Abstract
The human gut microbiome plays an essential role in maintaining human health including in degradation of dietary fibres and carbohydrates further used as nutrients by both the host and the gut bacteria. Previously, we identified a polysaccharide utilization loci (PUL) involved in sucrose and raffinose family oligosaccharide (RFO) metabolism from one of the most common Firmicutes present in individuals, Ruminococcus gnavus E1. One of the enzymes encoded by this PUL was annotated as a putative sucrose phosphate phosphorylase (RgSPP). In the present study, we have in-depth characterized the heterologously expressed RgSPP as sucrose 6F-phosphate phosphorylase (SPP), expanding our knowledge of the glycoside hydrolase GH13_18 subfamily. Specifically, the enzymatic characterization showed a selective activity on sucrose 6F-phosphate (S6FP) acting both in phosphorolysis releasing alpha-d-glucose-1-phosphate (G1P) and alpha-d-fructose-6-phosphate (F6P), and in reverse phosphorolysis from G1P and F6P to S6FP. Interestingly, such a SPP activity had never been observed in gut bacteria before. In addition, a phylogenetic and synteny analysis showed a clustering and a strictly conserved PUL organization specific to gut bacteria. However, a wide prevalence and abundance study with a human metagenomic library showed a correlation between SPP activity and the geographical origin of the individuals and, thus, most likely linked to diet. Rgspp gene overexpression has been observed in mice fed with a high-fat diet suggesting, as observed for humans, that intestine lipid and carbohydrate microbial metabolisms are intertwined. Finally, based on the genomic environment analysis, in vitro and in vivo studies, results provide new insights into the gut microbiota catabolism of sucrose, RFOs and S6FP.
Keywords: sucrose metabolism, sucrose phosphorylase, Ruminococcus gnavus E1, GH13_18, human gut microbiome
Data Summary
1. The Ruminococcus gnavus E1 sucrose 6F-phosphate phosphorylase (SPP) sequence is available from GenBank under accession number FQ790378 (see Fig. S5, available in the online version of this article).
2. All sequences, from the glycoside hydrolase GH13_18 subfamily used in the present study have been extracted from the CAZy database, URL: http://www.cazy.org.
3. Data used for the prevalence and abundance study were provided by the MetaHIT project (http://www.metahit.eu).
Impact Statement.
The human gut microbiome is considered an organ due to its key and specific functions in host metabolism, host protection and immune-system development. It is also able to modulate its own species composition to aid resiliency and to re-establish an intestinal stability. Developments in (meta)genome sequencing technologies and bioinformatic tools have now enabled scientists to study the microbiome's complex composition, its specific functions and the bacteria–host interactions. Recently, polysaccharide utilization loci (PUL) from Bacteroidetes have been investigated, highlighting their ability to use complex polysaccharide as a carbon source. By contrast, studies about gene clusters from Firmicutes are fewer. This paper will be of interest to those working in the fields of bacterial genomics and metabolism, human gut microbiology and CAZymes. For the first time, to the best of our knowledge, the ability of a strict anaerobe Firmicutes strain (i.e. Ruminococcus gnavus E1), found in more than 90 % of healthy humans, to metabolize the sucrose 6F-phosphate originating from plants via a glycoside hydrolase GH13_18 enzyme located in a PUL has been proposed, paving the way to interesting new pathways in sucrose, raffinose family oligosaccharides and sucrose 6F-phosphate catabolisms. More broadly, the Rgspp gene abundance and expression has been, respectively, studied in a human metagenomic library and in vivo with a high-fat diet, to figure out the link between lipid and carbohydrate metabolisms.
The human intestine is colonized by a complex, diverse and dynamic community of microorganisms, the so-called microbiome, which is in permanent interaction with the host [1]. It is now well established that the gut microbiome is of considerable interest for health. It is indeed involved in food degradation and assimilation, formation of bile salts, protection against pathogens, integrity of epithelial layer and immunity (for review see Sekirov et al., 2010 [1]), while a gut microbiome imbalance (i.e. dysbiosis) has been associated with several metabolic diseases, like obesity [2]. Among the many functions attributed to the intestinal microbiota, the metabolism of carbohydrates is of crucial importance.
Sucrose [α-d-glucopyranosyl-(1,2)-β-d-fructofuranoside], which is one of the most abundant soluble carbohydrates in plant tissues and in processed food, is usually degraded into glucose and fructose by a sucrase isomaltase, an intestinal membrane linked enzyme produced by the host, and leads by further processing to the generation of energy in the form of ATP (for a review see Gericke et al. [3]). Sucrose is also a structural component of sucrosyl-oligosaccharides, including fructans (FOS, e.g. 1-kestose, nystose and fructo-furanosyl nystose) and the raffinose family oligosaccharides (RFOs, e.g. raffinose, stachyose and verbascose), which are both important classes of carbohydrates in the plant kingdom [4]. Conversely to sucrose, FOS and RFOs are resistant to the action of human enzymes in the digestive tract, but they can be cleaved by microbial gut enzymes inducing a beneficial effect for the host's health [5]. For example, bacterial α-galactosidases hydrolyse RFOs to release d-galactose and sucrose, which can also be metabolized by the resident microbiota. This microbial degradation relies on intracellular sucrose phosphorylases (SPs; EC 2.4.1.7).
Based on the CAZy classification (www.cazy.org [6]), the glycoside hydrolase 13 family members act on substrates containing α-glucoside linkages and consist of about 30 different enzyme specificities including glycoside hydrolases, transferases, isomerases and phosphorylases, subdivided in 42 subfamilies. All members share a conserved structural scaffold with seven highly conserved regions despite their low overall sequence identity [7, 8]. SPs belong to the glycoside hydrolase GH family 13 subfamily 18 (GH13_18) [9]. The GH13_18 members are retaining enzymes, which reversibly catalyse the reaction between sucrose and inorganic phosphate to synthesize alpha-d-glucose-1-phosphate (G1P) and d-fructose. The products are then slotted into microbial glycolytic pathways such as glycolysis. Due to their ability to perform in vitro reverse phosphorolysis from G1P and a wide range of acceptor molecules [10], SPs are used for biotechnological purposes, to produce, for example, a moisturizing agent (Glycoin [11]), glucosylated flavonoids (including resveratrol and quercetin [12, 13]) and rare disaccharides (including kojibiose and nigerose [14, 15]).
To date, only a few GH13_18 members have been biochemically characterized [16–27]. In 2004, the first 3D structure of the SP from Bifidobacterium adolescentis was solved [20]. In the last few years, new specificities have been described for members of the GH13_18 family. In 2014, Verhaeghe et al. revealed for the first time a GH13_18 from Thermoanaerobacterium thermosaccharolyticum allowing phosphorolysis of the 6F phosphorylated sucrose (sucrose-6F-phosphate; S6FP), releasing G1P and alpha-d-fructose-6-phosphate (F6P) [25]. This enzyme was, thus, considered as a sucrose 6F-phosphate phosphorylase (SPP), which was the first and the only enzyme with this specificity reported so far. Furthermore, three members of the GH13_18 family were recently described as strict glucosylglycerate phosphorylases (GGaPs) [26]. The GGaPs from Meiothermus silvanus, Spirochaeta thermophila and Escherichia coli (i.e. MsGGaPs, StGGaPs and EcGGaPs, respectively) catalyse the reversible phosphorolysis of glucosylglycerate into G1P and d-glycerate. Very recently, an enzyme from Marinobacter adhaerens HP15 able to catalyse the reversible phosphorolysis of 2-O-α-d-glucosylglycerol has been characterized and named as glucosylglycerol phosphorylase (MaGGoP [27]).
Ruminococcus gnavus E1 is a Gram-positive anaerobic bacterium belonging to the phylum Firmicutes. As one of the 57 most common species, it is present in around 90 % of individuals [28]. First isolated from the faeces of a healthy human [29], Ruminococcus gnavus E1 fosters a high interest due to its abilities: (i) to produce antimicrobial peptides active against Clostridium perfringens [30–32], (ii) to produce a large panel of glycoside hydrolases acting on dietary [33–35] and (iii) host mucin-glycans [36]. Two loci putatively involved in sucrose metabolism have previously been identified (i.e. Rgaga1 and Rgaga2). Specifically, the Rgaga2 locus, including genes putatively encoding a regulator, a GH13_31, two phosphotransferase system (PTS) sequences and a GH32, has been proposed to be involved in extracellular and intracellular sucrose assimilation [35]. Similarly, Rgaga1 consists of six genes predicted to encode a transcriptional regulator (agaR), an ABC transporter (agaE, H and G), a SP (sucP) and a bifunctional α-galactosidase/sucrose kinase (RgAgaSK). RgAgaSK is the only biochemically characterized bi-modular enzyme encoded by this locus so far. It produces sucrose 6-phosphate (S6P) or releases corresponding monosaccharides from sucrose or from RFOs based on sucrose kinase and α-galactosidase activities [33].
Here, we used an integrative approach to better understand the function and the metabolic role of the SucP enzyme (hereafter renamed as RgSPP). Genomic environment analysis, in-depth biochemical characterization, analysis of gene prevalence in the human gut microbiome and in vivo studies highlighted the role of this enzyme in carbohydrate catabolism by the prominent gut bacterium Ruminococcus gnavus E1.
