Abstract
The human immunodeficiency virus type 1 (HIV-1) genomic RNA (vRNA) has two major fates during viral replication: to serve as the template for the major structural and enzymatic proteins, or to be encapsidated and packaged into assembling virions to serve as the genomic vRNA in budding viruses. The dynamic balance between vRNA translation and encapsidation is mediated by numerous host proteins, including Staufen1. During HIV-1 infection, HIV-1 recruits Staufen1 to assemble a distinct ribonucleoprotein complex promoting vRNA encapsidation and viral assembly. Staufen1 also rescues vRNA translation and gene expression during conditions of cellular stress. In this work, we utilized novel Staufen1−/− gene-edited cells to further characterize the contribution of Staufen1 in HIV-1 replication. We observed a marked deficiency in the ability of HIV-1 to dissociate stress granules (SGs) in Staufen1-deficient cells and remarkably, the vRNA repositioned to SGs. These phenotypes were rescued by Staufen1 expression in trans or in cis, but not by a dsRBD-binding mutant, Staufen1F135A. The mistrafficking of the vRNA in these Staufen1−/− cells was also accompanied by a dramatic decrease in viral production and infectivity. This work provides novel insight into the mechanisms by which HIV-1 uses Staufen1 to ensure optimal vRNA translation and trafficking, supporting an integral role for Staufen1 in the HIV-1 life cycle, positioning it as an attractive target for next-generation antiretroviral agents.
Keywords: HIV-1, Staufen1, stress granules, stress response to infection, viral genomic RNA (vRNA), vRNA trafficking
INTRODUCTION
In response conditions of cellular stress, including that of viral infection, the host cell reprograms its translational machinery to inhibit viral gene expression by assembling translationally silent ribonucleoprotein (RNP) complexes known as stress granules (SGs) (Anderson and Kedersha 2009; Thomas et al. 2011). Viruses are obligate intracellular parasites that utilize the host cell machinery to facilitate the expression of their own genes, and viral replication can be significantly decreased by an impediment to cellular mRNA translation. Hence, many viruses have developed mechanisms to circumvent the cellular stress response (for reviews, see Valiente-Echeverría et al. 2012; Poblete-Durán et al. 2016).
Two types of SGs that differ in their morphology, composition, and mechanisms of assembly have been described by Fujimura et al. (2012). In our previous work, we have shown that human immunodeficiency virus type 1 (HIV-1) disrupts canonical type I SG assembly in an eIF2α-phosphorylation (eIF2α-P) independent manner via an interaction between the amino-terminal domain of the capsid (CA) domain on the main HIV-1 structural protein pr55Gag and the host eukaryotic elongation factor 2 (eEF2) (Abrahamyan et al. 2010; Valiente-Echeverría et al. 2014). pr55Gag could also mediate the disassembly of preexisting SGs via an interaction with Ras GTPase-activating protein-binding protein 1 (G3BP1) (Valiente-Echeverría et al. 2014). Moreover, HIV-1 is also capable of blocking the assembly of type II, noncanonical SGs by reducing the amount of hypophosphorylated 4EBP1 associated with the 5′ cap, potentially through an interaction with its target, eIF4E (Cinti et al. 2016). We have also demonstrated that HIV-1 blocks the assembly of Type 1 SGs induced by HIV-1 nucleocapsid (NC) expression via the recruitment of the host dsRNA binding protein Staufen1 (Rao et al. 2018).
Staufen1 plays roles in various steps of the HIV-1 replication cycle, including Gag multimerisation, vRNA encapsidation, and viral assembly (Chatel-Chaix et al. 2004, 2007, 2008; Abrahamyan et al. 2010). A role for Staufen1 in viral encapsidation was first described when it was determined that Staufen1 is incorporated into virions in a vRNA-dependent manner (Mouland et al. 2000). Overexpression of Staufen1 was demonstrated to increase vRNA encapsidation several-fold, resulting in a significant impairment of viral infectivity (Mouland et al. 2000). The amino-terminal moiety of Staufen1 is also required for efficient Gag multimerisation (Chatel-Chaix et al. 2007). The third dsRNA binding domain (dsRBD3) of Staufen1 interacts specifically with the zinc fingers of the NC domain of Gag in an RNA-independent manner (Chatel-Chaix et al. 2008). This interaction results in the formation of high molecular weight, detergent insoluble, Staufen1-HIV-1-dependent RNPs (SHRNPs), also containing Gag, Upf1, and the vRNA, and many other viral and cellular proteins (Mallardo et al. 2003; Chatel-Chaix et al. 2004, 2007, 2008; Abrahamyan et al. 2010; Milev et al. 2012; Tosar et al. 2012). In the context of SHRNPs, Staufen1 likely interacts with the NC domain of Gag to enhance Gag assembly and vRNA packaging (Abrahamyan et al. 2010; Milev et al. 2010). The formation of this RNP also promotes vRNA encapsidation. The knockdown of Staufen1 has been shown to result in a significant reduction of viral infectivity (Chatel-Chaix et al. 2004). It was later described that during conditions of oxidative stress, HIV-1 can prevent the assembly of SGs, but promotes the assembly of SHRNPs (Abrahamyan et al. 2010). SHRNPs are distinct from both SGs and processing bodies (PBs), and their formation is also hypothesized to serve to prevent the degradation of the vRNA (Abrahamyan et al. 2010). Therefore, the formation of the SHRNPs contributes to the ability of HIV-1 in preventing SG assembly to ensure viral production (Abrahamyan et al. 2010).
In this work, we investigated the ability of HIV-1 to prevent the assembly of Type 1, arsenite (Ars)-induced SGs in novel HCT116 cells that are deficient of Staufen1, as generated by CRISPR-Cas9-mediated gene editing. We observed that Staufen1 knockout (SKO) resulted in impaired SG dissociation by HIV-1, rerouting of the viral genomic RNA to SGs, decreased viral gene expression and marked defects in viral production. These results highlight the importance of host protein Staufen1 in optimal vRNA trafficking and in the prevention of SG assembly for efficient virus production.
