Abstract
Pathologic grading plays a key role in prostate cancer risk stratification and treatment selection, traditionally assessed from systemic core needle biopsies sampled throughout the prostate gland. Multiparametric magnetic resonance imaging (mpMRI) has become a well-established clinical tool for detecting and localizing prostate cancer. However, both pathologic and radiologic assessment suffer from poor reproducibility among readers. Artificial intelligence (AI) methods show promise in aiding the detection and assessment of imaging-based tasks, dependent on the curation of high-quality training sets. This review provides an overview of recent advances in AI applied to mpMRI and digital pathology in prostate cancer which enable advanced characterization of disease through combined radiology-pathology assessment.
Prostate cancer is the sixth most common cancer worldwide and the second leading cause of cancer death in American men (1). Prostate cancer has a diverse range of risk based upon disease presentation, ranging from low-risk indolent to high-risk aggressive disease. Early detection and localization of prostate cancer at a treatable stage is important as excellent cancer-specific survival is expected for most locally confined diseases. The most common method to diagnose prostate cancer uses transrectal ultrasonography (TRUS) to sample twelve regions distributed throughout the prostate, most often recommended after an elevated prostate specific antigen (PSA) blood test or an abnormal digital rectal exam (2). Common treatments for patients with localized intermediate and high-risk disease include radical prostatectomy and radiation therapy, whereas active surveillance is commonly considered for patients harboring low-risk disease. Selecting among these treatment options is difficult due to the clinical and biologic heterogeneity of prostate cancer disease.
Clinical risk assessment is primarily determined by pathologic grade based on the Gleason grading system, as initially proposed in the 1960s and modified multiple times thereafter (3). While the Gleason grading system has limitations, it generally captures the full range of biologic aggressiveness and has historically correlated with the risk of recurrent cancer after definitive treatment (4). Gleason pattern 3 consists of well-formed, individual glands of various sizes. Cancers containing only Gleason pattern 3 (i.e., overall Gleason score assignment 3+3) are generally considered low risk. Gleason pattern 4 includes poorly formed, fused, and cribriform glands. Within the spectrum of intermediate-risk disease, the presence of cribriform pattern in radical prostatectomy specimens has shown association to poor surgical and clinical endpoints, including higher rates of extra-prostatic extension, positive surgical margins, biochemical recurrence, and cancer-specific mortality (5, 6). Gleason pattern 5 consists of high-risk histologic patterns including sheets of tumor, individual cells, and cords of cells. In general, tumors containing only Gleason 3+3 have a minimal risk of progression to metastatic cancer, while tumors with dominant Gleason patterns 4 and 5 cancers carry a higher likelihood of progression to metastatic disease and cancer-specific mortality. Prostate cancer is known to exhibit heterogeneous distribution of morphologies both within and across tumors of the same patient, with increasing number of observed morphologies in larger cancers of worsening grade (7). Due to this diversity, the final grade as reported by the Gleason system is based on the combination of the most dominant and second most dominant patterns observed. In some settings a tertiary pattern, otherwise defined as a minor component of high-grade cancer, is reported in combination with the overall Gleason score when a small population of high-grade glands are observed upon evaluation of radical prostatectomy specimens. Although it is central in risk assessment and patient management for localized prostate cancer, the Gleason scoring system is known to suffer from high variability across pathologists, and even within the same pathologist, due to the subjective nature of scoring assignment (8, 9).
Multiparametric magnetic resonance imaging (mpMRI) is well-established as an important aide for prostate cancer diagnosis. mpMRI provides high-contrast, high-resolution anatomical images of the prostate and pelvic regions using T2-weighted imaging, diffusion-weighted imaging (DWI), and dynamic contrast enhanced (DCE) imaging sequences for the detection of clinically significant prostate cancers (10). However, the combined assessment and interpretation of all mpMRI sequences requires a considerable level of expertise from radiologists. The current classification of abnormal regions on mpMRI imaging is defined by the Prostate Imaging Reporting and Detection System version 2 (PI-RADSv2) (11). This system, while widely accepted, is also prone to inter-reader variability, leading to variable sensitivity, specificity, and a wide range of cancer detection rates (12–14). Despite these shortcomings, mpMRI has shown practice-changing value for urologists to more accurately direct needle biopsies into suspicious regions of the prostate and superior performance in disease sampling compared with TRUS biopsy techniques (15–17). However, even with targeting based on mpMRI findings, biopsy sampling error and tumor heterogeneity leads to inaccurate pathologic characterization of final whole-organ assessment after surgery.
