Abstract
DNA methylation and Polycomb are key factors in the establishment of vertebrate cellular identity and fate. Here we report de novo missense mutations in DNMT3A, encoding the DNA methyltransferase DNMT3A, that cause microcephalic dwarfism, a hypocellular disorder of extreme global growth failure. Substitutions in the PWWP domain abrogate binding to the histone modifications H3K36me2/3, and alter DNA methylation in patient cells. Polycomb-associated DNA methylation canyons/valleys, hypomethylated domains encompassing developmental genes, become methylated with concomitant depletion of H3K27me3 and H3K4me3 bivalent marks. Such de novo DNA methylation occurs during differentiation of Dnmt3aW326R pluripotent cells in vitro, and is also evident in Dnmt3aW326R/+ dwarf mice. We therefore propose that the interaction of the DNMT3A PWWP domain with H3K36me2/3 normally limits DNA methylation of polycomb-marked regions. Our findings implicate the interplay between DNA methylation and polycomb at key developmental regulators as a determinant of organism size in mammals.
Introduction
Microcephalic dwarfism represents a group of conditions of profound size reduction in humans. These single gene disorders are distinguished from other forms of dwarfism by severity and morphology. Growth is globally impaired pre- and post-natally with proportionate scaling1. Reduced brain size in microcephalic dwarfism differentiates it from other forms of dwarfism and reflects an early developmental origin. We and others have discovered many microcephalic dwarfism genes to encode essential components of the cell cycle machinery, including replication licensing components2–5 and key mitotic proteins6–8. Mutations in these genes result in reduced cell number and consequently organism size1.
As cell number is also the major determinant of size differences between mammals9 and the molecular basis for many microcephalic dwarfism patients still remains to be defined, we performed whole-exome sequencing (WES) to identify novel genetic causes and inform understanding of size regulation.
Results
De novo mutations in DNMT3A causes microcephalic dwarfism
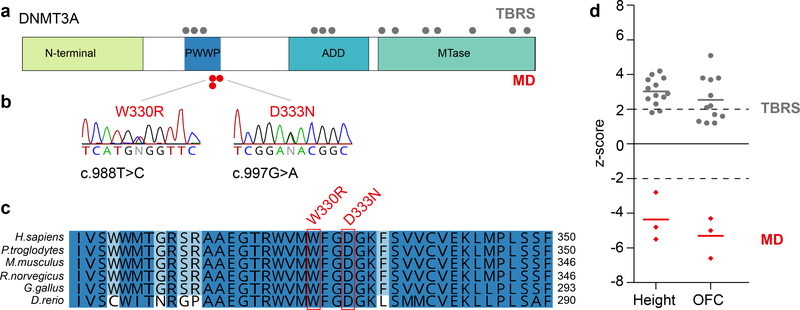
WES trio analysis of a microcephalic dwarfism family identified a de novo DNMT3A heterozygous mutation in the proband (NM_175629.2:c.988T>C, Fig. 1a,b and Supplementary Table 1). This resulted in the replacement of a tryptophan residue with an arginine at codon 330 (p.W330R) in the highly conserved PWWP domain of this DNA methyltransferase (Fig. 1c). NGS sequencing of our patient cohort then identified an unrelated patient with the same heterozygous de novo missense variant in DNMT3A (c.988T>C p.W330R, Supplementary Table 1). This substitution was not present in the GnomAD10 database suggesting it to be absent from the general population. The two individuals were phenotypically similar, exhibiting significant, proportionate reduction in head circumference and height (Supplementary Note, clinical synopsis). The shared clinical phenotype, in conjunction with independent de novo mutation of the same highly conserved residue, led us to conclude that these were pathogenic mutations. More recently, we ascertained a further microcephalic dwarfism patient with a de novo mutation in an adjacent codon (c.997G>A; p.D333N). Notably, the growth parameters of all three patients contrast markedly with those of previously reported patients with de novo germline missense and truncating loss of function DNMT3A mutations11,12, who have the reciprocal phenotype of macrocephalic overgrowth Tatton-Brown Rahman syndrome (TBRS, Fig. 1d). As DNMT3A haploinsufficiency causes overgrowth13, this suggested the c.988T>C and c.997G>A mutations to be genetic ‘gain of function’ mutations.
Fig. 1|. De novo mutations in DNMT3A cause microcephalic dwarfism.
a, Schematic of DNMT3A protein and domains. Position of microcephalic dwarfism (MD) mutations (red) and Tatton-Brown-Rahman syndrome (TBRS) overgrowth (grey) mutations (Tatton-Brown et al. 2014) (MTase, DNA methyltransferase domain) b, The heterozygous de novo c.988T>C mutation results in substitution of a Tryptophan residue (patient 1 and 2). The heterozygous de novo c.997G>A mutation results in substitution of an Aspartic acid residue (patient 3). Both residues are conserved in vertebrates c, and replaced with a physiochemically dissimilar residue: Arginine (p.W330R) and Asparagine (p.D333N) respectively. Sequence alignments, Clustal Omega. d, The W330R and D333N mutations cause extreme growth failure and microcephaly (red diamonds, n=3 independent patients), in direct contrast to DNMT3A overgrowth patients (grey circles, n=13 and n=12 patients, respectively for height and OFC). Height and head circumference (OFC) plotted as z-scores (s.d. for population mean adjusted for age and sex). Dashed lines at −2 and +2 s.d indicate 95% confidence interval for general population. Horizontal bars, mean values for respective patient groups. TBRS morphometric data reproduced from Tatton-Brown et al. 201411.
DNMT3AW330R is stably expressed
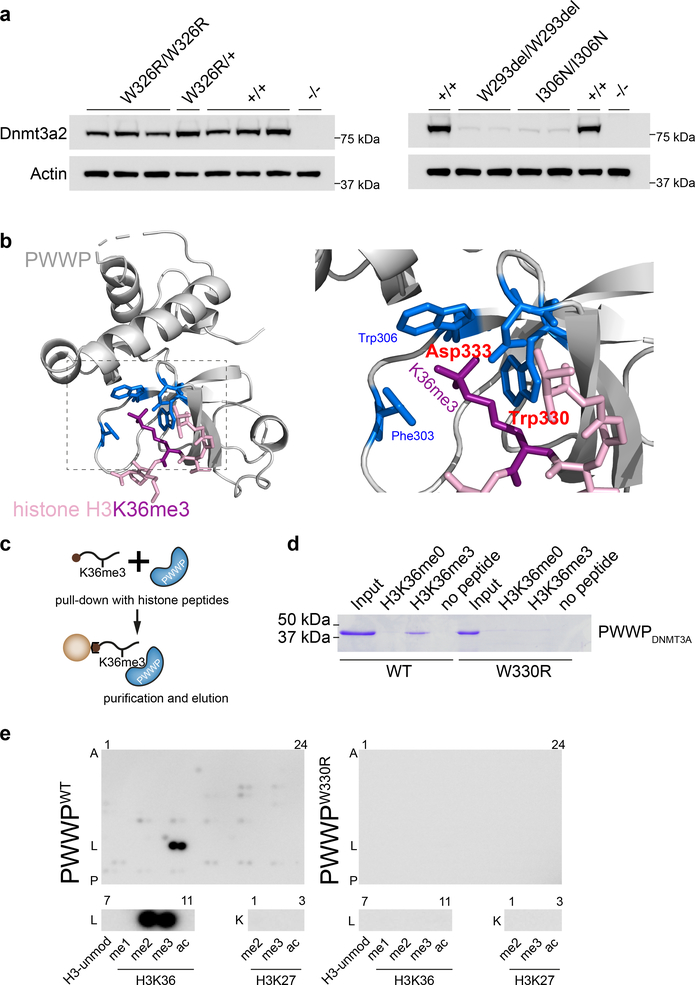
To model the consequences of the W330R substitution on DNMT3A stability we engineered mouse embryonic stem cells (mESCs) homozygous and heterozygous for the orthologous mutation, W326R, using CRISPR/Cas9-mediated homology-directed repair14. Immunoblotting of these lines established that the Dnmt3aW326R protein is stably expressed. In contrast, mESCs homozygous for the overgrowth PWWP mutations W293del and I306N (W297del and I310N in human, respectively), had markedly reduced Dnmt3a levels (Fig. 2a). We also generated recombinant wildtype and mutant human DNMT3A PWWP domains as GST-fusion proteins. While we were able to efficiently express and purify PWWPWT and PWWPW330R proteins, the overgrowth PWWPW297del and PWWPI310N proteins did not yield stable protein (Supplementary Fig. 1a). This supports the notion that the W330R mutation alters PWWP function, distinct from that of PWWP overgrowth mutations, which interfere with protein stability.
Fig. 2|. The W330R mutation impairs binding of di/tri-methylated H3K36.
a, Murine Dnmt3aW326R protein, containing the orthologous substitution to W330R, is stably expressed, in contrast to corresponding overgrowth PWWP mutations (W293del, I306N). Immunoblotting of cell lysates from CRISPR/Cas9 genome-edited mouse embryonic stem cells (mESC). Multiple independent cell lines, with genotypes as indicated. Representative of n=3 (WT, W326R lines) and n=2 (W293del, I306N) independent experiments. Immunoblots are cropped. b, Structural modelling of the PWWP domain predicts the W330R mutation to disrupt interaction with H3K36me3. The highlighted amino acids (blue) form a cage that binds trimethylated lysine 36 (purple). The amino acids altered in MD patients (tryptophan at codon 330 and aspartate at codon 333) are labelled in red. Backbones of PWWP and histone H3 N-terminal tail depicted in grey and pink respectively. c,d, Recombinant PWWPWT but not PWWPW330R protein binds H3K36me3 peptide. (c) Schematic of streptavidin pull-down of biotinylated histone peptides. (d) Coomassie stained gel of eluted protein from histone peptide pull-downs (cropped). Input, 9% of total protein. Histone peptide H3 (aa 21–44). H3K36me0 corresponding unmodified peptide. Representative of n=3 expts. e, PWWPW330R does not bind H3K36me2, H3K36me3 or other histone-tail modifications. MODified™ Histone Peptide Array representing 384 distinct or combinatorial histone modifications probed with recombinant PWWP proteins as indicated. Below, magnified insets of row L7–11 (histone 3 aa26–45) and K1–3 (histone 3 aa16–35) demonstrates that PWWPWT binds to H3K36me2 (L9) and H3K36me3 (L10), but PWWPW330R does not. Representative of n = 2 independent expts; see also Supplementary Fig. 1b.