RESULTS
HIV-1 has impaired ability to dissociate stress granules in Staufen1-deficient cells
Previous studies show that siRNA-mediated depletion of Staufen1 facilitates Ars-induced SG assembly while Staufen1 overexpression impairs SG assembly (Thomas et al. 2009). In HIV-1 expressing cells, Staufen1 prevents the assembly of Ars-induced SGs by promoting the assembly of SHRNPs (Abrahamyan et al. 2010). HIV-1 also disassembles SGs, irrespective of eIF2α phosphorylation status, by interacting with the eukaryotic elongation factor eEF2 via the capsid (CA) domain of the precursor structural protein, pr55Gag (referred to as Gag herein) (Valiente-Echeverría et al. 2014). In contrast, the carboxy-terminal NC domain of Gag, when expressed alone, induces eIF2α phosphorylation and SG assembly, but cannot be dissolved by CA. These observations suggest that the activities of the CA and NC domains control SG dynamics, likely to be important at several steps of HIV-1 replication. Nevertheless, we have shown that Staufen1 inhibits NC-induced SG assembly and rescues HIV-1 viral production (Rao et al. 2018). In this study, we therefore evaluated the ability of HIV-1 to dissociate Ars-induced SGs in novel, SKO cells, generated as described in Materials and Methods (S Hassine and L DesGroseillers, in prep.). These cells grow normally with no apparent change in phenotype. Parental HCT116 and SKO cells were either mock transfected or transfected with pNL4.3, a plasmid that expresses the full-length HIV-1 genome and expresses all auxiliary and regulatory viral genes as described in Materials and Methods. Twenty-four hours later, cells were left untreated or were briefly treated with Ars, and SGs were visualized by TIAR1 staining by indirect immunofluorescence for quantification. In wild-type (WT) HCT116 cells, Ars treatment resulted in the assembly of SGs in 82.9 (±13.1)% of mock-transfected cells (Fig. 1A,D). However, only 17.7 (±12.7)% of pNL4.3-transfected cells contained SGs, demonstrating that HIV-1 expression dissociates Ars-induced SGs in HCT116 cells (Fig. 1A,D). SGs were not observed in mock (pcDNA3.1 transfected) untreated cells, top rows in Fig. 1A,B). Similar to what was found in HCT116 cells, Ars treatment resulted in the assembly of SGs in the majority of cells (89.4 [±6.6]%) of mock-transfected SKO cells (Fig. 1B,D). SKO cells transfected with pNL4-3 however, were 62.9 (±11.6)% positive for SGs as a result of Ars treatment (Fig. 1B,C) indicating an impaired ability of HIV-1 to dissociate and block SG assembly. To determine whether reintroducing Staufen1 to SKO cells could rescue the HIV-1-induced SG blockade, we first transfected SKO cells with a Staufen1/YFP expression construct to rescue Staufen1 expression (Chatel-Chaix et al. 2004). Cells were treated with Ars 24 h later, and SGs were visualized by TIAR1 staining by indirect immunofluorescence. Comparable to the effects of HIV-1 expression in WT HCT116 cells, coexpression of Staufen1/YFP and pNL4.3 in SKO cells reverted to the HIV-1-mediated SG blockade phenotype where only 16.7 (±5.9)% of cells exhibited SGs (Fig. 1B, bottom row), indicating that Staufen1 is required for the SG blockade imposed by HIV-1.
FIGURE 1.

HIV-1 has impaired ability to dissociate Ars-induced SGs in SKO cells and is rescued by Staufen1 in trans and in cis. (A) HCT116 cells were transfected with pcDNA3.1 or proviral HIV-1 construct, pNL4.3. Twenty-four hours later, cells were either mock-treated or treated with 500 µM Ars for 1 h and then p24 (green) and TIAR1 (red) were detected by indirect immunofluorescence. Nuclei were stained with DAPI (blue). Scale bars are 10 µm. (B) Staufen1 gene-edited SKO cells were transfected with pcDNA3.1 (Mock), pNL4.3, or pNL4.3 and a Staufen1/YFP cDNA expression construct to rescue Staufen1 in trans. Twenty-four hours later, cells were either mock treated or treated with Ars for 1 h and p24 (green) and TIAR1 (red) were detected by indirect immunofluorescence. Nuclei were stained with DAPI (blue). Gag is pseudocolored green; inset shows Staufen1-YFP expression in yellow. Scale bars are 10 µm. (C) HCT116 cells or SKO cells (as indicated) were transfected either with pNL4.3, pNL4.3/Staufen1-HA, or pNL4.3/Staufen1F135A-HA, to rescue with WT or dsRBD3 mutant Staufen1, in cis. Twenty-four hours later, cells were either mock treated or treated with Ars for 1 h, and p24 (green) and eIF3 (pink) were detected by indirect immunofluorescence. Nuclei were stained with DAPI (blue). Scale bars are 10 µm. (D) Quantification of cells containing SGs from panels A–C. Error bars represent the standard deviation from three independent experiments with at least 100 cells counted per treatment. Asterisks represent statistically significant differences between groups (one-way ANOVA: [**] P ≤ 0.01; [****] P ≤ 0.001).
In the next series of experiments, we performed experiments with pNL4.3 and chimeric pNL4.3 constructs in which Staufen1 is inserted within the nef open reading frame to rescue Staufen1 or the Staufen1 mutant expression in cis. We stained with eIF3, which is found in type I but not type II SGs (Cinti et al. 2016). As shown in Figure 1C (top row), HIV-1 blocks type I SGs as shown previously. However, in SKO cells, HIV-1 is again unable to suppress SG assembly when cells are stressed with Ars. In a similar manner, when Staufen1 is rescued in cis using pNL4.3/Staufen1-HA, the HIV-1 imposed blockade of SGs is recovered to a similar level obtained with Staufen1/YFP expressed in trans (Fig. 1C, third row), but not when the dsRBD3 mutant, pNL4.3/Staufen1F135A-HA is used to rescue in cis (Fig. 1C, bottom row; Fig. 1D). Because Staufen1's ability to bind both Gag and RNA is dramatically impaired with this F135A mutant (Ramos et al. 2000; Chatel-Chaix et al. 2008), it is likely that these interactions are largely required to rescue the HIV-1-imposed SG phenotype. These results, compiled in Figure 1D, demonstrate that Staufen1 is recruited by HIV-1 to inhibit Ars-induced SG assembly via Staufen1's F135A region in its third dsRBD.
