liu Q., Lin Y.Z., Du C., Tang Y.D., Na L., Wang X., Zhou J.H. A unique evolution of the s2 gene of equine infectious anemia virus in hosts correlated with particular infection statuses. Viruses. 2014;6:4265–4279. doi: 10.3390/v6114265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ntziora F., Paraskevis D., Haida C., Magiorkinis E., Manesis E., Papatheodoridis G., Manolakopoulos S., Beloukas A., Chryssoy S., Magiorkinis G., et al. Quantitative detection of the m204v hepatitis B virus minor variants by amplification refractory mutation system real-time PCR combined with molecular beacon technology. J. Clin. Microbiol. 2009;47:2544–2550. doi: 10.1128/JCM.00045-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen Q., Belmonte I., Buti M., Nieto L., Garcia-Cehic D., Gregori J., Perales C., Ordeig L., Llorens M., Soria M.E., et al. New real-time-pcr method to identify single point mutations in hepatitis C virus. World J. Gastroenterol. 2016;22:9604–9612. doi: 10.3748/wjg.v22.i43.9604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Salazar-Gonzalez J.F., Bailes E., Pham K.T., Salazar M.G., Guffey M.B., Keele B.F., Derdeyn C.A., Farmer P., Hunter E., Allen S., et al. Deciphering human immunodeficiency virus type 1 transmission and early envelope diversification by single-genome amplification and sequencing. J. Virol. 2008;82:3952–3970. doi: 10.1128/JVI.02660-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Geng Q.M., Li H.P., Bao Z.Y., Liu Y.J., Zhuang D.M., Li L., Liu S.Y., Li J.Y. Indinavir resistance evolution in one human immunodeficiency virus type 1 infected patient revealed by single-genome amplification. Virol. Sin. 2010;25:316–328. doi: 10.1007/s12250-010-3122-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guinoiseau T., Moreau A., Hohnadel G., Ngo-Giang-Huong N., Brulard C., Vourc’h P., Goudeau A., Gaudy-Graffin C. Deep sequencing is an appropriate tool for the selection of unique hepatitis C virus (HCV) variants after single genomic amplification. PLoS ONE. 2017;12:e0174852. doi: 10.1371/journal.pone.0174852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Presloid J.B., Novella I.S. Rna viruses and rnai: Quasispecies implications for viral escape. Viruses. 2015;7:3226–3240. doi: 10.3390/v7062768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu F., Wu X., Li L., Zou Y., Liu S., Wang Z. Evolutionary characteristics of morbilliviruses during serial passages in vitro: Gradual attenuation of virus virulence. Comp. Immunol. Microbiol. Infect. Dis. 2016;47:7–18. doi: 10.1016/j.cimid.2016.05.007. [DOI] [PubMed] [Google Scholar]
- 30.Vignuzzi M., Stone J.K., Arnold J.J., Cameron C.E., Andino R. Quasispecies diversity determines pathogenesis through cooperative interactions in a viral population. Nature. 2006;439:344–348. doi: 10.1038/nature04388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martella V., Blixenkrone-Moller M., Elia G., Lucente M.S., Cirone F., Decaro N., Nielsen L., Banyai K., Carmichael L.E., Buonavoglia C. Lights and shades on an historical vaccine canine distemper virus, the rockborn strain. Vaccine. 2011;29:1222–1227. doi: 10.1016/j.vaccine.2010.12.001. [DOI] [PubMed] [Google Scholar]
- 32.Han X., Zhang P., Yu W., Xiang W., Li X. Amino acid mutations in the env gp90 protein that modify n-linked glycosylation of the chinese eiav vaccine strain enhance resistance to neutralizing antibodies. Virus Genes. 2016;52:814–822. doi: 10.1007/s11262-016-1382-2. [DOI] [PubMed] [Google Scholar]
- 33.Glebova O., Knyazev S., Melnyk A., Artyomenko A., Khudyakov Y., Zelikovsky A., Skums P. Inference of genetic relatedness between viral quasispecies from sequencing data. BMC Genomics. 2017;18:918. doi: 10.1186/s12864-017-4274-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.