Unique from many other primary cancer models, the surgical practice of radical removal of the entire prostate gland in localized disease allows for 1:1 spatial correspondence between radiologic imaging and pathologic imaging. This has led to a large body of literature characterizing tissue characteristics with functional imaging characteristics of mpMRI, most prominently using DWI imaging characteristics such as the apparent diffusion coefficient (ADC). Restriction of water molecules leads to differential signal properties of DWI, specifically reflecting the increased cellular density and nuclear-to-cytoplasmic ratio disturbed in cancerous tissues. In prostate cancer, several studies have established a negative relationship between ADC values and tumor Gleason score (18–20). However, the outright use of quantitative DWI characteristics for lesion detection is limited by the lack of clear distinction in ADC values of normal and tumor regions (21). Furthermore, the relationship between ADC and tumor-level Gleason score overall remains moderate, likely due to local heterogeneity in the proportion (density) and spatial distribution (sparsity) of cancerous cells within the tumor (22). Truong et al. (23) have reported preliminary evidence that advanced morphologic features, such as cribriform, influences the visibility of prostate lesions more so than tumors of the same grade with differing architectural patterns.
Overall, the complex heterogeneity of localized prostate cancer has led to variability in clinical practice for both radiology and pathology. The use of artificial intelligence (AI) applications in both radiology and pathology has increased substantially in the last decade (24), including more recently with the adoption of deep learning techniques applied to medical imaging. Here, we provide a summary of recent AI applications to radiology, pathology, and combined radiology-pathology characterization of localized prostate cancer where the intrinsic connection between histology and functional imaging in prostate cancer motivates the use of combined techniques for enhanced characterization.
Current uses of artificial intelligence in localized prostate cancer
Radiologic applications
In general, AI shows great promise in decreasing reader interpretation times, increasing performance of non-expert radiologists, and enabling large-scale screening practices without additional burden to radiologists. Applications of AI to prostate mpMRI are specifically expected to increase sensitivity of prostate cancer detection and decrease inter-reader variability (25). There are several examples of AI-based systems for prostate mp-MRI in the literature. In general, mp-MRI based AI tasks either focus on automated detection, aiming to localize suspicious regions within an image set, or automated diagnostic classification, aiming to predict the aggressiveness of a region of interest.
Traditional machine-learning based detection systems typically require several pre-processing steps, including extraction of prostate gland regions and quantitative metrics from MRI sequences. To date, several studies have evaluated in-house AI algorithms developed for automated detection on prostate cancer on mpMRI (26–31). Despite differences in terms of features used for voxel and candidate region classification, MRI modalities, and methods used for classification and multimodal fusion, these studies demonstrate robust detection on the order of 75% to 80% or more, within the range of reported radiologist performance (11). In a recent multi-reader, multi-institutional study, Gaur et al. (32) have shown in their multi-institution study that AI-based detection, from algorithm presented in (33), improved specificity in conjunction with PI-RADSv2 categorization as well as slightly improved radiologist efficiency, and they found an index lesion sensitivity for PIRADSv2 ≥3 of 78%. Here the greatest benefit was seen in the TZ where it helped moderately experienced readers to achieve 83.8% sensitivity with automated detection versus 66.9% with mpMRI alone. Litjens et al. (28) were able to demonstrate the combining AI-based prediction and PI-RADS improved both detection of cancer, as well as discrimination of clinically revelant (i.e., aggressive) disease.
While fewer studies have reported on the use of deep learning algorithms for automated detection of prostate cancer on mpMRI, early literature supports improved detection rates compared with previous works (34–36). Notably, Song et al. (37) report high diagnostic performance of 87% sensitivity in a cohort of 195 patients. Here, combination of automated detection and with expert radiologist PI-RADSv2 classification improved the detection of clinically significant cancer than with the radiologist alone. Multi-task algorithms, such as the method reported by Alkadi et al. (38), combine prostate gland segmentation for spatial context with lesion detection for improved performance. Yang et al. (39) report novel work demonstrating separate development and training on T2-weighted and ADC images improved detection performance when compared with methods learning across image sets. As the literature matures, it is expected that further increase in performance will be observed.
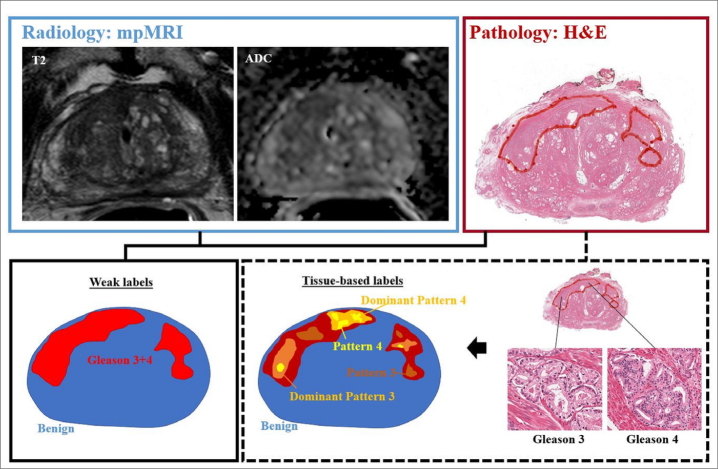
While these examples utilize various techniques and imaging features, they have all demonstrated promising ability to detect prostate cancer on mpMRI. However, they are inherently limited by training against a pathology validation set in which only a single Gleason score is assigned to each lesion, so called “weak labels”, that underestimate known intra-lesion heterogeneity (Fig.). This has resulted in underperformance of algorithms for certain pathologic grades (33). For these reasons, current AI algorithms are not ideally suited for intra-lesion spatial targeting of aggressive regions on mpMRI at biopsy. Ideally, voxel-based classifiers such as those previously described should be used to train the algorithm against a system that defines each “patch” of tumor in terms of its regional Gleason score to allow for more accurate spatial learning.