The HIV-1 viral RNA accumulates in stress granules induced in Staufen1-deficient cells
SGs are associated with silenced transcripts, and many viruses are known to subvert the function of these RNA granules to promote viral gene expression (Lloyd 2012). Since SGs were observed in HIV-1 expressing SKO cells (Fig. 1), we sought to determine whether these SGs contain sequestered, HIV-1 genomic RNA, vRNA. HCT116 and SKO cells were mock transfected and treated with or without Ars, or transfected with pNL.4.3 (Fig. 2A,B). Twenty-four hours later, the vRNA and SGs were visualized using combined fluorescence in situ hybridization and indirect immunofluorescence for TIAR1, respectively. In both cell lines, TIAR1 displayed a diffuse staining pattern and Ars-induced large, TIAR1+ SGs (Fig. 2A,B, top and middle rows). The vRNA presented as a regular, punctate staining pattern in untreated (without Ars), HIV-1-expressing HCT and SKO cells (Supplemental Fig. S1). As shown earlier, HIV-1 (identified herein as vRNA-expressing cells) suppressed SG assembly in Ars-treated HCT116 cells (Fig. 2A, bottom row). However, in the Ars-treated, pNL4.3-transfected SKO cells, the vRNA predominantly colocalized with the SG marker, TIAR1 (Fig. 2B, third row). Indeed, when SKO cells were transfected with pNL4.3-Staufen1-HA to rescue Staufen1 expression in cis, the HIV-1-mediated blockade to SG assembly was reestablished and vRNA was observed in the cytoplasm of cells (Fig. 2B, bottom row; Supplemental Fig. S1). Therefore, in the absence of Staufen1, the vRNA is mistrafficked and routed aberrantly to SGs.
FIGURE 2.

HIV-1 genomic RNA (vRNA) is sequestered in Ars-induced SGs in SKO cells. (A) HCT116 cells were transfected with pcDNA3.1 or pNL4.3. Twenty-four hours later, cells were either mock treated or treated with Ars for 1 h, and then HIV-1 genomic RNA (vRNA; red) and TIAR1 (cyan) were identified by coupled fluorescence in situ hybridization/immunofluorescence. Nuclei were stained with DAPI (blue). Scale bars are 10 µm. (B) SKO cells were transfected with pcDNA3.1, pNL4.3, or pNL4.3.-Staufen1-HA to rescue Staufen1 expression in cis. Twenty-four hours later, cells were either mock treated or treated with Ars for 1 h and then p24 (green) and TIAR1 (cyan) were detected by indirect immunofluorescence. Nuclei were stained with DAPI (blue). Scale bars are 10 µm.
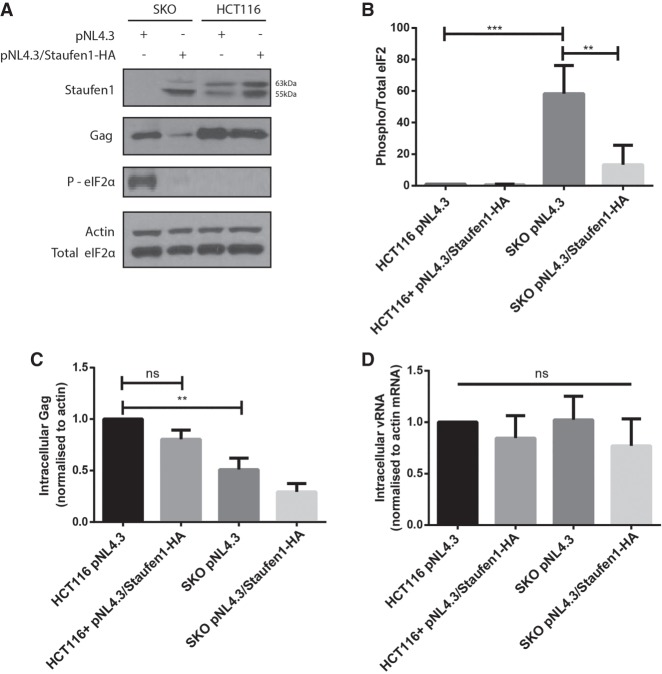
Staufen1 deficiency promotes eIF2α phosphorylation and a reduction in Gag
The phosphorylation of eIF2α is triggered by conditions of stress, including that induced by viral infection, resulting in a block to mRNA translation initiation and the assembly of SGs (Kedersha et al. 1999; Balachandran et al. 2000). Since SKO cells have an impaired ability to dissociate Ars-induced SGs, we evaluated the phosphorylation status of eIF2α in HIV-1-expressing cells, and its subsequent effect on viral gene expression. HCT116 and SKO cells were transfected with either pNL4.3 or pNL4.3-Staufen1-HA. Whole cell lysates were collected and the levels of phosphorylated eIF2α and Gag were quantified by western blotting at 24 h posttransfection. eIF2α was not phosphorylated in HCT116 cells transfected with expressing either of the proviral constructs, pNL4.3 or pNL4.3/Staufen1-HA (Fig. 3A,B), and no statistically significant differences in Gag expression levels were found either (Fig. 3A,C). However, robust phosphorylation of eIF2α was observed in the HIV-1-expressing SKO cells, and this was accompanied by a 51.0 (±11.2)% decrease in Gag expression (Fig. 3A–C). The coexpression of Staufen1 in cis and HIV-1 in the SKO background led to decreased eIF2α phosphorylation (Fig. 3A,B), but was however, unable to rescue Gag expression in these cells (Fig. 3A,C). To determine whether these effects on Gag expression were due to impaired translation of the vRNA or due to reduced vRNA levels, RT-qPCR was used to quantify vRNA isolated from cell lysates. No significant differences in the levels of intracellular vRNA were observed between HCT116 or SKO cells transfected with either pNL4.3 or pNL4.3/Staufen1-HA (Fig. 3D). While we did not determine cellular translation levels, Staufen1 was likely affecting HIV-1 gene expression at a posttranscriptional level since we found no significant differences in intracellular vRNA (Fig. 3D). Therefore, these results imply that the effects of Staufen1 deficiency on Gag levels are likely due to a posttranscriptional defect, and not a transcriptional one.
FIGURE 3.
SKO cells exhibit impaired intracellular vRNA expression. (A) HCT116 and SKO cells were transfected with either pNL4.3 or pNL4.3-Staufen1-HA, to rescue Staufen1 in cis. Twenty-four hours later, cell lysates were subjected to SDS-PAGE, immunoblotted, and probed to investigate eIF2α phosphorylation and Gag expression. (B) Densitometric quantification of phosphor(P)-eIF2α was determined by ImageJ analysis. Values presented in the graph are normalized against the total amount of eIF2α in the cell lysate and represent fold change with the HCT116 pNL4.3-transfected cells being arbitrarily set to one. Error bars represent the standard error of the mean from three independent experiments. Asterisks represent statistically significant differences between groups (one-way ANOVA: [**] P ≤ 0.01; [***] P ≤ 0.005). (C) Densitometry quantifications of Gag levels were determined by ImageJ analysis. Values presented in the graph are normalized against the total amount of Gag in the cell lysate and represent fold change with the HCT116 pNL4.3-transfected cells being arbitrarily set to one. Error bars represent the standard error of the mean from three independent experiments. Asterisks represent statistically significant difference between groups (one-way ANOVA: [**] P ≤ 0.01). (D) HIV-1 genomic RNA, vRNA, and levels from cells transfected as described above were measured by RT-PCR and normalized to the HCT116 pNL4.3-transfected condition that was arbitrarily set to one. Error bars represent the standard deviation from three independent experiments. ns, no statistical differences between the means.