The DNMT3AW330R substitution impairs binding to methylated H3K36
The PWWP-domain of DNMT3A binds post-translationally modified histone H3 that has been tri-methylated at Lysine 36 (H3K36me3)15,16. Tryptophan 330 is one of three aromatic amino acids that along with an aspartate residue (Asp333), form an aromatic cage around the methylated lysine17,18 (Fig. 2b). Structural modelling of the DNMT3AW330R substitution predicts that the arginine substitution substantially disrupts this interaction (interaction destabilization: 11.8 kcal/mol).
To test this experimentally, we performed pulldown experiments of histone tail peptides using GST-PWWP fusion proteins. Whereas PWWPWT interacted with an H3K36me3 modified histone-tail peptide but not the corresponding unmodified peptide, we did not detect an interaction of the mutant PWWPW330R with H3K36me3 (Fig. 2c,d). To confirm this and assess whether the W330R substitution conferred an alternative binding specificity on the PWWP domain, a peptide array containing 384 unique and combinatorial histone tail modifications was probed with recombinant protein. PWWPWT bound strongly to H3K36me3 and H3K36me2 as previously reported15,19. However, under the same experimental conditions, PWWPW330R did not bind to any histone modification represented on the array (Fig. 2e and Supplementary Fig. 1b).
The second mutation, p.D333N, is located at the aspartate residue that forms part of the cage surrounding H3K36me2/3 (Fig. 2b). As substitution of this residue is known to abrogate H3K36me2/3 binding15, we conclude that both the W330R and D333N substitutions are likely to impair DNMT3A’s binding of methylated H3K36.
As the N-terminal and ADD-domains of DNMT3A also mediate chromatin interactions20,21, this suggested that DNMT3AW330R and DNMT3AD333N proteins would have altered chromatin-binding specificity, which in turn could modify the pattern of DNA methylation in patient cells.
Increased DNA Methylation occurs at key developmental genes in patient cells
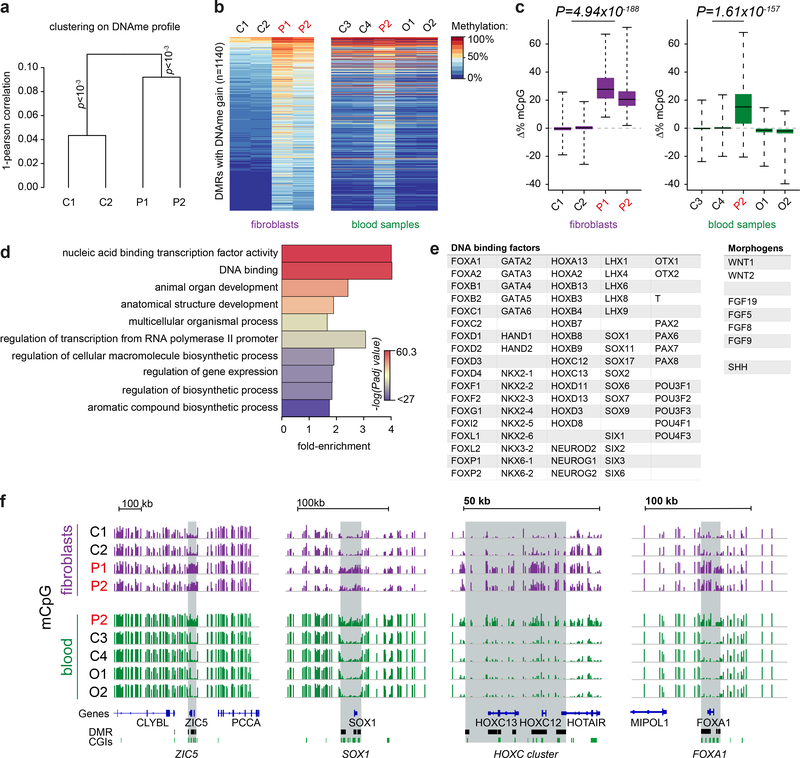
We therefore assessed the genome-wide distribution of DNA methylation in patient-derived fibroblasts using Illumina Infinium MethylationEPIC beadchips. Unsupervised hierarchical clustering established that dermal primary fibroblasts from DNMT3AW330R/+ microcephalic dwarfism patients had similar DNA methylation profiles, significantly distinct from those of healthy subjects (Fig. 3a, p<0.001 for each group). 1878 differentially methylated regions (DMRs) were common to both patients (Fig. 3b and Supplementary Fig. 2). Consistent with altered genomic targeting of DNMT3AW330R, the majority of DMRs were hypermethylated relative to controls (n=1140, Fig. 3b,c and Supplementary Table 2). Notably, the same regions of increased DNA methylation were also present in DNMT3AW330R/+ patient peripheral blood leukocytes (PBLs) indicating this to be a reproducible signature and not a consequence of in vitro culture22 (Fig. 3b,c). Furthermore, the same hypermethylated DMRs were also evident in patient P3’s PBLs (Supplementary Fig. 3). In contrast, DMRs hypomethylated in fibroblasts were not observed in patient PBLs (n=738; Supplementary Fig. 2a,b, Supplementary Fig. 3b,c and Supplementary Table 3). DNMT3AW330R hypermethylated DMRs were not evident in DNMT3A overgrowth patient PBLs, (Fig. 3b,c), and were also absent from pericentrin (PCNT) null patient fibroblasts indicating they were not a general consequence of microcephalic dwarfism (Supplementary Fig. 2c).
Fig. 3|. DNA methylation is increased at key developmental gene loci in patient cells.
a, DNA methylation in DNMT3AW330R/+ patient fibroblasts significantly differs from controls. Unsupervised Ward clustering based on Pearson correlations of all probes from Illumina EPIC arrays for n=2 independent patients and 2 independent controls. Pvclust, approximately-unbiased p-values using 1000 bootstraps. b,c, A methylation signature is evident in DNMT3AW330R patient cells across tissues, comprising 1140 sites of increased methylation. (b) Heat map of differentially methylated regions (DMRs) hypermethylated in patient fibroblasts and peripheral blood leukocytes (PBLs). P1, P2, patients (DNMT3AW330R/+); C1-C4 healthy controls; O1, O2, TBRS overgrowth patients. (c) Quantification of DNA methylation for DMRs (n=1140 DMRs) depicted in panel (b). Box, 25th-75th percentile; whiskers, full data range; centre line, median; Δ%mCpG, percent change of methylation relative to mean of control. p value, two-sided, paired Wilcoxon rank sum tests for mean of control probes vs mean patient probes. d, Gene ontology analysis of genes associated with hypermethylated DMRs. Top ten significant hits shown. Color indicates Benjamini-Hochberg adjusted FDR significance level, genes associated with DMR probes (n=907 genes) versus genes associated with all probes on the array (n=18159), two-sided Fisher’s exact test. e, Exemplars of DNA binding factors and morphogens associated with DMRs. f, Representative genome browser views of hypermethylated DMRs demonstrating increased DNA methylation at key developmental genes in microcephalic dwarfism patient samples. All tracks scaled 0–100% mCpG, DNA methylation. CGI, CpG islands.
Gene ontology analysis for the genes located closest to the hypermethylated DMRs demonstrated a striking association with transcription factors and developmental processes (Fig. 3d and Supplementary Table 4). Notably multiple Hox, lineage-specific transcription factors and morphogen genes were evident in the DMR gene list (Fig. 3e). Visual inspection of the DMRs established these regions to contain CpG islands (CGIs) and encompass genomic regions surrounding these developmental genes (Fig. 3f).
Hypermethylation of Polycomb-marked DNA methylation valleys in patient cells
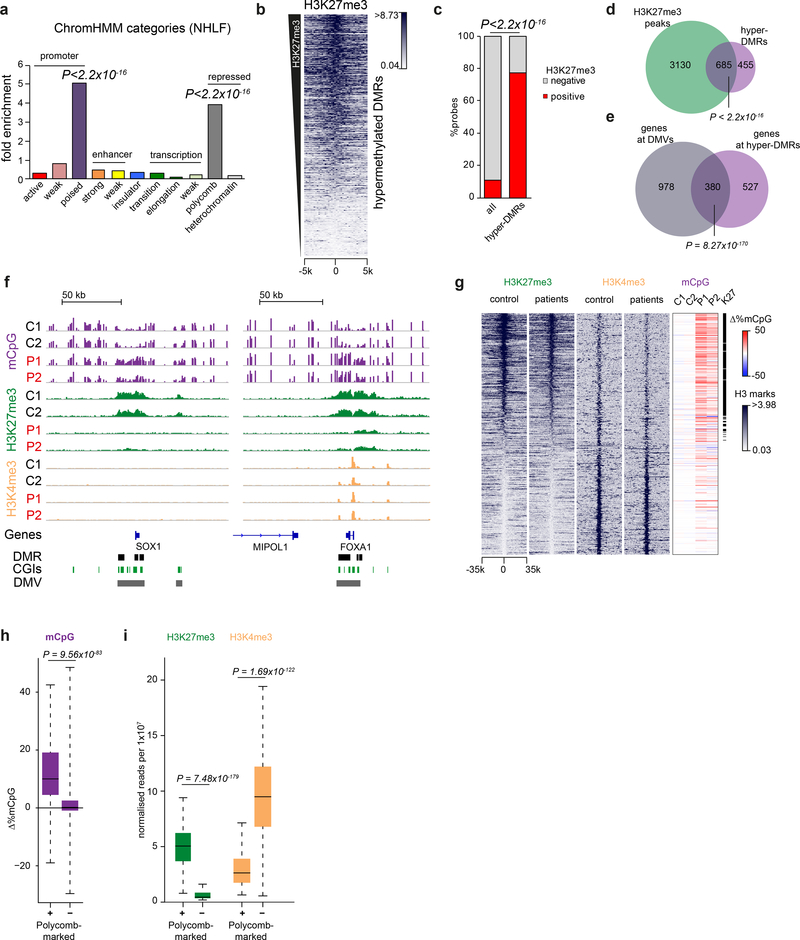
To understand the genomic context of the hypermethylated DMRs, we investigated their chromatin state by intersecting the DMRs with existing ChromHMM annotations for normal human lung fibroblasts (NHLF)23. The DMRs were significantly enriched for ‘Poised-Promoter’ and ‘Polycomb-Repressed’ ChromHMM categories (Fig. 4a), both of which are associated with Polycomb repressive complexes (PRCs). To directly address if these hypermethylated DMRs were Polycomb-marked regions in dermal primary fibroblasts, we next performed ChIP-seq for H3K27me3, the epigenetic signature of the Polycomb repressive complex 2 (PRC2)24,25. Significant enrichment for control fibroblast H3K27me3 peaks was seen at DMR sites (P < 2.2×10−16, Fig. 4b-d) confirming them to be normally marked by H3K27me3.