Figure.
Example mpMRI (T2-weighted and ADC images) with corresponding whole mount hematoxylin-eosin (H&E) section demonstrating two pathologically-defined tumors. Overall assessment in this patient resulted in Gleason grade 3+4 score assignment, resulting in weak labels derived from total tumor extent. However, detailed pathologic assessment demonstrates majority of Gleason pattern 4 is located in the anterior portion of the tumor, with predominately Gleason pattern 3 throughout remainder of the tumor field. Opportunities for improved spatial learning include density mapping of dominant pathologic grading and exclusion of non-cancerous structures within tumor field.
AI-enabled pathology-to-radiology correlation
To improve spatial labeling at the mp-MRI level, detailed pathologic information of each lesion would theoretically be required as input. Spatial annotation of this level is impractical due to the time-consuming nature of spatially annotating at the full resolution of digital pathology images, which can range up to 10 GB on the order of 100 000×100 000 pixels per section for whole mount prostate specimens (40). For these reasons, historical applications of AI techniques have included approaches for semantic segmentation of histology slides, such as automated detection and segmentation of all nuclei and tissue components to allow for further classification and/or analysis of diseased regions (41).
Commonly used machine learning workflows in prostate cancer rely heavily on semantic segmentation of epithelial, stromal, and lumen components on tissue slides due to the varying representation of each within normal and malignant structures (42). Gertych et al. (43) developed machine learning techniques for differentiating stroma and epithelial components, followed by classifier of benign and malignant regions using quantitative features derived from epithelial components. Gorelick et al. (44) reported improved classification of malignant tissue when including segmentation of lumen. Others have focused on segmentation of entire glandular structures (45), which have been used to enable morphology quantification for further classification of malignant tissues (46, 47). More recently, Li et al. (48) were able to demonstrate that an expectation-maximization approach to deep learning can provide improved gland segmentation in prostate cancer.
These applications have additionally shown promise for serving as pre-processing steps to enable more complex histopathologic spatial analysis with radiologic imaging. Kwak et al. (31) have shown that MRI signal characteristics are significantly associated with tissue composition density derived from automated tissue-level compartment segmentation of stroma, epithelium, epithelial nuclei, and lumen. In a similar analysis, McGarry et al. (49) shows that radiopathomic maps of epithelium and lumen density can be used to distinguish cancerous regions of varying grade. The use of AI for correlation between pathologic features and radiologic signature has shown that both histologic grade and mpMRI diffusion characteristics correlate more strongly with glandular components and crowding than with nuclear count and crowding (50–52). Chatterjee et al. (50) demonstrated that differences between cancers with Gleason patterns 3, 4, and 5 were also greater for the gland component volumes than for the cellularity metrics. Meanwhile, Hectors et al. (51) were able to establish a stronger relationship using advanced diffusion models mimicking human tissue. Importantly, establishing the relationship between tissue components and imaging signatures across these studies were achieved utilizing multiple different algorithms applied to pathologic data, providing strong evidence for distinct biophysical foundations for radiologic signatures and motivating utility and development of new imaging techniques (53).
Advanced pathologic applications
In several cancers, automated detection and quantification of tissue components demonstrate utility in predicting patient outcome. For example, nuclear and stromal features have shown correlation to patient prognosis in breast cancer (54, 55). Tumor infiltrating lymphocyte detection via deep learning models allows for spatial analysis of immune profiling, demonstrating independent prediction of treatment outcomes (56). However, these techniques so far have not demonstrated value in prostate cancer, where the major focus has turned to automated Gleason grading on digital pathologic assessment.
Despite increasing popularity of machine learning applications to complex medical imaging problems, automated algorithms for Gleason grading demonstrate limited success largely because of the subjectivity of the system (57–59). This includes sub-optimal prediction in binary classification of low- versus high-risk disease, with some studies showing highest error rates in the group of intermediate-to-high risk prostate cancers (57). However, these machine learning methods are highly sensitive to multiple pre-processing and feature extraction steps. Traditional machine learning methods have relied on building classifiers from handcrafted features derived in relatively homogeneous settings, such as those derived in tissue microarrays. This leads to lack of generalizability to whole slide images of heterogeneous tissue components. More recent advances in deep learning applications have shown promise for improved performance of prostate cancer grading algorithms.