Staufen1 deficiency results in impaired viral infectivity
Since reduced Gag was observed in the SKO cells (Fig. 3A,C), we aimed to determine whether this reduction was correlated with reduced virus production. HCT116 or SKO cells were either transfected with proviral constructs, pNL4.3 or pNL4.3/Staufen1-HA, and p24 levels from supernatants, as a measure of virus production, were quantified by enzyme-linked immunosorbent assay (ELISA). Coexpression of HIV-1 and Staufen1 in WT HCT116 cells displayed a 55.9 (±7.9)% reduction in virus relative to HIV-1 expression alone (Fig. 4A). However, the reduction of virus production from SKO cells was more dramatic, showing a decrease of 78.4 (±6.3)%, and 84.2 (±5.6)% from coexpression of HIV-1 and Staufen1 (using pNL4.3/Staufen1-HA) relative to HIV-1 expressing HCT116 cells (Fig. 4A). Since both the overexpression and knockdown of Staufen1 lead to increased vRNA encapsidation and reduced viral infectivity (Mouland et al. 2000; Chatel-Chaix et al. 2004; Abrahamyan et al. 2010), we sought to quantitate vRNA encapsidation in the released virus. For this, we used semiquantitative RT-qPCR to quantify vRNA from supernatants of either WT HCT116 or SKO cells transfected with pNL4.3 or pNL4.3/Staufen1-HA. The infectivity of viruses produced from these cells was measured using HIV-1 infectivity indicator TZM-bl cells. In agreement with our previous work (Mouland et al. 2000), Staufen1 overexpression in WT HCT116 cells caused a 2.1 (±0.3) fold increase in the vRNA/p24 ratio, a measure of viral encapsidation, relative to the HIV-1 expressing HCT116 cells, indicating that Staufen1 expression enhanced vRNA encapsidation (Fig. 4B). As compared to the WT HCT116 HIV-1 expressing cells, SKO cells exhibited a 3.8 (±0.7)-fold and 5.7 (±2.1)-fold increase in vRNA/p24 ratio in the HIV-1 expressing and HIV-1 and Staufen1-HA coexpressing cells (using pNL4.3/Staufen1-HA), respectively (Fig. 4B). These latter data are also in agreement with our previous observations of impaired infectivity of progeny virions upon Staufen1 knockdown (Chatel-Chaix et al. 2004). This correlated with a 62.5 (±0.1)%, 56.2 (±24.6)%, and 88.58 (±8.9)% decrease in virus infectivity from the supernatants from Staufen1-HIV-1 coexpressing HCT116 cells, of SKO cells either expressing HIV-1, and coexpressing Staufen-1 and HIV-1 (using pNL4.3/Staufen1-HA), respectively (Fig. 4C). Therefore, virus derived from SKO cells contain enhanced vRNA but exhibit largely decreased infectivity.
FIGURE 4.
The viruses generated from SKO cells have impaired infectivity. HCT116 and SKO cells were transfected with either pNL4.3 or pNL4.3.-Staufen1-HA to rescue Staufen1 in cis. The amount of p24 in the supernatant of transfected cells was measured by p24 ELISA. Error bars represent the standard deviation from three independent experiments. Asterisks represent statistically significant difference between groups (one-way ANOVA: [****] P ≤ 0.001). (B) vRNA levels from cells transfected as described above were measured by RT-PCR and normalized to the amount of virus in the supernatant as measured by p24 ELISA. Error bars represent the standard deviation from three independent experiments. Asterisks represent statistically significant differences between groups (one-way ANOVA: [**] P ≤ 0.01). (C) The infectivity of the virus in the supernatants of cells transfected as described above was quantified by X-Gal staining assay and normalized to the amount of virus in the supernatant as measured by p24 ELISA. Error bars represent the standard deviation from three independent experiments. Asterisks represent statistically significant difference between groups (one-way ANOVA: [**] P ≤ 0.01).
DISCUSSION
This work demonstrates an integral requirement for Staufen1 in the process of HIV-1-mediated SG disassembly and for efficient viral gene expression. We show that HIV-1 has an impaired ability to dissociate Ars-induced SGs in SKO cells. We also demonstrate by mutational analysis that this effect is mediated by the RNA- and Gag- binding activity of the third dsRBD of Staufen1 (Fig. 1). We provide novel evidence of the vRNA is rerouted and mistrafficked to SGs in the SKO cells, when exposed to Ars-induced oxidative stress (Fig. 2). We also demonstrate that HIV-1 expression in SKO cells results in a massive phosphorylation of eIF2α and an impairment of viral gene expression, increased vRNA encapsidation, and defects in infectivity of the viral progeny generated from these cells (Figs. 3, 4). Importantly, this work is the first report demonstrating that HIV-1 expression results in the phosphorylation of the translation initiation factor eIF2α during conditions of Staufen1 deficiency, indicating that Staufen1 is responsible for preventing eIF2α phosphorylation during HIV-1 infection. Our previous work demonstrates that HIV-1 is able to inhibit Ars-induced SGs via the recruitment of eEF2 by capsid (CA) domain of Gag and that Cyclophilin A stabilizes this interaction (Valiente-Echeverría et al. 2014). However, in the absence of Staufen1, HIV-1 is unable to prevent Ars-induced SGs, indicating an important role for Staufen1 in HIV-1-mediated SG disassembly and the implication of multiple host factors in countering this stress response. Therefore, in HCT116 cells Staufen1 plays crucial roles in preventing SG assembly, likely by promoting the assembly of SHRNPs or a particular type of RNA granule (Abrahamyan et al. 2010), resulting in the proper trafficking of the vRNA, its encapsidation, and the generation of infectious virus particles. It will be of value to further characterize the effects of Staufen1 deficiency in HIV-1-tropic cell types.