Fig. 4|. DNA methylation is increased at polycomb-marked DNA methylation valleys.
a, Hypermethylated DMRs in DNMT3AW330R patient cells are significantly enriched at poised promoters and polycomb-repressed regions. Plotted, enrichment of chromatin state categories as identified in normal human lung fibroblasts (NHLF) by ChromHMM in patient hypermethylated DMRs. P-values for each enriched category, two-sided Fisher’s exact test hyper-DMR probes (n=10871 probes) vs all probes (n=403348). (ChromHMM: software annotating Chromatin state by a Hidden Markov Model)57. b-d, H3K27me3 sites in control dermal fibroblasts correlate with hypermethylated DMRs in patient cells. (b) Heat map of normalised H3K27me3 ChIP-seq reads in control fibroblasts (mean of C1, C2) centred on DMRs, ranked by DMR mean H3K27me3 levels. Scale indicates normalised read counts. Window size, 250 bp. (c,d) Quantification of H3K27me3 enrichment at hypermethylated DMRs. (c) Percentage of Infinium array probes overlapping H3K27me3 peaks in control fibroblasts (red, mean of C1 and C2). All, all probes on the array (n=403348 probes). Hyper-DMRs, probes within hypermethylated DMRs (n=10871). p-value, two-sided Fisher’s exact test. (d) Venn diagram displaying overlap of hypermethylated DMRs (n=1140) with H3K27me3 peaks (n=3815) in controls. p value, two-sided Fisher’s exact test. (e) Genes associated with hypermethylated DMRs (n=907 genes), significantly overlap genes associated with DMVs (n=1,358). Two-sided Fisher’s exact test, genes associated with hyper-DMRs vs all genes represented on array. (f) Increased methylation is distributed across H3K27me3 regions, but excluded from H3K4me3 peaks. Representative IGV genome browser views. For all tracks: DNA methylation (magenta, scale 0–100%), H3K27me3 (green, scale 0–4 scaled read counts per 107 reads), H3K4me3 (yellow, scale 0–8 scaled read counts per 107 reads) in control (C1, C2) and patient (P1, P2) dermal fibroblasts. DNA methylation data for SOX1 and FOXA1 (Fig. 3f) are shown again for comparison with H3K27me3 and H3K4me3. g, Polycomb-marked DNA methylation valleys (DMVs) are hypermethylated in DNMT3AW330R/+ patients. Shown, heat maps of n=1,152 DMVs26 of normalised H3K27me3/K4me3 read counts for control (C1,C2 mean) and patient (P1,P2 mean) fibroblasts, centred on DMVs and ranked by mean H3K27me3 levels in controls. Δ%mCpG, percent change of DMV methylation relative to mean of controls. Window size, 500bp. h, i, Quantification of data shown in panel g. (h) Polycomb-marked DMVs exhibit increased methylation in patient cells, while non-polycomb associated regions do not. Y-axis indicates mean difference between patients and controls: 0, no change; >0 increased in patients; <0 decreased in patients. (i) Polycomb-marked DMVs with increased methylation in patient cells, exhibit lower levels of H3K4me3 in controls (C1, C2 mean). Box, 25th-75th percentile; (h) whiskers, full data range; (i) whiskers, 1.5x interquartile range; centre line, median. Polycomb marked DMV definition, see methods. (p-values in h,i, two-sided Wilcoxon rank sum tests, polycomb positive (+) (n=524) versus negative (−) (n=628) DMVs).
Notably the regions of increased DNA methylation in patient cells were not confined to CGIs and often extended over tens of kilobases of genomic sequence (Fig. 3f). Their extent and location were reminiscent of ordinarily hypomethylated domains, that have been termed ‘DNA methylation valleys’ (DMVs)26,27, ‘DNA methylated canyons’28 or ‘broad non-methylated islands’29. These have been demonstrated to be evolutionary conserved regions, often associated with Polycomb-regulated developmental genes. Subsequent analysis confirmed a significant overlap between genes within reported DMVs26 and genes associated with DNMT3AW330R/+ hypermethylated DMRs (P = 8.3×10−170, Fig 4e).
Comparison of H3K27me3-marked DMVs in control fibroblasts with those lacking H3K27me3, established that the polycomb-associated DMVs were specifically hypermethylated in patients (P = 9.6×10−83, Fig. 4f-h). Subsequent H3K4me3 ChIP-seq showed that hypermethylated DMVs also contained H3K4me3 peaks (Fig. 4f), consistent with ChromHMM ‘poised-promoter’ predictions (Fig. 4a). DMVs without Polycomb marks exhibited higher levels of H3K4me3 (Fig. 4 g,i), consistent with transcriptionally active loci.
In DNMT3AW330R/+ patient fibroblasts, H3K27me3 levels were reduced at hypermethylated DMRs and H3K27me3 marked DMVs (Fig. 4f,g, Supplementary Fig. 4a-f). However, levels of the H3K27me3 methyltransferase EZH2 were normal in patient fibroblasts, and total cellular levels of H3K27me3-marked histones were unchanged when assessed by mass-spectroscopy (Supplementary Fig. 4g-i). Therefore, reduction in H3K27me3 was likely the result of DNMT3A-mediated DNA methylation inhibiting PRC2 binding/activity30,31. H3K4me3 levels were also reduced at hypermethylated DMRs and H3K27me3 marked DMVs (Supplementary Fig. 5a-e), significantly more than at other H3K4me3 peaks in the genome, consistent also with this reduction being a secondary consequence of DNA hypermethylation.
We therefore concluded that the W330R mutation is associated with hypermethylation of Polycomb-marked DMVs in patient cells, impacting on bivalent histone marks and modifying the chromatin state at key developmental regulators.
H3K36me3 and H3K27me3 histone modifications are usually mutually exclusive32,33, and strongly anti-correlated genome-wide34. To confirm this was also the case for DMRs and DMVs we performed H3K36me3 ChIP Rx-seq, and indeed few H3K36me3 ChIP-seq reads were present in DMVs in control and patient cells, with no enrichment over ChIP input seen (Supplementary Fig. 6a,b). Furthermore, hypermethylated DMRs in both control and patient fibroblasts were substantially depleted for H3K36me3 ChIP-seq peaks, when compared to all Infinium array probe sites (Supplementary Fig. 6c).
Hypermethylation at Polycomb marked loci occurs upon differentiation of DNMT3AW326R pluripotent stem cells
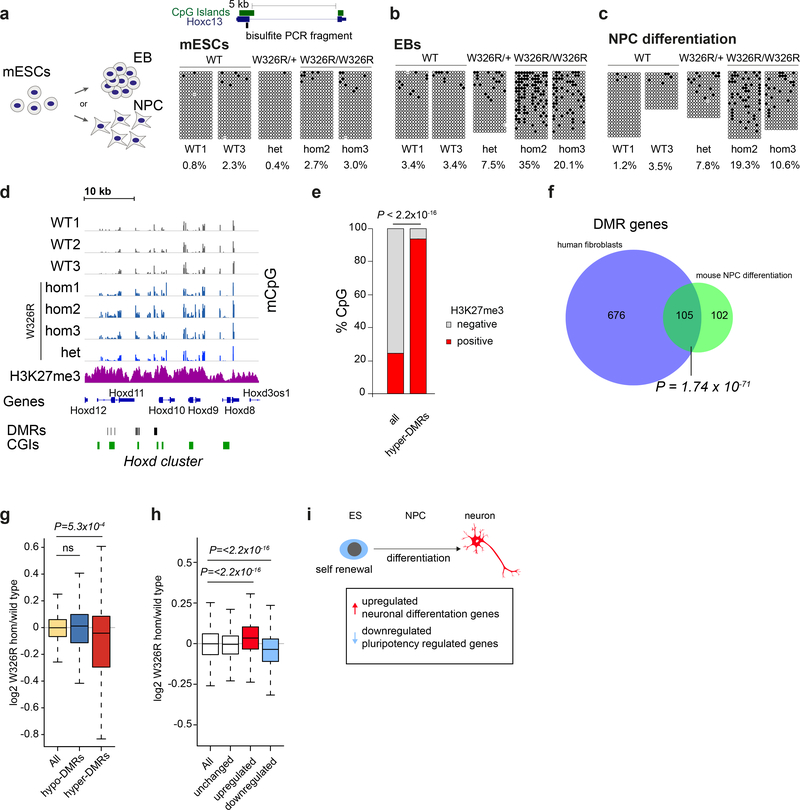
As large scale de novo DNA methylation occurs during early embryogenesis35, we reasoned that the increased methylation detected in patient fibroblasts and leukocytes was likely to have developmental origins. We addressed this possibility using the previously generated Dnmt3aW326R mES cell lines, containing the W330R-orthologous murine mutation (Fig. 2a). However, bisulfite sequencing of the promoter CpG islands of Hoxc13, Sox1 and Foxa1 (loci we had established to have increased methylation in patient cells), demonstrated similar low levels of DNA methylation in wild-type, DNMT3AW326R/+ and DNMT3AW326R/W326R ES cells (Fig. 5a and Supplementary Fig. 7a,b). Nevertheless, upon differentiation to embryoid bodies (EBs), DNA hypermethylation became evident in DNMT3AW326R/+ and DNMT3AW326R/W326R cells relative to controls (Fig. 5b). To exclude skewing of lineage fate in EBs as a confounding explanation for altered methylation, directed differentiation of mESCs to neural progenitor cells (NPCs)36 was also performed. This also demonstrated increased methylation in DNMT3AW326R cells (Fig. 5c). Furthermore, Reduced Representation Bisulfite Sequencing (RRBS)37 established that such methylation occurred at many Polycomb-marked loci in neurally-differentiated cells (Fig. 5d,e). 342 hypermethylated DMRs were detected in DNMT3AW326R/W326R cells relative to wild-type controls (Supplementary Table 5). These regions were significantly enriched for H3K27me3 ChIP-seq peaks derived from a wildtype neural-progenitor differentiation dataset38 (P < 2.2×10−16, Fig. 5e). As well, 105 of 207 DMR-associated genes overlapped orthologous gene loci for hypermethylated patient fibroblast DMRs (P = 1.7×10−71 Fisher’s exact test, Fig. 5f). Therefore, we conclude that the W326R substitution causes methylation at Polycomb-marked developmental genes from early stages of cell fate specification and differentiation in vitro.