Work by Nir et al. (60) evaluating multiple machine and deep learning methods demonstrate best results of 92% accuracy in cancer detection and 79% accuracy in classification of low and high grade prostate cancers when trained on tissue micro-array data, though agreement with expert pathologists was moderately low and overall system accuracy suffered when validated in an external dataset of whole-mount images. Other preliminary works differentiating low-to-intermediate (Gleason Grade ≤ 3+4) from unfavorable intermediate-to-high risk (Gleason Grade ≥ 4+3) only achieved 75% classification accuracy (61). Improved classification was found when performance was evaluated in a set of well-curated tissue microarrays, where Arvanti et al. (62) reported 70% overall sensitivity in classifying tissues as benign, Grade 3, Grade 4, or Grade 5. Here, Grade 5 achieved the highest per-class recall of 88% in contrast to 58% in Grade 4. Despite this, the deep learning model was able to achieve high agreement with pathologist interpretations (kappa, 0.71–0.75). High level annotations derived within 40 patients were used to build a custom network for both detection and grading of prostate cancer, demonstrating 99% epithelial detection accuracy and 71%–79% accuracy for low- and high-grade disease (63). In the largest study to date, Nagpal et al. (64) have developed and validated their deep learning algorithm against a cohort of 29 board-certified pathologists in Gleason scoring whole-slide images from prostatectomy patients and was able to provide more accurate quantitation of Gleason patterns and better risk stratification for prediction of biochemical recurrence. Even still, performance remains 70% for Gleason grading.
Remaining challenges for AI applications in pathology include stain normalization and color matching (65). Heterogeneity in these properties are underexplored due to the small nature of most datasets currently available. As noted, both machine learning and deep learning suffer in the cohort of intermediate-risk prostate cancers. However, given that morphology and classification pattern tasks vary widely across differing tumor types, direct application of algorithms developed in other tissue types may not translate. Computationally, reading in the entire image is prohibitively expensive and limits most AI applications to patch-based techniques. As technology continues to increase computing power, new opportunities for image-based tasks at full-scale resolution will become available.
Future opportunities for integrated techniques, radiogenomics, and beyond
Currently, both radiology and pathology are limited in their ability to detect and classify intermediate prostate cancers, which represent a critical point in clinical treatment decision-making. Here, opportunities for new integrated techniques combining imaging modalities for outcome prediction could be readily developed as datasets mature. However, as discussed, there is a lack of high-quality annotations and most models are constructed using weakly labeled data ignoring tissue heterogeneity. Improving performance of pathologic classifiers can serve as the basis for high-resolution radiologic labels, whereby pixel-wise predication of prostate cancers can replace current weak labels. Furthermore, the promise of integrated techniques comes with the idea that improved pathologic prediction and grading can lead to improved radiologic prediction.
While outside the scope of this review, new development of AI frameworks demonstrate that proof-of-principle integration of diverse data is feasible. Methods for coupling weak and strong labeled data demonstrated the highest level discrimination of low risk (Gleason ≤ 7) from high risk (Gleason ≥ 8) in digital pathology assessment of prostate cancer specimens (62). Combined machine learning and deep learning techniques for longevity prediction show proof-of-principle for combining multi-dimensional data for outcome prediction (66). Ensemble-based methods of cascaded deep learning algorithms can be readily used to evaluate the utility of combined prediction of radiologic and pathologic prediction for patient outcome or prediction of disease risk, including molecular characterization. Recently, novel integration of pathologic features and genomic characteristics for prediction of cancer outcomes further demonstrates a role for AI methods in integrated precision medicine initiatives (67).
In the combination of multi-modality multi-dimensional data, pathologic imaging and evaluation represents an ideal intermediary step to enable further radiogenomic associations. Radiogenomics, i.e., the correlation of radiographic signatures with genomic features, has been evaluated for prostate cancer in several small studies (68). Following radiogenomic work in glioma by Smedley et al. (69), there is an opportunity to apply deep learning for discovery-based prediction within radiogenomic frameworks. Recently published work demonstrates widespread transcriptome heterogeneity within prostate cancer (70). This formative work by Berglund et al. (71) demonstrates differential gene expression across regions of the tumor microenvironment, particularly in peripheral vs. more central regions of the tumor, of a single patient. However, the degree to which this results in differential growth patterns remains largely unexplored. Preliminary work correlating spatially distinct mpMRI normal and suspicious regions with whole exome sequencing additionally demonstrate heterogeneity within prostate lesions.
Conclusion
In conclusion, there exists a clinical need in prostate cancer for improvement in standardized assessment and characterization on pathologic and radiologic interpretation. The intimate connection between histology and functional imaging characteristics creates a unique opportunity for AI applications to improve detection, classification, and overall prognostication. As demonstrated across AI literature, the need for mature datasets with high quality annotations are necessary to continue advancements in this field.
Main points.
Recent studies show artificial intelligence (AI) has potential to aide in detection and classification of prostate cancer on radiology and pathology imaging.
Current radiology AI applications suffer from training data that does not reflect the heterogeneity in tissue composition observed on pathology.
Pathology-based AI applications enable high quality radiology-pathology correlation and have the potential to serve as input to train improved radiology-based AI algorithms.
Footnotes
Financial disclosure
This project has been funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under Contract No. HHSN261200800001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Conflict of interest disclosure
The authors declared no conflicts of interest.