In HIV-1-expressing cells, the vRNA has two main fates: to serve as a template for mRNA translation and synthesis of the major HIV-1 structural and enzymatic proteins Gag and GagPol, and to be packaged into progeny, budding viruses. The majority of the vRNA is not captured for encapsidation, but rather serves other roles toward generating viral proteins, or acts as a cofactor for viral assembly (Kaye and Lever 1999; Butsch and Boris-Lawrie 2000, 2002; Poon et al. 2002). The regulation of the fate of the vRNA, that is, whether it is translated or encapsidated, is reported to be determined by conformational switches in secondary structure of the vRNA (Abbink and Berkhout 2003). The vRNA long-distance interaction (LDI) secondary structure is used for translation, while the branched multiple hairpin (BMH) secondary structure is used for genome encapsidation (Abbink and Berkhout 2003). Binding of the NC protein to the vRNA is believed to cause a rearrangement from the LDI to the BMH conformation (Huthoff and Berkhout 2001). Staufen1 interacts with the NC domain of Gag to promote vRNA encapsidation via the assembly of SHRNPS that putatively serve as a scaffold for vRNA packaging. During Staufen1 overexpression in WT HCT116 cells, the virus generated from HIV-1 expressing cells contained increased levels of vRNA (Fig. 4B), yet had decreased viral infectivity (Fig. 4C), in agreement with previous studies showing that an overexpression of Staufen1 also results in enhanced vRNA encapsidation and decreased infectivity of progeny virions (Mouland et al. 2000). This further supports a role for Staufen1 in vRNA encapsidation via the assembly of SHRNPs.
SHRNPs are unable to assemble in cells lacking Staufen1. Enhanced vRNA encapsidation was observed in the SKO cells, and was accompanied by a phosphorylation of eIF2α (Figs. 3A, 4B). These data are similar to previous reports in which siRNA-mediated depletion of Staufen1 also resulted in enhanced vRNA encapsidation and defects in viral infectivity (Abrahamyan et al. 2010). Therefore, since SHNRPs are unable to assemble in SKO cells, the vRNA is mistrafficked and assembles in distinct RNPs that promote excessive vRNA encapsidation, resulting in the generation of defective progeny virus. The association of RNP assembly to packaging has been intimated to be the case for the Ty particle of retroelements (Beliakova-Bethell et al. 2006; Beckham and Parker 2008). More recently, it was demonstrated that vRNA packaging is initiated by the targeting of Gag to a subset of host RNA granules containing unspliced HIV-1 RNA (Barajas et al. 2018). During HIV-2 infection, the HIV-2 genomic RNA accumulates in SGs in the absence of active translation prior to being selected for packaging by the HIV-2 Gag polyprotein (Soto-Rifo et al. 2014). This report demonstrates that HIV-1 requires Staufen1, both to suppress SG assembly and to avert vRNA trafficking to visible, type I SGs. It is likely that the vRNA routes to poorly characterized but HIV-1–specific RNP granules (e.g., SHRNPs) for optimal vRNA packaging.
A deficiency of Staufen1 also has marked effects on vRNA translation. The decrease in Gag expression and reduced viral release observed in the SKO cells could be the result of the mistrafficking of the intracellular vRNA, with more of the total vRNA becoming encapsidated instead of being translated. Moreover, the impaired cellular translation as a result of eIF2α phosphorylation, along with the impaired ability of HIV-1 to inhibit the cellular stress response in the SKO cells, could also contribute to reduced viral gene expression and viral release. Recent work demonstrates that the recruitment of RNAs to SGs is dependent on the disengagement of ribosomes from the mRNA (Khong and Parker 2018). The amino-terminal region of Staufen1 binds to ribosomal subunits to stabilize polysomes on translating mRNAs, including the HIV-1 vRNA (Thomas et al. 2009; Rao et al. 2018). Therefore, in the absence of Staufen1, the stability of the ribosomes on the vRNA might be compromised, resulting in the mistrafficking of the vRNA into SGs rather than their translation. This further highlights the role of Staufen1 in vRNA translation (Dugre-Brisson et al. 2005; Abrahamyan et al. 2010; Rao et al. 2018).
Rescue of Staufen1 expression in SKO cells resulted in the reestablishment of the ability of HIV-1 to dissociate Ars-induced SGs and a reduction in eIF2α phosphorylation (Figs. 1, 3A), supporting a role for Staufen1 in translational derepression (Abrahamyan et al. 2010; Rao et al. 2018). Staufen1 also competes with the interferon (IFN)-inducible host protein PKR and prevents PKR-mediated eIF2α-phosphorylation and subsequent translational shutdown, highlighting another role for Staufen1 in translational derepression (Elbarbary et al. 2013; Dixit et al. 2016). However, Staufen1 rescue was unable to restore intracellular Gag expression and the amount of virus released into the supernatant (Figs. 3A, 4A). Rescue of Staufen1 led to increased vRNA encapsidation and a decrease in viral infectivity, further demonstrating that the balance of vRNA translation and encapsidation is a tightly regulated process that requires conserved levels of constitutively expressed Staufen1 in host cells (Huthoff and Berkhout 2001).
Overall, this work highlights the integral role of Staufen1 in the generation of infectious HIV-1 particles, in routing the genomic RNA in the cell and sheds light on the dynamic intracellular interplay between Staufen1, the vRNA and Gag toward the regulation of viral gene expression.
MATERIALS AND METHODS
Cell culture and transfection conditions
Staufen1 knockout (SKO) cells were generated by transfecting HCT116 cells with a plasmid encoding GFP, Cas9, and sgRNA targeting exon 5 of the Staufen1 gene (Horizon Discovery) and using Lipofectamine 3000 (Life Technologies/Thermo Fisher). Forty-eight hours posttransfection, GFP positive cells were sorted by FACS, and single cells were expanded into colonies in 96-well plates. Loss of Staufen1 expression in cells was monitored by western blotting using anti-Staufen1 antibody and by Polymerase chain reaction (PCR). Cells were maintained in McCoy's Media (Life Technologies/Thermo Fisher) supplemented with 1% penicillin/streptomycin and 10% fetal bovine serum (Hyclone). SKO cells grew normally with no apparent phenotype (S Hassine and L DesGroseillers, in prep.). Cells were transfected with 1 µg of total DNA per 4 × 105 cells using JetPrime (PolyPlus transfections) according to the manufacturer's instructions. Twenty-four hours after transfection, cells were fixed or lysed. Cells were treated with 500 μM arsenite (NaAs02; Sigma-Aldrich) for 1 h (Cinti et al. 2017).