Methods
Materials
Oligonucleotides, Zeocin, Geneticin, ampicillin, kanamycin and all restriction DNA modifying enzymes (except DNA polymerase) were purchased from Invitrogen. Culture media were from BD Difco. PrimeSTAR HS DNA polymerase for PCRs was from Takara. E. coli DH5α (supE44, hsdR17, recA1, endA1, gyrA96, thi-1, relA1) was used for the DNA procedures (Invitrogen). Sodium phosphate and potassium phosphate were purchased from Sigma–Aldrich, sodium acetate was from Calbiochem and sodium hydroxide was from Fischer Scientific. Ultracel system and Ultracel PES membrane were from Millipore. The protein assay reagent was from Bio-Rad. Standard diet (SD) and high-fat (HF) diet were provided by SAFE Laboratories.
Cloning, expression and purification of RgSPP
The Rgspp gene (accession no. FQ790378) was cloned from the genomic DNA of Ruminococcus gnavus E1 and amplified as described by Bruel et al. in 2011 with PCR reaction primers (Table S1) [33]. Recombinant Rgspp, with a C-terminal (His)6-tag, was synthesized from E. coli BL21(DE3) grown in LB broth containing 50 mg ampicillin l−1 as an overnight culture at 20 °C. Bacterial lysis was carried out with Cell Disruptor (Constant System) in the binding buffer for affinity chromatography (20 mM MOPS buffer pH 7.4, 40 mM imidazole) at 1.37 kbar. After centrifugation at 1500 g, 30 min at 4 °C, the soluble fraction was loaded onto a Ni-NTA column and the recombinant protein was eluted with 125 mM imidazole in MOPS buffer pH 7.4. Fractions containing proteins were pooled, dialysed against HEPES buffer (50 mM, pH 7.0) and concentrated to 5 mg ml−1. The concentrated proteins were injected onto a Sephacryl S200 (26/60) size exclusion column (SEC) with flow rate of 1 ml min−1 and purified to near homogeneity (>90 %). The purity of the protein was checked by SDS-PAGE (12 %) [37], with an overall yield of 50 mg (l of culture)−1. The protein concentration was determined using the Bio-Rad protein assay kit with BSA as the standard.
Enzyme assays
Phosphorolytic activity and kinetic parameters
Phosphorylase activity was measured using two different methods. In the first one, G1P concentration was indirectly quantified with 0.90 µM enzyme, by following, at 340 nm, the coupled production of NADPH from NADP+ and G1P by phosphoglucomutase and glucose-6-phosphate dehydrogenase [17]. In the second method, substrate and product concentrations were determined by High-Performance Anion-Exchange Chromatography coupled with Pulsed-Amperometric Detection (HPAEC-PAD). The phosphorylase activity assays were performed over 18 h using 0.18 µM enzyme. The protocol and gradient used are the same as described by Lafond et al. in 2011, except that buffer A was composed of 5 mM NaOAc and 80 mM NaOH [38]. Prior to being analysed, samples were inactivated by heating for 5 min at 70 °C. In both cases, reactions were realized with 20 mM sucrose or S6FP in 100 mM phosphate buffer pH 6.0 at 40 °C. The samples were injected on a Dionex system equipped with a GP40 gradient pump, an ED40 pulsed amperometric detector, an AS3500 auto-sampler (Thermo-Electron) and a CarboPac PA-100 analytical column at 25 °C (250×4 mm).
Specific activity
The specificity toward different substrates was analysed using 10 mM G1P as donor and 10 mM acceptor, i.e. d-fructose, d-glucose, d-galactose, d-xylose, d-leucrose, d-isomaltulose, d-fructose 6-phosphate, d-glucose 6-phosphate, d-galactose 6-phosphate, d-glucose 1,6-diphosphate, d-fructose 6-diphosphate, and 100 mM maltotetraose and maltoheptaose. All reactions were monitored in 50 mM MOPS buffer pH 6.0 at 40 °C, for 18 h, and with 0.18 or 0.64 µM enzyme.
Kinetic parameters
Kinetic parameters were determined using the same conditions described previously with production of NADPH and S6FP as the substrate in 100 mM sodium phosphate buffer, pH 6.0, at 40 °C and by incubating 0.1 µM enzyme with seven different concentrations of S6FP (1.0 to 20 mM) in a reaction volume of 500 µl. Km and kcat values were calculated using the nonlinear regression Michaelis–Menten equation.
Effect of temperature and pH on enzyme activity
Optimal pH for RgSPP was determined using the coupled method with S6FP in 50 mM sodium phosphate buffer in a pH range of 5.0 to 8.0 at 45 °C and with 0.92 µM enzyme. Optimal temperatures were also determined using the same enzymatic assay in 50 mM sodium phosphate buffer pH 6.0 and temperatures ranging from 20 to 80 °C.
Caecal content (CC) collection and extraction of RNA
The in vivo assays included an experimental model of infection (C57BL6 gnotobiotic mice) subcontracted to Germ Free Animals Facility ANAXEM platform INRA, UMR 1319 Micalis, France. Animal experiments were performed according to the guidelines of the French Ethics Committee, i.e. agreement number A78-322-6 for mice maintained for 10 days on a control diet and agreement number A78-718 for mice maintained for 2 months on a HF diet (Table S2). C57BL6 mouse strains are established models for diet-induced obesity [39]. Mice were reared in Trexler type isolators (LaCalhène), fed ad libitum with commercial diets sterilized by gamma ray and supplied with sterile autoclaved drinking water. Male mice (8–9 weeks old) sourced by ANEXEM and maintained on SD were inoculated with Ruminococcus gnavus E1 (0.5 ml late log-phase culture at 108 cells ml−1), by intra-gastric route, on 3 consecutive days in order to generate Ruminococcus gnavus E1 monoxenic mice. Then mice were randomly divided into two separate groups. The first group of Ruminococcus gnavus E1 monoxenic mice (n=10) was assigned for 10 days on a SD to obtain an effective colonization of the digestive tract by Ruminococcus gnavus E1, according to Graziani et al. [40]. The second group (n=10) was assigned for 2 months on a HF diet containing 34.9/100 g fat, 26/100 g carbohydrate and 26/100 g protein provided by SAFE Laboratories (Table S2). The C57BL6 mouse strain has proved particularly useful as the mice readily gain weight when fed HF diets, and are more susceptible to obesity and glucose intolerance when fed HF diets for at least 2 months [41–43]. After 10 days (i.e. for the SD) or 2 months (i.e. for the HF diet), individual faecal samples were collected and bacterial counts estimated. Colonization by Ruminococcus gnavus E1 was controlled and analysed by plating serial dilutions of suspensions obtained from ground colon in an anaerobic chamber using brain-heart infusion media, whereas the analyses of bacterial c.f.u. were performed with the CC samples. Body weight was also monitored every 2 weeks and food consumption was monitored weekly. Then, the animals were sacrificed, and faeces and the CCs were collected in 1 ml Tri-Reagent (Molecular Research Center) and frozen at −80 °C until RNA extraction was carried out.
DNA isolation and PCR analysis of Ruminococcus gnavus E1
For each analysis, chromosomal DNA was isolated from 100 to 120 mg mouse faeces, using the Fast DNA SPIN kit for faeces (MP). Amplification of 16S rDNA was performed using oligonucleotide primers 16F8 and 16R1541, which corresponded to bacterial 16S rRNA gene conserved sequences (from positions 8 to 1541 on the E. coli 16S rRNA). The PCR conditions used were: DNA (50 ng); annealing at 60 °C (20 s), polymerization at 72 °C (30 s) and denaturation at 94 °C (20 s). Amplification reactions (30 cycles) were carried out in a Mastercycler Nexus GX2 (Eppendorf). Resulting DNA fragments were then sequenced.
RNA isolation and quantitative Reverse Transcription-PCR analysis of the faeces and the CCs
Two hundred milligrams fresh material (faeces/CCs) were used for total RNA extraction according to the protocol described by Doré et al. and cleaned up with the RNeasy mini kit (Qiagen) [44]. RNA was spectrophotometrically quantified (260 nm) and purity assessed by the A260/A280 ratio using a NanoDrop 2000c UV-Vis spectrophotometer (Thermo Fischer Scientific). Reverse transcription of 6.25 ng RNA was performed with qScript cDNA (Quantabio) and 50 ng random primers in a 20 µl volume at 42 °C for 1 h with reverse transcriptase. All procedures were according to the manufacturer’s instructions. PCR was performed in the presence of SYBR green reagent with a Light Cycler 480 (Roche Technology) and carried out with 25 ng cDNA in 20 µl containing 10 µl PCR master mix reagent and 0.5 µM each primer. The sequences of the primers corresponding to the genes in mice were designed using the Universal Probe Library assay design centre (Roche) and are reported in Table S1. Thermal cycling conditions were as follows: 5 min denaturation at 94 °C; followed by 45 cycles of 10 s at 94 °C, 10 s at 60 °C and 10 s at 72 °C. Data were collected using the Light Cycler 480 software (Platform AVB, iSm2, Marseille, France).