Fig. 5|. Hypermethylation of polycomb-marked regions is observed on differentiation of Dnmt3aW326R pluripotent stem cells.
a-e, DNA methylation at DMRs occurs during cellular differentiation to embryoid bodies (EBs) and neural progenitor cells (NPCs) in CRISPR/Cas9-edited Dnmt3aW326R mESCs. Bisulfite sequencing of the Hoxc13 locus of (a) LIF/serum maintained mESCs, (b) after 9 days differentiation to EBs and (c) after 9 days neural induction to NPCs. For EBs and NPC differentiation, representative of n=2 independent experiments each. Blocks, independent cell lines; open and closed circles, unmethylated and methylated CpGs, respectively; dots, undetermined methylation status; columns CpG sites; rows individual sequences. Total percentage methylation calculated per sample. (d) Genome browser view of RRBS DNA methylation profiles after 9 days neural differentiation. Tracks, independent wild type (dark grey), and Dnmt3aW326R (blue) cell lines. Neural precursor cell H3K27me3 data (magenta) from published ChIP-seq dataset38. DNA methylation (scale 0–80%, all tracks). (e) Hypermethylated DMRs are enriched for H3K27me3 peaks in wildtype NPCs. Percentage of CpGs observed in RRBS overlapping with H3K27me3 peaks. H3K27me3 data from wild type NPC ChIP-seq dataset38. All, all CpGs observed (n=1178718 CpGs). Hyper-DMRs, CpGs within hypermethylated DMRs (3117). P-value, two-sided Fisher’s exact test. f, Hypermethylated gene loci in Dnmt3aW326R NPCs substantially overlap those in patient cells. Venn diagram of orthologous genes (human n=781; mouse n=207) associated with respective DMRs. P-value, two-sided Fisher’s exact test. (g) Reduced expression for genes associated with hyper-DMRs is evident during NPC differentiation. RNA-seq data for NPC differentiation experiment from panel d. (n=3 wild-type clones, n=3 Dnmt3aW326R/W326R clones). log2 CPM ratios of Dnmt3aW326R/W326R versus wildtype at 9 day NPC differentiation plotted. Box, 25th-75th percentile; whiskers, 1.5x interquartile range from box; centre line, median. Two-sided Wilcoxon rank sum test, All genes with coverage in RRBS (n=12620 genes) vs genes associated with hypo-DMRs (n=169) or hyper-DMRs (n=161). h-i, Neurogenic gene transcription bias in Dnmt3aW326R/W326R NPCs. (h) log2 CPM ratios of genes for Dnmt3aW326R/W326R versus wild-type 9 day-differentiated NPCs. All, all genes n=13,022; and gene sets, upregulated (n=3,864 genes), unchanged (n=3,516) and downregulated genes (n=3,281) during differentiation from mESCs to neurons. Box, 25th-75th percentile; whiskers, 1.5x interquartile range from box; centre line, median. Two-sided Wilcoxon rank sum test, log2 W326R/WT for All vs up or downregulated gene sets. (i) Schematic: Gene sets defined on basis of published dataset of mESC differentiation to terminally differentiated neurons38. Downregulated and upregulated gene sets defined as those genes with reduced and increased transcripts respectively in neurons relative to ES cells. The downregulated set therefore contains pluripotency-related genes (light blue) and the upregulated set, neuronal differentiation genes (light red).
Neurogenic gene expression bias in Dnmt3aW326R NPCs
To understand the transcriptional consequences of DMR hypermethylation, we next performed RNA-seq on Dnmt3aW326R/W326R NPCs and DNMT3AW330R/+ fibroblasts. We found a significant downregulation of transcription of genes associated with hypermethylated DMRs, whereas transcript levels at hypomethylated DMRs were unchanged (Fig. 5g and Supplementary Fig. 8a-c). We reasoned that many of the DMR/DMV-associated genes are transcription factors, that would consequently perturb developmental transcriptional networks. Prior work has demonstrated differentiation to be impaired in Dnmt3a-deficient NPCs and hematopoietic stem cells (HSCs) with enhanced expression of multipotency/stem cell genes and decreases in differentiation/neurogenic gene transcripts 31,39,40. As W326R is a ‘gain of function’ mutation, we postulated that a reciprocal transcriptional phenotype would be evident in Dnmt3aW326R/W326R NPCs. Accordingly, we examined two gene sets, representing genes that are respectively up and down-regulated during differentiation of mESCs to terminally-differentiated neurons38 (Fig. 5h). In line with our expectation, Dnmt3aW326R/W326R NPCs demonstrated a transcriptional bias towards expression of neurogenic genes at the expense of genes normally expressed in the pluripotent state. This suggests that hypermethylation of DMV/DMRs could lead to a skewing of stem/progenitor cells towards differentiation away from self-renewal.
Dnmt3aW326R/+ mice have reduced brain size and body weight
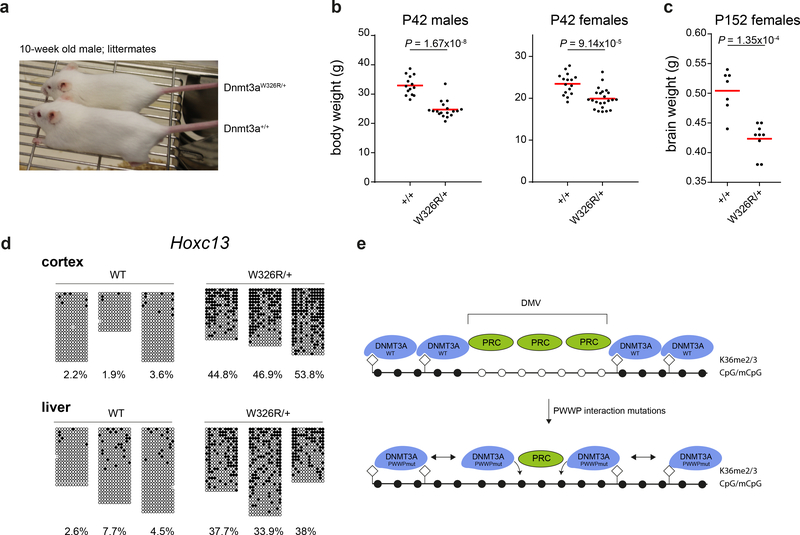
Finally, we generated a Dnmt3aW326R/+ mouse using CRISPR/Cas9-mediated homology directed repair (Supplementary Fig. 9a,b) to provide an in vivo model. Recapitulating the patient growth restriction phenotype, Dnmt3aW326R/+ mice were viable, healthy and morphologically unremarkable, but were proportionately small with significantly reduced body and brain weight (Fig. 6a-c, Supplementary Fig 9c,d). Furthermore, bisulfite sequencing of cerebral cortex and liver provided in vivo confirmation of hypermethylation at polycomb-regulated regions, with substantial methylation observed at the Hoxc13 and Sox1 loci in Dnmt3aW326R/+ mice (Fig. 6d,Supplementary Fig. 9e). Furthermore, RRBS analysis confirmed that genome-wide, NPC hypermethylated DMRs were hypermethylated in the Dnmt3aW326R/+ mouse cortex (Supplementary Fig. 9f-h).
Fig. 6|. Dnmt3aW326R/+ mice have reduced brain size and body weight, alongside hypermethylation of developmental genes.
a, 10-week old Dnmt3aW326R/+ mouse next to wild-type littermate. (b) Body weight for 6 week-old Dnmt3aW326R/+ mice compared to wild type littermates. Males, n=14 wildtype and n=18 Dnmt3aW326R/+ animals. Females, n= 16 wildtype and n=23 Dnmt3aW326R/+ animals. (c) Brain weight of female Dnmt3aW326R/+ mice compared to wild type litter mates at 5 months of age. n=7 wildtype and 9 Dnmt3aW326R/+ animals. h,i P-values, two-tailed t-test. Horizontal bar, mean weight per group. (d) Locus-specific (Hoxc13) bisulfite sequencing for cortex and liver samples from Dnmt3aW326R/+ and wild-type littermates (n=3/group; female, age 8 weeks). e, Proposed model linking disruption of the H3K36me2/3<--> PWWP interaction with DMV DNA methylation. WT-DNMT3A is normally targeted to H3K36me2 and H3K36me3, marks present widely in the genome32,58, but rarely coexist with H3K27me3 32,33. This limits availability of free-DNMT3A to bind at other locations. When the PWWP-H3K36me2/3 interaction is disrupted, sufficient free DNMT3A is available to methylate genomic DNA at DMVs. Enzymatic activity of DNMT3A and DNA methylation impair PRC2 chromatin binding30,31, leading to secondary loss of H3K27me3. Notably, the long isoform of DNMT3A (DNMT3A1) localises to the edge of Polycomb domains21,43. When mutated it is therefore well placed to methylate these regions. DNMT3A1 is also the major isoform expressed after ESC differentiation21, potentially explaining timing of hypermethylation. Filled circles methylated CpG, open circles unmethylated CpG. Diamonds, H3K36me2/3 modified histones.