Plasmids
pNL4.3, a proviral HIV-1 clone that encodes the full-length RNA genome, the structural and enzyme proteins Gag, Gag/Pol, envelope, the complete set of auxiliary (Vpr, Vpu, Vif) and regulatory genes (tat, rev, nef), was obtained from the NIH AIDS reagent program. The Staufen1 cDNA expression construct pCMV-Staufen1-YFP used to rescue Staufen1 in trans was previously described (Chatel-Chaix et al. 2004). pNL4.3/Staufen1-HA and pNL4.3/Staufen1F135A-HA were previously described (Chatel-Chaix et al. 2004) and contain either the WT Staufen1 or dsRBD3 mutant cDNA fused to the HA epitope tag in place of the Nef coding sequence. pcDNA3.1 used in mock conditions was purchased from Invitrogen.
Antibodies
Rabbit anti-Staufen1 antiserum generated to the full-length recombinant protein was produced and purified at the McGill University Cell Imaging and Analysis Network, and was used at 1:1000 for western blotting; mouse anti-p24 (clone 183-H12-5C) (NIH AIDS Reference and Reagent Program) was used to identify Gag at 1:400 for indirect immunofluorescence and at 1:10,000 for western blotting; and goat anti-TIAR1 (Santa-Cruz Biotechnology) was used for indirect immunofluorescence microscopy at 1:500; rabbit anti-phospho-eIF2α (Ser51) (Cinti et al. 2016) (Cell Signaling Technology) was used for indirect immunofluorescence microscopy at 1:200 and for western blotting at 1:1000; goat anti-eIF3b (Santa-Cruz Biotechnology) 1:250, as described by Valiente-Echeverría et al. (2014); mouse anti-eIF2α (Cell Signaling Technology) and mouse anti-actin (Abcam) were used for western blotting at 1:1000 and at 1:10,000, respectively. Horseradish peroxidase-conjugated secondary antibodies were purchased from Rockland Immunochemicals, AlexaFluor secondary antibodies were purchased from Life Technologies, and were used as recommended by the manufacturers.
Immunofluorescence, fluorescence in situ hybridization, and imaging analyses
Following transfection, cells were washed once in Dulbecco's phosphate-buffered saline (DPBS) (Thermo Fisher Scientific) and fixed with 4% paraformaldehyde for 20 min. Cells were then washed with DPBS, incubated in 0.1 M glycine for 10 min, washed with DPBS, incubated in 0.2% Triton X-100 for 5 min, and washed in DPBS. A digoxigenin-labeled RNA probe targeting the vRNA was synthesized in vitro in the presence of digoxigenin-labeled UTP (Roche) as described by Vyboh et al. (2012). To stain the vRNA, cells were DNase (Invitrogen) treated for 15 min (25 U per coverslip), then incubated in hybridization solution for 16–18 h at 42°C (50% formamide, 1 mg/mL tRNA, 2× SSPE, 5× Denharts, 5 U RNaseOut [Invitrogen], 50 ng probe). Cells were then incubated in 50% formamide for 15 min at 42°C and incubated twice in 2× SSPE for 5 min each at 42°C. Cells were briefly washed in PBS before being blocked in 1× blocking solution (Roche). Primary antibodies were applied for 1 h at 37°C and then washed for 10 min in DPBS followed by secondary antibodies for 1 h. Cells were washed for 20 min in DPBS before being mounted on glass slides using ProLong Gold Antifade Reagent with DAPI (Life Technologies). Negative isotype-matched antibodies were used to control staining specificity. Confocal laser scanning microscopy was performed using a Leica DM16000B microscope equipped with a WaveFX spinning disk confocal head (Quorum Technologies), and images were acquired with a Hamamatsu ImageEM EM-charge coupled device camera. Scanning was performed and digitized at a resolution of 1024 × 1024 pixels. Filter sets and laser wavelengths were described earlier (Monette et al. 2011; Valiente-Echeverría et al. 2014). Image processing and analyses were performed by Imaris software (version 8.4.1 Bitplane/Andor) or by MetaXpress software (Molecular Devices). All imaging experiments were performed at least three times. The observed phenotypes were representative of n > 50 cells per condition from each experiment. SGs were defined as large G3BP1 or T-cell-restricted intracellular antigen-related protein (TIAR1) foci measuring >0.5 μm, and a cell was deemed as SG positive when it exhibited at least three or more SGs (Gilks et al. 2004).
Western blotting
Cells were collected following transfection, washed with DPBS (Corning), and lysed in ice-cold lysis buffer (100 mM NaCl, 10 mM Tris, pH 7.5, 1 mM EDTA, 0.5% Nonidet P-40, protease and phosphatase inhibitor cocktail [Roche]). Cell lysates were quantified by the Bradford assay (Bio-Rad), and 20 µg of lysates were denatured in Laemmli sample buffer and incubated for 5 min at 95°C. The proteins were separated by SDS-PAGE and transferred onto nitrocellulose membranes (Bio-Rad). Membranes were blocked with 5% nonfat milk in Tris-buffered saline pH 7.4 and 0.5% Tween 20 (TBST) and then incubated with primary antibodies. Following washes using TBST, the membranes were incubated with horseradish peroxidase conjugated secondary antibodies (Rockland Immunochemicals) and detected using Western Lightning Plus-ECL reagent (Perkin-Elmer). Signal intensity and densitometry analyses were performed using ImageJ (NIH).
Nucleic acid extraction, reverse transcription, and PCR analysis
Intracellular and supernatant RNA extraction were performed using TRIzol and TRIzol LS Reagent, respectively (Thermo Fisher Scientific) following manufacturer's instructions. cDNA was obtained using the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems). cDNA and primers were then added to GoTaq Green Master Mix (Promega). GAPDH was amplified using the primers GAPDH_1 forward 5′-TGACCACAGTCCATGCCATC-3′ and GAPDH_1 reverse 5′-ATGATGTTCTGGAGAGCCCC-3′, and HIV-1 vRNA using the primers pNL4-3_1 forward 5′-GGGAGCTAGAACGATTCGCA-3′ and pNL4-3_1 reverse 5′-GGATGGTTGTAGCTGTCCCA-3′. The PCR products were visualized on a 1% agarose gel by staining the DNA with RedSafe Nucleic Acid Staining Solution (iNtRON). Signals were captured using a Gel Doc System and intensities were normalized to the GAPDH signal.
Quantification of virus in supernatants
Forty-eight hours posttransfection, cell culture supernatants were harvested and passed through a 0.2 µm filter (VWR) to remove cellular debris and centrifuged at 20,000 rpm for 1 h. The pellet containing the virus was resuspended in 200 µL RPMI and the levels of p24 were determined by ELISA (Perkin-Elmer).