Cycle thresholds were normalized to rpoB or gyrB levels and fold changes were calculated against the normalized control of induction and untreated values when applicable. The relative quantification of glycosyltransferase and gene expression was performed using the comparative ∆∆Ct method [45]. Each sample was treated in triplicate to ensure statistical significance of the analysis. The P value was determined by t-test. The significance P values shown are at least <0.005. As a control, additional reactions were performed using, as a template, a Reverse Transcription mixture without enzyme, a Reverse Transcription mixture without RNA and chromosomal DNA.
In silico analysis
The gene model from locus RUGNEv3_61221 of Ruminococcus gnavus E1 (GenBank accession no. FQ790378.1) was downloaded from the Microbial Genome Annotation and Analysis Platform (MaGE, accessible via https://www.genoscope.cns.fr/) and translated to the corresponding protein sequence (GenBank accession no. CCA61958.1), herein referred to as RgSPP. Eighteen hundred other amino acid sequences of bacterial GH13_18 members were extracted from the public version of the CAZy database [6]. The RgSPP sequence was submitted to the Phyre2 server [46] in order to generate a 3D structural model and visualized using PyMOL software (PyMOL Molecular Graphics System, version 2.0; Schrödinger). The SignalP 4.1 server was used to determine the presence and location of protein signal peptide cleavage sites in the RgSPP amino acid sequence [47]. Multiple alignment of characterized GH13_18 members was performed using mafft einsi advanced mode [48]. The secondary structure of AAO33821 from Bifidobacterium adolescentis based on its solved crystal structure (PDBID: 1R7A) was analysed using ESPript (http://espript.ibcp.fr) [49]. Where present, signal sequences and additional modules were removed to isolate the catalytic modules for bioinformatics analysis. A multiple sequence alignment was produced using the Muscle program [50], and the evolutionary history was inferred using the neighbour-joining method [51]. The evolutionary distances were computed using the Poisson correction method [52] and are in the following units: the number of amino acid substitutions per site. The analysis involved 1065 amino acid sequences. All ambiguous positions were removed for each sequence pair. There was a total of 987 positions in the final dataset. Evolutionary analyses were conducted in mega6 software [53].
Cluster analysis was based on the neighbour-joining method with the closely related bacterium Ruminococcus gnavus E1 as the out-group root. Synteny blocks were analysed by the MaGE platform, which allowed the comparison of coding sequencespredicted from the genomic DNA of Ruminococcus gnavus E1 to those predicted from genomic DNA present in the PkGDB (Prokaryotic Genome DataBase) and the National Center for Biotechnology Information RefSeq database (a collection of raw sequences from whole-genome sequencing). Beyond a simple sequence comparison, this interface allows the analysis of the synteny between two chromosomes.
Gene prevalence and abundance analysis
Sequences of the GH13_18 referenced in the CAZy database in September 2018 were searched by blastp analysis (E value=0, identity ≥90 %) in the translated catalogue of 9.9 million reference genes constructed using gut metagenome sequences of 1267 subjects from 3 continents (USA, China, Europe): 139 USA HMP samples; 760 European faecal samples from the MetaHIT project; 368 Chinese faecal samples [54]. The microbial gene richness of GH13_18 in human gut was assessed by recovering the prevalence and occurrence frequency data [54] of homologous sequence of the catalogue assigned to GH13_18 from the 9.9 gene frequency matrix in the 1267 subjects (http://meta.genomics.cn/meta/dataTools).
Results and Discussion
Cloning, heterologous expression and functional characterization of RgSPP
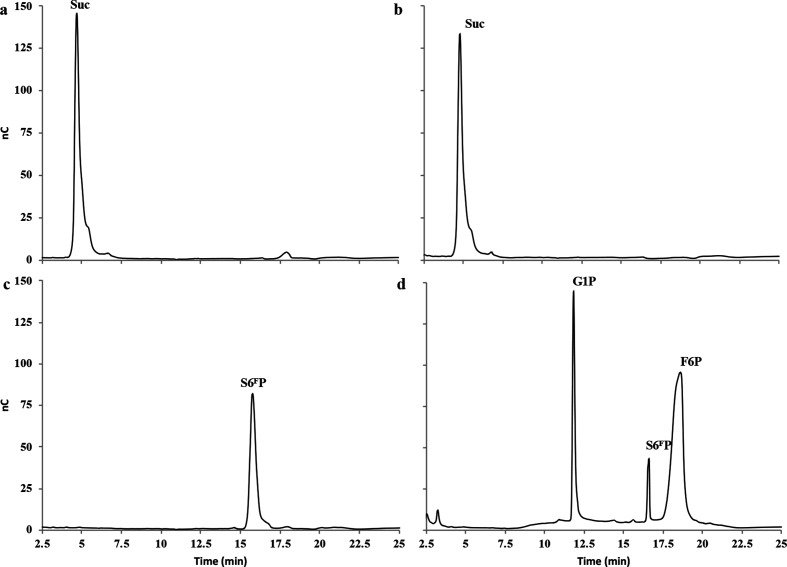
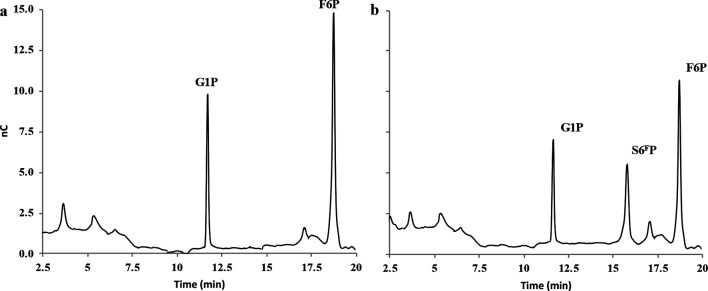
In order to perform biochemical and molecular studies, the RgSPP-encoding gene was cloned into the pOPINE vector, and heterologously expressed in E. coli BL21. After affinity purification using immobilized metal affinity chromatography (Ni-NTA, Fig. S1a) and an additional SEC (S200; Fig. S1b), the recombinant protein tagged with a C-terminal (His)6-tag showed on SDS-PAGE an apparent molecular mass of 55 kDa (Fig. S2), which is in agreement with its theoretical molecular mass (56 165 Da). SEC analysis indicated that RgSPP is a monomer in solution (Fig. S1b). The purified RgSPP was first assayed against sucrose and S6FP in the presence of inorganic phosphate, in order to determine its phosphorolysis activity. G1P and F6P were released only from S6FP, indicating that the enzyme is a SPP and not a SP (Figs 1 and S3). In order to accurately determine the substrate specificity of RgSPP, different carbohydrates and phosphorylated carbohydrates were then considered as acceptors for the reverse phosphorolysis reaction in the presence of G1P as glycosyl donor (Table 1). The results showed that the RgSPP only used the F6P as acceptor. Considering this S6FP synthetic reaction (Fig. 2), RgSPP displayed a specific activity of 0.22 U mg−1. All in all, these data obtained by HPAEC-PAD, demonstrate that RgSPP is a highly specific enzyme, able to reversely convert S6FP+Pi into G1P+F6P (Figs 1 and 2). Previously, this activity was reported only once for a member of the GH13_18 family, from Thermoanaerobacterium thermosaccharolyticum (TtSPP), which is a thermophilic obligate anaerobe [25]. However, SPP activity has never been described within the human gut microbiota so far.
Fig. 1.
HPAEC-PAD profiles of the RgSPP reaction products obtained after 24 h in the presence of 20 mM sucrose (b) and S6FP (d) and Pi (phosphorolysis). Controls without enzyme are displayed for sucrose (a) and S6FP (c).
Table 1. RgSPP activity on various substrates.
| Reaction | Substrate | Relative activity (%) |
|---|---|---|
| Phosphorolysis* | Sucrose 6F-phosphate | 100 |
| Sucrose | nd | |
| Synthesis† | d-Fructose-6-phosphate | 100 |
| d-Fructose | nd | |
| d-Glucose | nd | |
| d-Galactose | nd | |
| d-Mannose | nd | |
| d-N-Acetylgalactosamine | nd | |
| d-Xylose | nd | |
| Maltotetraose | nd | |
| Maltoheptaose | nd |
nd, Not detected.
*At 40 °C with 100 mM phosphate buffer at pH 6.5 and 20 mM donor.
†At 40 °C with 50 mM MOPS buffer at pH 6.5, 10 mM donor (α-d-glucose 1-phosphate) and 10 mM acceptor.
Fig. 2.
HPAEC-PAD profiles of the RgSPP reaction products obtained after 24 h in the presence of 100 µM G1P and F6P (reverse phosphorolysis). (a) Negative control and (b) assay with 0.18 µM RgSPP.