Discussion
Here we report widespread DNA hypermethylation at Polycomb-regulated regions resulting from a gain of function mutation in DNMT3A. As such genomic regions contain key developmental genes, classical patterning defects might be expected, but, surprisingly, the DNMT3AW330R mutation instead causes an extreme growth disorder.
Unexpectedly our findings suggest that the DNMT3A PWWP domain limits DNA methylation at Polycomb-regulated regions. DNMT3A has been previously shown to counter H3K27 tri-methylation in vivo, with wild-type (but not catalytically dead) DNMT3A opposing PRC2 binding in neural stem cells31. In patient cells, it is therefore likely that altered binding specificity of DNMT3AW330R leads to it methylating polycomb-associated DMRs and DMVs, with a secondary reduction occurring in H3K27me3 due to impaired binding of PRC2 to methylated DNA30,31.
Biochemically, binding to H3K36me2/3 is abrogated in the PWWPW330R mutant. How then could this impaired interaction with H3K36me2/3 connect with DNA methylation of H3K27me3 regions? We favour a model where widespread distribution of H3K36me2/3 leads to wild-type DNMT3A being targeted to many genomic sites and limiting its availability to non-preferred sites such as polycomb-associated regions (Fig 6e). Consistent with this model, we see low levels of H3K36me3 at DMRs and DMVs, explained by H3K36me2/3 rarely co-existing with H3K27me3 on histones32,33. As well, genome-wide, H3K36me3 is strongly anticorrelated with H3K27me3, and low levels of H3K36me2 correlated with increased H3K27me334. Furthermore, H3K36me2 is actively removed by KDM2A from unmethylated CpG regions41 and Nsd1-mediated H3K36me2 methylation has recently been shown to restrict deposition of H3K27me3 34.
In our model we propose that disruption of PWWP-H3K36me2/3 interactions in patient cells would increase availability of DNMT3AW330R to interact with DNA in polycomb regions (Fig 6e), increasing the possibility of DNA methylation, consequently impairing PRC2 binding and polycomb-domain integrity. Alternative explanations are also possible. For instance, the PWWPWT-H3K36me2/3 interaction may normally be required for enzymatic activity, whereas DNMT3AW330R may be permissive for DNA methylation without the interaction; or the PWWP domain may mediate non-histone interactions critical to restrict it from Polycomb-marked loci. Further studies, including assessment of DNMT3AW330R localization by ChIP-seq to determine genomic distribution, will be important in distinguishing between these possibilities. Nonetheless, our findings establish the DNMT3A PWWP domain as a factor countering methylation of key developmental loci, one that may act alongside Tet enzymes42,43 and FBXL1044 to ensure their hypomethylation.
Previously identified microcephalic dwarfism genes impair cell proliferation to reduce cell number and organism size1, so how might this mutation in DNMT3A act? Both DNA methyltransferases and Polycomb can impart heritable transcriptionally repressive epigenetic marks. However, while DNA promoter methylation is considered to stably silence genes45, Polycomb repression is potentially reversible, maintaining plasticity of gene expression and enabling robust switching to gene activation in response to developmental cues46,47. Dnmt3a loss in hematopoietic stem cells leads to expanded stem cell numbers at a cost to differentiated progeny39. Likewise, Dnmt3a null neural stem cells have markedly reduced neurogenic potential31. Conversely, loss of the PRC2 H3K27me3 methyltransferase, Ezh2, from cortical progenitors impairs self-renewal promoting premature neuronal differentiation48, and here we observe a transcriptional bias away from pluripotency towards differentiation in Dnmt3aW326R/W32R NPCs (Fig 5.). Hence, gain of function DNMT3A mutations might increase cellular differentiation leading to premature depletion of stem/progenitor cell pools and reduce final cell numbers in tissues and consequently organism size (Supplementary Fig. 8d).
Like DNMT3A, haploinsufficiency of H3K36 methyltransferases NSD1 and SETD2, cause macrocephalic overgrowth13,49,50. Mutations in genes encoding EZH2, and EED subunits of Polycomb complexes also cause overgrowth51–53 and PHC1 mutation results in microcephalic dwarfism54. While DNA methylation and Polycomb-repression are thought of as mutually antagonistic and exclusive processes at specific loci, our findings linking H3K27, H3K36 and DNA methylation, suggest a yet to be defined common developmental mechanism for these syndromes. Furthermore, given NSD1, DNMT3A and EZH2 are both height QTLs and somatically mutated in cancer55,56, the interplay between Polycomb and DNA methylation has wider relevance both to neoplastic processes and physiological regulation of human size, that warrants further investigation.
Materials and Methods
Research subjects
Genomic DNA from affected children and family members was extracted using standard protocols. Informed, written consent was obtained from all participating families. The study was approved by the Scottish Multicentre Research Ethics Committee (04:MRE00/19) and the Institutional Review Board of Cincinnati Children’s Hospital Medical Center (Protocol#2014–5919). All relevant ethical regulations were followed. Genotypes of TBRS patients were as follows: O1: DNMT3A – heterozygous c.1936G>C p.Gly464Arg; O2: DNMT3A – heterozygous c.2086del p.Gln696ArgFsTer9.
Exome sequencing
Exome sequencing of patients 1 and 3 was performed by Edinburgh Genomics and Cincinnati Children’s Hospital Sequencing Core Facility respectively as described previously59,60. Patient 2 was sequenced by Illumina MiSeq using a custom targeted capture (SureSelect, Agilent Technologies) targeting DNMT3A and other primordial dwarfism/microcephaly genes. Confirmatory Sanger sequencing was performed on all affected individuals and their parents. Primers listed in Supplementary Table 6. Further details see Supplementary Note.
Cell culture
Primary fibroblast cell lines were maintained at 3% O2 in Dulbecco’s modified Eagle’s medium (DMEM; Life Technologies) supplemented with 10% FBS and 5% penicillin-streptomycin antibiotics or in AmnioMAX-C100 (Life Technologies). HeLa cells, a kind gift from G. Stewart (Birmingham) originally obtained from ATCC, were maintained in Dulbecco’s modified Eagle’s medium (DMEM; Life Technologies) supplemented with 10% FBS and 5% penicillin-streptomycin antibiotics. E14 Tg2a IV mESCs were cultured on 0.1% gelatine coated dishes and maintained in Glasgow’s Minimum Essential Medium (GMEM; Life Technologies) supplemented with 10% FBS (HyClone), 1 mM Sodium Pyruvate (Sigma); 1x MEM non-essential amino acids (Sigma), 2 mM L-Glutamine, 5% penicillin-streptomycin antibiotics, 0.001% β-mercaptoethanol (Sigma) and leukemia inhibitory factor. Details for differentiation protocols see Supplementary Note.
Generation of CRISPR/Cas9 edited mESCs
Guide RNAs were designed using the optimized CRISPR design webtool (http://crispr.mit.edu/) with corresponding oligonucleotides cloned into pSpCas9(BB)-2A-GFP or pSpCas9n(BB)-2A-GFP (kind gift from Feng Zhang, Addgene Plasmids pX458:#48138, pX461:#48140)14. ssDNA oligonucleotides (ssODN, IDT Ultramers) repair template sequences for homology directed repair listed in Supplementary Table 6. Two independent CRISPR/Cas9 strategies were employed to generate the W326R mutation in the clones used in this study: each using different gRNAs, and either Cas9-nickase (nCas9) or wildtype Cas9 respectively. Vectors containing guide RNA sequences were transfected together with single stranded DNA oligonucleotides using FuGENE HD transfection reagent (Promega). GFP-positive cells were selected by FACS (FACSAriaII, FACSDiva Software Version 6.1.3, Becton-Dickinson) 48 hours after transfection and plated at clonal density. Individual colonies were grown up and validated by Sanger sequencing.
Immunoblot analysis and antibodies
Whole cell extracts for mESCs, human primary Fibroblasts and HeLa cells were obtained by sonication in UTB buffer (8 M urea, 50 mM Tris, pH 7.5, 150 mM β-mercaptoethanol) and analyzed by SDS-PAGE using 4–12% NuPage Bis-Tris Protein gels (Life Technologies) and transferred onto nitrocellulose membrane. Immunoblotting was performed using antibodies to Dnmt3a (Novus Biologicals NB120–13888; 1:500), EZH2 (Cell Signaling #5246S; 1:1000) and Actin (Sigma A2066; 1:5000). Images acquired with ImageQuant LAS 4000. Uncropped images in Supplementary Fig. 11.
RNA interference
EZH2 was targeted with 40nM of an ON-TARGETplus Human siRNA SMARTpool (L-004218–00-0005, Dharmacon) and cells harvested 48 hours after transfection with RNAiMAX (Thermo Fisher).
RT-PCR and RNA-sequencing
RNA was extracted using the RNeasy kit (QIAGEN) according to manufacturer instructions with DNAseI (QIAGEN) treatment. For RT-PCR cDNA was generated using SuperScript III Reverse Transcriptase (Invitrogen) and random primers (Promega). Primers for RT-PCR listed in Supplementary Table 6.