Infectivity assay
Viral titers of cell supernatants were quantified using the X-gal staining assay in TZM-bl cells as described in Xing et al. (2016). Briefly, dilutions of supernatants of each condition were added to TZM-bl cells seeded onto 96-well plates (Corning). After 48 h, cells were fixed with 1% paraformaldehyde, washed, and treated with X-Gal for the detection of β-galactosidase by counting blue TZM-bl cells.
Statistical analysis
All data presented are representative of at least three independent experiments and are presented as the mean ± standard deviation (SD). A P-value ≤0.05 in one-way ANOVA test was considered statistically significant, as indicated in the figure legends. GraphPad Prism 6 (GraphPad Software Inc.) was used for statistical analyses and graph creation.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
We thank Meijuan Niu for expert technical expertise, Maureen Oliviera, Llinca Ibanescu, Cesar Collazos, and Bonnie Spira for p24 ELISAs and the National Institutes of Health Reference and Reagent program for antibodies and reagents used in this study. This work was supported by grant MOP-56974 from the Canadian Institutes of Health Research (CIHR) (to A.J.M.) and by grant MOP-229979 from the CIHR to L.D.G. S.R. and R.A. were supported in part by The Canadian HIV Cure Enterprise Team grant HIG-133050 (to A.J.M.) from the CIHR in partnership with the Canadian Foundation for HIV-1/AIDS Research and International AIDS Society. The funders had no role in study design, data collection, and interpretation, or the decision to submit the work for publication.
Author contributions: S.R. and A.J.M. conceived the study and designed experiments. S.H. and L.D.G. generated the Staufen1−/− HCT116 cell line and provided methodological expertise for edited cell lines. S.R. conducted experiments shown in Figures 1A,B, 2A, 3, and 4; A.M. conducted experiments shown in Figures 1C,D, and Supplemental Figure S1; R.A. conducted experiments in Figure 2B; S.R., S.H., A.M., L.D., and A.J.M. drafted the manuscript; and S.R., A.M., and A.J.M. revised the manuscript. All authors read and approved the final manuscript.
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.069351.118.
REFERENCES
- Abbink TE, Berkhout B. 2003. A novel long distance base-pairing interaction in human immunodeficiency virus type 1 RNA occludes the Gag start codon. J Biol Chem 278: 11601–11611. 10.1074/jbc.M210291200 [DOI] [PubMed] [Google Scholar]
- Abrahamyan L, Chatel-Chaix L, Ajamian L, Milev M, Monette A, Clément JF, Song R, Lehmann M, DesGroseillers L, Laughrea M, et al. 2010. Novel Staufen1 ribonucleoproteins prevent formation of stress granules but favour encapsidation of HIV-1 genomic RNA. J Cell Sci 123: 369–383. 10.1242/jcs.055897 [DOI] [PubMed] [Google Scholar]
- Anderson P, Kedersha N. 2009. RNA granules: post-transcriptional and epigenetic modulators of gene expression. Nat Rev Mol Cell Biol 10: 430–436. 10.1038/nrm2694 [DOI] [PubMed] [Google Scholar]
- Balachandran S, Roberts PC, Brown LE, Truong H, Pattnaik AK, Archer DR, Barber GN. 2000. Essential role for the dsRNA-dependent protein kinase PKR in innate immunity to viral infection. Immunity 13: 129–141. 10.1016/S1074-7613(00)00014-5 [DOI] [PubMed] [Google Scholar]
- Barajas BC, Tanaka M, Robinson BA, Phuong DJ, Chutiraka K, Reed JC, Lingappa JR. 2018. Identifying the assembly intermediate in which Gag first associates with unspliced HIV-1 RNA suggests a novel model for HIV-1 RNA packaging. PLoS Pathog 14: e1006977 10.1371/journal.ppat.1006977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckham CJ, Parker R. 2008. P bodies, stress granules, and viral life cycles. Cell Host Microbe 3: 206–212. 10.1016/j.chom.2008.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beliakova-Bethell N, Beckham C, Giddings TH Jr, Winey M, Parker R, Sandmeyer S. 2006. Virus-like particles of the Ty3 retrotransposon assemble in association with P-body components. RNA 12: 94–101. 10.1261/rna.2264806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butsch M, Boris-Lawrie K. 2000. Translation is not required to generate virion precursor RNA in human immunodeficiency virus type 1-infected T cells. J Virol 74: 11531–11537. 10.1128/JVI.74.24.11531-11537.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butsch M, Boris-Lawrie K. 2002. Destiny of unspliced retroviral RNA: ribosome and/or virion? J Virol 76: 3089–3094. 10.1128/JVI.76.7.3089-3094.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatel-Chaix L, Clement JF, Martel C, Beriault V, Gatignol A, DesGroseillers L, Mouland AJ. 2004. Identification of Staufen in the human immunodeficiency virus type 1 Gag ribonucleoprotein complex and a role in generating infectious viral particles. Mol Cell Biol 24: 2637–2648. 10.1128/MCB.24.7.2637-2648.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatel-Chaix L, Abrahamyan L, Frechina C, Mouland AJ, DesGroseillers L. 2007. The host protein Staufen1 participates in human immunodeficiency virus type 1 assembly in live cells by influencing pr55Gag multimerization. J Virol 81: 6216–6230. 10.1128/JVI.00284-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatel-Chaix L, Boulay K, Mouland AJ, Desgroseillers L. 2008. The host protein Staufen1 interacts with the Pr55Gag zinc fingers and regulates HIV-1 assembly via its N-terminus. Retrovirology 5: 41 10.1186/1742-4690-5-41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cinti A, Le Sage V, Ghanem M, Mouland AJ. 2016. HIV-1 Gag blocks selenite-induced stress granule assembly by altering the mRNA cap-binding complex. mBio 7: e00329 10.1128/mBio.00329-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cinti A, Le Sage V, Milev MP, Valiente-Echeverría F, Crossie C, Miron MJ, Panté N, Olivier M, Mouland AJ. 2017. HIV-1 enhances mTORC1 activity and repositions lysosomes to the periphery by co-opting Rag GTPases. Sci Rep 7: 5515 10.1038/s41598-017-05410-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixit U, Pandey AK, Mishra P, Sengupta A, Pandey VN. 2016. Staufen1 promotes HCV replication by inhibiting protein kinase R and transporting viral RNA to the site of translation and replication in the cells. Nucleic Acids Res 44: 5271–5287. 10.1093/nar/gkw312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dugre-Brisson S, Elvira G, Boulay K, Chatel-Chaix L, Mouland AJ, DesGroseillers L. 2005. Interaction of Staufen1 with the 5′ end of mRNA facilitates translation of these RNAs. Nucleic Acids Res 33: 4797–4812. 