The biochemical properties of RgSPP were then investigated for S6FP phosphorolysis. The optimum pH was 6.0 (Fig. S4a). This value corresponds to that obtained with the other characterized members of the GH13_18 family (between 6.0 and 6.5), which are, as described further, all bacterial phosphorylases. This is in agreement with the physiological role of these enzymes, which is to perform intracellular phosphorolysis of oligosaccharides. The optimal temperature was 45 °C, a value surprisingly higher than that met in the human gut (Fig. S4b). Such thermoactive enzymes have already been found in the human gut microbiome, and could be due to the ability of gut microbes to colonize various habitats [55]. In addition, fridge storage cannot be considered because the purified protein was not stable at 4 °C after 3 days, whereas RgSPP was stable for at least 4 weeks at −20 °C (data not shown). RgSPP phosphorolysis Km and kcat values are summarized in Table 2. This enzyme is 112 times more efficient for S6FP phosphorolysis than TtSPP [25]. Moreover, it is noteworthy that RgSPP is strictly specific for S6FP, in contrast to TtSPP, which also displays high activity on sucrose.
Table 2. RgSPP kinetic parameters determined on sucrose 6F-phosphate.
| Reaction | Enzyme |
Km
(mM) |
kcat
(s−1) |
kcat/Km
(mM−1 s−1) |
Reference |
|---|---|---|---|---|---|
| Phosphorolysis | RgSPP | 1.70 | 740 | 435.3 | Present work |
| TtSPP | 12.7 | 82.6 | 6.5 | [23] |
Sequence analysis
Based on sequence alignment, RgSPP belongs to glycoside hydrolase family GH13 subfamily 18 according to the CAZy classification (http://www.cazy.org/ [6]). SignalP failed to predict a signal peptide suggesting a cytoplasmic subcellular localization [47].
Multiple sequence alignment for all the characterized members of the GH13_18 family with RgSPP was performed (Fig. S5). The deduced amino acid sequence (492 aa) was compared to other characterized SP, GGaP and GGoP sequences, and revealed about 32 to 35 % identities to the characterized SPs from Leuconostoc mesenteroides spp. (BAA14344.1 [17], ABS59292.1 [21], AAX33736.1 [23]), Streptococcus mutans (CAA30846.1 [16]), Bifidobacterium adolescentis (AAO33821.1 [20]), Bifidobacterium longum spp. (AAO84039.1 [19], BAF62433.1 [22]), Lactobacillus reuteri (AGK37834.1 [24]), Pelomonas saccharophila (AAD40317.1 [18]); 34 % to TtSPP from Thermoanaerobacterium thermosaccharolyticum (ADL69407.1 [25]); 27 and 28 % to MsGGaP and MaGGoP (ADH62582.1 and ADP98617.1 [26, 27]), respectively (Fig. S5). Previously Verhaeghe et al., in 2014, proposed that the residue H344 in TtSPP is present in all SPP [25]. The mutation of this residue H344 to Tyr led to a decreased ratio of activity on phosphorylated fructose over fructose, suggesting a crucial role of this residue in phosphate binding. However, in RgSPP, the equivalent residue for H344 in TtSPP is a tyrosine (Y355), as observed for all the others characterized members of the subfamily, including the GGaPs and GGoPs, suggesting that the S6FP specificity might not be restricted to this residue. This observation is corroborated by the observation of Y344 – the homologous residue in SP from Bifidobacterium adolescentis (AAO33821, PDBID 1R7A) – which has a side chain pointing out of the overall structure is unlikely involved in enzyme activity [20]. Nevertheless, we can speculate that the hydroxyl function could form an H bond or even a phosphoester bridge with the phosphate carried by the C6 of the fructose residue to stabilize the substrate into the catalytic pocket.
Phylogenetic, synteny analysis and metabolic pathways
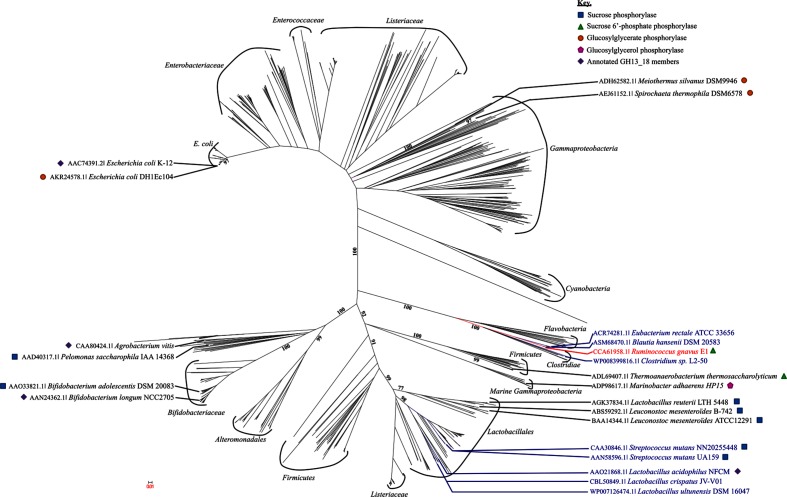
In order to gain more insight in the phylogenetic diversity of the GH13_18 subfamily, a phylogenetic tree was reconstructed, using sequences classified in the CAZy database (Fig. 3) [6]. GH13_18 enzymes are distributed in diverse phylogenetic groups, such as lactic acid bacteria or cyanobacteria from different ecosystems like soil, marine and human gastrointestinal microbiota. The present phylogenetic tree reveals different clades; seven containing characterized SP, SPP, GGaP or GGoP enzymes (e.g. Bifidobacteriaceae, Lactobacilliales and Thermoanaerobacteria [10, 25–27, 56]). Franceus et al., in 2017, proposed a phylogenetic tree of the GH13_18 subfamily with two major clusters, one with all SP and SPP enzymes and the other one with enzymes exhibiting GGaP activity, but this was without taking into account the identification of the new MaGGoP that is interlocked between the SP and SPP enzymes [26, 27]. Consequently, GH13_18 activity cannot be predicted from its clade status. Here, we observed that RgSPP belongs to the Clostridiae clade constituted by only gut bacteria (e.g. Blautia, Clostridium, Eubacterium) (Fig. 3) despite the fact that RgSPP and TtSPP, which shared the same substrate specificity, clustered in two different but close clades. It is noteworthy that most of the GH13_18 were found in the lactic acid bacteria group, known for their diverse benefits for humans (e.g. probiotic, oral health, etc.), underlying the importance of such activities.
Fig. 3.
Phylogenic tree of subfamily GH13_18. The tree includes all the GH13_18 listed in the CAZy database. Members sharing syntenies with Rgspp are indicated in dark blue. Enzyme specificity is indicated in the key.
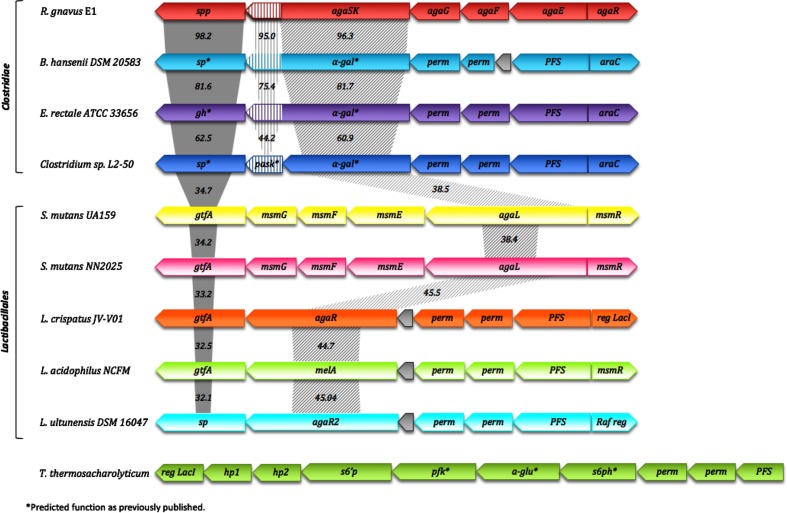
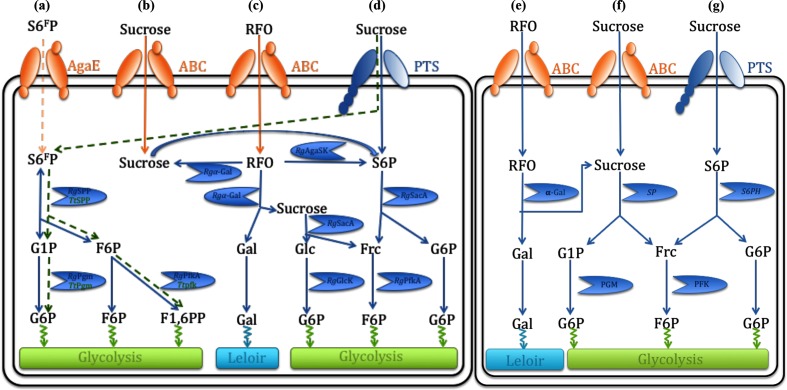
As previously mentioned, the Rgspp gene is located in the Rgaga1 locus previously described by Bruel et al., in 2011 [33], of which the organization is similar to (i) the Rgaga2 locus suggested to play essential roles in the metabolism of dietary sucrose and oligosaccharides [35], and (ii) polysaccharide utilization loci (PUL) as defined by Terrapon et al., in 2015 [57], but with an ABC transporter rather than a TonB-dependent transporter/SusD family lipoprotein-encoding gene pair specific from the Bacteroidetes [58]. In order to figure out the accurate role of such Gram-positive PUL in the human gut microbiome, we performed a synteny study based on all the prokaryotic strains with characterized GH13_18 enzymes to date (Fig. 4), coupled with a deep mining of the Ruminococcus gnavus E1 genome, that led us to propose a new model of sucrose, S6FP and RFO metabolic pathways (Fig. 5a–g). The clustering alignment showed for the four first sequences found in the Clostridiae clade of the phylogenetic tree, a quite high conservation of this classical Gram-positive PUL organization [58], where spp genes are always in a position downstream of those encoding bi-functional enzymes presenting kinase and α-galactosidase activities. Moreover, the different ABC-transporter- and PTS-encoding genes (i.e. agaG, agaF and agaE for the Ruminococcus gnavus E1 cluster, and perm and PFS for Blautia hansenii, Eubacterium rectale and Clostidium spL2-50) and the response regulator genes (i.e. araC), located in a position upstream of the kinase-α-gal gene are also conserved. The comparison between these clusters showed that the spp, kinase and α-gal gene sequences are highly conserved, with at least 62.5, 44.2 and 60.9 % identities, respectively, whereas identities were less important with enzymes originally from the Lactobacillale clade (Fig. 