For RNA-sequencing, random primed cDNA from poly-A selected RNA was converted into an Illumina sequencing library and single-end 50bp reads generated on an Illumina HiSeq machine (GATC Biotech Konstanz, Germany).
RNA-seq data were aligned to the genome using bowtie 2 (v2.3.1). Further data processing details see Supplementary Note. Alignment statistics are provided in Supplementary Table 7 and summaries of the data are shown in Supplementary Fig. 8a,b.
Structural Modelling
The impact of W330R on the interaction with H3K36me3 was modelled with FoldX61 using the crystal structure of the DNMT3B PWWP domain bound to H3K36me3 (PDB ID: 5CIU). The change in interaction energy caused by the equivalent W263R mutation in DNMT3B was calculated with the AnalyseComplex function. Since the H3K36me3 binding site is highly conserved between DNMT3A and DNMT3B, including full conservation of all the aromatic residues involved in binding highlighted in Figure 2, this suggests that W330R would also disrupt the interaction.
Generation of recombinant PWWP protein
PWWP domain of DNMT3A was expressed in E.coli and purified using standard methods, documented in the Supplementary Note.
Histone peptide pull downs
20 μg of purified recombinant GST-PWWP fusion protein and 2000 pmol of histone H3 biotinylated peptides (AnaSpec peptides; AS-64440 and AS-64441) were diluted in interaction buffer (50 mM Tris/HCl pH8.0, 100 mM NaCl, 2 mM EDTA, 0.1% Triton X-100 freshly supplemented with 0.5 mM DTT, 0.2 mM PMSF and 1x protease inhibitor cocktail, Roche)15. Reactions were incubated overnight under rotation at 4°C. MyOne T1 streptavidin beads (Life Technologies) were added to the reactions and rotated for 4h at 4°C, followed by three washes with interaction buffer. 20 μl of sample loading buffer (50 mM Tris pH6.8, 20% Glycerol, 20% SDS, 625 mM β-mercaptoethanol, bromphenolblue) were added to beads, boiled for 5 min and eluted proteins separated on 15% SDS-PAGE and visualised with Coomassie Blue R250.
Peptide arrays
Peptide arrays were processed following manufacturer instructions for the MODified Histone Peptide Arrays (Active Motif). In brief, arrays were blocked and washed with buffers provided. 10nM or 100nM wildtype or W330R DNMT3A GST-tagged PWWP protein was diluted in interaction buffer (100 mM KCl, 20 mM Hepes pH7.5, 1 mM EDTA, 0.1 mM DTT, 10% glycerol)15 and incubated overnight at 4°C on an orbital shaker. Protein-peptide interactions were detected with an antibody directed against the GST-tag (GE Healthcare 27–4577-01; 1:5000) with subsequent ECL-based detection. c-Myc mouse monoclonal antibody (1:2000, Active-Motif). Images acquired using ImageQuant LAS 4000.
Infinium® MethylationEPIC BeadChip
Fibroblast genomic DNA extracted using the DNeasy Blood & Tissue Kit (QIAGEN). DNA was bisulfite converted using the EZ DNA Methylation kit (Zymo Research, Infinium assay protocol). Infinium® MethylationEPIC BeadChip performed according to manufacturer instructions by Edinburgh WTCRF. The Bioconducter package minfi (v1.22.1) was used to process raw Infinium idat files (ssNoob method)62,63. For further details, see Supplementary Note. Overall summaries of Infinium methylation data are shown in Supplementary Figure 10a,b and d.
Chromatin immunoprecipitation and sequencing
Cross-linked chromatin immunoprecipitation was adapted from previous publications 64,65, further detailed in the Supplementary Note. For H3K27me3 single-end 50bp reads were generated on an Illumina HiSeq machine (GATC Biotech Konstanz, Germany). For H3K4me3, H3K36me3 ChIP-Rx and H3K27me3 ChIP-Rx single-end 75bp reads were generated on an Illumina NextSeq 550 machine (WTCRF Edinburgh, UK).
ChIP-seq read quality assessment and alignment was performed as for RNA-seq. For ChIP Rx-seq, reads were aligned to a combination of the hg19 and dm6 genomes using the same settings. Multi-mapping reads excluded as for RNA-seq. Additionally, PCR duplicates excluded using SAMBAMBA (v0.5.9)66. Sequencing statistics are shown in Supplementary Table 8. For further analysis details see Supplementary Note.
Histone acid extraction and histone PTM detection by mass spectrometry
Histones were acid extracted as previously described32 with minor modifications and LC MS/MS analyses were performed on an Orbitrap Fusion Lumos coupled to Dionex Ultimate3000RSLCnano UHPLC system. For further details see Supplementary Note.
Bisulfite PCR sequencing
Genomic DNA was isolated using the DNeasy Blood & Tissue Kit (QIAGEN) or Phenol-Chloroform extraction. 250–500ng DNA was bisulfite converted with the EZ DNA Methylation-Lightning Kit (Zymo Research) according to manufacturer instructions. Converted DNA was eluted twice in 10 μl elution buffer. Bisulfite PCR primer sequences provided in Supplementary Table 6. Products were amplified using FastStart PCR Master Mix (Roche), purified using the QIAquick PCR purification Kit (QIAGEN) and subcloned into pGEMT-easy (Promega). Individual bacterial colonies were sanger sequenced using M13 sequencing primers, analysed using BISMA67 and results formatted with the BiQ Analyzer Diagrams tool68. In two independent experiments of NPC/EB differentiation, the following cell lines were used: For EB experiments: WT3, hom2 (n=2); WT1, hom3, het (n=1). For NPC, WT1, hom2, hom3, het (n=2); WT2,WT3, hom1 (n=1).
Reduced Representation Bisulfite Sequencing (RRBS)
Genomic DNA isolated with DNeasy Blood & Tissue Kit (QIAGEN) or Nucleon BACC2 Genomic DNA Extraction Kit (illustra) and quantified by Qubit (Invitrogen). DNA from mouse cortex samples were concentrated using Agencourt AMPure XP technology. 200ng of purified DNA samples (for NPC differentiation: DNA from experiment depicted in Fig.5c; for mouse cortexes in Fig. 6d, Supplementary Fig. 9e) were processed using the Ovation RRBS Methyl-Seq system kit (NuGen Technologies) according to instructions with modifications documented in the Supplementary Note. RRBS sequencing was aligned and processed using Bismark (v0.16.3)69.
Processed RRBS files were assessed for conversion efficiency based on the proportion of methylated reads mapping to the λ genome spike-in (>99.5% in all cases, Supplementary Table 9) and processed in R to call DMRs. Alignment statistics provided in Supplementary Table 9. BigWigs were generated from RRBS data using CpGs with coverage ≥5. BigWigs for Patient 3 and Control 3 were generated only from CpGs with coverage ≥5 in both samples to facilitate visual comparison (shown Figure S3d). Overall summaries of RRBS data are shown in Figure S10c, f-h. Mean methylation in each sample was calculated as the weighted mean across all CpGs observed on autosomes irrespective of coverage (methylated coverage/total coverage).
Generation of Dnmt3aW326R mice
A template for in vitro transcription was prepared by PCR, using the pX458-based plasmid containing the Dnmt3a-targeting gRNA sequence, a T7-tagged gRNA specific forward primer and a universal reverse primer (sequences Supplementary Table 6), PCR product purified by QIAquick PCR Purification (QIAGEN) and gRNA was produced by in vitro transcription (NEB HiScribe T7 High Yield RNA Synthesis Kit) using 1 μg of PCR product, and purified using the RNeasy Mini Kit (QIAGEN). Transgenic mice were generated by cytoplasmic injection of gRNA (25 ng/μl), Cas9 mRNA (50 ng/μl; L-6125; TriLink Biotechnologies) and ssODN repair template (150 ng/μl) into B6CBAF1/J single cell embryos. All resulting pups were screened by PCR amplification and sanger sequencing of the targeted region (primer sequences, Supplementary Table 6). F0 males were crossed with CD-1 females to establish germline transmission. F1 Dnmt3aW326R/+ males were crossed with CD-1 females and F2 offspring used for phenotyping and tissue collection (investigators were blinded to genotypes). Mouse studies were approved by the University of Edinburgh animal welfare and ethical review board (AWERB) and conducted according to UK Home Office regulations under a UK Home Office project license.
Statistical analysis
Statistical testing was performed using R v3.4.2 and GraphPad PRISM 6. Tests used indicated in figure legends. All tests were two-sided, unless otherwise stated. Further details of specific analyses provided in the relevant methods sections, below and Supplementary Note.
Hierarchical clustering
Clustering was performed on processed Infinium Beta probe values using R (Pearson correlation distance and Ward method). Cluster significance was tested using the CRAN package ‘pvclust’ (v2.0.0).
Differentially methylated region identification
Windows of 5 contiguous probes or CpGs were used to identify DMRs for Infinium and RRBS data respectively. For Infinium arrays DMRs were called on the basis of ≥3 probes in a window having a difference in Beta value of at least 0.1 between each individual patient sample and each of the two control fibroblast lines, changed in the same direction, and with no CpG ≥1000bp from its neighbours in the window. For this analysis we also only considered probes showing the same difference in both the patient 1 replicates. Overlapping or contiguous DMR windows were merged. For enrichment analyses, the set of DMR probes was compared to a genome-wide background control set of ‘All’ probes, derived from genomic regions spanning genomically-contiguous probes that fulfilled the same distance threshold criteria (ie. CpG ≤1000bp from neighbours). DMR methylation level, defined as the mean Beta value of all probes located in the DMR. Fibroblast DMRs are provided in Supplementary Tables 2 and 3.