References
- 1.Grönberg H. Prostate cancer epidemiology. Lancet. 2003;361:859–864. doi: 10.1016/S0140-6736(03)12713-4. [DOI] [PubMed] [Google Scholar]
- 2.Costa DN, Pedrosa I, Donato F, Jr, Roehrborn CG, Rofsky NM. MR imaging–transrectal US fusion for targeted prostate biopsies: implications for diagnosis and clinical management. Radiographics. 2015;35:696–708. doi: 10.1148/rg.2015140058. [DOI] [PubMed] [Google Scholar]
- 3.Gleason DF. Classification of prostatic carcinomas. Cancer Chemother Rep. 1966;50:125–128. [PubMed] [Google Scholar]
- 4.Epstein JI, Egevad L, Amin MB, et al. The 2014 International Society of Urological Pathology (ISUP) Consensus Conference on Gleason Grading of Prostatic Carcinoma: Definition of Grading Patterns and Proposal for a New Grading System. Am J Surg Pathol. 2016;40:244–252. doi: 10.1097/PAS.0000000000000530. [DOI] [PubMed] [Google Scholar]
- 5.Kweldam CF, Wildhagen MF, Steyerberg EW, Bangma CH, van der Kwast TH, van Leenders GJ. Cribriform growth is highly predictive for postoperative metastasis and disease-specific death in Gleason score 7 prostate cancer. Mod Pathol. 2015;28:457–464. doi: 10.1038/modpathol.2014.116. [DOI] [PubMed] [Google Scholar]
- 6.Iczkowski KA, Torkko KC, Kotnis GR, et al. Digital quantification of five high-grade prostate cancer patterns, including the cribriform pattern, and their association with adverse outcome. Am J Clin Pathol. 2011;136:98–107. doi: 10.1309/AJCPZ7WBU9YXSJPE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aihara M, Wheeler TM, Ohori M, Scardino PT. Heterogeneity of prostate cancer in radical prostatectomy specimens. Urology. 1994;43:60–67. doi: 10.1016/S0090-4295(94)80264-5. [DOI] [PubMed] [Google Scholar]
- 8.Allsbrook WC, Mangold KA, Johnson MH, Lane RB, Lane CG, Epstein JI. Interobserver reproducibility of Gleason grading of prostatic carcinoma: general pathologist. Hum Pathol. 2001;32:81–88. doi: 10.1053/hupa.2001.21134. [DOI] [PubMed] [Google Scholar]
- 9.Egevad L, Ahmad AS, Algaba F, et al. Standardization of Gleason grading among 337 European pathologists. Histopathology. 2013;62:247–256. doi: 10.1111/his.12008. [DOI] [PubMed] [Google Scholar]
- 10.Ploussard G, Epstein JI, Montironi R, et al. The contemporary concept of significant versus insignificant prostate cancer. Eur Urol. 2011;60:291–303. doi: 10.1016/j.eururo.2011.05.006. [DOI] [PubMed] [Google Scholar]
- 11.Weinreb JC, Barentsz JO, Choyke PL, et al. PI-RADS Prostate Imaging - Reporting and Data System: 2015, Version 2. Eur Urol. 2016;69:16–40. doi: 10.1016/j.eururo.2015.08.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rosenkrantz AB, Ginocchio LA, Cornfeld D, et al. Interobserver reproducibility of the PI-RADS Version 2 Lexicon: a multicenter study of six experienced prostate radiologists. Radiology. 2016;280:793–804. doi: 10.1148/radiol.2016152542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mehralivand S, Bednarova S, Shih JH, et al. Prospective evaluation of PI-RADS™ version 2 using the International Society of Urological Pathology Prostate Cancer Grade Group System. J Urol. 2017;198:583–590. doi: 10.1016/j.juro.2017.03.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Valerio M, Donaldson I, Emberton M, et al. Detection of clinically significant prostate cancer using magnetic resonance imaging-ultrasound fusion targeted biopsy: a systematic review. Eur Urol. 2015;68:8–19. doi: 10.1016/j.eururo.2014.10.026. [DOI] [PubMed] [Google Scholar]
- 15.Siddiqui MM, Rais-Bahrami S, Turkbey B, et al. Comparison of MR/ultrasound fusion-guided biopsy with ultrasound-guided biopsy for the diagnosis of prostate cancer. JAMA. 2015;313:390–397. doi: 10.1001/jama.2014.17942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Borkowetz A, Platzek I, Toma M, et al. Direct comparison of multiparametric magnetic resonance imaging (MRI) results with final histopathology in patients with proven prostate cancer in MRI/ultrasonography-fusion biopsy. BJU Int. 2016;118:213–220. doi: 10.1111/bju.13461. [DOI] [PubMed] [Google Scholar]
- 17.Kasivisvanathan V, Rannikko AS, Borghi M, et al. MRI-targeted or standard biopsy for prostate-cancer diagnosis. N Engl J Med. 2018;378:1767–1777. doi: 10.1056/NEJMoa1801993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gibbs P, Liney GP, Pickles MD, Zelhof B, Rodrigues G, Turnbull LW. Correlation of ADC and T2 measurements with cell density in prostate cancer at 3.0 Tesla. Invest Radiol. 2009;44:572–576. doi: 10.1097/RLI.0b013e3181b4c10e. [DOI] [PubMed] [Google Scholar]