4). Although, synteny is observed with clusters from different genomes belonging to the Lactobacillale clade where the α-gal gene is still conserved, the kinase gene is not. Interestingly, the absence of kinase genes in the Lactobacillale clusters appears to be correlated with the presence of a sp gene as described for the five GH13_18 characterized enzymes from Streptococcus mutans, Lactobacillus crispatus, Lactobacillus acidophilus and Lactobacillus ultunensis [59–62] suggestive of different sucrose and RFO metabolic pathways (Fig. 5). Indeed, the absence of a kinase gene led to (i) an inability to phosphorylate sucrose or RFOs to S6P contrary to the Ruminococcus gnavus E1 clade via SK-α-Gal enzymes (Figs. 4 and 5b–d) and (ii) confirmed the involvement of the PTS for the phosphorylation of sucrose in S6P, and the subsequent hydrolysis in Frc and G6P by a sucrose 6-phosphate hydrolase (see RgSacA accession no. CCG93499.1; E.C. 3.2.1.26 and S6PH in Fig. 5d, g, respectively). Fructose is phosphorylated afterwards by a fructokinase to F6P (RgFruk, accession no. EDN78042.1; E.C. 2.7.1.11; Fig. 5d, g). Moreover, the conservation of the α-gal gene shows the ability to remove the galactosyl units from the RFOs, which then will be managed by the metabolic pathway of Leloir for the Ruminococcus gnavus E1 and Lactobacillale bacteria (Fig. 5c, e). Finally, considering the two clusters carrying the only two SPP characterized to date, i.e. RgSPP and TtSPP, we observed that surprisingly there is no synteny between the two organisms (Fig. 4). Nevertheless, it appeared that there is a real relationship between the SPP activity and the presence of a kinase catalytic domain in the same cluster (i.e. AgaSK in the Rgaga1 cluster and PFK in the Ttspp cluster). Strengthening the previous hypothesis from Verhaeghe et al., in 2014 [25], where sucrose is phosphorylated in S6FP during translocation by a PTS; here, we propose a translocation of the S6FP via the AgaE PTS component. Then, S6FP could be metabolized by RgSPP to produce on one hand G1P, which is subsequently transformed by a phosphoglucomutase (RgPgm accession no. WP_101882627.1; E.C. 5.4.2.2) into G6P, on the other hand F6P could end up as intermediate in the glycolytic pathway or could be phosphorylated first by RgPfka yielding Fructose 1,6-di-phosphate (F1,6PP) (Fig. 5a).
Fig. 4.
Genomic environment of the Rgspp gene. The values indicate the percentage identities between spp or sp, kinase and α-gal genes.
Fig. 5.
Hypothetical sucrose, sucrose 6-phosphate and sucrose 6F-phosphate metabolic pathways in the Ruminococcus gnavus E1, T. thermolyticum and Lactobacillales bacteria (adapted from Bruel et al. and Verhaeghe et al. [24, 32]). Pathways for uptake and catabolism in the Ruminococcus gnavus E1 clade of (a) S6FP, (b) and (d) sucrose, and (c) RFO, and in the lactobacillales bacteria of (e) RFO, and (f) and (g) sucrose. The T. thermolyticum S6F P (a) metabolic pathway proposed by Verhaeghe et al., in 2014, appears in green, whereas the sucrose pathways follow the (f) and (g) models. RgSPP, sucrose 6F-phosphorylase; RgAgaSK, α-galactosidase/sucrose kinase; Rgα-Gal, α-galactosidase; RgPfkA, 6-phosphofructokinase; RgPgm, phosphoglucomutase; RgSacA, sucrose 6P hydrolase; RgGlcK, glucose kinase; SP, sucrose phosphorylase; S6PH, sucrose 6-phospho-hydrolase; AgaE and ABC, ABC transporters; PTS, phosphoenolpyruvate-dependent sugar phosphotransferase system; S6FP, sucrose 6F-phosphate; S6P, sucrose 6-phosphate; RFO, raffinose family oligosaccharide.
Finally, no synteny has been identified between the Rgaga1 cluster and the GGaP and GGoP genomic environments recently characterized by Franceus et al., in 2017 and 2018, respectively [26, 27], suggesting that this PUL organization is linked to the sucrose, S6FP and RFO metabolic pathways [33, 35].
Prevalence in the human gut microbiota
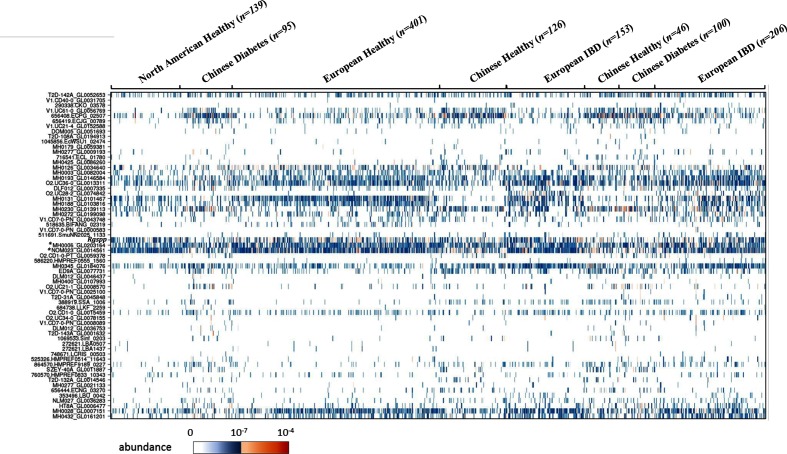
Among the GH13_18 sequences referenced in the CAZy database, 954 shared at least 90 % sequence identity with 63 sequences of the human gut metagenomic gene catalogue established from 1267 subjects [54]. Their abundance and prevalence in the various cohorts are presented in Fig. 6 and values are summarized in Table S3.
Fig. 6.
Prevalence and abundance of the human gut GH13_18 gene homologues found in the human gut reference catalogue (1267 individuals from diverse geographical origins and with different health status). Connections between GH13_18 sequences and sequences from the catalogue are indicated in Table S3. The target Rgspp shares, respectively, 97.8 and 98.4 % identities with the sequences MH0006_GL0203164 (*) and NOM023_GL0014561 (*) of the catalogue.
By investigating the prevalence of the GH13_18 homologues in individuals, we observed that there are differences due to geographical origin rather than an impact of the health status of the individuals. Indeed, the genes from E. coli strains corresponding to the sequences V1.UC61-0_GL0056769, 656408.ECPG_02507 and 656419.ECJG_00789 of the catalogue were more abundant in the Chinese subjects, together with the gene ADQ02266.1 (100 % identity with the sequence MH0230_GL0139113 of the catalogue) from the Bifidobacterium longum subsp. longum BBMN68 strain [63]. This strain, described as a health-promoting strain, was previously isolated from the faeces of a healthy Chinese centenarian. By contrast, other genes were less abundant in Chinese subjects, such as sequence AAO33821.1 from Bifidobacterium adolescentis DSM 20083 (99.8 % identity with MH0131_GL0101467), sequence CBL13462.1 from Roseburia intestinalis (97.6 % identity with 72-stool_revised_C812172_1_gene66964) and sequence ADC85157.1 from Bifidobacterium animalis subsp. lactis BB-12 (100 % identity with O2.UC28-2_GL0074842), independently of the clinical status. Besides, sequence AFJ26298.1 from Streptococcus parasanguinis (98.6 % identity with MH0003_GL0082004), sequence ACD98776.1 from Bifidobacterium longum DJO10A (99.6 % identity with MH0432_GL0161201) and to a lesser extent sequence AOL09592.1 from Bifidobacterium longum 35 664 (99 % identity with MH0193_GL0146584) were poorly represented in Chinese and USA subjects compared to European ones. It is noteworthy that only two sequences appear as biomarkers of the European subjects whose clinical status was intestinal bowel disease (IBD) [28]. One is the homologue sequence of ADC85157.1 from Bifidobacterium animalis subsp. lactis BB-12 (100 % identity with O2.UC28-2_GL0074842), which is in agreement with the study of Hao et al., published in 2011, where the administration of Bifidobacterium animalis subsp. lactis BB-12 in combination with other probiotics to 32 patients with ulcerative colitis tended to increase remission [63, 64]. The other one, which is highly prevalent (found in 45 % of individuals), is the sequence CED94009.1 from Romboutsia ilealis CRIB (98.8 % identity with MH0345_GL0184076). These results provide new insights about the presence of homologous sequences from Romboutsia ilealis related to IBD. Indeed, Gerritsen et al. were not able to draw conclusions about the correlations between prevalence and/or abundance of Romboutsia ilealis and specific human diseases likely due to the limited number of available human datasets [65].