For RRBS data DMRs were called using a binomial linear model to test for a difference in the proportion of methylated reads for each CpG in the homozygous mutant samples versus controls. CpGs showing significant differences were then identified as those with Benjami-Hochberg adjusted p-values <0.05. DMRs were then called in a manner similar to that used for the Infinium arrays but using a distance threshold of 200bp. A control set of CpG sites that were within the distance threshold was used as a background control set of ‘All’ CpGs for enrichment analyses. Only CpGs where coverage was ≥10 in all samples were considered for DMR calling (1,516,046 CpGs). For RRBS DMR methylation level was defined as the weighted mean methylation level (methylated coverage/total coverage) from all CpGs observed within the DMR region irrespective of coverage. NPC DMRs are provided in Supplementary Tables 5 and 10.
Enrichment of DMRs in ChromHMM segmentations
Infinium probes were mapped to existing ChromHMM annotations70 using the BEDtools intersect function (v2.27.1)71. Identical ChromHMM labels were merged for analysis. To test for enrichment of an annotation, a Fisher’s exact test was performed for number of DMR probes against number of control probes.
DNA methylation valley analysis
Previously reported DMVs26 were mapped to hg19 using the UCSC liftover tool and merged DMV regions from all 5 cell-types determined using the Bedtools merge function. DMV methylation level, defined as mean Beta value of all probes present in the DMV. DMVs were then mapped to their closest genes using ChIPpeakAnno (details Supplementary Note). Using DMR and control gene lists (as defined in GO analysis section), DMR enrichment at DMVs was tested by Fisher’s exact test, comparing the proportion of DMR-associated genes that were DMV genes with the proportion of control genes that were DMV genes.
Analysis of histone modifications at DMRs, DMVs and ChIP-seq peaks
Non-overlapping windows of 250bp (for DMRs and ChIP-seq peaks) or 500bp (for DMVs) were defined centred on each region of interest, with ChIP-seq read counts/window calculated using BEDtools’ coverage function. Read counts were scaled to counts per 10 million based on total number of mapped reads/sample and divided by the input read count to provide a normalised read counts. To prevent windows with zero reads in the input sample generating a normalised count of infinity, an offset of 0.5 was added all windows prior to scaling and input normalisation. Regions where coverage was 0 in all samples were removed from the analysis. ChIP-Rx was analysed similarly before samples were scaled using a normalisation factor generated from the number of reads mapping to the spike-in Drosophila genome. Reads mapping to the Drosphila genome in each ChIP and input sample were first scaled to reads per 1×107. The scaling factor was then calculated as the ratio of the scaled Drosophila reads in two ChIP samples over their respective ratio from the input samples, ie scaling factor, S, for sample n compared to reference sample ref: Sn=(dRPTMChIP-n/dRPTMChIP-ref)/(dRPTMIN-n/dRPTMIN-ref), where dRPTM = Drosophila Reads per 1×107 for ChIP and input, IN, runs respectively (modified from published method to take account of the presence of an input sample)65. To statistically test differences in histone modification levels, normalised read depths across DMRs/DMVs were compared using a Wilcoxon rank sum test. H3K27me3-marked DMVs defined as those containing a H3K27me3 ChIP-seq peak replicated in both control fibroblast lines. H3K27me3 and H3K4me3 peaks used for quantitative analysis were defined by merging peaks called in the two controls (using Bedtools merge). Only autosomal peaks overlapping those called in both control samples and containing Infinium probes in the background control set from DMR calling were used for analysis. The subsets of these peaks overlapping DMRs were defined using Bedtools intersect. The change in histone modification levels within regions of interest was defined as the log2 ratio of the mean mutant normalised read count over control normalised read count. The profile of H3K36me3 at DMVs was generated by calculating normalised read counts in 10 scaled windows across DMVs together with 500bp windows extending 10Kb up- and down-stream of each DMV. Colour scales for ChIP-seq heatmaps range from the minimum to the 90% quantile of the normalised read count for the reference dataset in each set of heatmaps.
Analysis of enrichment of histone modifications at DMRs
BEDtools intersect was used to overlap DMR probes with histone modification peaks. % DMR probes mapping to peaks was tested against % background control probes mapping to peaks using a Fishers exact test. A similar strategy was applied for mouse RRBS DMRs, testing DMR CpG versus background control CpG sites.
RNAseq analysis
To analyse RNAseq, the number of reads mapping to each ENSEMBL annotated gene (human: Release 75/GCRh37, mouse: Release 91/mm10) was calculated using the featureCounts module of the subread aligner (v1.5.2)72. Only reads mapping to exons were considered. Gene read counts were then analysed using EdgeR (v3.18.1)73 with Trimmed Mean of M-values (TMM) normalization74. The log2 normalised counts per million (CPM) and log2 ratios calculated by EdgeR were then subject to further analysis. Only genes where CPM was >1 in ≥2 samples and that are annotated as protein coding in ENSEMBL were considered for analysis (human fibroblasts, 11,963 genes; mouse NPCs, 13,022 genes). To generate lists of genes differentially regulated in published data of mES cells differentiated to terminally differentiated neurons38, similar pre-processing was applied (resulting in data for 14,780 genes). Differential expression was then called using an F-test of a generalised linear model fitted to the data taking account of sample batch using EdgeR. Up and down regulated genes were called as those with Benjamini-Hochberg corrected FDR < 0.01 and a log-fold change >|1| (4,147 and 4,067 genes respectively). Genes unchanged in the analysis were defined as those with Benjamini-Hochberg corrected FDR > 0.05 (4,067 genes).
Reporting Summary
Further information on experimental design is available in the Reporting Summary.
Data availability
The human next-generation sequencing data used in the manuscript are available on request from the relevant Data Access Committee from the European Genome–Phenome Archive (EGA). The exome data is available under the accession EGAS00001003231. Human RNA-seq, RRBS and ChIP-seq under the accession EGAS00001003232. The data are not publicly available to ensure protection of patient sequence data confidentiality through controlled access. Processed data files and mouse RNA-seq/RRBS are available in GEO under accession GSE120558.
Supplementary Material
Acknowledgements:
We are grateful to families and clinicians for their involvement and participation. We would like to thank W. Bickmore, R. Meehan, N. Hastie, T.Baubec and I. Adams for helpful discussions. G. Kelsey for discussion of unpublished data. P. Madapura, G. Taylor, L. Duthie and R. Illingworth for technical advice, E. Freyer, A. Meynert, IGMM FACS and Sequencing Cores, CBS, WTCCB mass-spectroscopy facility and the WTCRF for technical support. A.P.J. is supported by the Medical Research Council UK (MRC, U127580972) and the European Research Council (ERC), through ERC Starter Grant 281847; and now by the European Union’s Horizon 2020 research and innovation programme ERC Advanced Grant (grant agreement No: 788093). D.S. is a Cancer Research UK Career Development Fellow (reference C47648/A20837), and work in his laboratory is also supported by a Medical Research Council University grant to the MRC Human Genetics Unit. J.M. is supported by a Medical Research Council Career Development Award (MR/M02122X/1). P.H. was supported by a fellowship within the Postdoc-Program of the German Academic Exchange Service (DAAD). V. Hwa is supported by funding from NIH NICHHD R01HD078592. T.A. is supported by Wellcome Trust funding to R.C.A. (200885). J.R. is supported by the Wellcome Trust through a Senior Research Fellowship (103139) and a multi-user equipment grant (108504). The Wellcome Centre for Cell Biology is supported by core funding from the Wellcome Trust (203149).
Footnotes
Competing Interests:
The authors declare no competing interests.