- 19.Tamada T, Sone T, Jo Y, et al. Apparent diffusion coefficient values in peripheral and transition zones of the prostate: comparison between normal and malignant prostatic tissues and correlation with histologic grade. J Magn Reson Imaging. 2008;28:720–726. doi: 10.1002/jmri.21503. [DOI] [PubMed] [Google Scholar]
- 20.Turkbey B, Shah VP, Pang Y, et al. Is apparent diffusion coefficient associated with clinical risk scores for prostate cancers that are visible on 3-T MR images? Radiology. 2011;258:488–495. doi: 10.1148/radiol.10100667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Klotz L, Vesprini D, Sethukavalan P, et al. Longterm follow-up of a large active surveillance cohort of patients with prostate cancer. J Clin Oncol. 2015;33:272–277. doi: 10.1200/JCO.2014.55.1192. [DOI] [PubMed] [Google Scholar]
- 22.Hambrock T, Somford DM, Huisman HJ, et al. Relationship between apparent diffusion coefficients at 3.0-T MR imaging and Gleason grade in peripheral zone prostate cancer. Radiology. 2011;259:453–461. doi: 10.1148/radiol.11091409. [DOI] [PubMed] [Google Scholar]
- 23.Truong M, Hollenberg G, Weinberg E, Messing EM, Miyamoto H, Frye TP. Impact of Gleason subtype on prostate cancer detection using multiparametric magnetic resonance imaging: correlation with final histopathology. J Urol. 2017;198:316–321. doi: 10.1016/j.juro.2017.01.077. [DOI] [PubMed] [Google Scholar]
- 24.Hosny A, Parmar C, Quackenbush J, Schwartz LH, Aerts HJWL. Artificial intelligence in radiology. Nat Rev Cancer. 2018;18:500–510. doi: 10.1038/s41568-018-0016-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Greer MD, Lay N, Shih JH, et al. Computer-aided diagnosis prior to conventional interpretation of prostate mpMRI: an international multi-reader study. Eur Radiol. 2018;28:4407–4417. doi: 10.1007/s00330-018-5374-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hambrock T, Vos PC, Hulsbergen-van de Kaa CA, Barentsz JO, Huisman HJ. Prostate cancer: computer-aided diagnosis with multiparametric 3-T MR imaging--effect on observer performance. Radiology. 2013;266:521–530. doi: 10.1148/radiol.12111634. [DOI] [PubMed] [Google Scholar]
- 27.Lemaitre G, Marti R, Rastgoo M, Meriaudeau F. Computer-aided detection for prostate cancer detection based on multi-parametric magnetic resonance imaging. Conf Proc IEEE Eng Med Biol Soc. 2017;2017:3138–3141. doi: 10.1109/EMBC.2017.8037522. [DOI] [PubMed] [Google Scholar]
- 28.Litjens GJ, Barentsz JO, Karssemeijer N, Huisman HJ. Clinical evaluation of a computer-aided diagnosis system for determining cancer aggressiveness in prostate MRI. Eur Radiol. 2015;25:3187–3199. doi: 10.1007/s00330-015-3743-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Metzger GJ, Kalavagunta C, Spilseth B, et al. Detection of prostate cancer: quantitative multiparametric MR imaging models developed using registered correlative histopathology. Radiology. 2016;279:805–816. doi: 10.1148/radiol.2015151089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peng Y, Jiang Y, Antic T, Giger ML, Eggener SE, Oto A. Validation of quantitative analysis of multiparametric prostate MR images for prostate cancer detection and aggressiveness assessment: a cross-imager study. Radiology. 2014;271:461–471. doi: 10.1148/radiol.14131320. [DOI] [PubMed] [Google Scholar]
- 31.Kwak JT, Xu S, Wood BJ, et al. Automated prostate cancer detection using T2-weighted and high-b-value diffusion-weighted magnetic resonance imaging. Med Phys. 2015;42:2368–2378. doi: 10.1118/1.4918318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gaur S, Lay N, Harmon SA, et al. Can computer-aided diagnosis assist in the identification of prostate cancer on prostate MRI? a multi-center, multi-reader investigation. Oncotarget. 2018;9:33804–33817. doi: 10.18632/oncotarget.26100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lay N, Tsehay Y, Greer MD, et al. Detection of prostate cancer in multiparametric MRI using random forest with instance weighting. J Med Imaging (Bellingham) 2017;4:024506. doi: 10.1117/1.JMI.4.2.024506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu L, Tian Z, Zhang Z, Fei B. Computer-aided detection of prostate cancer with MRI: technology and applications. Acad Radiol. 2016;23:1024–1046. doi: 10.1016/j.acra.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sumathipala Y, Lay N, Turkbey B, Smith C, Choyke PL, Summers RM. Prostate cancer detection from multi-institution multiparametric MRIs using deep convolutional neural networks. J Med Imaging (Bellingham) 2018;5:044507. doi: 10.1117/1.JMI.5.4.044507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mehrtash A, Sedghi A, Ghafoorian M, et al. Classification of clinical significance of MRI prostate findings using 3D convolutional neural networks. Proc SPIE Int Soc Opt Eng. 2017;10134:101342A. doi: 10.1117/12.2277123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Song Y, Zhang YD, Yan X, et al. Computer-aided diagnosis of prostate cancer using a deep convolutional neural network from multiparametric MRI. J Magn Reson Imaging. 2018;48:1570–1577. doi: 10.1002/jmri.26047. [DOI] [PubMed] [Google Scholar]