The target RgSPP sequence and its homologues from Eubacterium rectale (ACR74281.1) present about 98 % identity with the sequences MH0006_GL0203164 and NOM023_GL0014561 of the catalogue. Both sequences are the most prevalent genes of the family and were found in 67.9 and 72.1 % of the individuals, respectively. They are distributed in the various cohorts, to a lesser extent within the Chinese cohort, whatever their clinical status. Similarly, the homologous sequences from Blautia (ASM68470.1, 97.8 % identity with MH0028_GL0007151) and Clostridium (WP_008399816, 98.4 % identity with MH0126_GL0034640) present a relatively high prevalence and are found in 50.9 and 29.7 % of individuals, respectively. Moreover, the sequence AAO33821.1 from Bifidobacterium adolescentis DSM 20083 (99.8 % identity with MH0131_GL0101467), encoding a characterized SP [20], presents a prevalence of about 45.5 % with a specific geographical distribution among European and USA cohorts independently of the clinical status. The other homologous sequences known to encode characterized SPs, whose homologues were also associated within the catalogue sequences, were found in less than 5 % of the individuals. Interestingly, the sequence AKR24578.1 from E. coli DH1Ec104 (99.4 % identity with 656408.ECPG_02507), which was characterized as a GGaP [26], is present in about 42.4 % of the individuals with a clear abundance in the Chinese cohorts independently of the clinical status. In total, only 17 sequences were found in more than 20 % of the subjects. For most of them it was possible to discriminate the geographical origin of the individuals, with differential abundances in the Chinese cohorts compared to the European and/or USA ones, suggesting that these differences are not related to the clinical status of the individuals but most likely to diet, indicative of the presence of sucrose-related compounds in their food intakes.
HF diet influence on Rgspp mRNA expression in monoxenic mice
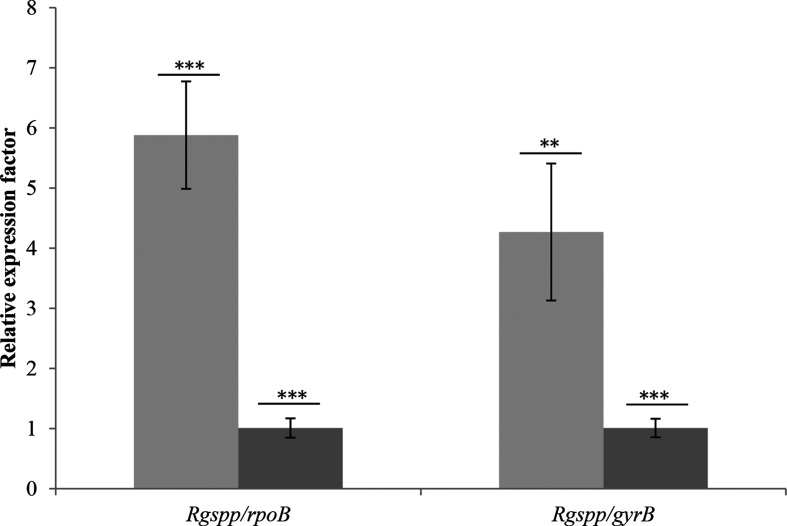
Although the abundance of the Rgspp sequence in the gut microbiome was not found specifically correlated to IBD or diabetes status but mostly to diet, we wanted to check whether diet could affect the expression of the Rgspp gene and, more globally, carbohydrate metabolism. It is indeed well known that, for example, a HF diet affects the composition of the gut microbiota, together with the host metabolism [66]. However, the influence of this diet on bacterial metabolism is poorly studied. In 2014, Daniel et al. fed mice with HF diet for 12 weeks [67]. Metaproteomic and metabolomic data obtained, targeting 1760 bacterial proteins and 86 annotated metabolites, revealed distinct HF-diet-specific profiles demonstrating the impact of diet on the microbial metabolism. In the present study, we used two different trials composed of monoxenic mice inoculated with Ruminococcus gnavus E1. In trial one, mice were fed with a SD and in trial two with a HF diet (Table S2), which contained five times less sucrose. As shown in Fig. 7, a fivefold increase in Rgspp mRNA was observed for the HF diet group, suggesting a close relationship between lipid and sucrose metabolism, and confirming the impact of the HF diet on the gut microbiome previously highlighted by Daniel et al. in 2014 [67].
Fig. 7.
Expression of the RgSPP-encoding gene in monoxenic mice fed with different diets. Dark grey, SD (n=10); light grey, HF diet (n=10). Two different housekeeping genes were used to normalize the experiment, rpoB (left) and gyrB (right). P values were determined using Student's t-test. **, P<0.005; ***, P<0.001.
Conclusion
It is well-know that in plants the biosynthesis of sucrose occurs in the cytosol of the leaves, the two last steps being: (i) UDP-glucose+fructose-6P<>sucrose 6F-phosphate+UDP catalysed by a sucrose phosphate synthase; and (ii) sucrose 6F-phosphate+H2O<>sucrose+Pi catalysed by a SPP. We can assume that during consumption of plant dietary constituents, a part of S6FP is ingested, reaching the distal intestinal compartment without any modification (Fig. 5a). At this point, we can suggest a translocation of the S6FP across the Ruminococcus gnavus E1 cell wall to the cytosolic compartment, via the ABC transporter AgaE. S6FP will be then hydrolyses by RgSPP releasing G1P and F6P, considered as metabolic intermediates of microbial carbohydrate catabolism. Nevertheless, more investigation looks to be necessary to confirm these hypotheses.
Data Bibliography
Additional sequences and data used in this study were obtained from the following sources.
Verhaeghe T, Aerts D, Diricks M, Soetaert W, Desmet T. The quest for a thermostable sucrose phosphorylase reveals sucrose 6’-phosphate phosphorylase as a novel specificity. Appl Microbiol Biotechnol 2014;98:7027–7037.
Bruel L, Sulzenbacher G, Cervera Tison M, Pujol A, Nicoletti C, Perrier J et al. α-Galactosidase/sucrose kinase (AgaSK), a novel bifunctional enzyme from the human microbiome coupling galactosidase and kinase activities. J Biol Chem 2011;286:40814–40823.
Franceus J, Pinel D, Desmet T. Glucosylglycerate phosphorylase, an enzyme with novel specificity involved in compatible solute metabolism. Appl Environ Microbiol 2017;83:e01434-17.
Franceus J, Decuyper L, D’Hooghe M, Desmet T. Exploring the sequence diversity in glycoside hydrolase family 13_18 reveals a novel glucosylglycerol phosphorylase. Appl Microbiol Biotechnol 2018;102:3183–3191.
Qin J, Li R, Raes J, Arumugam M, Burgdorf KS et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 2010;464:59–65.
Supplementary Data
Funding information
This work was co-funded, in part, by Aix-Marseille Université, CNRS and the Ministère Français de l’Enseignement Supérieur et de la Recherche, and by the European Union’s framework programme Horizon 2020 (MSCA-IF-2015 707457, CaTSYS).
Author contributions
The author’s contribution were as follow: AT was involved in study design, data acquisition, data interpretation and manuscript writing; LB was involved in biochemical and enzymatic analysis; EL was involved the prevalence analysis; CN was involved in the qPCR assays; DN was involved in the tree design; BH was involved in the sequences extraction from CAZy database; GV in study design, data interpretation and manuscript writing; JP designed and piloted the in vivo assays; TG was the co-coordinator of the study and was involved in the data interpretation and manuscript writing. ML was the co-coordinator of the study and was involved in the manuscript writing and editing. All authors read and approved the final manuscript.
Acknowledgements
We are grateful to the Analyse et Valorisation de la Biodiversité platform for access to the qPCR thermocycler.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Ethical statement
Animal experiments were performed according to the guidelines of the French Ethics Committee, i.e. agreement number A78-322-6 for mice maintained for 10 days on a control diet and agreement number A78-718 for mice maintained for 2 months on a HF diet.