Accession Codes:
DNMT3A –NM_175629.2
EGA: WES data: EGAS00001003231; RNAseq/RRBS/ChIPseq EGAS00001003232
GEO: awaiting accession codes
References:
- 1.Klingseisen A & Jackson AP Mechanisms and pathways of growth failure in primordial dwarfism. Genes and Development 25, 2011–2024 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bicknell LS et al. Mutations in the pre-replication complex cause Meier-Gorlin syndrome. Nat Genet 43, 356–9 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bicknell LS et al. Mutations in ORC1, encoding the largest subunit of the origin recognition complex, cause microcephalic primordial dwarfism resembling Meier-Gorlin syndrome. Nat Genet 43, 350–5 (2011). [DOI] [PubMed] [Google Scholar]
- 4.Guernsey DL et al. Mutations in origin recognition complex gene ORC4 cause Meier-Gorlin syndrome. Nat Genet 43, 360–4 (2011). [DOI] [PubMed] [Google Scholar]
- 5.Burrage LC et al. De Novo GMNN Mutations Cause Autosomal-Dominant Primordial Dwarfism Associated with Meier-Gorlin Syndrome. Am J Hum Genet 97, 904–13 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rauch A et al. Mutations in the pericentrin (PCNT) gene cause primordial dwarfism. Science 319, 816–9 (2008). [DOI] [PubMed] [Google Scholar]
- 7.Griffith E et al. Mutations in pericentrin cause Seckel syndrome with defective ATR-dependent DNA damage signaling. Nat Genet 40, 232–6 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martin CA et al. Mutations in PLK4, encoding a master regulator of centriole biogenesis, cause microcephaly, growth failure and retinopathy. Nat Genet 46, 1283–92 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Conlon I & Raff M Size control in animal development. Cell 96, 235–44 (1999). [DOI] [PubMed] [Google Scholar]
- 10.Lek M et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature 536, 285–91 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tatton-Brown K et al. Mutations in the DNA methyltransferase gene DNMT3A cause an overgrowth syndrome with intellectual disability. Nat Genet 46, 385–8 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tlemsani C et al. SETD2 and DNMT3A screen in the Sotos-like syndrome French cohort. J Med Genet (2016). [DOI] [PubMed] [Google Scholar]
- 13.Okamoto N, Toribe Y, Shimojima K & Yamamoto T Tatton-Brown-Rahman syndrome due to 2p23 microdeletion. Am J Med Genet A 170A, 1339–42 (2016). [DOI] [PubMed] [Google Scholar]
- 14.Ran FA et al. Genome engineering using the CRISPR-Cas9 system. Nat Protoc 8, 2281–2308 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dhayalan A et al. The Dnmt3a PWWP domain reads histone 3 lysine 36 trimethylation and guides DNA methylation. J Biol Chem 285, 26114–20 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sankaran SM, Wilkinson AW, Elias JE & Gozani O A PWWP Domain of Histone-Lysine N-Methyltransferase NSD2 Binds to Dimethylated Lys-36 of Histone H3 and Regulates NSD2 Function at Chromatin. J Biol Chem 291, 8465–74 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qin S & Min J Structure and function of the nucleosome-binding PWWP domain. Trends Biochem Sci 39, 536–47 (2014). [DOI] [PubMed] [Google Scholar]
- 18.Rondelet G, Dal Maso T, Willems L & Wouters J Structural basis for recognition of histone H3K36me3 nucleosome by human de novo DNA methyltransferases 3A and 3B. J Struct Biol 194, 357–67 (2016). [DOI] [PubMed] [Google Scholar]
- 19.Kungulovski G et al. Application of histone modification-specific interaction domains as an alternative to antibodies. Genome Res 24, 1842–53 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Du J, Johnson LM, Jacobsen SE & Patel DJ DNA methylation pathways and their crosstalk with histone methylation. Nat Rev Mol Cell Biol 16, 519–32 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Manzo M et al. Isoform-specific localization of DNMT3A regulates DNA methylation fidelity at bivalent CpG islands. EMBO J 36, 3421–3434 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meissner A et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature 454, 766–70 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ernst J et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature 473, 43–9 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cao R et al. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science 298, 1039–43 (2002). [DOI] [PubMed] [Google Scholar]
- 25.Kuzmichev A, Jenuwein T, Tempst P & Reinberg D Different EZH2-containing complexes target methylation of histone H1 or nucleosomal histone H3. Mol Cell 14, 183–93 (2004). [DOI] [PubMed] [Google Scholar]
- 26.Xie W et al. Epigenomic analysis of multilineage differentiation of human embryonic stem cells. Cell 153, 1134–48 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li Y et al. Genome-wide analyses reveal a role of Polycomb in promoting hypomethylation of DNA methylation valleys. Genome Biol 19, 18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jeong M et al. Large conserved domains of low DNA methylation maintained by Dnmt3a. Nat Genet 46, 17–23 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Long HK et al. Epigenetic conservation at gene regulatory elements revealed by non-methylated DNA profiling in seven vertebrates. Elife 2, e00348 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bartke T et al. Nucleosome-interacting proteins regulated by DNA and histone methylation. Cell 143, 470–84 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu H et al. Dnmt3a-dependent nonpromoter DNA methylation facilitates transcription of neurogenic genes. Science 329, 444–8 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sidoli S et al. Middle-down hybrid chromatography/tandem mass spectrometry workflow for characterization of combinatorial post-translational modifications in histones. Proteomics 14, 2200–11 (2014). [DOI] [PubMed] [Google Scholar]
- 33.Yuan W et al. H3K36 methylation antagonizes PRC2-mediated H3K27 methylation. J Biol Chem 286, 7983–9 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Streubel G et al. The H3K36me2 Methyltransferase Nsd1 Demarcates PRC2-Mediated H3K27me2 and H3K27me3 Domains in Embryonic Stem Cells. Mol Cell 70, 371–379 e5 (2018). [DOI] [PubMed] [Google Scholar]
- 35.Smallwood SA & Kelsey G De novo DNA methylation: a germ cell perspective. Trends Genet 28, 33–42 (2012). [DOI] [PubMed] [Google Scholar]
- 36.Pollard SM, Benchoua A & Lowell S Neural stem cells, neurons, and glia. Methods Enzymol 418, 151–69 (2006). [DOI] [PubMed] [Google Scholar]
- 37.Meissner A et al. Reduced representation bisulfite sequencing for comparative high-resolution DNA methylation analysis. Nucleic Acids Res 33, 5868–77 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tippmann SC et al. Chromatin measurements reveal contributions of synthesis and decay to steady-state mRNA levels. Mol Syst Biol 8, 593 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Challen GA et al. Dnmt3a is essential for hematopoietic stem cell differentiation. Nat Genet 44, 23–31 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jeong M et al. Loss of Dnmt3a Immortalizes Hematopoietic Stem Cells In Vivo. Cell Rep 23, 1–10 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Blackledge NP et al. CpG Islands Recruit a Histone H3 Lysine 36 Demethylase. Molecular Cell 38, 179–190 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wiehle L et al. Tet1 and Tet2 Protect DNA Methylation Canyons against Hypermethylation. Mol Cell Biol 36, 452–61 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gu T et al. DNMT3A and TET1 cooperate to regulate promoter epigenetic landscapes in mouse embryonic stem cells. Genome Biol 19, 88 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Boulard M, Edwards JR & Bestor TH FBXL10 protects Polycomb-bound genes from hypermethylation. Nat Genet 47, 479–85 (2015). [DOI] [PubMed] [Google Scholar]
- 45.Goll MG & Bestor TH Eukaryotic cytosine methyltransferases. Annu Rev Biochem 74, 481–514 (2005). [DOI] [PubMed] [Google Scholar]
- 46.Voigt P, Tee WW & Reinberg D A double take on bivalent promoters. Genes Dev 27, 1318–38 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Klose RJ, Cooper S, Farcas AM, Blackledge NP & Brockdorff N Chromatin sampling--an emerging perspective on targeting polycomb repressor proteins. PLoS Genet 9, e1003717 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pereira JD et al. Ezh2, the histone methyltransferase of PRC2, regulates the balance between self-renewal and differentiation in the cerebral cortex. Proc Natl Acad Sci U S A 107, 15957–62 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kurotaki N et al. Haploinsufficiency of NSD1 causes Sotos syndrome. Nat Genet 30, 365–6 (2002). [DOI] [PubMed] [Google Scholar]
- 50.Luscan A et al. Mutations in SETD2 cause a novel overgrowth condition. J Med Genet 51, 512–7 (2014). [DOI] [PubMed] [Google Scholar]
- 51.Tatton-Brown K et al. Germline mutations in the oncogene EZH2 cause Weaver syndrome and increased human height. Oncotarget 2, 1127–33 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gibson WT et al. Mutations in EZH2 cause Weaver syndrome. Am J Hum Genet 90, 110–8 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cohen AS et al. A novel mutation in EED associated with overgrowth. J Hum Genet 60, 339–42 (2015). [DOI] [PubMed] [Google Scholar]
- 54.Awad S et al. Mutation in PHC1 implicates chromatin remodeling in primary microcephaly pathogenesis. Hum Mol Genet 22, 2200–13 (2013). [DOI] [PubMed] [Google Scholar]
- 55.Tatton-Brown K et al. Mutations in Epigenetic Regulation Genes Are a Major Cause of Overgrowth with Intellectual Disability. Am J Hum Genet 100, 725–736 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wood AR et al. Defining the role of common variation in the genomic and biological architecture of adult human height. Nat Genet 46, 1173–86 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ernst J & Kellis M Chromatin-state discovery and genome annotation with ChromHMM. Nat Protoc 12, 2478–2492 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Methods only references:
- 58.Barski A et al. High-resolution profiling of histone methylations in the human genome. Cell 129, 823–37 (2007). [DOI] [PubMed] [Google Scholar]
- 59.Murray JE et al. Extreme growth failure is a common presentation of ligase IV deficiency. Hum Mutat 35, 76–85 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.de Bruin C et al. An XRCC4 splice mutation associated with severe short stature, gonadal failure, and early-onset metabolic syndrome. J Clin Endocrinol Metab 100, E789–98 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Guerois R, Nielsen JE & Serrano L Predicting changes in the stability of proteins and protein complexes: a study of more than 1000 mutations. J Mol Biol 320, 369–87 (2002). [DOI] [PubMed] [Google Scholar]
- 62.Triche TJ Jr., Weisenberger DJ, Van Den Berg D, Laird PW & Siegmund KD Low-level processing of Illumina Infinium DNA Methylation BeadArrays. Nucleic Acids Res 41, e90 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fortin JP, Triche TJ Jr. & Hansen KD Preprocessing, normalization and integration of the Illumina HumanMethylationEPIC array with minfi. Bioinformatics 33, 558–560 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Illingworth RS, Holzenspies JJ, Roske FV, Bickmore WA & Brickman JM Polycomb enables primitive endoderm lineage priming in embryonic stem cells. Elife 5(2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Orlando DA et al. Quantitative ChIP-Seq normalization reveals global modulation of the epigenome. Cell Rep 9, 1163–70 (2014). [DOI] [PubMed] [Google Scholar]
- 66.Tarasov A, Vilella AJ, Cuppen E, Nijman IJ & Prins P Sambamba: fast processing of NGS alignment formats. Bioinformatics 31, 2032–4 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rohde C, Zhang Y, Reinhardt R & Jeltsch A BISMA--fast and accurate bisulfite sequencing data analysis of individual clones from unique and repetitive sequences. BMC Bioinformatics 11, 230 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bock C et al. BiQ Analyzer: visualization and quality control for DNA methylation data from bisulfite sequencing. Bioinformatics 21, 4067–8 (2005). [DOI] [PubMed] [Google Scholar]
- 69.Krueger F & Andrews SR Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics 27, 1571–2 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ernst J & Kellis M ChromHMM: automating chromatin-state discovery and characterization. Nat Methods 9, 215–6 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Quinlan AR & Hall IM BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–2 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Liao Y, Smyth GK & Shi W featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–30 (2014). [DOI] [PubMed] [Google Scholar]
- 73.Robinson MD, McCarthy DJ & Smyth GK edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–40 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Robinson MD & Oshlack A A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol 11, R25 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The human next-generation sequencing data used in the manuscript are available on request from the relevant Data Access Committee from the European Genome–Phenome Archive (EGA). The exome data is available under the accession EGAS00001003231. Human RNA-seq, RRBS and ChIP-seq under the accession EGAS00001003232. The data are not publicly available to ensure protection of patient sequence data confidentiality through controlled access. Processed data files and mouse RNA-seq/RRBS are available in GEO under accession GSE120558.