- 38.Alkadi R, Taher F, El-Baz A, Werghi N. A Deep Learning-based approach for the detection and localization of prostate cancer in T2 magnetic resonance images. J Digit Imaging. 2018 doi: 10.1007/s10278-018-0160-1. doi: 10.1007/s10278-018-0160-1. Published online 30 November 2018 [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang X, Liu C, Wang Z, et al. Co-trained convolutional neural networks for automated detection of prostate cancer in multi-parametric MRI. Med Image Anal. 2017;42:212–227. doi: 10.1016/j.media.2017.08.006. [DOI] [PubMed] [Google Scholar]
- 40.Kwak JT, Sankineni S, Xu S, et al. Correlation of magnetic resonance imaging with digital histopathology in prostate. Int J Comput Assist Radiol Surg. 2016;11:657–666. doi: 10.1007/s11548-015-1287-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stotzka R, Männer R, Bartels PH, Thompson D. A hybrid neural and statistical classifier system for histopathologic grading of prostatic lesions. Anal Quant Cytol Histol. 1995;17:204–218. [PubMed] [Google Scholar]
- 42.Rashid S, Fazli L, Boag A, Siemens R, Abolmaesumi P, Salcudean SE. Separation of benign and malignant glands in prostatic adenocarcinoma. Med Image Comput Comput Assist Interv. 2013;16:461–468. doi: 10.1007/978-3-642-40760-4_58. [DOI] [PubMed] [Google Scholar]
- 43.Gertych A, Ing N, Ma Z, et al. Machine learning approaches to analyze histological images of tissues from radical prostatectomies. Comput Med Imaging Graph. 2015;46:197–208. doi: 10.1016/j.compmedimag.2015.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gorelick L, Veksler O, Gaed M, et al. Prostate histopathology: learning tissue component histograms for cancer detection and classification. IEEE Trans Med Imaging. 2013;32:1804–1818. doi: 10.1109/TMI.2013.2265334. [DOI] [PubMed] [Google Scholar]
- 45.Singh M, Kalaw EM, Giron DM, Chong KT, Tan CL, Lee HK. Gland segmentation in prostate histopathological images. J Med Imaging (Bellingham) 2017;4:027501. doi: 10.1117/1.JMI.4.2.027501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nguyen K, Sarkar A, Jain AK. Prostate cancer grading: use of graph cut and spatial arrangement of nuclei. IEEE Trans Med Imaging. 2014;33:2254–2270. doi: 10.1109/TMI.2014.2336883. [DOI] [PubMed] [Google Scholar]
- 47.Peng Y, Jiang Y, Eisengart L, Healy MA, Straus FH, Yang XJ. Computer-aided identification of prostatic adenocarcinoma: Segmentation of glandular structures. J Pathol Inform. 2011;2:33. doi: 10.4103/2153-3539.83193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li J, Speier W, Ho KC, et al. An EM-based semi-supervised deep learning approach for semantic segmentation of histopathological images from radical prostatectomies. Comput Med Imaging Graph. 2018;69:125–133. doi: 10.1016/j.compmedimag.2018.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McGarry SD, Hurrell SL, Iczkowski KA, et al. Radio-pathomic maps of epithelium and lumen density predict the location of high-grade prostate cancer. Int J Radiat Oncol Biol Phys. 2018;101:1179–1187. doi: 10.1016/j.ijrobp.2018.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chatterjee A, Watson G, Myint E, Sved P, McEntee M, Bourne R. Changes in epithelium, stroma, and lumen space correlate more strongly with gleason pattern and are stronger predictors of prostate ADC changes than cellularity metrics. Radiology. 2015;277:751–762. doi: 10.1148/radiol.2015142414. [DOI] [PubMed] [Google Scholar]
- 51.Hectors SJ, Semaan S, Song C, et al. Advanced diffusion-weighted imaging modeling for prostate cancer characterization: correlation with quantitative histopathologic tumor tissue composition-a hypothesis-generating study. Radiology. 2018;286:918–928. doi: 10.1148/radiol.2017170904. [DOI] [PubMed] [Google Scholar]
- 52.Hill DK, Heindl A, Zormpas-Petridis K, et al. Non-invasive prostate cancer characterization with diffusion-weighted MRI: insight from in silico studies of a transgenic mouse model. Front Oncol. 2017;7:290. doi: 10.3389/fonc.2017.00290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chatterjee A, Bourne RM, Wang S, et al. Diagnosis of prostate cancer with noninvasive estimation of prostate tissue composition by using hybrid multidimensional MR imaging: a feasibility study. Radiology. 2018;287:864–873. doi: 10.1148/radiol.2018171130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lu C, Romo-Bucheli D, Wang X, et al. Nuclear shape and orientation features from H&E images predict survival in early-stage estrogen receptor-positive breast cancers. Lab Invest. 2018;98:1438–1448. doi: 10.1038/s41374-018-0095-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Beck AH, Sangoi AR, Leung S, et al. Systematic analysis of breast cancer morphology uncovers stromal features associated with survival. Sci Transl Med. 2011;3:108–113. doi: 10.1126/scitranslmed.3002564. [DOI] [PubMed] [Google Scholar]