Footnotes
Abbreviations: CC, caecal content; F6P, alpha-d-fructose-6-phosphate; GGaP, glucosylglycerate phosphorylase; G1P, alpha-d-glucose-1-phosphate; GGoP, glucosylglycerol phosphorylase; HF, high fat; HPAEC-PAD, high-performance anion-exchange chromatography coupled with pulsed-amperometric detection; IBD, intestinal bowel disease; PTS, phosphotransferase system; PUL, polysaccharide utilization loci; RFO, raffinose family oligosaccharide; SD, standard diet; SEC, size exclusion column; SP, sucrose phosphorylase; SPP, sucrose 6F-phosphate phosphorylase.
All supporting data, code and protocols have been provided within the article or through supplementary data files. Five supplementary figures and three supplementary tables are available with the online version of this article.
References
- 1.Sekirov I, Russell SL, Antunes LC, Finlay BB. Gut microbiota in health and disease. Physiol Rev. 2010;90:859–904. doi: 10.1152/physrev.00045.2009. [DOI] [PubMed] [Google Scholar]
- 2.Boulangé CL, Neves AL, Chilloux J, Nicholson JK, Dumas ME. Impact of the gut microbiota on inflammation, obesity and metabolic disease. Genome Med. 2016;8 doi: 10.1186/s13073-016-0303-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gericke B, Amiri M, Naim HY. The multiple roles of sucrase-isomaltase in the intestinal physiology. Mol Cell Pediatr. 2016;3 doi: 10.1186/s40348-016-0033-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van den Ende W. Multifunctional fructans and raffinose family oligosaccharides. Front Plant Sci. 2013:247. doi: 10.3389/fpls.2013.00247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Collins S, Reid G. Distant site effects of ingested prebiotics. Nutrients. 2016;8:523. doi: 10.3390/nu8090523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lombard V, Golaconda Ramulu H, Drula E, Coutinho PM, Henrissat B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014;42:D490–D495. doi: 10.1093/nar/gkt1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Janeček Š, Svensson B, MacGregor EA. α-Amylase: an enzyme specificity found in various families of glycoside hydrolases. Cell Mol Life Sci. 2014;71:1149–1170. doi: 10.1007/s00018-013-1388-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.MacGregor EA, Janeček Š, Svensson B. Relationship of sequence and structure to specificity in the α-amylase family of enzymes. Biochim Biophys Acta. 2001;1546:1–20. doi: 10.1016/S0167-4838(00)00302-2. [DOI] [PubMed] [Google Scholar]
- 9.Stam MR, Danchin EG, Rancurel C, Coutinho PM, Henrissat B. Dividing the large glycoside hydrolase family 13 into subfamilies: towards improved functional annotations of alpha-amylase-related proteins. Protein Eng Des Sel. 2006;19:555–562. doi: 10.1093/protein/gzl044. [DOI] [PubMed] [Google Scholar]
- 10.Aerts D, Verhaeghe TF, Roman BI, Stevens CV, Desmet T, et al. Transglucosylation potential of six sucrose phosphorylases toward different classes of acceptors. Carbohydr Res. 2011;346:1860–1867. doi: 10.1016/j.carres.2011.06.024. [DOI] [PubMed] [Google Scholar]
- 11.Goedl C, Schwarz A, Mueller M, Brecker L, Nidetzky B. Mechanistic differences among retaining disaccharide phosphorylases: insights from kinetic analysis of active site mutants of sucrose phosphorylase and alpha,alpha-trehalose phosphorylase. Carbohydr Res. 2008;343:2032–2040. doi: 10.1016/j.carres.2008.01.029. [DOI] [PubMed] [Google Scholar]
- 12.de Winter K, Verlinden K, Křen V, Weignerová L, Soetaert W, et al. Ionic liquids as cosolvents for glycosylation by sucrose phosphorylase: balancing acceptor solubility and enzyme stability. Green Chem. 2013;15:1949–1955. doi: 10.1039/c3gc40449h. [DOI] [Google Scholar]
- 13.de Bruyn F, van Brempt M, Maertens J, van Bellegem W, Duchi D, et al. Metabolic engineering of Escherichia coli into a versatile glycosylation platform: production of bio-active quercetin glycosides. Microb Cell Fact. 2015;14:138. doi: 10.1186/s12934-015-0326-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Verhaeghe T, de Winter K, Berland M, de Vreese R, D'Hooghe M, et al. Converting bulk sugars into prebiotics: semi-rational design of a transglucosylase with controlled selectivity. Chem Commun. 2016;52:3687–3689. doi: 10.1039/C5CC09940D. [DOI] [PubMed] [Google Scholar]
- 15.Kraus M, Görl J, Timm M, Seibel J. Synthesis of the rare disaccharide nigerose by structure-based design of a phosphorylase mutant with altered regioselectivity. Chem Commun. 2016;52:4625–4627. doi: 10.1039/C6CC00934D. [DOI] [PubMed] [Google Scholar]
- 16.Russell RR, Mukasa H, Shimamura A, Ferretti JJ. Streptococcus mutans gtfA gene specifies sucrose phosphorylase. Infect Immun. 1988;56:2763–2765. doi: 10.1128/iai.56.10.2763-2765.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koga T, Nakamura K, Shirokane Y, Mizusawa K, Kitao S, et al. Purification and some properties of sucrose phosphorylase from Leuconostoc mesenteroides. Agric Biol Chem. 1991;55:1805–1810. doi: 10.1271/bbb1961.55.1805. [DOI] [PubMed] [Google Scholar]
- 18.Trethewey RN, Fernie AR, Bachmann A, Fleischer-Notter H, Geigenberger P, et al. Expression of a bacterial sucrose phosphorylase in potato tubers results in a glucose-independent induction of glycolysis. Plant Cell Environ. 2001;24:357–365. doi: 10.1046/j.1365-3040.2001.00679.x. [DOI] [Google Scholar]
- 19.Kim M, Kwon T, Lee HJ, Kim KH, Chung DK, et al. Cloning and expression of sucrose phosphorylase gene from Bifidobacterium longum in E. coli and characterization of the recombinant enzyme. Biotechnol Lett. 2003;25:1211–1217. doi: 10.1023/A:1025035320983. [DOI] [PubMed] [Google Scholar]
- 20.Sprogøe D, van den Broek LA, Mirza O, Kastrup JS, Voragen AG, et al. Crystal structure of sucrose phosphorylase from Bifidobacterium adolescentis. Biochemistry. 2004;43:1156–1162. doi: 10.1021/bi0356395. [DOI] [PubMed] [Google Scholar]
- 21.Lee J-H, Yoon S-H, Nam S-H, Moon Y-H, Moon Y-Y, et al. Molecular cloning of a gene encoding the sucrose phosphorylase from Leuconostoc mesenteroides B-1149 and the expression in Escherichia coli. Enzyme Microb Technol. 2006;39:612–620. doi: 10.1016/j.enzmictec.2005.11.008. [DOI] [Google Scholar]
- 22.Nishimoto M, Kitaoka M. Identification of the putative proton donor residue of lacto-N-biose phosphorylase (EC 2.4.1.211) Biosci Biotechnol Biochem. 2007;71:1587–1591. doi: 10.1271/bbb.70064. [DOI] [PubMed] [Google Scholar]
- 23.Lee J-H, Moon Y-H, Kim N, Kim Y-M, Kang H-K, et al. Cloning and expression of the sucrose phosphorylase gene from Leuconostoc mesenteroides in Escherichia coli. Biotechnol Lett. 2008;30:749–754. doi: 10.1007/s10529-007-9608-y. [DOI] [PubMed] [Google Scholar]
- 24.Teixeira JS, Abdi R, Su MS, Schwab C, Gänzle MG. Functional characterization of sucrose phosphorylase and scrR, a regulator of sucrose metabolism in Lactobacillus reuteri. Food Microbiol. 2013;36:432–439. doi: 10.1016/j.fm.2013.07.011. [DOI] [PubMed] [Google Scholar]
- 25.Verhaeghe T, Aerts D, Diricks M, Soetaert W, Desmet T. The quest for a thermostable sucrose phosphorylase reveals sucrose 6'-phosphate phosphorylase as a novel specificity. Appl Microbiol Biotechnol. 2014;98:7027–7037. doi: 10.1007/s00253-014-5621-y. [DOI] [PubMed] [Google Scholar]
- 26.Franceus J, Pinel D, Desmet T. Glucosylglycerate phosphorylase, an enzyme with novel specificity involved in compatible solute metabolism. Appl Environ Microbiol. 2017;83::e01434-17. doi: 10.1128/AEM.01434-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Franceus J, Decuyper L, D'Hooghe M, Desmet T. Exploring the sequence diversity in glycoside hydrolase family 13_18 reveals a novel glucosylglycerol phosphorylase. Appl Microbiol Biotechnol. 2018;102:3183–3191. doi: 10.1007/s00253-018-8856-1. [DOI] [PubMed] [Google Scholar]
- 28.Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ramare F, Nicoli J, Dabard J, Corring T, Ladire M, et al. Trypsin-dependent production of an antibacterial substance by a human Peptostreptococcus strain in gnotobiotic rats and in vitro. Appl Environ Microbiol. 1993;59:2876–2883. doi: 10.1128/aem.59.9.2876-2883.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dabard J, Bridonneau C, Phillipe C, Anglade P, Molle D, et al. Ruminococcin A, a new lantibiotic produced by a Ruminococcus gnavus strain isolated from human feces. Appl Environ Microbiol. 2001;67:4111–4118. doi: 10.1128/AEM.67.9.4111-4118.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pujol A, Crost EH, Simon G, Barbe V, Vallenet D, et al. Characterization and distribution of the gene cluster encoding RumC, an anti-Clostridium perfringens bacteriocin produced in the gut. FEMS Microbiol Ecol. 2011;78:405–415. doi: 10.1111/j.1574-6941.2011.01176.x. [DOI] [PubMed] [Google Scholar]