10.1093/nar/gki794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbarbary RA, Li W, Tian B, Maquat LE. 2013. STAU1 binding 3′ UTR IRAlus complements nuclear retention to protect cells from PKR-mediated translational shutdown. Genes Dev 27: 1495–1510. 10.1101/gad.220962.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimura K, Sasaki AT, Anderson P. 2012. Selenite targets eIF4E-binding protein-1 to inhibit translation initiation and induce the assembly of non-canonical stress granules. Nucleic Acids Res 40: 8099–8110. 10.1093/nar/gks566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilks N, Kedersha N, Ayodele M, Shen L, Stoecklin G, Dember LM, Anderson P. 2004. Stress granule assembly is mediated by prion-like aggregation of TIA-1. Mol Biol Cell 15: 5383–5398. 10.1091/mbc.e04-08-0715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huthoff H, Berkhout B. 2001. Two alternating structures of the HIV-1 leader RNA. RNA 7: 143–157. 10.1017/S1355838201001881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaye JF, Lever AM. 1999. Human immunodeficiency virus types 1 and 2 differ in the predominant mechanism used for selection of genomic RNA for encapsidation. J Virol 73: 3023–3031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha NL, Gupta M, Li W, Miller I, Anderson P. 1999. RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2 α to the assembly of mammalian stress granules. J Cell Biol 147: 1431–1442. 10.1083/jcb.147.7.1431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khong A, Parker R. 2018. mRNP architecture in translating and stress conditions reveals an ordered pathway of mRNP compaction. J Cell Biol 217: 4124–4140. 10.1083/jcb.201806183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd RE. 2012. How do viruses interact with stress-associated RNA granules? PLoS Pathog 8: e1002741 10.1371/journal.ppat.1002741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallardo M, Deitinghoff A, Muller J, Goetze B, Macchi P, Peters C, Kiebler MA. 2003. Isolation and characterization of Staufen-containing ribonucleoprotein particles from rat brain. Proc Natl Acad Sci 100: 2100–2105. 10.1073/pnas.0334355100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milev MP, Brown CM, Mouland AJ. 2010. Live cell visualization of the interactions between HIV-1 Gag and the cellular RNA-binding protein Staufen1. Retrovirology 7: 41 10.1186/1742-4690-7-41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milev MP, Ravichandran M, Khan MF, Schriemer DC, Mouland AJ. 2012. Characterization of staufen1 ribonucleoproteins by mass spectrometry and biochemical analyses reveal the presence of diverse host proteins associated with human immunodeficiency virus type 1. Front Microbiol 3: 367 10.3389/fmicb.2012.00367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monette A, Panté N, Mouland AJ. 2011. HIV-1 remodels the nuclear pore complex. J Cell Biol 193: 619–631. 10.1083/jcb.201008064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouland AJ, Mercier J, Luo M, Bernier L, DesGroseillers L, Cohen EA. 2000. The double-stranded RNA-binding protein Staufen is incorporated in human immunodeficiency virus type 1: evidence for a role in genomic RNA encapsidation. J Virol 74: 5441–5451. 10.1128/JVI.74.12.5441-5451.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poblete-Durán N, Prades-Pérez Y, Vera-Otarola J, Soto-Rifo R, Valiente-Echeverría F. 2016. Who regulates whom? An overview of RNA granules and viral infections. Viruses 8: E180 10.3390/v8070180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon DT, Chertova EN, Ott DE. 2002. Human immunodeficiency virus type 1 preferentially encapsidates genomic RNAs that encode Pr55Gag: functional linkage between translation and RNA packaging. Virology 293: 368–378. 10.1006/viro.2001.1283 [DOI] [PubMed] [Google Scholar]
- Ramos A, Grünert S, Adams J, Micklem DR, Proctor MR, Freund S, Bycroft M, St Johnston D, Varani G. 2000. RNA recognition by a Staufen double-stranded RNA-binding domain. EMBO J 19: 997–1009. 10.1093/emboj/19.5.997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao S, Cinti A, Temzi A, Amorim R, You JC, Mouland AJ. 2018. HIV-1 NC-induced stress granule assembly and translation arrest are inhibited by the dsRNA binding protein Staufen1. RNA 24: 219–236. 10.1261/rna.064618.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soto-Rifo R, Valiente-Echeverría F, Rubilar PS, Garcia-de-Gracia F, Ricci EP, Limousin T, Décimo D, Mouland AJ, Ohlmann T. 2014. HIV-2 genomic RNA accumulates in stress granules in the absence of active translation. Nucleic Acids Res 42: 12861–12875. 10.1093/nar/gku1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas MG, Martinez Tosar LJ, Desbats MA, Leishman CC, Boccaccio GL. 2009. Mammalian Staufen 1 is recruited to stress granules and impairs their assembly. J Cell Sci 122: 563–573. 10.1242/jcs.038208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas MG, Loschi M, Desbats MA, Boccaccio GL. 2011. RNA granules: the good, the bad and the ugly. Cell Signal 23: 324–334. 10.1016/j.cellsig.2010.08.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tosar LJ, Thomas MG, Baez MV, Ibanez I, Chernomoretz A, Boccaccio GL. 2012. Staufen: from embryo polarity to cellular stress and neurodegeneration. Front Biosci (Schol Ed) 4: 432–452. 10.2741/s277 [DOI] [PubMed] [Google Scholar]
- Valiente-Echeverría F, Melnychuk L, Mouland AJ. 2012. Viral modulation of stress granules. Virus Res 169: 430–437. 10.1016/j.virusres.2012.06.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valiente-Echeverría F, Melnychuk L, Vyboh K, Ajamian L, Gallouzi IE, Bernard N, Mouland AJ. 2014. eEF2 and Ras-GAP SH3 domain-binding protein (G3BP1) modulate stress granule assembly during HIV-1 infection. Nat Commun 5: 4819 10.1038/ncomms5819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vyboh K, Ajamian L, Mouland AJ. 2012. Detection of viral RNA by fluorescence in situ hybridization (FISH). J Vis Exp: e4002 10.3791/4002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing L, Wang S, Hu Q, Li J, Zeng Y. 2016. Comparison of three quantification methods for the TZM-bl pseudovirus assay for screening of anti-HIV-1 agents. J Virol Methods 233: 56–61. 10.1016/j.jviromet.2016.03.008 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.