- 56.Saltz J, Gupta R, Hou L, et al. Spatial organization and molecular correlation of tumor-infiltrating lymphocytes using deep learning on pathology images. Cell Rep. 2018;23:181–193. doi: 10.1016/j.celrep.2018.03.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rezaeilouyeh H, Mollahosseini A, Mahoor MH. Microscopic medical image classification framework via deep learning and shearlet transform. J Med Imaging (Bellingham) 2016;3:044501. doi: 10.1117/1.JMI.3.4.044501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tabesh A, Teverovskiy M, Pang HY, et al. Multi-feature prostate cancer diagnosis and Gleason grading of histological images. IEEE Trans Med Imaging. 2007;26:1366–1378. doi: 10.1109/TMI.2007.898536. [DOI] [PubMed] [Google Scholar]
- 59.Sparks R, Madabhushi A. Explicit shape descriptors: novel morphologic features for histopathology classification. Med Image Anal. 2013;17:997–1009. doi: 10.1016/j.media.2013.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nir G, Hor S, Karimi D, et al. Automatic grading of prostate cancer in digitized histopathology images: Learning from multiple experts. Med Image Anal. 2018;50:167–180. doi: 10.1016/j.media.2018.09.005. [DOI] [PubMed] [Google Scholar]
- 61.Zhou N, Fedorov A, Fennessy F, Kikinis R, Gao Y. Large scale digital prostate pathology image analysis combining feature extraction and deep neural network. 2017. arXiv [Internet] Cited March 2019. Available from: https://arxiv.org/abs/1705.02678.
- 62.Arvaniti E, Fricker KS, Moret M, et al. Automated Gleason grading of prostate cancer tissue microarrays via deep learning. Sci Rep. 2018;8:12054. doi: 10.1038/s41598-018-30535-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li W, Li J, Sarma KV, et al. Path R-CNN for prostate cancer diagnosis and gleason grading of histological images. IEEE Trans Med Imaging. 2019:945–954. doi: 10.1109/TMI.2018.2875868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nagpal K, Foote D, Liu Y, et al. Development and validation of a deep learning algorithm for improving gleason scoring of prostate cancer. 2018. arXiv [Internet] Cited March 2019. Available from: https://arxiv.org/abs/1811.06497. [DOI] [PMC free article] [PubMed]
- 65.Janowczyk A, Basavanhally A, Madabhushi A. Stain normalization using Sparse AutoEncoders (StaNoSA): application to digital pathology. Comput Med Imaging Graph. 2017;57:50–61. doi: 10.1016/j.compmedimag.2016.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Oakden-Rayner L, Carneiro G, Bessen T, Nascimento JC, Bradley AP, Palmer LJ. Precision radiology: predicting longevity using feature engineering and deep learning methods in a radiomics framework. Sci Rep. 2017;7:1648. doi: 10.1038/s41598-017-01931-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mobadersany P, Yousefi S, Amgad M, et al. Predicting cancer outcomes from histology and genomics using convolutional networks. Proc Natl Acad Sci USA. 2018;115:E2970–E2979. doi: 10.1073/pnas.1717139115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Stoyanova R, Pollack A, Takhar M, et al. Association of multiparametric MRI quantitative imaging features with prostate cancer gene expression in MRI-targeted prostate biopsies. Oncotarget. 2016;7:53362–53376. doi: 10.18632/oncotarget.10523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Smedley NF, Hsu W. Using deep neural networks for radiogenomic analysis. Proc IEEE Int Symp Biomed Imaging. 2018;2018:1529–1533. doi: 10.1109/ISBI.2018.8363864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Berglund E, Maaskola J, Schultz N, et al. Spatial maps of prostate cancer transcriptomes reveal an unexplored landscape of heterogeneity. Nat Commun. 2018;9:2419. doi: 10.1038/s41467-018-04724-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jamshidi N, Margolis DJ, Raman S, Huang J, Reiter RE, Kuo MD. Multiregional radiogenomic assessment of prostate microenvironments with multiparametric MR imaging and DNA whole-exome sequencing of prostate glands with adenocarcinoma. Radiology. 2017;284:109–119. doi: 10.1148/radiol.2017162827. [DOI] [PMC free article] [PubMed] [Google Scholar]