- 32.Crost EH, Ajandouz EH, Villard C, Geraert PA, Puigserver A, et al. Ruminococcin C, a new anti-Clostridium perfringens bacteriocin produced in the gut by the commensal bacterium Ruminococcus gnavus E1. Biochimie. 2011;93:1487–1494. doi: 10.1016/j.biochi.2011.05.001. [DOI] [PubMed] [Google Scholar]
- 33.Bruel L, Sulzenbacher G, Cervera Tison M, Pujol A, Nicoletti C, et al. α-Galactosidase/sucrose kinase (AgaSK), a novel bifunctional enzyme from the human microbiome coupling galactosidase and kinase activities. J Biol Chem. 2011;286:40814–40823. doi: 10.1074/jbc.M111.286039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aguilera M, Rakotoarivonina H, Brutus A, Giardina T, Simon G, et al. Aga1, the first alpha-galactosidase from the human bacteria Ruminococcus gnavus E1, efficiently transcribed in gut conditions. Res Microbiol. 2012;163:14–21. doi: 10.1016/j.resmic.2011.10.005. [DOI] [PubMed] [Google Scholar]
- 35.Cervera-Tison M, Tailford LE, Fuell C, Bruel L, Sulzenbacher G, et al. Functional analysis of family GH36 α-galactosidases from Ruminococcus gnavus E1: insights into the metabolism of a plant oligosaccharide by a human gut symbiont. Appl Environ Microbiol. 2012;78:7720–7732. doi: 10.1128/AEM.01350-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Crost EH, Tailford LE, Le Gall G, Fons M, Henrissat B, et al. Utilisation of mucin glycans by the human gut symbiont Ruminococcus gnavus is strain-dependent. PLoS One. 2013;8:e76341. doi: 10.1371/journal.pone.0076341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 38.Lafond M, Tauzin A, Desseaux V, Bonnin E, Ajandouz E-H, et al. GH10 xylanase D from Penicillium funiculosum: biochemical studies and xylooligosaccharide production. Microb Cell Fact. 2011;10:20. doi: 10.1186/1475-2859-10-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.West DB, Boozer CN, Moody DL, Atkinson RL. Dietary obesity in nine inbred mouse strains. Am J Physiol. 1992;262:R1025–R1032. doi: 10.1152/ajpregu.1992.262.6.R1025. [DOI] [PubMed] [Google Scholar]
- 40.Graziani F, Pujol A, Nicoletti C, Dou S, Maresca M, et al. Ruminococcus gnavus E1 modulates mucin expression and intestinal glycosylation. J Appl Microbiol. 2016;120:1403–1417. doi: 10.1111/jam.13095. [DOI] [PubMed] [Google Scholar]
- 41.Rossmeisl M, Rim JS, Koza RA, Kozak LP. Variation in type 2 diabetes-related traits in mouse strains susceptible to diet-induced obesity. Diabetes. 2003;52:1958–1966. doi: 10.2337/diabetes.52.8.1958. [DOI] [PubMed] [Google Scholar]
- 42.Rabot S, Membrez M, Bruneau A, Gérard P, Harach T, et al. Germ-free C57BL/6J mice are resistant to high-fat-diet-induced insulin resistance and have altered cholesterol metabolism. Faseb J. 2010;24:4948–4959. doi: 10.1096/fj.10-164921. [DOI] [PubMed] [Google Scholar]
- 43.Faith JJ, McNulty NP, Rey FE, Gordon JI. Predicting a human gut microbiota's response to diet in gnotobiotic mice. Science. 2011;333:101–104. doi: 10.1126/science.1206025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Doré J, Sghir A, Hannequart-Gramet G, Corthier G, Pochart P. Design and evaluation of a 16S rRNA-targeted oligonucleotide probe for specific detection and quantitation of human faecal Bacteroides populations. Syst Appl Microbiol. 1998;21:65–71. doi: 10.1016/S0723-2020(98)80009-X. [DOI] [PubMed] [Google Scholar]
- 45.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 46.Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc. 2015;10:845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Petersen TN, Brunak S, von Heijne G, Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods. 2011;8:785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 48.Katoh K, Frith MC. Adding unaligned sequences into an existing alignment using MAFFT and LAST. Bioinformatics. 2012;28:3144–3146. doi: 10.1093/bioinformatics/bts578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Robert X, Gouet P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014;42:W320–W324. doi: 10.1093/nar/gku316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 52.Zuckerkandl E, Pauling L. Evolutionary divergence and convergence in proteins. In: Bryson V, Vogel HJ, editors. Evolving Genes and Proteins. New York: Academic Press; 1965. pp. 97–166. (editors) [Google Scholar]
- 53.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li J, Jia H, Cai X, Zhong H, Feng Q, et al. An integrated catalog of reference genes in the human gut microbiome. Nat Biotechnol. 2014;32:834–841. doi: 10.1038/nbt.2942. [DOI] [PubMed] [Google Scholar]
- 55.Tasse L, Bercovici J, Pizzut-Serin S, Robe P, Tap J, et al. Functional metagenomics to mine the human gut microbiome for dietary fiber catabolic enzymes. Genome Res. 2010;20:1605–1612. doi: 10.1101/gr.108332.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Goedl C, Nidetzky B. Sucrose phosphorylase harbouring a redesigned, glycosyltransferase-like active site exhibits retaining glucosyl transfer in the absence of a covalent intermediate. Chembiochem. 2009;10:2333–2337. doi: 10.1002/cbic.200900429. [DOI] [PubMed] [Google Scholar]
- 57.Terrapon N, Lombard V, Gilbert HJ, Henrissat B. Automatic prediction of polysaccharide utilization loci in Bacteroidetes species. Bioinformatics. 2015;31:647–655. doi: 10.1093/bioinformatics/btu716. [DOI] [PubMed] [Google Scholar]
- 58.Sheridan PO, Martin JC, Lawley TD, Browne HP, Harris HM, et al. Polysaccharide utilization loci and nutritional specialization in a dominant group of butyrate-producing human colonic Firmicutes. Microb Genom. 2016;2:e000043. doi: 10.1099/mgen.0.000043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ferretti JJ, Huang TT, Russell RR. Sequence analysis of the glucosyltransferase A gene (gtfA) from Streptococcus mutans Ingbritt. Infect Immun. 1988;56:1585–1588. doi: 10.1128/iai.56.6.1585-1588.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Altermann E, Russell WM, Azcarate-Peril MA, Barrangou R, Buck BL, et al. Complete genome sequence of the probiotic lactic acid bacterium Lactobacillus acidophilus NCFM. Proc Natl Acad Sci USA. 2005;102:3906–3912. doi: 10.1073/pnas.0409188102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ojala T, Kuparinen V, Koskinen JP, Alatalo E, Holm L, et al. Genome sequence of Lactobacillus crispatus ST1. J Bacteriol. 2010;192:3547–3548. doi: 10.1128/JB.00399-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sun Z, Harris HM, McCann A, Guo C, Argimón S, et al. Expanding the biotechnology potential of lactobacilli through comparative genomics of 213 strains and associated genera. Nat Commun. 2015;6:8322. doi: 10.1038/ncomms9322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hao Y, Huang D, Guo H, Xiao M, An H, et al. Complete genome sequence of Bifidobacterium longum subsp. longum BBMN68, a new strain from a healthy Chinese centenarian. J Bacteriol. 2011;193:787–788. doi: 10.1128/JB.01213-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wildt S, Nordgaard I, Hansen U, Brockmann E, Rumessen JJ. A randomised double-blind placebo-controlled trial with Lactobacillus acidophilus La-5 and Bifidobacterium animalis subsp. lactis BB-12 for maintenance of remission in ulcerative colitis. J Crohns Colitis. 2011;5:115–121. doi: 10.1016/j.crohns.2010.11.004. [DOI] [PubMed] [Google Scholar]
- 65.Gerritsen J, Hornung B, Renckens B, van Hijum S, Martins dos Santos VAP, et al. Genomic and functional analysis of Romboutsia ilealis CRIBT reveals adaptation to the small intestine. PeerJ. 2017;5:e3698. doi: 10.7717/peerj.3698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Murphy EA, Velazquez KT, Herbert KM. Influence of high-fat diet on gut microbiota: a driving force for chronic disease risk. Curr Opin Clin Nutr Metab Care. 2015;18:515–520. doi: 10.1097/MCO.0000000000000209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Daniel H, Gholami AM, Berry D, Desmarchelier C, Hahne H, et al. High-fat diet alters gut microbiota physiology in mice. ISME J. 2014;8:295–308. doi: 10.1038/ismej.2013.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.